Summary
Despite being of vital importance for seed establishment and grain quality, starch degradation remains poorly understood in organs such as cereal or legume seeds. In cereals, starch degradation requires the synergetic action of different isoforms of α‐amylases. Ubiquitous overexpression of TaAmy2 resulted in a 2.0–437.6‐fold increase of total α‐amylase activity in developing leaf and harvested grains. These increases led to dramatic alterations of starch visco‐properties and augmentation of soluble carbohydrate levels (mainly sucrose and α‐gluco‐oligosaccharide) in grain. Interestingly, the overexpression of TaAMY2 led to an absence of dormancy in ripened grain due to abscisic acid (ABA) insensitivity. Using an allosteric α‐amylase inhibitor (acarbose), we demonstrated that ABA insensitivity was due to the increased soluble carbohydrate generated by the α‐amylase excess. Independent from the TaAMY2 overexpression, inhibition of α‐amylase during germination led to the accumulation of soluble α‐gluco‐oligosaccharides without affecting the first stage of germination. These findings support the hypotheses that (i) endosperm sugar may overcome ABA signalling and promote sprouting, and (ii) α‐amylase may not be required for the initial stage of grain germination, an observation that questions the function of the amylolytic enzyme in the starch degradation process during germination.
Keywords: amylase, wheat, sucrose, starch, degradation, germination, dormancy
Significance Statement
This study described the application of plant biotechnology to provide evidence as to whether elevated levels of α‐amylase in the mature grain can affect germination positively or negatively. These findings support the hypotheses that (i) endosperm sugar may overcome abscisic acid signalling and promote sprouting, and (ii) α‐amylase may not be required for the initial stage of grain germination.
INTRODUCTION
α‐Amylases (EC 3.2.1.1) have been omnipresent throughout evolution, from archaebacteria to humans and of course, the plant kingdom. The α‐amylase family (GH13 family) represents the largest group of the glycoside hydrolase families among the carbohydrate active enzymes (CAZy database, http://www.cazy.org/index.html) (Regina et al., 2004). α‐Amylases are endo‐hydrolases catalysing the cleavage of α‐1,4‐glucan linkages of complex carbohydrate structures such as starch, glycogen, or related oligo‐ and polysaccharides (Majzlova et al., 2013). In plants, α‐amylases are produced for three primary purposes: (i) to ensure the degradation of transitory starch in vegetative tissue providing the energy required for plant metabolic function and growth at night (MacNeill et al., 2017), (ii) to respond to biotic stress by removing any internal source of carbohydrate for the pathogen in a ‘scorched‐earth’ strategy (Andersen et al., 2018), and (iii) to provide the energy required by the plant during grain development and germination processes to complete its reproductive life cycle (Guzmán‐Ortiz et al., 2018). For example, Arabidopsis thaliana contains three isoforms: AtAMY1, AtAMY2, and AtAMY3 (Yu et al., 2005). In barley, four α‐amylase categories have been identified, HvAMY1–HvAMY4 (Radchuk et al., 2009; Rogers, 1985), whereas in rice, at least 10 α‐amylase genes are clustered into five hybridization groups and three subfamilies (OsAMY1, AsAMY2, and OsAMY3) (Damaris et al., 2019; Nanjo et al., 2004). All subfamilies in both barley and rice have been demonstrated to be expressed at different grain developmental stages and in various tissues (Damaris et al., 2019; Karrer et al., 1991).
In wheat, four isoforms of α‐amylase have been identified to date (Barrero et al., 2013; Mieog et al., 2017). The two major α‐amylases, TaAMY1 and TaAMY2 have been primarily identified and isolated based on their isoelectric point (pI) (Ainsworth et al., 1985). TaAMY1 has a high pI α‐amylase encoded by multigene families located on the long arm of chromosome 6. TaAMY1 has been described as the germinating α‐amylase due to its accumulation in the aleurone layer both at late maturity and during the first stage of germination (Mieog et al., 2017; Nishikawa et al., 1981). The α‐amylase 2 (TaAMY2) families have been located on the long arm of the group 7 chromosomes. This so‐called ‘green’ α‐amylase has been detected in the pericarp of the developing grain during the first 15 days after anthesis and appears at a later stage of germination (Gale et al., 1983; Kozo et al., 1988). The TaAmy3 and TaAmy4 loci have been mapped to chromosome 5 and 4 respectively (Baulcombe et al., 1987; Mieog et al., 2017). The role and function of TaAmy4 remains to be elucidated despite some preliminary results suggested co‐expression with TaAmy1 (Mieog et al., 2017). While a recent study suggested that both TaAMY3 and TaAMY4 were non‐functional during evolution (Ju et al., 2019), studies on the impact of TaAmy3 overexpression in wheat grain have demonstrated clear impact on starch and end‐product functionality (Ral et al., 2016, 2018; Whan et al., 2014).
Wheat is one of the world’s three major food crops in food security with global wheat production in 2019 estimated at 765 million tonnes, making it the second most‐produced cereal after maize (FAO Stat: http://www.fao.org/worldfoodsituation/csdb/en/). Owing to its economic significance, wheat TaAMY1 and TaAMY2 have been heavily characterized due to their involvement in two of the most severe quality defects of the industry; late maturity α‐amylase and pre‐harvest sprouting (PHS) (for review Mares and Mrva, 2014). Both have altered α‐amylase expression, dormancy, germination, and/or grain functionality (Biddulph et al., 2008; Kondhare et al., 2012, 2015). During germination, the synthesis of α‐amylase mRNA and efficiency of translation is accelerated by gibberellins (GA) (Higgins et al., 1976) and strongly inhibited by abscisic acid (ABA) (Ho and Varner, 1975; Jacobsen, 1973). ABA is the key hormone regulating dormancy and preventing germination in the seed and directly influences PHS (Gubler et al., 2005). The effects of the GA‐insensitivity alleles on late maturity α‐amylase expression suggest a role for the germination hormone in the enzyme synthesis (Barrero et al., 2013).
Until recently, the physiological properties of the various types of α‐amylase on starch in the grain and the potential impacts of α‐amylase accumulation on grain development, starch accumulation, functionality, and germination were largely unknown (Mieog et al., 2017). One of the aims of this study was to assess the functional properties of an uncharacterized wheat α‐amylase to date. This study describes an approach where one of the main wheat α‐amylases (TaAmy2) was overexpressed ubiquitously in wheat, and its consequence on grain properties and starch functionality was studied. The impact of elevated levels of TaAMY2 on germination also highlights a new development on the general mechanism through which α‐amylase acts on starch degradation during grain germination.
RESULTS
Generation of TaAmy2 overexpression line (UA2OE)
To investigate the mode of action and the impact of TaAMY2 on native starch in the grain we developed TaAmy2 overexpressing transgenic wheat plants in the variety Fielder (spring wheat cultivar). The TaAmy2 cDNA cassette was placed under the control of the Zea mays ubiquitin promoter to ensure TaAmy2 gene expression throughout the entire plant lifespan and more specifically during grain development and germination (Figure S1).
Twelve hygromycin resistant TaAmy2 overexpression (UA2OE) wheat T0 plants were selected by polymerase chain reaction (PCR) using construct‐specific primers (Table S1) following Agrobacterium tumefaciens‐mediated transformation of immature embryos. Quantitative real‐time PCR detected between one and 11 insertion events at T1 generation. Three lines were selected for further characterization including one independent event with potentially one insertion number (FC 127‐3), two events with multiple insertion numbers (FC127‐7 and FC127‐12) and a negative control (transformed with no insert) (NC) (Table S2.1–S2.3). Copy number was monitored from generations T1 to T4. The UA2OE line FC127‐3 has a stable copy number with two copies detected across several generations whereas FC127‐7 and FC127‐12 have various high copy numbers (Table S2). Relative quantification on total protein extracted from individual grains and comparison of TaAMY2 peptides, using mass spectrometry analysis, solely identified TaAMY2 peptides in the positive lines (Figure S2 and S3). Relative quantitation and comparison of TaAMY2 peptides on dry grain detected five TaAMY2 peptides in the UA2OE compared with NC. No TaAMY1 and TaAMY3 target peptides were detected in either the positive lines or NCs.
All three independent events showed strong elevated levels of α‐amylase activity (Table S3). In the T4 grains, copy number, total α‐amylase activity, and starch damage were assessed. The copy number was significantly correlated using lienar Spearman correlation with α‐amylase activity and damaged starch percentage at 0.772 and 0.764, respectively (Figure S4 and Table S3). The soluble sugar exhibited a low correlation (0.377) with the copy number. In addition, all test lines showed a significant increase in the percentage of damaged starch compared with their NCs.
All three lines FC127‐3, FC127‐7, and FC127‐12 were selected as the representative homozygous lines labelled UA2OE3.1, UA2OE7.2, and UA2OE12.1 for ubiquitin TaAmy2 overexpression, together with NC. The grains from T4 and T5 generations were used for further characterization. Compared with the [NC, 0.11 ceralpha unit (CU) g−1 wholemeal], all UA2OE lines showed a significant increase in total α‐amylase activity of 114.3–437.6‐fold (Figure 1a) ranging from 12.84 to 49.12 CU·g−1 wholemeal and a specific elevation in damaged starch percentage of 5.9–7.9‐fold compared with NC with 1.29% ± 0.7 SE (Figure 1b).
Figure 1.
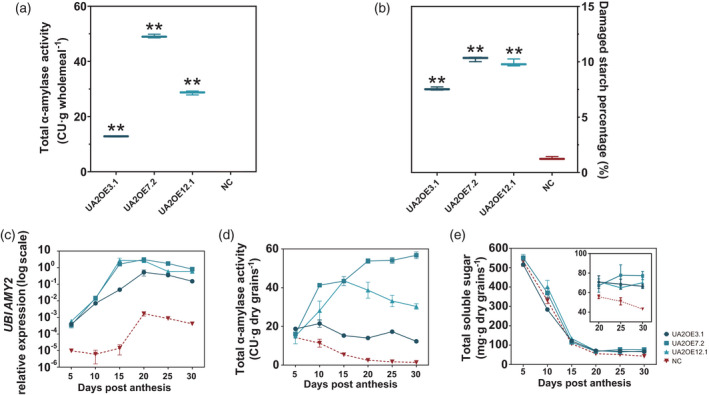
Total α‐amylase, damaged starch percentage and engineered TaAMY2 profile of UA2OE (blue) and negative control (NC) (red) in mature and developing grains. (a) Total α‐amylase activity is expressed in ceralpha units (CU) of per gram wholemeal and (b) damaged starch is expressed in percentage dry weight. (c) TaAMY2 of construct transcript level (relative to geometric mean of TaActin, Ta.14126.1.S1_at, and Ta.7894.3.A1_at) seed development in whole grain; (d) total α‐amylase activity and (e) total soluble sugar of three UA2OE lines (blue) and NC (red) during grain development. One‐way ANOVA was used for statistical analysis. Asterisks indicate significant differences between the UA2OE lines and NC (**P < 0.01). Values are the mean and standard error of (a,b) three technical replicates and (c–e) three biological replicates.
Overexpressed TaAmy2 profile during grain development
The specific expression level of the ubiquitin TaAmy2 was assessed during grain development using real‐time PCR relative to TaActin, Ta.14126.1.S1_at, and Ta.7894.3.A1_at using the approach of Mieog et al. (2017) (Figure 1c). Relative expression of TaAmy2 in NC remained at a very low level at all time points and despite a slight increment after 15 days post‐anthesis (DPA) demonstrating the specificity of our assay for TaAmy2. In the three UA2OE lines, the elevated TaAmy2 transgene expression increased from 5 days DPA and throughout the grain development reaching its maximum between 15 and 20 DPA (Figure 1c). The expression level of TaAmy1 and TaAmy3 showed no significant difference between UA2OE lines and NC (Figure S5).
The relative expression of exogenous TaAmy2 was elevated during grain development in the UA2OE lines and translated into an elevated level of total α‐amylase activity throughout grain development. In NC, the activity was highest at 5 DPA (14.26 CU g−1 dry grain) before slowly decreasing to a very low level at 30 DPA (1.36 CU g−1 dry grain) (Figure 1d). In the three UA2OE lines, the total α‐amylase activity increased significantly compared with NC. In UA2OE3.1, total activity remained at a stable level ranging from 12.30 to 21.48 CU g−1 dry grain during grain development. In UA2OE7.2, the activity gradually increased from 16.09 to 56.57 CU g−1 dry grain throughout grain development. In UA2OE12.1, total α‐amylase activity peaked at 15 DPA (38.72 CU g–1 dry grain) and thereafter, slightly decreased to reach 30.3 CU g−1 dry grain at 30 DPA. At the beginning of grain development, no significant increase was observed in total soluble sugar, but from 20 DPA, the level of soluble sugar seemed to be slightly but significantly higher in all three UA2OE lines compared with NC (Figure 1e).
No impact on transitory starch
While the TaAMY2 sequence did not contain a chloroplast targeting peptide, the impact of TaAMY2 overexpression on leaf transitory starch metabolism was assessed to ensure that transitory starch metabolism was not impacted. Total α‐amylase activity, leaf starch content and total soluble sugars were measured on leaf tissue sampled from plants in the vegetative stage (jointing stage) grown in glasshouse under a 16 h light/8 h dark regime. The three UA2OE lines showed a significant elevated total α‐amylase activity with 5.23, 0.43, and 1.3 CU g−1 dry leaf, respectively, at sunrise and with 5.95, 0.44, and 2.59 CU g−1 dry leaf at sunset while the control line did not show any significant α‐amylase activity (Figure S6A). The three UA2OE lines and NC exhibited similar starch and soluble sugar patterns at sunset and sunrise (Figure S6B and S6C). Visual observation did not detect any changes in plant morphology or development.
TaAMY2 overexpression impact on grain composition
Several differences highlighted the impact of TaAMY2 overexpression on mature grain composition. Overall, the thousand grain weight was significantly reduced (6.1–12.8% reduction to the NC) (Table S4). Compared with NC (54.35 ± 0.65%), all three UA2OE lines showed a significant reduction (by 5.27–7.90%) in total starch content (Figure 2a). All three UA2OE lines had 3.3–4.3‐fold increase of total soluble sugar in mature grains compared with NC (Figure 2b). Several types of soluble sugar were significantly higher in all three UA2OE lines. For instance, the total α‐gluco‐oligosaccharides showed the highest increase by 37.7–53.2‐fold with 38.9, 53.5 and 54.8 mg g−1 wholemeal, respectively, compared with NC with 0.6 mg g−1 wholemeal (Figure 2c). Free glucose and sucrose levels were elevated by 9.3–16.0‐fold (P = 3.1 × 10−9, Figure 2d) and 2.8–3.2‐fold (P < 5.59 × 10−8, Figure 2e) respectively. Free fructose remained at a very low level but still showed a significant increase (P = 0.0003) by 2.4–3.0‐fold with an average of 0.99 mg g−1 wholemeal in three UA2OE lines compared with NC with 0.4 mg g−1 wholemeal (Figure 2f). Total fructan remained the main carbohydrate after starch in the mature wheat grains and showed a significant (P = 0.0002) increase in all three UA2OE lines by 2–3‐fold compared with NC (Figure 2g).
Figure 2.
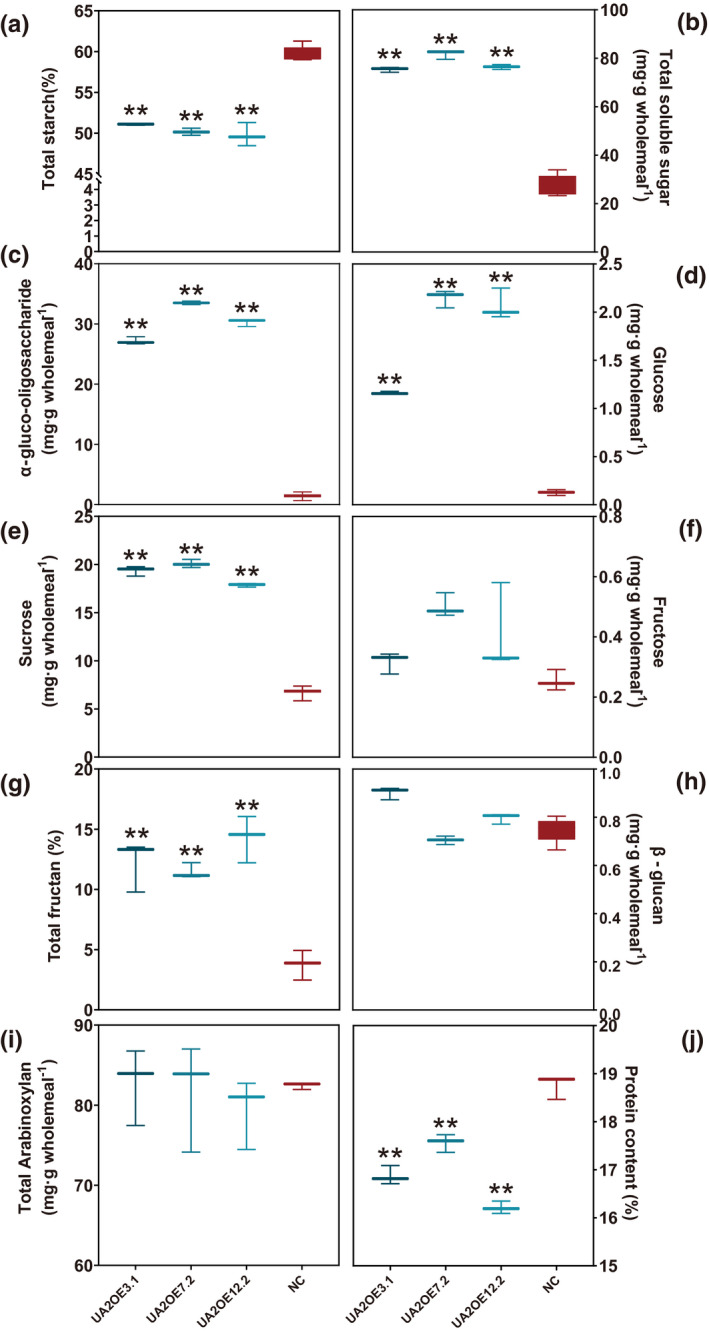
Grain compositional analysis of three UA2OE lines and negative control (NC) at maturity. Total starch (a), fructan (g) and protein (j) are expressed in percentage dry weight. Total soluble carbohydrate (b), α‐gluco‐oligosaccharide (c), glucose (d), sucrose (e), fructose (f), β‐glucan (h), and total arabinoxylan (i) are expressed in milligram per gram wholemeal. Three biological replicates were performed. One‐way ANOVA was used for statistical analysis. Asterisks indicate significant differences between the two treatments (*P < 0.05; **P < 0.01).
In addition, variation in β‐glucan and total arabinoxylan contents (Figure 2h,I) were detected, though not significantly different from NC. Protein content in the three UA2OE lines showed a significant (P < 1.38 × 10−6) decrease with 16.2–17.5% compared with NC with 18.7% (Figure 2j).
Impact on starch granule morphology, structural, and rheological properties
A range of measurements and observations including microscopy techniques, granule size distribution, amylose content, and chain length distribution were used to investigate whether the elevated level of TaAMY2 impacted the morphology and/or starch structure (Figure 3 and Figure S7).
Figure 3.
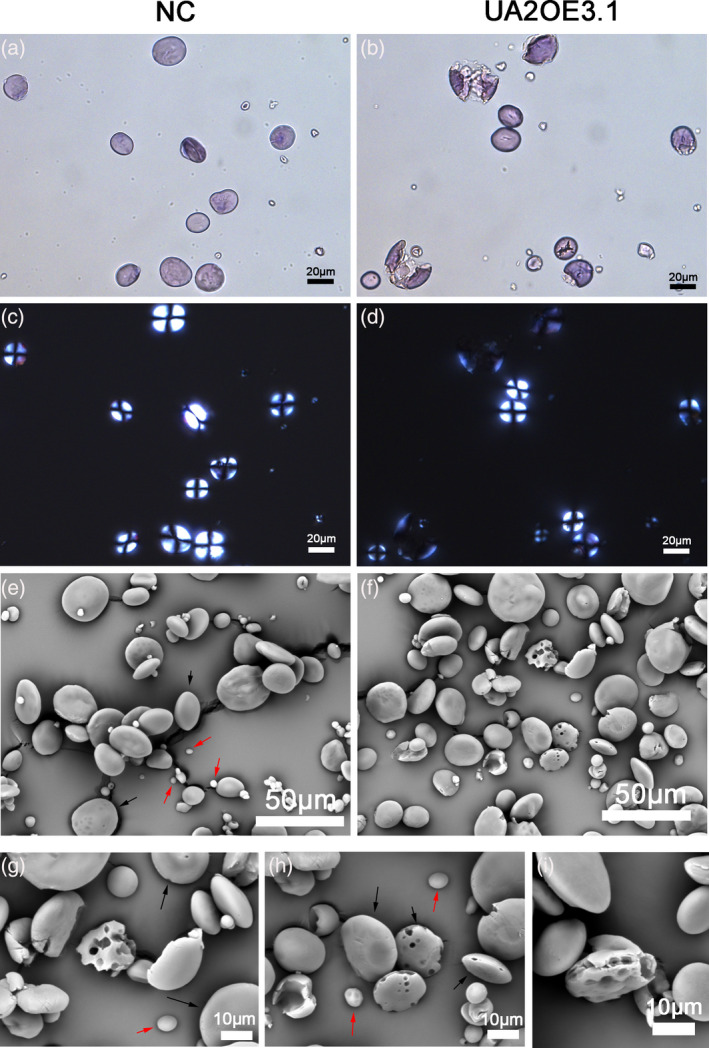
Impact of UA2OE on starch granules in mature grains. Light micrographs of isolated starch stained with iodine (a, b), polarized light micrographs (c, d), and scanning electron micrographs of isolated starch granules (e–i) from negative control (NC) (a, c, e) and UA2OE3.1 (b, d, f, g–i). A‐type granules (starch granule diameter >10 μm) and B‐type granules (starch granule diameter <10 μm) are indicated by black arrows and red arrows, respectively.
Light microscopy revealed clear morphological differences between the starch granules of UA2OE and NC lines. While most of the A granules in the NC showed a classical lenticular shape and the B granules were spherical (Figure 3a), most starch granules from UA2OE showed signs of partial to almost total digestion (Figure 3b). Polarizing and brightfield images revealed that most unbroken starch granules in UA2OE and all starch granules in the NC displayed the typical ‘Maltese cross’ associated with starch crystallinity (Figure 3c,d), whereas partially digested starch granules from UA2OE showed a very slight response to polarized light (Figure 3d). The morphological characteristics of starch granules were also examined by scanning electron microscopy (SEM). Starch granules in these NC lines displayed a smooth surface with no sign of damage (Figure 3e). In the UA2OE lines, while many starch granules seemed to remain unaffected, several starch granules showed signs of damage including pores, loss of structural integrity and altered shapes overall. The SEM of starch granules from UA2OE3.1 in mature grains showed many pin holes on the surface of granules (Figure 3f–i) similar to those observed following enzymatic hydrolysis of barley starch during malting and in germinating wheat (Faltermaier et al., 2014). SEM analysis of UA2OE3.1 and NC from 15 to 30 DPA indicated that the presence of pin holes appears from 20 to 25 DPA suggesting that degradation only occurs toward the end of the starch filling process (Figure S7). These results confirmed the significant increase of soluble carbohydrates seen from 20 DPA.
The three UA2OE lines and NC showed no significant difference for their amylose content (P = 0.079) or for their particle size distribution analysis (Figure S8a,b). The amylopectin chain length distribution difference was performed revealing no significant differences on chain length distribution with or without β‐amylase pre‐treatment (Figure S8c,d).
The Rapid Visco Analyser was used to investigate the pasting properties of wholemeal flour from UA2OE lines in the presence or absence of an enzyme inhibitor (AgNO3) (Figure S9 and Table 1). In the absence of silver nitrate, NC had low peak and final viscosity values at 1277.7 and 684.3 centipoises (cP) on average respectively. The profiles of the UA2OE were very different compared with NC, with even lower peaks (71.7 ± 0.3, 42 ± 4, and 47.7 ± 0.3, respectively) and final viscosities (29.7 ± 0.9, 22.5 ± 0.5, and 26.3 ± 1.2, respectively) near the limit of detection. When α‐amylase inhibitor was added, all the UA2OE lines and NC profiles increased significantly although the RVA profile of UA2OE lines was still slightly lower compared with NC.
Table 1.
Rapid Visco Analyser profile and gelatinization properties of UA2OE and NC
| Samples | Rapid Visco Analyser | Gelatinization endotherm | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Peak viscosity (cP) | Trough viscosity (cP) | Breakdown (cP) | Final viscosity (cP) | Setback (cP) | Onset temp. | Peak temp. | Conclusion temp. | ∆H (J·g−1) | |
| NC | 1277.7 ± 5.5 | 291.7 ± 2.9 | 986 ± 8.4 | 684.3 ± 4.9 | 392.7 ± 2.2 | 60.6 ± 0.1 | 63.9 ± 0.3 | 70.3 ± 0.5 | 6.3 ± 0.2 |
| NC + AgNO3 | 3946.7 ± 141.3 | 1421 ± 70.2 | 2525.7 ± 77.3 | 3019.3 ± 114.4 | 1598.3 ± 46.3 | ||||
| UA2OE3.1 | 71.7 ± 0.3** | 22.7 ± 0.3** | 49 ± 0** | 29.7 ± 0.9** | 7 ± 1.2** | 61.2 ± 0.5 | 64.2 ± 0.5 | 69.5 ± 0.6 | 6.0 ± 0.3 |
| UA2OE3.1 + AgNO3 | 2541.7 ± 57.3** | 1016.3 ± 16.2** | 1525.3 ± 41.6** | 2145 ± 16.2** | 1128.7 ± 1.3** | ||||
| UA2OE7.2 | 42 ± 4** | 11.5 ± 0.5** | 30.5 ± 3.5** | 22.5 ± 0.5** | 11 ± 1** | 61.5 ± 0.4 | 64.6 ± 0.3 | 71.3 ± 0.5 | 5.7 ± 0.1 |
| UA2OE7.2 + AgNO3 | 2294 ± 113.2** | 1167.3 ± 89.7** | 1126.7 ± 119.5** | 2128.7 ± 94.6** | 961.3 ± 65.1** | ||||
| UA2OE12.1 | 47.7 ± 0.3** | 19.3 ± 0.9** | 28.3 ± 0.7** | 26.3 ± 1.2** | 7 ± 0.6** | 61.6 ± 0.3 | 64.5 ± 0.4 | 70.3 ± 0.4 | 6.8 ± 0.3 |
| UA2OE12.1 + AgNO3 | 2190 ± 7.2** | 1030 ± 86.2** | 1160 ± 93.4** | 1996.3 ± 35.4** | 966.3 ± 55.8** | ||||
Rapid Visco Analyser profiles of UA2OE and negative control (NC) in presence or absence of the Silver Nitrate (α‐amylase inhibitor). Viscosity is expressed in Centipoise (cP). Temperatures are expressed in °C in gelatinization endotherm. Enthalpy of gelatinization (∆H) is expressed in joules per g (J g−1). Results displayed are the mean of three independent assays. Asterisks indicate significant differences between UA2OE and NC (*P < 0.05; **P < 0.01).
Differential scanning calorimetry analysis performed on purified starch revealed that the UA2OE lines had no significant difference on onset (To), peak (Tp), conclusion (Tc) temperatures, and enthalpy (ΔH) compared with NC (Table 1).
Reduced dormancy in TaAMY2 overexpression line
Germination assays with grains harvested at physiological maturity and 4 weeks after‐physiological maturity from the mother plant were performed to determine the influence of TaAMY2 overexpression in dormancy retention and germination. NC at harvest‐ripe showed dormancy with 73.3% ± 3.3 SE of grains showing coleorhiza emergence (CE) at the end of the 10‐day experiment while the grains of all three UA2OE lines showed a low level of dormancy with CE at 100% after 6 days (Figure 4a). Four weeks after physiological maturity, NC lost dormancy and showed a similar germination pattern with 100% at 4 days post‐imbibition (DPI) (Figure 4b).
Figure 4.
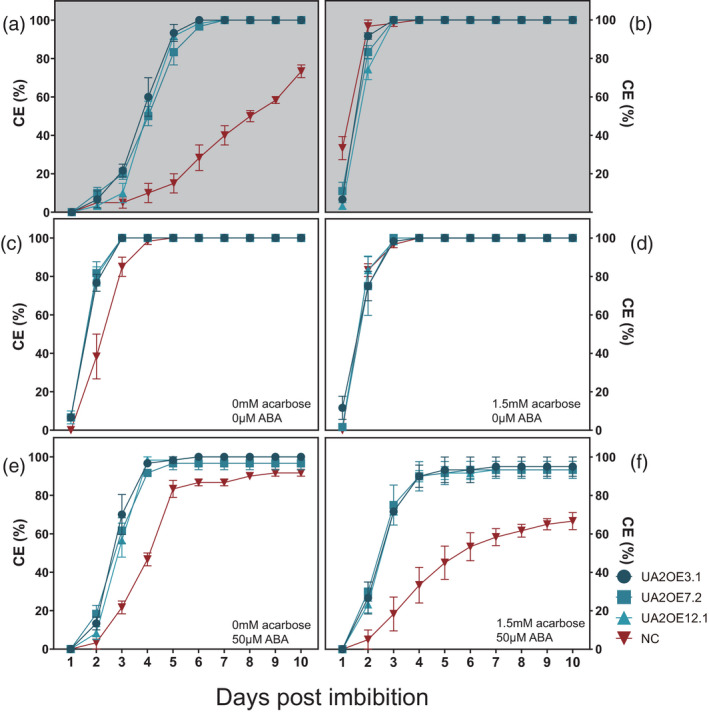
Coleorhiza emergence (CE) in whole grains and effects of abscisic acid (ABA) and acarbose on half grain germination. CE for whole grains at physiological maturity (a) and 4 weeks after‐physiological maturity (b) in whole grains (grey background). CE on half grains at ripening (c), in presence of 1.5 mm acarbose (d, f) and 50 μm ABA (e, f). Germination rate comparisons between three UA2OE lines (blue) and negative control (red). UA2OE3.1, UA2OE7.2, UA2OE12.1, and NC were indicated as circle (dark blue), square (blue), triangle (light blue) and inverted triangle (red), respectively. Error bars represent standard error of three replicate plates.
TaAMY2 overexpression overcomes ABA‐induced dormancy in half grains
To determine whether the lack of dormancy in the three UA2OE lines was due to ABA insensitivity, different concentrations of ABA were applied to halved grains of both UA2OE and NC. Cutting grains in half broke the dormancy and caused rapid germination to 100% CE in all three UA2OE lines and NC within 3–4 days (Figure 4c). ABA resistance was tested by the addition of ABA at incremental concentrations (0, 20, 50, 100, and 150 μm). The addition of ≥50 μm of exogenous ABA was able to restore partially some dormancy in NC. However, the addition of 50 μm of exogenous ABA was unable to induce dormancy in all three UA2OE lines (Figure 4e). All three lines showed a similar germination rate response in the presence of increased concentrations of ABA (100 and 150 μm) compared with NC (Figure S10a,b) suggesting that TaAMY2 overexpression led to ABA insensitivity.
Two clear phenotypes emerged from the dry grain characterization that could be associated with grain dormancy and ABA insensitivity, (i) the presence of elevated levels of overexpressed TaAMY2, and (ii) the presence of elevated levels of soluble sugars. To confirm whether the ABA insensitivity was directly caused by the elevated presence of an active α‐amylase in dry grain, we performed in parallel germination experiments using acarbose, an allosteric inhibitor of α‐amylase.
The presence of acarbose had no observed effect on the germination of non‐dormant UA2OE lines and NC in both whole (Figure S10c,d) or half grains (Figure 4d,f). The UA2OE half grains showed a high level of CE reaching 100% after 7 days of germination whereas NC still maintained considerable dormancy with CE at 66.6% ± 4.4 SE after 10 days in the presence of acarbose and ABA (Figure 4f). We could therefore conclude that α‐amylase activity was not the direct cause of the ABA resistance. Moreover, the presence of an exogenous α‐amylase inhibitor did not affect the germination in either the UAMY2OE lines or the NC, but it seemed that the combination of acarbose and ABA had a synergistic effect on increasing dormancy in NC.
The only other remarkable phenotype was the presence of elevated levels of soluble sugar. It was reasonable to assume that this presence of elevated soluble sugar could provide the energy required for the grain to trigger germination.
Impact of TaAMY2 overexpression on soluble sugar profile during germination
We decided to monitor different soluble carbohydrate profiles to see how the presence of elevated levels of TaAMY2 could affect sugar accumulation during germination. Acarbose was also added to investigate whether inhibition of α‐amylase activity in grains after imbibition affected the soluble sugar accumulation. In the absence of acarbose, NC showed a very low α‐amylase activity at 2 DPI, rapidly increasing to 78.7 CU g−1 wholemeal at 6 DPI (Figure 5a). However, all three UA2OE lines showed an initial decrease of total α‐amylase activity to reach levels below 15 CU g−1 wholemeal, until 3 DPA before subsequently increasing to 57.4, 84.3, and 62.8 CU g−1 wholemeal, respectively, at 6 or 7 DPI. The acarbose concentration of 1.5 mm inhibited most of the α‐amylase with activity reduced to levels below 1 CU g−1 in all three UA2OE lines and NC (Figure 5a).
Figure 5.
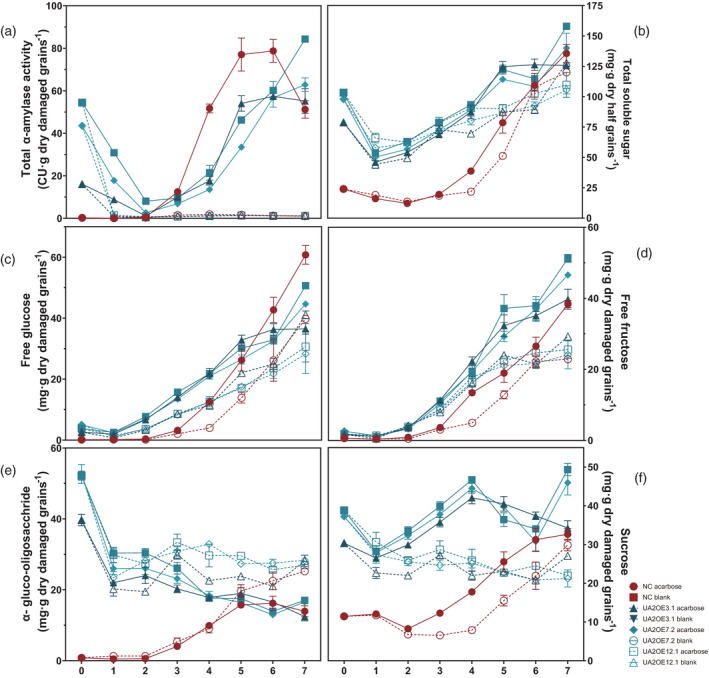
Effects of acarbose on α‐amylase activity and total soluble sugar accumulation during germination. Three UA2OE lines, UA2OE3.1 (triangle, dark blue), UA2OE7.2 (square, blue), and UA2OE (rhomb, wathet blue), and NC (circle, red) half grains during germination in the absence (plain and solid) and presence of 1.5 mm acarbose (dash and hollow) for 7 days. Total α‐amylase activity is expressed in CU·mg−1 dry tissue (a), total soluble sugar is expressed in mg·g−1 dry damaged grains (b), free glucose (c), free fructose (d), α‐gluco‐oligosaccharide (e) and sucrose (f). Values are expressed as mean ± SE.
Total soluble sugars were measured throughout the germination experiment (Figure 5b–f). In NC, the total soluble sugar dropped slightly prior to 2 DPI from 23.9 mg g−1 DW to 12.1 mg·g−1 DW in the presence or absence of acarbose (Figure 5b). From 4 DPI, the presence of acarbose reduced the overall amount of soluble sugar by 15.4 mg g−1 DW but incrementally kept the same profile indirectly confirming the impact of acarbose on α‐amylase activity in the germinating grain. All three UA2OE profiles were drastically different to NC. Total soluble sugar was sharply reduced at 1 DPI by an average of 40.4 mg g−1 DW but still remained at a higher level than NC, then slowly increased to reach a level similar to NC at 7 DPI. The soluble sugar level was not affected by acarbose until 4 DPI, but then its accumulation slowed down. Free glucose and fructose displayed a similar profile that incrementally increased during germination (Figure 5c,d). In the absence of acarbose, all three UA2OE lines maintained relatively higher glucose and fructose contents compared with NC until 5 DPI. In the presence of acarbose, all UA2OE lines and NC showed similar and increasing accumulation profiles over time but glucose and fructose levels were lower in the presence of acarbose from 3 DPI.
The α‐gluco‐oligosaccharide content was quite intriguing. In the absence of acarbose, the NC showed a slow building of α‐gluco‐oligosaccharide across the 7 DPI (from 0.8 to 13.9 mg g−1 DW) while all three UA2OE lines displayed a consistent decrease of α‐gluco‐oligosaccharides from 39.7 ± 1.6, 52.2 ± 1.3, and 52.7 ± 2.7 5 mg g−1 DW to 12.4 ± 2.0, 17.0 ± 0.3 and 16.6 ± 8.0 mg g−1 DW (Figure 5e). In the presence of acarbose, the NC showed a strong increase of α‐gluco‐oligosaccharides reaching 25.2 mg g−1 DW at 7 DPI. The three UA2OE lines on the other hand, showed a decrease from 39.7 ± 1.6, 52.2 ± 1.3 and 52.7 ± 2.7 5 mg·g−1 DW to reach about 27 mg·g−1 DW at 7 DPI reaching similar levels to those seen in the NC with acarbose. All three UA2OE lines and NC displayed higher levels of α‐gluco‐oligosaccharides in general at 7 DPI when acarbose was added.
Finally, sucrose content showed a distinct pattern in the NC and the three UA2OE lines. In the absence of acarbose, NC showed a slight decrease until 2 DPA before rising to its highest level at 7 DPA (Figure 5f). The three UA2OE lines decreased from 30.4 ± 0.6, 38.8 ± 0.4, and 37.2 ± 0.7 mg·g−1 DW to about 28 mg·g−1 DW at 1 DPI then increased, reaching a peak of 42.1 ± 1.5, 46.7 ± 0.3, and 44.5 ± 1.7 mg·g−1 DW at 4 DPI. In the presence of acarbose, the level of sucrose is generally lower in all UA2OE lines and NC. The presence of acarbose reduced the overall amount of sucrose in both NC and UA2OE lines.
DISCUSSION
Impact of TaAMY2 overexpression on starch granule properties
The overexpression of TaAMY2 led to a 114.3–437.6‐fold increase in total α‐amylase activity in dry grain. This resulted in the appearance of pin holes, pores, and an overall alteration in starch granule morphology. This was associated with a quantifiable reduction of grain weight, increase in the proportion of damaged starch granules and an elevated level of soluble carbohydrates, including α‐gluco‐oligosaccharides, sucrose, fructan, and glucose. However, it is interesting to note that while increase in total α‐amylase activity could be detected from early grain development, the increase of soluble carbohydrate was only significant after 20 DPA. Our first hypothesis argues that this later occurrence of soluble carbohydrate is an indication of starch degradation at a later stage of grain development. This was confirmed by the presence of visible signs of degradation only from 25 DPA. The absence of starch digestion, at least until the end of the starch filling process, may be an indication of different localization between the engineered TaAMY2 enzyme and its substrate. During grain fill, starch in cereal grain is produced in amyloplasts. TaAMY2 is named the ‘green’ α‐amylase and its role is to fuel grain cellularization in early development (Radchuk et al., 2009), but there has been no indication in the literature to date of TaAMY2 being plastid targeted. During maturation, the grain triggers a rapid net water loss and the membrane of the desiccated amyloplasts degrades (Pepler et al., 2006). If the location of TaAMY2 is ectopic of the plastid, the degradation of the granule could only occur when the lipid membrane surrounding the amyloplast breaks down after 20 DPA giving rise to an increase in soluble sugars and visible signs of degradation from 20 to 25 DPA.
Consequently, rheological properties were significantly altered with the total absence of viscosity peaks on the RVA profile and a significant increase in gelatinization temperature. While these phenotypes are consistent with those observed when TaAMY3 was overexpressed (Ral et al., 2016; Whan et al., 2014), some specificities would suggest a different modus operandi between TaAMY2 and TaAMY3. The UA2OE grain displayed a wider distribution of pores, pins, and degradation marks across the granules while most of the pores in the case of TaAMY3 overexpression were located on the wheat starch granule groove (Whan et al., 2014). Li et al. (2012) suggested that α‐amylases were able to aim towards specific weak points on the surface of the crystalline structure such as amorphous or non‐homogeneous regions. Descriptions of the starch‐binding domains among the cereal α‐amylases highlighted marked differences regarding their carbohydrate‐binding domain (Janecek et al., 2013, 2014; Mieog et al., 2017). Structural analysis of the high and low pI α‐amylase in barley demonstrated that the low pI α‐amylase had an additional sugar‐binding domain or ‘sugar tong’ (Tranier et al., 2005). It was speculated that this sugar tong enables the α‐amylase to bind and break the highly branched sugars generated from preliminary degradation caused by the high pI α‐amylase. Thus, it is plausible that, in excess, TaAMY2 has a stronger ability to bind to the surface of the granule thereby creating more damage than previously described for other isoforms.
The augmentation of pores led to a clear increase in the proportion of damaged starch and consequently resulted in an almost total absence of viscosity profile. The presence of damaged starch and pin holes would accelerate water absorption and increase accessibility to additional hydrolases during the process (Johnston et al., 2019). The addition of silver nitrate, an enzyme inhibitor, however significantly restored the viscosity profile. These results confirmed parallel findings from Bhattacharya and Corke (1996) and Ral et al. (2016); while some of the starch damage occurred before grain maturity, the low viscosity was the result of α‐amylase activity during the hydrated environment of the test rather than starch damage existing before analysis. The absence of changes in gelatinization properties of the isolated starch determined by DSC tends to confirm it.
Overexpressing TaAMY2 does not participate in degradation of starch in developing leaves
Higher levels of α‐amylase activity were detected in the leaves of young UA2OE plants both at sunrise and sunset but without any significant impact on transitory starch degradation (Figure S6). The level of transitory starch and soluble sugar during the circadian cycle remained unchanged. While we cannot rule out that the overexpressed enzyme may accumulate outside of the chloroplast due to the lack of a chloroplast targeting peptide, it is unlikely the excess of TaAMY2 had any significant detrimental effect on carbon allocation and plant development in our transgenic lines. These results are consistent with the absence of a major role of α‐amylase in transitory starch degradation described in A. thaliana. Knock‐out mutants of AtAMY1 and AtAMY2 and tDNA insertion mutant of the plastidial AtAMY3 did not show any impact on starch breakdown (Lloyd et al., 2005; Yu et al., 2005). The role of AtAMY3 in starch degradation was only discovered when several mutations including inactivation of iso‐amylase (ISA3) and β‐amylase (BAM1) and AtAMY3 were combined (Streb et al., 2012).
TaAMY2 overexpression affects dormancy causing ABA insensitivity
Seed dormancy is defined as a hibernation‐like state where seeds do not germinate even under optimum conditions (Bewley, 1997). Dormancy is also a very important physiological and economical trait for cereal. Strong dormancy reduces plant emergence thus impacting yield, while absence of dormancy causes seed germination on the mother plant due to environmental conditions such as heavy rainfall and before ripening known as PHS (for review Rodríguez et al., 2015). Overexpressing TaAMY2 resulted in grains with almost total absence of dormancy before any after‐ripening. The presence of readily available α‐amylase in UA2OE mature grains would suggest a faster germination rate once dormancy is broken down rather than an absence of dormancy. Historically, absence of dormancy is caused by an alteration of the plant hormone antagonistic interaction between ABA involved in the dormancy induction and GA promoting dormant grain germination (Zeevaart and Creelman, 1988). The experiments germinating UA2OE and NC half seeds in the presence of exogenous ABA and/or acarbose were designed to identify the possible mechanisms underlying the absence of dormancy. Damaging the mature grain is known to break coat imposed dormancy while addition of incremental levels of ABA would demonstrate any potential ABA insensitivity (Bykova et al., 2011; Chen et al., 2010). The ABA addition restored some level of dormancy in NC but had very limited impact on UA2OE grain germination profile suggesting some form of ABA insensitivity within our transgenic lines.
The addition of acarbose, an allosteric inhibitor of glycoside hydrolases (specifically, α‐glucosidase enzymes and α‐amylase) (Konishi et al., 1994; Oudjeriouat et al., 2003) was designed to confirm whether the elevated level of α‐amylase was the direct cause of the ABA insensitivity. Acarbose almost completely inhibited the total α‐amylase activity during the 7 DPI but failed to have any impact on grain germination thus suggesting a limited impact of α‐amylase on the ABA resistance. The synergistic effect of acarbose and ABA on increasing NC dormancy while having no effect on UAMY2OE lines could be another indication that the cause of ABA insensitivity was not directly due to α‐amylase activity during germination but occurred before imbibition with acarbose. This result was consistent with Lin et al. (2008) who found no significant correlation between germinating barley α‐amylase activity and germination rate. While, α‐amylase synthesis in the aleurone layer is critical for seed germination in cereal grains and is strongly regulated by ABA (negatively) and GA (positively), α‐amylase is not required for breaking dormancy (Nee et al., 2017). When dormancy is broken down and germination occurs, GA is secreted from the embryo into the aleurone layer (Smith and Hooley, 2002) promoting the expression of α‐amylases (Gubler et al., 1995). The α‐amylase production is completed during the first 24–48 h following imbibition to mobilize the energy from starch (Radchuk et al., 2009) and to ensure seedling development. However, isolated embryos are able to germinate in tissue culture conditions (Du et al., 2018). It was therefore reasonable to assume that ABA resistance was caused by the second marked phenotype: the excess of soluble carbohydrate.
However, high α‐amylase levels in developing grain could strongly increase starch degradation and indirectly affect seed germination (Yang et al., 2014). The accumulation of α‐amylase at the end of grain development could promote the degradation of starch into soluble sugars at levels that may affect dormancy (De Laethauwer et al., 2013). Exogenous sugars are able to overcome the germination inhibition of ABA in whole seeds of A. thaliana (Finkelstein and Lynch, 2000; Garciarrubio et al., 1997) and sugar can overcome dormancy promoted by osmotic stress in wheat (Hu et al., 2012). More recently, Du et al. (2018) described a mutation in PHS8 in Rice affecting Starch Debranching Enzyme ISA1. Inactivation of ISA1 led to high concentrations of soluble sugars (mainly sucrose and glucose) suppressing the expression of OsABI3 and OsABI5 (two transcription factors mediating the effect of ABA) and reducing sensitivity to ABA. The elevated sugars provided by the activity of TaAMY2 in UA2OE endosperm seems to have produced the same response.
What is the role of α‐amylase in starch degradation during germination?
The analysis of the soluble sugar accumulation profiles during germination in the presence and absence of acarbose aimed to understand whether the accumulation of soluble sugar during development was sufficient to trigger germination. The analysis of the acarbose effect on α‐amylase activity and total soluble sugar on both NC and UA2OE has yielded many results allowing us also to speculate on the α‐amylase role during germination and TaAMY2 modus operandi. TaAMY2 was shown to be expressed after TaAMY1 at the later stage of grain germination (Mieog et al., 2017). As shown in the NC without acarbose, total amylase activity increases significantly from 3 DPI when the enzymatic arsenal required from starch degradation has been released, including endogenous α‐amylase.
From imbibition to 4 DPI when overexpressed TaAMY2 is the only active α‐amylase, the total sugar accumulation profile was markedly different for the UA2OE lines compared with the NC. The elevated amount of soluble sugars generated before grain maturation by the excess of TaAMY2 were slowly metabolized. These differences were driven mainly by the α‐gluco‐oligosaccharides and sucrose. While not being the by‐product of starch degradation, the presence of elevated sucrose could be a by‐product of the increased glucose and fructose content (Whan et al., 2014). Edelman et al. (1959) found that glucose in the scutellum of barley grains is absorbed from the endosperm and converted into sucrose, which is subsequently transported to the seedling. Similar production could be at play in response to the elevated concentration of sucrose raw material.
Irrespective of the presence of an overexpressed TaAMY2, the effect of acarbose on the starch degradation mechanism is noteworthy. When acarbose was added, the entire set of analysed soluble sugars decreased in both UA2OE lines and NC apart from the α‐gluco‐oligosaccharide content. The α‐gluco‐oligosaccharide in UA2OE lines maintain the same profile but with a higher general level while in NC, the increase was markedly higher, particularly from 5 DPI. This accumulation of α‐gluco‐oligosaccharide coupled to a decrease in glucose, fructose, and sucrose content when acarbose was added suggests a strong involvement of α‐amylase in general to the conversion of α‐gluco‐oligosaccharides for subsequent breakdown to disaccharides. It has been demonstrated that TaAMY2 like the barley HvAMY1 presents an additional sugar‐binding domain (SBS) or sugar tong (Robert et al., 2003). It has been hypothesized that this extra SBS makes these α‐amylases more prone to degrade highly branched polysaccharides and particularly glucosyl residues near branched points (Cockburn et al., 2015). As TaAMY2 expression follows TaAMY1 occurrence, it is plausible that the role of TaAMY2 would be to complement the starch degradation partially initiated by TaAMY1 and specifically targeting highly branched oligosaccharides. Comparative studies on the impact of overexpressing TaAMY1 or TaAMY2 on sugar accumulation profiles during germination would provide interesting information on the respective role of TaAMY1 and TaAMY2 during starch degradation in cereals.
According to Lloyd and Kötting (2016), the starch degradation pathway to glucose proceeds via concerted and sequential actions of the following enzymes: α‐amylase, Limit dextrinase, β‐amylase, and Maltase producing branched glucans and linear α‐gluco‐oligosaccharides, linear glucans, maltose and glucose respectively. However, the accumulation of α‐gluco‐oligosaccharides when acarbose is used would suggest a more complex role of α‐amylase in this degradation cascade. New perspectives on the role of starch‐degrading enzymes during grain germination are likely to emerge from more targeted investigations.
EXPERIMENTAL PROCEDURES
Vector construction and wheat transformation
A wheat TaAmy2 was amplified from cDNA prepared from developing grain at 5 DPA from the variety Chara. The ORF of TaAmy2 (nt 1–1314) with strong similarity with GenBank accession no. KY368736 (Figure S1a). Sequence comparison using the Wheat EnsemblPlant search engine showed cDNA translated protein sequence displayed a 99% identity with Chinese Spring TaAMY2 TraesCS7D02G380400 located on chromosome 7D. TaAmy2 cassette was cloned into pUbiIRcasNOT (Figure S1b) in sense orientation using a gateway system and under control of the Z. mays ubiquitin promoter (Christensen and Quail, 1996) according to the methods described in Whan et al. (2014). Transformation was carried out on immature embryos using the method of Richardson et al. (2014).
Plant growth, rearing, and sampling
All plants were grown in glasshouses at CSIRO Agriculture and Food, Canberra, Australia Capital Territory, Australia, under natural light with no additional artificial lighting on a diurnal temperature cycle of 14/20°C. Different generations of GM plants and their NCs were grown in glasshouses at CSIRO Black Mountain science and Innovation Park, Canberra, ACT, Australia as described in Ral et al. (2012). Plants were watered automatically at a rate equivalent of 10 mm of water every 3 days. For molecular analysis and vegetative characterization, biological tissue samples were collected from three individual plants for each event grown simultaneously in the same glasshouse.
Developing grains samples (dissected where appropriate) and other tissue samples were snap‐frozen in liquid nitrogen and stored at −80°C until time of analysis.
For grain compositional analysis, grains from several T4 plants for each independent event and NC were grown simultaneously in the same glasshouse. Stability of the copy number were assessed by PCR and grains were harvested, bulked, and processed for analysis.
Grain milling and starch extraction
Wheat grain from the UA2OE lines and their negative segregant NCs, were milled into wholemeal flour using an UDY Cyclone Sample Mill (UDY Corporation, Fort Collins, Co., USA) fitted with a 0.5‐mm screen without any previous moisture conditioning. For each line, the grains from two to three individuals were combined and used as a biological replicate for deeper analysis.
Starch isolation followed the method of Regina et al. (2004) with some modifications. Starch slurry was filtered through a 200‐µm sieve first and a 100‐µm sieve. Tailings were removed with 90% Percoll (v/v) before the final water washes, centrifugation, and freeze drying.
Thousand grain weights
One thousand grains were determined in triplicate for each line using a Contador Seed Counter (Graintec Scientific, Queensland, Australia) then weighed.
Construct copy number
For genomic DNA extraction, vegetative plant material was obtained from multiple generations of plants and construct copy numbers were determined as described in Mieog et al. (2013) with some modifications. The NOS terminator was used as the PCR selection marker and wheat Epsilon Cyclase (EC) from genome A (EC A) was used as the reference gene (Table S1). The real‐time PCR reactions were run in PikoReal96 PCR (Thermo, Finland) and included 10 μl Sensimix SYBR green with fluorescein, 5 μl primer mix and 5 μl DNA template (5–10 ng/μl).
Real‐time quantitative PCR
For gene transcript levels, developing grains were obtained from T4 plants and put into liquid nitrogen rapidly then store at −80°C. RNA was isolated using the NucleSpin RNA kit (Scientifix, Clayton, Australia) and RNA was purified following the manufacturer’s protocol. Quantitative reverse transcription‐PCR was performed following the protocol described in Mieog et al. (2017). For normalization, three reference genes, TaActin, Ta.14126.1.S1_at, and Ta.7894.3.A1_at, were used as internal control genes (Ji et al., 2011; Long et al., 2010) and the level of target gene expression was calculated using the E▵▵ C t method (Michael, 2001) against the geometric mean of three internal reference genes. The primers of quantitative reverse transcription‐PCR were listed in Table S1. For expression of TaAmy2, primers were designed to be specific for the TaAmy2 sequence of the construct.
Protein extraction and digestion
Triplicate biological samples taken from wholemeal flour for α‐amylase TaAMY2 overexpression positive lines (UA2OE; A1, A2, A3) and negative segregant control lines (UA2NS; A4N, A5N, A6N) were prepared and digested as described in Whan et al. (2014) (Table S5).
Liquid chromatography–tandem mass spectometry analysis and development of multiple reaction monitoring assay for α‐amylase (TaAMY2)
The digested peptides (4 μl) from the positive overexpression lines (UA2OE; A1, A2, A3) were chromatographically separated following the method described in Colgrave et al. (2014), Colgrave et al. (2017), Whan et al. (2014), and MacLean et al. (2010). The detailed method is described in Methods S1.
Iodometric estimation of amylose content
Amylose content was measured following small‐scale iodine adsorption method on flour as described in Regina et al. (2004) with a SPECTROstar Nano Microplate Reader (BMG LABTECH, Mornington, Australia) at 620 nm. Standard samples containing amylose ranging from 0% to 100% were used to generate a standard curve. The absorbance of the test samples was converted to percentage amylose using a regression equation derived from the standard samples. All positive and negative lines of UA2OE were analysed in triplicate.
α‐Amylase assay
α‐Amylase activity was determined for 10 mg wholemeal or leaves samples and/or two to eight ground developing grain samples. The 96‐well plate adapted cerealpha kit (Megazyme International Ireland, Bray Business Park, Bray, Co. Wicklow, Ireland) was used according to Newberry et al. (2018). Results displayed are the mean of three independent assays of three biological replicates.
Carbohydrate measurement
Freeze‐dried samples from either dried, developing grains, or leaf tissues were ground using a hammer mill (ESPE GmbH & Co. KG, Seefeld, Germany). Triplicate 10 mg aliquots were extracted three times in 400 μl boiling 80% (v/v) ethanol each.
Total soluble sugar was measured using 2% anthrone in 70% sulphuric acid (Whan et al., 2014) and compared with a standard curve established using the anthrone solution with a glucose gradient prepared with a 1 mg·ml−1 glucose stock solution. For the test samples, 10 μl of sugar extract was added to 1.5 ml screw‐capped tubes with 0.7 ml anthrone reagent and boiled for 10 min. Then, 200‐μl aliquots were transferred in triplicate to a flat‐bottomed 96‐well plate and absorbance measured at 630 nm with a SPECTROstar Nano Microplate Reader.
Sucrose was measured according to Birnberg and Brenner (1984) and adapted to a microplate assay. Assay buffer (PH 7) contained 0.01 m of KH2PO4, 0.01 m of K2HPO4, 1 mm of MgSO4, and 0.5 mm NADP. To 180 μl of assay buffer, 1 U of glucose‐6‐phosphate dehydrogenase (EC 1.1.1.49; Roche, Switzerland) and 0.2 U of phosphoglucomutase (EC 5.4.2.2; Roche) were added together with 20 μl of sample extract. The reactions were initiated with 0.25 U of sucrose phosphorylase (SucP, EC 2.4.1.7; Sigma Aldrich) and incubated at 37°C to completion. Absorbance was measured at 340 nm and the sucrose content was determined with a sucrose standard curve.
Free fructose and glucose were determined in a two‐step reaction sequence with changes in absorbance monitored at 340 nm as described by Campbell et al. (1999) with some modifications. Assay buffer contained 0.1 m Tris‐HCl, 5 mm MgSO4 (pH 8.1), 1 mm ATP, and 0.5 mm NADP. To 180 µl of assay buffer, 1 U hexokinase (EC 2.7.1.1; Roche) and 2 U glucose‐6‐phosphate dehydrogenase (Roche) were added together with 10 µl of sample extract. The reactions were incubated at 37°C and free glucose determined upon completion. Then 1.5 U phosphoglucose isomerase (EC 5.3.1.9; Roche) was added and free fructose determined as for free glucose using standard curves.
Total starch and ethanol soluble α‐gluco‐oligosaccharides were measured using the Total Starch assay kit from Megazyme according to the manufacturer’s protocol. Before total starch measurements, wholemeal samples were washed twice with 80% ethanol (v/v) to remove soluble sugars. In addition, before measuring α‐gluco‐oligosaccharides, 200 μl ethanol extract was dried and then resuspended in the assay buffer.
Fructan was measured using the Megazyme Fructan HK Kit. Total fructan was extracted from wholemeal using distilled water, and was described by Verspreet et al. (2012) and in triplicate. The total fructan was measured according to the Fructan HK Procedure (Megazyme; AOAC method 999.03 and AACC Method 32.32.01).
β‐Glucan was measured using the mixed‐linkage β‐glucan assay kit (Megazyme International Ireland, Ltd.), which permits the analysis of small‐scale samples (20 mg) by its procedure, with some modification. Three technical replicates were performed on all samples.
Total arabinoxylan was measured according to Sun et al. (2020). Triplicate 20 mg wholemeal samples were extracted by incubating at 100°C for 30 min in 1 ml 0.5 m H2SO4. The PGR reagent mixture contained 0.3 g phloroglucinol, 1.2 ml absolute ethanol, 27.5 ml acetic acid, 0.55 ml concentrated hydrochloric acid, and 0.3 ml glucose (7 mg·ml−1 in stock). Then, 100 μl of appropriately diluted sample or standard was added to 0.5 ml of PGR and incubated at 100°C for 25 min. Absorbances were determined at 552 and 510 nm and used to calculate total arabinoxylan.
Starch granule size distribution
The starch granule size distribution was measured using UA2OE and NC by a Malvern Mastersizer 3000 with the Hydro MV wet sample dispersion unit (Malvern Instruments, Ltd., Malvern, Worcs, UK). The concentration of the measured particles was 0.05 mg·ml−1 within the range of the instrument’s specifications. The B granule content was taken as the proportion of granules with a diameter of <10.1 μm.
Amylopectin chain length distribution
Sample preparation was adapted from O’Shea et al. (1998). Amylopectin chain‐length distribution was analysed by capillary electrophoresis using an Agilent 7100 CE with ZETALIF 488nm Laser Induced Fluorescence Detector and Agilent OpenLAB CDS ChemStation software (Agilent Technologies, Mulgrave, Australia Pty Ltd.).
Differential scanning calorimetry
The differential scanning calorimetry analysis of starch from the UA2OE positive and control lines was carried out using a Perkin DSC 8000 (PerkinElmer, Pty Ltd, Melbourne, Australia). Starch and water were premixed at ratio of 1:2 and approximately 50 mg weighed into a DSC pan, which was sealed and left to equilibrate overnight. This analysis followed the protocol described in Ral et al. (2008). The displayed results are the mean of three independent assays.
Pasting properties
Starch pasting properties were analysed using RVA 4500 (Perten Instruments Australia Pty Ltd, Sydney, NSW, Australia). A 9% starch suspension was equilibrated at 50°C for 2 min, heated to 95°C for 6 min, maintained at temperature for 4 min, cooled to 50°C for 4 min, and finally maintained at 50°C for 5 min. A constant rotating paddle speed (160 rpm) was used throughout the analysis.
Damaged starch
The damaged starch percentage was determined on 10‐mg wholemeal samples. The Starch Damage Assay Kit (Megazyme) was used, which followed a 96‐well plate format that adapted the manufacturer’s protocol, and with appropriate dilutions. The displayed results are the mean of three technical replicates for all UA2OE and negative lines.
Protein content
The total protein of the wholemeal flour was determined by the Dumas combustion method using DuMaster Buchi D‐480 (Switzerland). Two milligrams of wholemeal were used per assay and three technical replicates were performed for three UA2OE lines and NC. The factor of 5.8 was used to convert nitrogen amounts into crude protein content.
Microscopy
Purified starch granules were stained with iodine and imaged under bright field (BF) and polarized light using a Leica DM6B light microscope (Leica Microsystems, Wetzlar, Germany) with Leica DMC 4500 digital camera and LASX software.
The developing grains (15, 20, 25, and 30 DPA) were fixed in a modified FAA solution (Liu et al., 1993) for 24 h. The FAA solution contained 50% ethanol, 5% formaldehyde, 6% acetic acid, and 5% glycerol. Fixed samples were washed in 70% (v/v) ethanol for 2 h. The grains were cut into 1‐mm slices using a razor blade and freeze‐dried. Purified starch granules and grain slices were sputter coated with gold using an EMITECH 5500X and visualized in a Zeiss EVO LS 15 extended pressure SEM at 20 kV. Images were processed using Photoshop CS6.
Germination assay
Germination assays were performed as described by Jacobsen et al. (2013). Grains (T5 generation) from T4 plants were harvested at physiological maturity and 4 weeks after physiological maturity from the mother plant. The protocol for harvest followed Gubler et al. (2008) except for details outlined as follows. Harvested grain was dried at 37°C for 24 h with low humidity and were then stored at −20°C. Grains were imbibed on 9‐cm plastic Petri dishes containing 0.3% agarose (w/v) and MilliQ water. Plates were incubated at 20°C in dark. CE was counted every 24 h over the time of the experiment.
ABA (species ‘(±)’; Sigma‐Aldrich; CAS. no. 14375‐45‐2) was prepared by dissolving 7.93 mg of dry powder in 260 µl of absolute ethanol followed by dilution with sterilized MilliQ water to a 2‐mm stock solution concentration. ABA resistance was tested by addition of ABA (0, 20, 50, 100, and 150 μm) from a 2‐mm stock solution to sterilized 0.3% agarose (w/v and adjusted at pH 6) according to Kondhare et al. (2012). Acarbose (Sigma‐Aldrich; CAS. no. 56180‐94‐0) (Oudjeriouat et al., 2003) was added to sterilized 0.3% agarose to give the final desired concentration of acarbose (0 μm, 200 μm, 500 μm, 1 mm, and 1.5 mm) from a 2 mmol·ml−1 stock solution. The acarbose concentration of 1.5 mm was the minimum concentration to inhibit α‐amylase completely in wheat grains. The germination experiment included either three replicates of 20 whole grains of UA2OE and NC per 9 cm Petri dishes and placed crease down or 20 halved grains placed cut side down in 9 cm Petri dishes with 0.3% agarose containing acarbose and/or of ABA.
Statistical analysis
Statistical analysis was carried out with IBM SPSS Statistics (IBM Corp., Armonk, NY, USA). The ANOVA test was performed on all data to indicate significant differences. Figures were drafted by OriginPro 9.1 and GraphPad Prism 8.0.
CONFLICT OF INTERESTS
The authors declare that they have no competing interests.
Supporting information
Figure S1. Schematic representation of the pAMY2OEUBI construct for wheat transformation.
Figure S2. Relative quantitation and comparison of TaAMY2 peptides using mass spectrometry.
Figure S3. TaAMY2 sequence with mapped tryptic peptides (bold, underlined).
Figure S4. Correlation between copy number (T3 plants) and total α‐amylase activity (T4 grains, Ceralpha Units·gram−1 wholemeal).
Figure S5. Expression level of TaAmy1 (a) and TaAmy3 (b) in developing grains.
Figure S6. α‐Amylase activity, total starch and total soluble carbohydrate in vegetative stage leaf.
Figure S7. Effect of TaAMY2 overexpression on starch morphology during grain development SEM of representative grain cross sections from NC (a–d) and UA2OE3.1 (e–h) during grain development.
Figure S8. Amylose content and Amylopectin structure in three UA2OE lines and NC.
Figure S9. Rapid Visco Analyser comparison between three UA2OE lines and NC wholemeal.
Figure S10. Coleorhiza emergence (CE) comparisons of whole grains in presence (a) or absence (b) of 1.5 mm acarbose.
Table S1. Primer pairs used for copy number (qPCR) or gene expression (RT‐qPCR).
Table S2. Analysis of copy number from three independent transformation events in different generations.
Table S3. Spearman correlation coefficients between copy number, α‐amylase activity, soluble sugar and damaged starch.
Table S4. Thousand Kernel Weight (TKW) of TaAMY2 overexpression lines and NC.
Table S5. The information of α‐amylase protein extraction.
Table S6. Multiple reaction monitoring transitions of TaAMY2 peptides.
Methods S1. LC‐MS/MS analysis and development of multiple reaction monitoring (MRM) assay for α‐amylase (taAMY2).
ACKNOWLEDGEMENTS
The authors acknowledge Jose Barrero and Ross Dennis for input into experimental designs and helpful discussions; Christine Konik‐Rose, Phil Hands, Vivien Rolland, and Rosengela Devilla for their technical assistance. Financial support for the research was provided by the CSIRO Research Office and the Chinese Scholarship Council mobility program.
References
- Ainsworth, C.C. , Doherty, P. , Edwards, K.G.K. , Martienssen, R.A. & Gale, M.D. (1985) Allelic variation at α‐amylase loci in hexaploid wheat. Theoretical and Applied Genetics, 70, 400–406. [DOI] [PubMed] [Google Scholar]
- Andersen, E.J. , Ali, S. , Byamukama, E. , Yen, Y. & Nepal, M.P. (2018) Disease resistance mechanisms in plants. Genes (Basel), 9, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero, J.M. , Mrva, K. , Talbot, M.J. , White, R.G. , Taylor, J. , Gubler, F. et al. (2013) Genetic, hormonal, and physiological analysis of late maturity alpha‐amylase in wheat. Plant Physiology, 161, 1265–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D.C. , Huttly, A.K. , Martienssen, R.A. , Barker, R.F. & Jarvis, M.G. (1987) A novel wheat alpha‐amylase gene (alpha‐Amy3). Molecular and General Genetics, 209, 33–40. [DOI] [PubMed] [Google Scholar]
- Bewley, J.D. (1997) Seed germination and dormancy. The Plant Cell, 9, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya, M. & Corke, H. (1996) Selection of desirable starch pasting properties in wheat for use in white salted or yellow alkaline noodles. Cereal Chemistry, 73, 721–728. [Google Scholar]
- Biddulph, T.B. , Plummer, J.A. , Setter, T.L. & Mares, D.J. (2008) Seasonal conditions influence dormancy and preharvest sprouting tolerance of wheat (Triticum aestivum L.) in the field. Field Crops Research, 107, 116–128. [Google Scholar]
- Birnberg, P.R. & Brenner, M.L. (1984) A one‐step enzymatic assay for sucrose with sucrose phosphorylase. Analytical Biochemistry, 142, 556–561. [DOI] [PubMed] [Google Scholar]
- Bykova, N.V. , Hoehn, B. , Rampitsch, C. , Banks, T. , Stebbing, J.A. , Fan, T. et al. (2011) Redox‐sensitive proteome and antioxidant strategies in wheat seed dormancy control. Proteomics, 11, 865–882. [DOI] [PubMed] [Google Scholar]
- Campbell, J.A. , Hansen, R.W. & Wilson, J.R. (1999) Cost‐effective colorimetric microtiter plate enzymatic assays for sucrose, glucose and fructose in sugarcane tissue extracts. Journal of the Science of Food and Agriculture, 79, 232–236. [Google Scholar]
- Chen, S.Y. , Chien, C.T. , Baskin, J.M. & Baskin, C.C. (2010) Storage behavior and changes in concentrations of abscisic acid and gibberellins during dormancy break and germination in seeds of Phellodendron amurense var. wilsonii (Rutaceae). Tree Physiology, 30, 275–284. [DOI] [PubMed] [Google Scholar]
- Christensen, A.H. & Quail, P.H. (1996) Ubiquitin promoter‐based vectors for high‐level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Research, 5, 213–218. [DOI] [PubMed] [Google Scholar]
- Cockburn, D. , Nielsen, M.M. , Christiansen, C. , Andersen, J.M. , Rannes, J.B. , Blennow, A. et al. (2015) Surface binding sites in amylase have distinct roles in recognition of starch structure motifs and degradation. International Journal of Biological Macromolecules, 75, 338–345. [DOI] [PubMed] [Google Scholar]
- Colgrave, M.L. , Byrne, K. & Howitt, C.A. (2017) Liquid chromatography‐mass spectrometry analysis reveals hydrolyzed gluten in beers crafted to remove gluten. Journal of Agriculture and Food Chemistry, 65, 9715–9725. [DOI] [PubMed] [Google Scholar]
- Colgrave, M.L. , Goswami, H. , Blundell, M. , Howitt, C.A. & Tanner, G.J. (2014) Using mass spectrometry to detect hydrolysed gluten in beer that is responsible for false negatives by ELISA. Journal of Chromatography A, 1370, 105–114. [DOI] [PubMed] [Google Scholar]
- Damaris, R.N. , Lin, Z. , Yang, P. & He, D. (2019) The rice alpha‐amylase, conserved regulator of seed maturation and germination. International Journal of Molecular Sciences, 20, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laethauwer, S. , De Riek, J. , Stals, I. , Reheul, D. & Haesaert, G. (2013) α‐Amylase gene expression during kernel development in relation to pre‐harvest sprouting in wheat and triticale. Acta Physiologiae Plantarum, 35, 2927–2938. [Google Scholar]
- Du, L. , Xu, F. , Fang, J. , Gao, S. , Tang, J. , Fang, S. et al. (2018) Endosperm sugar accumulation caused by mutation of PHS8/ISA1 leads to pre‐harvest sprouting in rice. The Plant Journal, 95, 545–556. [DOI] [PubMed] [Google Scholar]
- Edelman, J. , Shibko, S.I. & Keys, A.J. (1959) The role of the scutellum of cereal seedlings in the synthesis and transport of sucrose. Journal of Experimental Botany, 10, 178–189. [Google Scholar]
- Faltermaier, A. , Waters, D. , Becker, T. , Arendt, E. & Gastl, M. (2014) Common wheat (Triticum aestivum L.) and its use as a brewing cereal – a review. Journal of the Institute of Brewing, 120, 1–15. [Google Scholar]
- Finkelstein, R.R. & Lynch, T.J. (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. The Plant Cell, 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M.D. , Law, C.N. , Chojecki, A.J. & Kempton, R.A. (1983) Genetic control of α‐amylase production in wheat. Theoretical and Applied Genetics, 64, 309–316. [DOI] [PubMed] [Google Scholar]
- Garciarrubio, A. , Legaria, J.P. & Covarrubias, A.A. (1997) Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta, 203, 182–187. [DOI] [PubMed] [Google Scholar]
- Gubler, F. , Hughes, T. , Waterhouse, P. & Jacobsen, J. (2008) Regulation of dormancy in barley by blue light and after‐ripening: effects on abscisic acid and gibberellin metabolism. Plant Physiology, 147, 886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F. , Kalla, R. , Roberts, J.K. & Jacobsen, J.V. (1995) Gibberellin‐regulated expression of a Myb gene in barley aleurone cells: evidence for Myb transactivation of a high‐pI alpha‐amylase gene promoter. The Plant Cell, 7, 1879–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F. , Millar, A.A. & Jacobsen, J.V. (2005) Dormancy release, ABA and pre‐harvest sprouting. Current Opinion in Plant Biology, 8, 183–187. [DOI] [PubMed] [Google Scholar]
- Guzmán‐Ortiz, F.A. , Castro‐Rosas, J. , Gómez‐Aldapa, C.A. , Mora‐Escobedo, R. , Rojas‐León, A. , Rodríguez‐Marín, M.L. et al. (2018) Enzyme activity during germination of different cereals: a review. Food Reviews International, 35, 177–200. [Google Scholar]
- Higgins, T.J.V. , Zwar, J.A. & Jacobsen, J.V. (1976) Gibberellic acid enhances the level of translatable mRNA for α‐amylase in barley aleurone layers. Nature, 260, 166–169. [Google Scholar]
- Ho, D.T.H. & Varner, J.E. (1975) Mode of action of abscisic‐acid in aleurone layers. Plant Physiology, 56, 44.16659255 [Google Scholar]
- Hu, M. , Shi, Z. , Zhang, Z. , Zhang, Y. & Li, H. (2012) Effects of exogenous glucose on seed germination and antioxidant capacity in wheat seedlings under salt stress. Plant Growth Regulation, 68, 177–188. [Google Scholar]
- Jacobsen, J.V. (1973) Interactions between gibberellic acid, ethylene, and abscisic acid in control of amylase synthesis in barley aleurone layers. Plant Physiology, 51, 198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, J.V. , Barrero, J.M. , Hughes, T. , Julkowska, M. , Taylor, J.M. , Xu, Q. et al. (2013) Roles for blue light, jasmonate and nitric oxide in the regulation of dormancy and germination in wheat grain (Triticum aestivum L.). Planta, 238, 121–138. [DOI] [PubMed] [Google Scholar]
- Janecek, S. , Svensson, B. & MacGregor, E.A. (2013) Alpha‐amylase – an enzyme present in various sequence‐based glycoside hydrolase families. FEBS Journal, 280, 546. [Google Scholar]
- Janecek, S. , Svensson, B. & MacGregor, E.A. (2014) Alpha‐amylase: an enzyme specificity found in various families of glycoside hydrolases. Cellular and Molecular Life Sciences, 71, 1149–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, X. , Dong, B. , Shiran, B. , Talbot, M.J. , Edlington, J.E. , Hughes, T. et al. (2011) Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiology, 156, 647–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, R. , Martin, J.M. , Vetch, J.M. , Byker‐Shanks, C. , Finnie, S. & Giroux, M.J. (2019) Controlled sprouting in wheat increases quality and consumer acceptability of whole‐wheat bread. Cereal Chemistry, 96, 866–877. [Google Scholar]
- Ju, L. , Deng, G. , Liang, J. , Zhang, H. , Li, Q. , Pan, Z. et al. (2019) Structural organization and functional divergence of high isoelectric point alpha‐amylase genes in bread wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.). BMC Genetics, 20, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer, E.E. , Litts, J.C. & Rodriguez, R.L. (1991) Differential expression of α‐amylase genes in germinating rice and barley seeds. Plant Molecular Biology, 16, 797–805. [DOI] [PubMed] [Google Scholar]
- Kondhare, K.R. , Farrell, A.D. , Kettlewell, P.S. , Hedden, P. & Monaghan, J.M. (2015) Pre‐maturity α‐amylase in wheat: the role of abscisic acid and gibberellins. Journal of Cereal Science, 63, 95–108. [Google Scholar]
- Kondhare, K.R. , Kettlewell, P.S. , Farrell, A.D. , Hedden, P. & Monaghan, J.M. (2012) Effects of exogenous abscisic acid and gibberellic acid on pre‐maturity α‐amylase formation in wheat grains. Euphytica, 188, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi, Y. , Okamoto, A. , Takahashi, J. , Aitani, M. & Nakatani, N. (1994) Effects of bay m 1099, an alpha‐glucosidase inhibitor, on starch metabolism in germinating wheat seeds. Bioscience, Biotechnology, and Biochemistry, 58, 135–139. [DOI] [PubMed] [Google Scholar]
- Kozo, N. , YoshihikoF, U. & Satoru, K. (1988) Genetic studies of α‐amylase isozymes in wheat. VI. Variation and differentiation in tetraploid wheat. Japanese Journal of Genetics, 63, 425–434. [Google Scholar]
- Li, C.Y. , Li, C. , Lu, Z.X. , Li, W.H. & Cao, L.P. (2012) Morphological changes of starch granules during grain filling and seed germination in wheat. Starch – Stärke, 64, 166–170. [Google Scholar]
- Lin, R. , Horsley, R.D. & Schwarz, P.B. (2008) Associations between caryopsis dormancy, α‐amylase activity, and pre‐harvest sprouting in barley. Journal of Cereal Science, 48, 446–456. [Google Scholar]
- Liu, C.M. , Xu, Z.H. & Chua, N.H. (1993) Auxin polar transport is essential for the establishment of bilateral symmetry during early plant embryogenesis. The Plant Cell, 5, 621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, J.R. , Kossmann, J. & Ritte, G. (2005) Leaf starch degradation comes out of the shadows. Trends in Plant Science, 10, 130–137. [DOI] [PubMed] [Google Scholar]
- Lloyd, J.R. & Kötting, O. (2016) Starch biosynthesis and degradation in plants. In: eLS. Chichester: John Wiley & Sons, Ltd., pp. 1–10. [Google Scholar]
- Long, X.Y. , Wang, J.R. , Ouellet, T. , Rocheleau, H. , Wei, Y.M. , Pu, Z.E. et al. (2010) Genome‐wide identification and evaluation of novel internal control genes for Q‐PCR based transcript normalization in wheat. Plant Molecular Biology, 74, 307–311. [DOI] [PubMed] [Google Scholar]
- MacLean, B. , Tomazela, D.M. , Shulman, N. , Chambers, M. , Finney, G.L. , Frewen, B. et al. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics, 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeill, G.J. , Mehrpouyan, S. , Minow, M.A.A. , Patterson, J.A. , Tetlow, I.J. & Emes, M.J. (2017) Starch as a source, starch as a sink: the bifunctional role of starch in carbon allocation. Journal of Experimental Botany, 68, 4433–4453. [DOI] [PubMed] [Google Scholar]
- Majzlova, K. , Pukajova, Z. & Janecek, S. (2013) Tracing the evolution of the alpha‐amylase subfamily GH13_36 covering the amylolytic enzymes intermediate between oligo‐1,6‐glucosidases and neopullulanases. Carbohydrate Research, 367, 48–57. [DOI] [PubMed] [Google Scholar]
- Mares, D.J. & Mrva, K. (2014) Wheat grain preharvest sprouting and late maturity alpha‐amylase. Planta, 240, 1176–1178. [DOI] [PubMed] [Google Scholar]
- Michael, W.P. (2001) A new mathematical model for relative quantification in real‐time RT–PCR. Nucleic Acids Research, 29, 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieog, J.C. , Howitt, C.A. & Ral, J.P. (2013) Fast‐tracking development of homozygous transgenic cereal lines using a simple and highly flexible real‐time PCR assay. BMC Plant Biology, 13, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieog, J.C. , Janeček, Š. & Ral, J.‐P. (2017) New insight in cereal starch degradation: identification and structural characterization of four α‐amylases in bread wheat. Amylase, 1, 35–49. [Google Scholar]
- Nanjo, Y. , Asatsuma, S. , Itoh, K. , Hori, H. & Mitsui, T. (2004) Proteomic identification of alpha‐amylase isoforms encoded by RAmy3B/3C from germinating rice seeds. Bioscience, Biotechnology, and Biochemistry, 68, 112–118. [DOI] [PubMed] [Google Scholar]
- Nee, G. , Xiang, Y. & Soppe, W.J. (2017) The release of dormancy, a wake‐up call for seeds to germinate. Current Opinion in Plant Biology, 35, 8–14. [DOI] [PubMed] [Google Scholar]
- Newberry, M. , Zwart, A.B. , Whan, A. , Mieog, J.C. , Sun, M. , Leyne, E. et al. (2018) Does late maturity alpha‐amylase impact wheat baking quality? Frontiers in Plant Science, 9, 1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa, B.K. , Furuta, Y. , Hina, Y. & Yamada, T. (1981) Genetic studies of α‐amylase isozymes in wheat. IV. Genetic analyses in hexaploid wheat. Japanese Journal of Genetics, 56, 345–385. [Google Scholar]
- O’Shea, M.G. , Samuel, M.S. , Konik, C.M. & Morell, M.K. (1998) Fluorophore‐assisted carbohydrate electrophoresis (FACE) of oligosaccharides: efficiency of labelling and high‐resolution separation. Carbohydrate Research, 307, 1–12. [Google Scholar]
- Oudjeriouat, N. , Moreau, Y. , Santimone, M. , Svensson, B. , Marchis‐Mouren, G. & Desseaux, V. (2003) On the mechanism of α‐amylase: acarbose and cyclodextrin inhibition of barley amylase isozymes. European Journal of Biochemistry, 270, 3871–3879. [DOI] [PubMed] [Google Scholar]
- Pepler, S. , Gooding, M.J. & Ellis, R.H. (2006) Modelling simultaneously water content and dry matter dynamics of wheat grains. Field Crops Research, 95, 49–63. [Google Scholar]
- Radchuk, V.V. , Borisjuk, L. , Sreenivasulu, N. , Merx, K. , Mock, H.P. , Rolletschek, H. et al. (2009) Spatiotemporal profiling of starch biosynthesis and degradation in the developing barley grain. Plant Physiology, 150, 190–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ral, J.P. , Bowerman, A.F. , Li, Z. , Sirault, X. , Furbank, R. , Pritchard, J.R. et al. (2012) Down‐regulation of Glucan, Water‐Dikinase activity in wheat endosperm increases vegetative biomass and yield. Plant Biotechnology Journal, 10, 871–882. [DOI] [PubMed] [Google Scholar]
- Ral, J.P. , Cavanagh, C.R. , Larroque, O. , Regina, A. & Morell, M.K. (2008) Structural and molecular basis of starch viscosity in hexaploid wheat. Journal of Agricultural and Food Chemistry, 56, 4188–4197. [DOI] [PubMed] [Google Scholar]
- Ral, J.‐P.‐F. , Sun, M. , Mathy, A. , Pritchard, J.R. , Konik‐Rose, C. , Larroque, O. & et al. (2018) A biotechnological approach to directly assess the impact of elevated endogenous α‐amylase on Asian white‐salted noodle quality. Starch – Stärke, 70, 1700089. [Google Scholar]
- Ral, J.P. , Whan, A. , Larroque, O. , Leyne, E. , Pritchard, J. , Dielen, A.S. et al. (2016) Engineering high alpha‐amylase levels in wheat grain lowers Falling Number but improves baking properties. Plant Biotechnology Journal, 14, 364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina, A. , Kosar‐Hashemi, B. , Li, Z. , Rampling, L. , Cmiel, M. , Gianibelli, M.C. et al. (2004) Multiple isoforms of starch branching enzyme‐I in wheat: lack of the major SBE‐I isoform does not alter starch phenotype. Functional Plant Biology, 31, 591. [DOI] [PubMed] [Google Scholar]
- Richardson, T. , Thistleton, J. , Higgins, T.J. , Howitt, C. & Ayliffe, M. (2014) Efficient Agrobacterium transformation of elite wheat germplasm without selection. Plant Cell, Tissue and Organ Culture, 119, 647–659. [Google Scholar]
- Robert, X. , Haser, R. , Gottschalk, T.E. , Ratajczak, F. , Driguez, H. , Svensson, B. et al. (2003) The structure of barley α‐amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding. Structure, 11, 973–984. [DOI] [PubMed] [Google Scholar]
- Rodríguez, M.V. , Barrero, J.M. , Corbineau, F. , Gubler, F. & Benech‐Arnold, R.L. (2015) Dormancy in cereals (not too much, not so little): about the mechanisms behind this trait. Seed Science Research, 25, 99–119. [Google Scholar]
- Rogers, J.C. (1985) Two barley α‐amylase gene families are regulated differently in aleurone cells. Journal of Biological Chemistry, 260, 3731–3738. [PubMed] [Google Scholar]
- Smith, S.J. & Hooley, R. (2002) An increase in the speed of response, and sensitivity, of Avena fatua aleurone layers and protoplasts to gibberellin. Journal of Plant Physiology, 159, 355–360. [Google Scholar]
- Streb, S. , Eicke, S. & Zeeman, S.C. (2012) The simultaneous abolition of three starch hydrolases blocks transient starch breakdown in Arabidopsis. Journal of Biological Chemistry, 287, 41745–41756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J. , Xu, F. & Lu, J. (2020) A glycoside hydrolase family 62 A‐L‐arabinofuranosidase from Trichoderma reesei and its applicable potential during mashing. Foods, 9, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranier, S. , Deville, K. , Robert, X. , Bozonnet, S. , Haser, R. , Svensson, B. et al. (2005) Insights into the ‘pair of sugar tongs’ surface binding site in barley α‐amylase isozymes and crystallization of appropriate sugar tongs mutants. Biologia, 60, 37–46. [Google Scholar]
- Verspreet, J. , Pollet, A. , Cuyvers, S. , Vergauwen, R. , Van den Ende, W. , Delcour, J.A. et al. (2012) A simple and accurate method for determining wheat grain fructan content and average degree of polymerization. Journal of Agriculture and Food Chemistry, 60, 2102–2107. [DOI] [PubMed] [Google Scholar]
- Whan, A. , Dielen, A.S. , Mieog, J. , Bowerman, A.F. , Robinson, H.M. , Byrne, K. et al. (2014) Engineering alpha‐amylase levels in wheat grain suggests a highly sophisticated level of carbohydrate regulation during development. Journal of Experimental Botany, 65, 5443–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Liu, Y. , Pu, Z. , Zhang, L. , Yuan, Z. , Chen, G. et al. (2014) Molecular characterization of high pI α‐amylase and its expression QTL analysis in synthetic wheat RILs. Molecular Breeding, 34, 1075–1085. [Google Scholar]
- Yu, T.S. , Zeeman, S.C. , Thorneycroft, D. , Fulton, D.C. , Dunstan, H. , Lue, W.L. et al. (2005) Alpha‐amylase is not required for breakdown of transitory starch in Arabidopsis leaves. Journal of Biological Chemistry, 280, 9773–9779. [DOI] [PubMed] [Google Scholar]
- Zeevaart, J.A.D. & Creelman, R.A. (1988) Metabolism and physiology of abscisic acid. Annual Review of Plant Physiology and Plant Molecular Biology, 39, 439–473. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Schematic representation of the pAMY2OEUBI construct for wheat transformation.
Figure S2. Relative quantitation and comparison of TaAMY2 peptides using mass spectrometry.
Figure S3. TaAMY2 sequence with mapped tryptic peptides (bold, underlined).
Figure S4. Correlation between copy number (T3 plants) and total α‐amylase activity (T4 grains, Ceralpha Units·gram−1 wholemeal).
Figure S5. Expression level of TaAmy1 (a) and TaAmy3 (b) in developing grains.
Figure S6. α‐Amylase activity, total starch and total soluble carbohydrate in vegetative stage leaf.
Figure S7. Effect of TaAMY2 overexpression on starch morphology during grain development SEM of representative grain cross sections from NC (a–d) and UA2OE3.1 (e–h) during grain development.
Figure S8. Amylose content and Amylopectin structure in three UA2OE lines and NC.
Figure S9. Rapid Visco Analyser comparison between three UA2OE lines and NC wholemeal.
Figure S10. Coleorhiza emergence (CE) comparisons of whole grains in presence (a) or absence (b) of 1.5 mm acarbose.
Table S1. Primer pairs used for copy number (qPCR) or gene expression (RT‐qPCR).
Table S2. Analysis of copy number from three independent transformation events in different generations.
Table S3. Spearman correlation coefficients between copy number, α‐amylase activity, soluble sugar and damaged starch.
Table S4. Thousand Kernel Weight (TKW) of TaAMY2 overexpression lines and NC.
Table S5. The information of α‐amylase protein extraction.
Table S6. Multiple reaction monitoring transitions of TaAMY2 peptides.
Methods S1. LC‐MS/MS analysis and development of multiple reaction monitoring (MRM) assay for α‐amylase (taAMY2).


