Abstract
Aims
CADASIL, the most prevalent hereditary cerebral small vessel disease, is caused by cysteine‐altering NOTCH3 variants (NOTCH3 cys ) leading to vascular NOTCH3 protein aggregation. It has recently been shown that variants located in one of NOTCH3 protein epidermal growth‐factor like repeat (EGFr) domains 1–6, are associated with a more severe phenotype than variants located in one of the EGFr domains 7–34. The underlying mechanism for this genotype–phenotype correlation is unknown. The aim of this study was to analyse whether NOTCH3 cys variant position is associated with NOTCH3 protein aggregation load.
Methods
We quantified vascular NOTCH3 aggregation in skin biopsies (n = 25) and brain tissue (n = 7) of CADASIL patients with a NOTCH3 cys EGFr 1–6 variant or a EGFr 7–34 variant, using NOTCH3 immunohistochemistry (NOTCH3 score) and ultrastructural analysis of granular osmiophilic material (GOM count). Disease severity was assessed by neuroimaging (lacune count and white matter hyperintensity volume) and disability (modified Rankin scale).
Results
Patients with NOTCH3 cys EGFr 7–34 variants had lower NOTCH3 scores (P = 1.3·10−5) and lower GOM counts (P = 8.2·10−5) than patients with NOTCH3 cys EGFr 1–6 variants in skin vessels. A similar trend was observed in brain vasculature. In the EGFr 7–34 group, NOTCH3 aggregation levels were associated with lacune count (P = 0.03) and white matter hyperintensity volume (P = 0.02), but not with disability.
Conclusions
CADASIL patients with an EGFr 7–34 variant have significantly less vascular NOTCH3 aggregation than patients with an EGFr 1–6 variant. This may be one of the factors underlying the difference in disease severity between NOTCH3 cys EGFr 7–34 and EGFr 1–6 variants.
Keywords: CADASIL, GOM deposit, NOTCH3 aggregation, NOTCH3 score, NOTCH3 variant position
The position of the pathogenic NOTCH3 variant in CADASIL patients is correlated with mutant NOTCH3 protein aggregation levels within the vessel wall in skin. The fact that NOTCH3 EGFr 7–34 variants aggregate less may explain the milder CADASIL phenotype associated with these EGFr 7–34 variants.
Abbreviations
- CADASIL
cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy
- EGFr
epidermal growth factor‐like repeat
- EM
electron microscopy
- GOM
granular osmiophilic material
- IQR
interquartile range
- NOTCH3 cys
NOTCH3 cysteine‐altering missense variant
- NOTCH3ECD
NOTCH3 protein's ectodomain
INTRODUCTION
Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) is a hereditary small vessel disease caused by NOTCH3 variants. 1 Pathogenic NOTCH3 variants in CADASIL are almost exclusively missense variants which lead to a cysteine amino acid change in one of the 34 epidermal growth factor‐like repeat (EGFr) domains of the NOTCH3 protein's ectodomain (NOTCH3ECD) 2 and are associated with increased NOTCH3ECD multimerization and aggregation. 3 , 4 Vascular pathological hallmarks of CADASIL are granular NOTCH3ECD staining of the vessel wall and the presence of granular osmiophilic material (GOM) deposits on electron microscopy (EM). 3 This vessel wall pathology is not only present in brain vasculature, but is also observed in other tissues, including skin. 5 , 6 , 7 , 8
Recently, we showed that the position of the NOTCH3 cysteine‐altering missense variant (NOTCH3 cys ) is an important modifier of disease severity: NOTCH3 cys variants in EGFr domains 7–34 are associated with a later onset of stroke, higher survival and a lower white matter hyperintensity (WMH) MRI lesion load than NOTCH3 cys variants in EGFr domains 1–6. 9 , 10 , 11 , 12 The molecular mechanisms underlying the NOTCH3 EGFr variant position effect are unknown.
As vascular NOTCH3ECD aggregation is widely held to be one of the key drivers of CADASIL pathogenesis, we hypothesised that EGFr 7–34 variants have a lower aggregation propensity than EGFr 1–6 variants. Here, we compared NOTCH3ECD aggregation between patients with NOTCH3 cys EGFr 7–34 variants and those with NOTCH3 cys EGFr 1–6 variants, by performing quantitative analysis of NOTCH3ECD staining and GOM deposits in skin biopsies. Also, we assessed end‐stage CADASIL brain vessels in patients with a NOTCH3 cys EGFr 7–34 variant and in patients with an EGFr 1–6 variant.
MATERIALS AND METHODS
Patient selection
Patients were selected from a prospective CADASIL cohort study at the Leiden University Medical Center, The Netherlands. All patients were members of Dutch CADASIL pedigrees registered in the Dutch CADASIL registry. All study participants gave written informed consent and all participated in the study between May 2019 and December 2020. All procedures performed in studies involving human participants were in accordance with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This study was approved by the medical ethics committee of the Leiden University Medical Center (P18.164). For the current study, 12 patients with a NOTCH3 cys EGFr 1–6 variant and 13 patients with a NOTCH3 cys EGFr 7–34 variant were selected from the database for analysis of their skin biopsy. The selection criteria were (1) age between 50 and 60 years; (2) for each NOTCH3 cys variant group (EGFr 1–6 versus EGFr 7–34), half of the patients fulfilled the pre‐defined criterium of a severe phenotype (‘EGFr 1‐6 severe’ group and ‘EGFr 7‐34 severe’ group) and half fulfilled the criterium of a mild phenotype (‘EGFr 1‐6 mild’ group and ‘EGFr 7‐34 mild’ group). A severe phenotype was defined as ≥3 lacunes on MRI; a mild phenotype was defined as 0–2 lacunes (Table 1). Healthy controls shown not to have the familial NOTCH3 cys variant were selected from the CADASIL2000 study and the 18‐year follow‐up of that cohort (approved by the Leiden University Medical Center medical ethics committee under P80.89 and P17.170, respectively). 13 , 14
TABLE 1.
Patient characteristics by EGFr group and disease severity
| Location of NOTCH3 cys variant | EGFr 1–6 | EGFr 1–6 | EGFr 7–34 | EGFr 7–34 | P value a |
|---|---|---|---|---|---|
| Phenotype | Severe | Mild | Severe | Mild | |
| N | 6 | 6 | 6 | 7 | |
| Age, mean (sd) | 55.5 (2.2) | 56.0 (2.8) | 56.0 (3.4) | 54.4 (3.2) | n.s. |
| Female, n (%) | 1 (17%) | 4 (67%) | 2 (33%) | 5 (71%) | n.s. |
| NOTCH3 cys variant (n) | R110C (1) | R141C (1) | R544C (1) | C568Y (1) | |
| R141C (1) | R169C (1) | C568Y (1) | R578C (5) | ||
| C144F (1) | R207C (3) | R578C (2) | G667C (1) | ||
| R153C (2) | C222Y (1) | C1015R (1) | |||
| R182C (1) | R1076C (1) | ||||
| Smoking, n (%) | 3 (50%) | 3 (50%) | 3 (50%) | 5 (71%) | n.s. |
| Hypertension, n (%) | 1 (17%) | 2 (33%) | 4 (67%) | 1 (14%) | n.s. |
| Hypercholesterolemia, n (%) | 2 (33%) | 1 (17%) | 3 (50%) | 1 (14%) | n.s. |
| mRS, n (%) | 0.005 b | ||||
| 0–1 | 0 (0%) | 5 (83%) | 2 (33%) | 6 (86%) | |
| ≥2 | 6 (100%) | 1 (17%) | 4 (67%) | 1 (14%) | |
| Prior clinical stroke, n (%) | 5 (83%) | 0 (0%) | 3 (50%) | 0 (0%) | 0.004 b |
| Presence of lacunes, n (%) | 6 (100%) | 0 (0%) | 6 (100%) | 0 (0%) | <0.001 b |
| Fazekas dwm, n (%) | <0.001 c | ||||
| 0–2 | 0 (0%) | 0 (0%) | 0 (0%) | 7 (100%) | |
| 3 | 6 (100%) | 6 (100%) | 6 (100%) | 0 (0%) |
Note: n: number; sd: standard deviation; n.s.: not significant.
P value for overall differences between patient groups (Chi‐square; two‐way ANOVA).
Statistical significance attributed to differences between severe and mild patients. No statistically significant difference between severely affected patients with a NOTCH3 cys EGFr 1–6 versus EGFr 7–34 variant, nor between mildly affected patients with an EGFr 1–6 versus EGFr 7–34 variant.
No statistically significant difference between EGFr 1–6 severe group and EGFr 7–34 severe group, but the EGFr 1–6 mild group had higher Fazekas deep white matter (dwm) scores than the EGFr 7–34 mild group (P = 0.001).
In addition to the 25 selected patients for analysis of skin biopsies, we included skin and brain material from six deceased CADASIL patients available in our centre, who had given written informed consent for autopsy and the use of their material for CADASIL research. Four patients had an EGFr 1–6 variant (deceased at 57, 58, 59 and 65 years of age) and two patients had an EGFr 7–34 variant (deceased at 69 and 84 years of age).
Study procedure
All patients included in the study followed an identical, one‐day study protocol with clinical assessment, brain MRI, neuropsychological test battery, skin biopsy and blood withdrawal. Clinical assessment included history of clinical stroke, functional ability status according to the modified Rankin Scale (mRS) and the presence of cardiovascular risk factors. Smoking was defined as current or past smoking. Individuals using an antihypertensive agent or with a previous diagnosis of hypertension (>140/90 mmHg) were considered to have hypertension. Individuals with non‐fasting levels of LDL‐c > 3.5 mmol/L or total cholesterol >6.5 mmol/L or with a previous diagnosis of hypercholesterolaemia were considered to have hypercholesterolaemia. Neuroimaging was performed on a Philips Medical Systems 3 Tesla MR. Lacunes were assessed and counted according to the STRIVE criteria. 15 White matter hyperintensities were semi‐quantitatively scored using the Fazekas scale for deep white matter. 16 A mask for subcortical grey matter structures and brain stem was generated from FLAIR images and used for WMH classification with Brain Intensity AbNormality Classification Algorithm (BIANCA) with settings optimised for CADASIL populations. 17 , 18 WMH volumes were calculated using FMRIB Software Library (FSL) v6.0.4, Analysis Group, FMRIB, Oxford, UK, using individually selected WMH thresholds.
NOTCH3ECD immunohistochemistry
Skin punch biopsies were taken from the lateral upper arm and processed to formalin‐fixed paraffin‐embedded tissue blocks. Post‐mortem material from skin and brain were also formalin‐fixed and paraffin‐embedded in tissue blocks. Per individual, two sections with a thickness of 5 μm were pretreated with proteinase K, washed three times with PBS and stained for 2 hours at room temperature with the primary mouse anti‐NOTCH3ECD antibody (clone 1E4, Millipore, dilution 1:1000, RRID:AB_2890101). The secondary antibody (rabbit‐anti‐mouse/biotin, Dako, dilution 1:200, RRID:AB_2636929) was incubated for 1 hour at room temperature, followed by development using the Avidin‐Biotin Complex (ABC Elite Kit, Vector, RRID:AB_2336810) for 30 min. Further staining with 3,3′‐diaminobenzidine (DAB, Sigma Aldrich) and the collection of full‐colour, full‐focus images was performed as described previously. 19 Harris' haematoxylin (diluted 1:3 for 5 s, Merck) was used as co‐staining reagent. Inner and outer boundaries of vessel walls were manually drawn. The NOTCH3ECD positive area, with a typical granular staining pattern, was determined using a Colour Threshold (Hue 0–50; Saturation 0–255; Brightness 0–175) in ImageJ, and expressed as percentage of vessel wall area (Figure 1C). We defined the NOTCH3 score 20 as the average of the 10 vessels with the highest NOTCH3ECD positive area per individual. The NOTCH3 score based on the average of these 10 vessels was found to be representative of the NOTCH3 score based on the average of all vessels, both in skin (median of 24 vessels were assessed per individual, range 11–55)(r = 0.97, R 2 = 0.95, P = 1.1·10−5, Supporting Information S1A) and brain (median of 22 vessels per individual, range 20–34)(r = 0.99, R 2 = 0.98, P = 5.2·10−5, Supporting Information S1B). In one individual (EGFr 1–6 group, severe phenotype), the NOTCH3 score could not be obtained due to insufficient tissue for immunohistochemistry.
FIGURE 1.
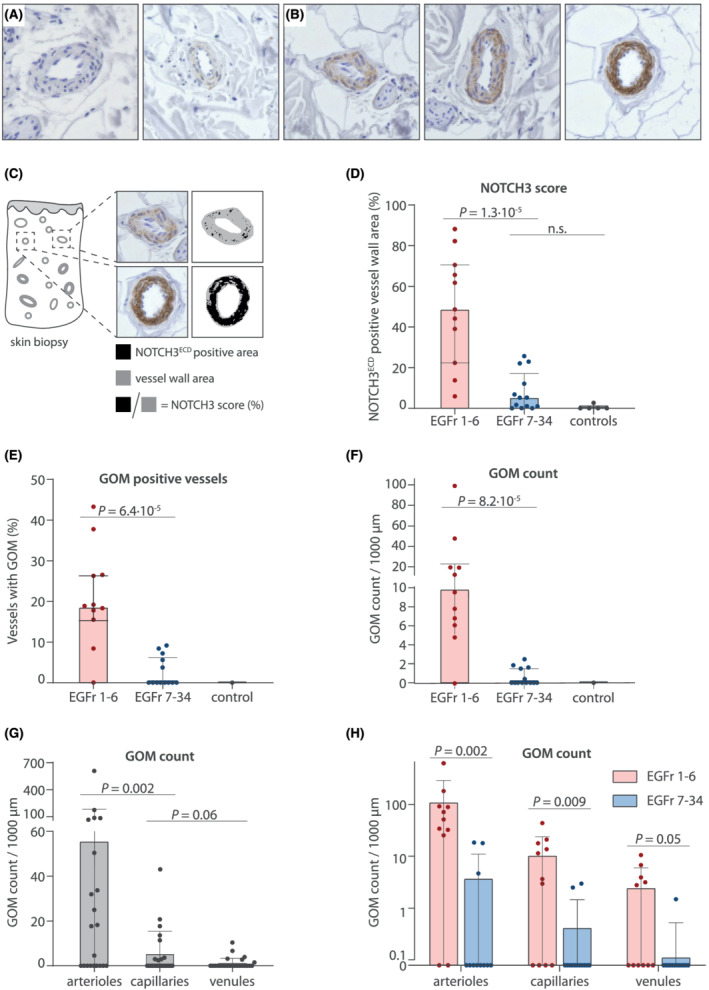
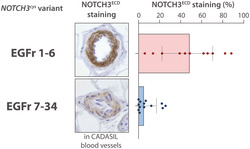
NOTCH3 score and GOM load are significantly lower in CADASIL patients with NOTCH3 cys variants in EGFr 7–34 in skin biopsies. (A,B) representative images of NOTCH3 immunostaining on skin vessels of (A) controls and (B) CADASIL patients. Controls show a negative NOTCH3 staining, whilst CADASIL patients show a positive and granular NOTCH3 staining. (C) NOTCH3 score: The vessel wall area positive for granular NOTCH3ECD staining is expressed as the percentage of the total vessel wall area. (D) Patients with NOTCH3 cys variants located in EGFr 7–34 have lower NOTCH3 scores than patients with EGFr 1–6 variants (median NOTCH3 score 5.2% [IQR 11.1] versus 48.6% [48.2], P = 1.3·10−5). (E,F) patients with NOTCH3 cys variants located in EGFr 7–34 have less GOM‐positive vessels than patients with EGFr 1–6 variants (median 0.0% [IQR 5.6] versus 18.8% [10.9], P = 6.4·10−5) and a lower overall GOM count (median 0.0 GOM/1000 μm [IQR 1.6] versus 9.8 [15.2], P = 8.2·10−5). (G) GOM deposits are more prevalent in arterioles than in capillaries and venules (median 11.2 GOM/1000 μm [IQR 50.6], 0.0 [3.3] and 0.0 [0.7], respectively). (H) The difference in GOM count between the EGFr groups was also observed in arterioles, capillaries and venules separately. To facilitate interpretation, data is plotted on log10‐axis and represent mean ± standard deviation in H. In D‐G, data represent median ± interquartile range. P values are calculated from transformed variables
Ultrastructural analysis of GOM
Skin biopsy processing for ultrastructural analysis was performed as described previously. 19 The presence of GOM deposits in blood vessel walls was evaluated by two independent observers who were blinded for NOTCH3 cys variant position and phenotype (GG and MNC). One skin biopsy of a control was included as a reference for normal ultrastructural vessel wall morphology, to ensure that the observers were only counting GOM deposits and not structures that might be similar to GOM deposits. GOM were identified and counted in a median of 16 vessels (range 7–27) for each individual. Blood vessels were further categorised into capillaries based on a lumen diameter of <7 μm. All other vessels were manually categorised by two independent observers into arterioles and venules based on ultrastructural hallmarks, that is, the shape of the mural cells (block‐like for arteriole; spindle‐like for venule) and completeness of the mural cell layer around the vessel. 21 In one individual (EGFr 1–6 group, mild phenotype), GOM could not be assessed as the tissue processed for electron microscopy did not contain any vessels.
Statistical analysis
Differences in patient characteristics were analysed using an independent samples t‐test or Chi‐square test. In all analyses, each data point reflects one individual (i.e., GOM count of one patient reflects the average of the GOM count in all analysed vessels of that patient). NOTCH3 score was square root transformed and GOM count was cubic root transformed to obtain a plausible normal distribution. One‐way ANOVA analyses were performed on transformed variables to assess the effect of EGFr group and vessel type. Two‐way interaction ANOVA analyses were performed on transformed variables to assess the overall effect of EGFr group; disease severity; and the interaction (i.e., whether the effect of disease severity was significantly different between the two EGFr groups). Two‐way interaction ANOVA was also used to assess the effect of cardiovascular risk factors. Post‐hoc pairwise comparisons were Bonferroni adjusted for multiple comparisons. All statistical analyses were two‐sided tests with a threshold for statistical significance of 0.05, using the IBM SPSS Statistics software, version 25.
RESULTS
Patient characteristics
Baseline characteristics of the study population are shown in Table 1. Disease severity and neuroimaging lesion load did not differ between the ‘EGFr 1‐6 severe’ and the ‘EGFr 7‐34 severe’ group, or between the ‘EGFr 1‐6 mild’ and ‘EGFr 7‐34 mild’ group, except for white matter hyperintensity lesion load. There was no significant difference in sex and cardiovascular risk factors between groups.
NOTCH3 cys EGFr 7–34 variants are associated with a lower NOTCH3 score and GOM load
Patients with a NOTCH3 cys EGFr 7–34 variant had a much lower NOTCH3 score than patients with an EGFr 1–6 variant (median NOTCH3 score 5.2% [IQR 11.1] versus 48.6% [48.2], P = 1.3·10−5; Figure 1A–D). Moreover, almost half (6/13) of patients with an EGFr 7–34 variant had no granular NOTCH3ECD staining at all in any of the skin vessels, whereas granular NOTCH3ECD staining was observed in all patients with an EGFr 1–6 variant. GOM deposits were also much less abundant in patients with an EGFr 7–34 variant compared to those with an EGFr 1–6 variant, in terms of both amount of GOM positive vessels (median 0.0% [IQR 5.6] versus 18.8% [10.9], P = 6.4·10−5; Figure 1E) and GOM count (median 0.0 GOM/1000 μm [IQR 1.6] versus 9.8 [15.2], P = 8.2·10−5; Figure 1F). The association between NOTCH3 cys variant position and NOTCH3 score and GOM count remained statistically significant after correction for cardiovascular risk factors and sex (NOTCH3 score P = 6.2·10−5, GOM count P = 2.5·10−4). Strikingly, the majority of individuals (8/13) with an EGFr 7–34 variant had no GOM deposits at all, whereas almost all individuals (10/11) with an EGFr 1–6 variant had GOM. NOTCH3 scores were highest for the most N‐terminal NOTCH3 cys variants (Supplementary Information S4). There was no difference in NOTCH3 score between NOTCH3 cys variants leading to a gain versus a loss of a cysteine residue (data not shown).
Overall, GOM deposits were most abundant in arterioles, but they were also observed in capillaries and even in venules (Figure 1G). The difference in GOM count between individuals with EGFr 1–6 and EGFr 7–34 variants was observed in all three vessel types (Figure 1H, Supporting Information S2 and S3).
Analysis of post‐mortem CADASIL brain tissue showed that the two individuals with an EGFr 7–34 variant had lower NOTCH3 scores in brain vessels than the four individuals with an EGFr 1–6 variant (Figure 2 A,B). No statistics could be performed for the brain NOTCH3 score due to the sample size. In the skin vessels of the deceased CADASIL patients, the NOTCH3 score was also lower in individuals with an EGFr 7–34 variant than in those with an EGFr 1–6 variant (Figure 2B), and skin and brain NOTCH3 scores in these individuals were correlated (r = 0.84, P = 0.02, Figure 2C).
FIGURE 2.
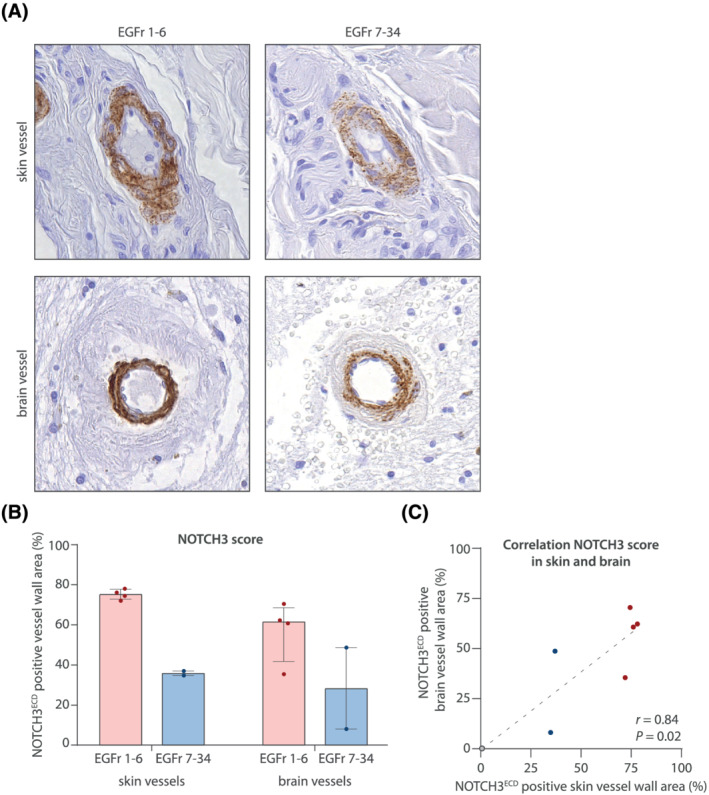
NOTCH3 cys variants in EGFr 7–34 are also associated with a lower NOTCH3 score in brain. (A) Example vessels from skin and brain tissue of a deceased CADASIL patient with a NOTCH3 cys variant in EGFr 1–6 and a deceased patient with a NOTCH3 cys variant in EGFr 7–34. (B) In both skin and brain vessels, the NOTCH3 score was lower in the EGFr 7–34 group compared to the EGFr 1–6 group. Data represent median ± interquartile range. (C) Correlation between the NOTCH3 score in skin and brain
Association of disease severity with NOTCH3 score and GOM count
Next, we assessed whether NOTCH3 score and GOM count in skin biopsies differed between mild and severe patients. Disease severity was not significantly associated with either NOTCH3 score or GOM count (P = 0.06 and P = 0.06, respectively)(Figure 3A,B). However, the ‘EGFr 7‐34 mild’ patient group did have a lower NOTCH3 score than the ‘EGFr 7‐34 severe’ patient group (median 1.0% [IQR 5.2] versus 14.5% [17.7], P = 0.051 in a post‐hoc analysis) and a lower GOM count (median 0.0 GOM/1000 μm [IQR 0.0] versus 0.0 [4.7], P = 0.28 in a post hoc analysis), although this difference was not statistically significant. In the EGFr 7–34 group, skin NOTCH3 score was significantly correlated with lacune count (r = 0.67, P = 0.03) and WMH volume (r = 0.70, P = 0.02), but not with disability (r = 0.24, P = 0.42)(Figure 3C–E).
FIGURE 3.
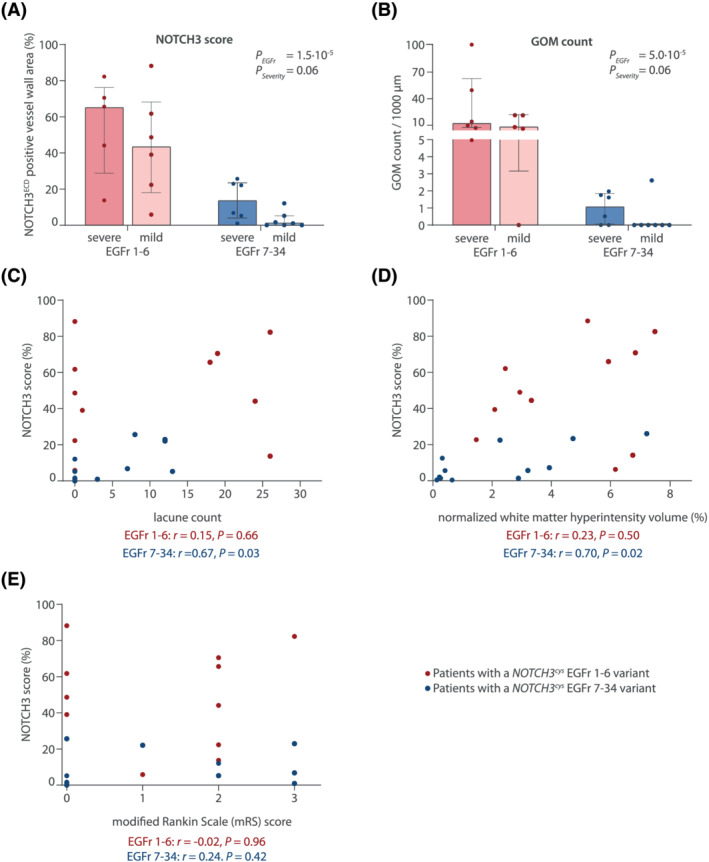
Association between NOTCH3 score, GOM load and disease severity. (A) NOTCH3 score and (B) GOM count were higher in patients with a severe phenotype compared to those with a mild phenotype (P = 0.06 for both NOTCH3 score and GOM after correction for EGFr group). This was mainly attributable to differences between mild and severe patients in the EGFr 7–34 group. Data represent median ± interquartile range. (C,D,E) scatter plot showing the relation between NOTCH3 score and lacune count (C), white matter hyperintensity volume (D) and modified Rankin scale score (E)
DISCUSSION
In this study, we show that vascular NOTCH3ECD aggregation load is lower in CADASIL patients with a NOTCH3 cys EGFr 7–34 variant than in patients with a NOTCH3 cys EGFr 1–6 variant. This finding suggests that there is a difference in aggregation properties between EGFr 1–6 and EGFr 7–34 NOTCH3cys mutant proteins. 3 , 4 , 22 A lower vascular NOTCH3ECD aggregation load may be one of the factors underlying the later and milder disease onset in CADASIL patients with an EGFr 7–34 variant compared to patients with an EGFr 1–6 variant. 9 , 10 , 11
EGFr domains 1–6 are more exposed to extracellular matrix proteins, some of which have been shown to co‐aggregate with NOTCH3ECD, such as HTRA1, LTBP‐1, TIMP3, and vitronectin, 23 , 24 , 25 which may explain why mutant EGFr 1–6 proteins are more prone to aggregate. Alternatively, aggregation properties may be related to the distance of the NOTCH3 cys variant from the recently identified non‐enzymatic cleavage site between EGFr 1 and 2. 26 Potentially in line with this, we observed a trend towards an association between NOTCH3 score and the distance from this cleavage site for variants located in EGFr domains 1–6.
Interestingly, the association between NOTCH3ECD aggregation and EGFr mutation position was much stronger than the association between NOTCH3ECD aggregation and disease severity. There was an association, however, between NOTCH3 score and MRI lesion load (lacune count and WMH volume), but this was only seen in the EGFr 7–34 group. Larger studies, including CADASIL patients with variable disease severity as well as asymptomatic individuals with an incidental NOTCH3 cys variant in population databases, are needed to further clarify the association between NOTCH3ECD aggregation in skin and disease severity.
Notably, most individuals in the ‘EGFr 7‐34 mild’ group had very low NOTCH3 scores and only one had GOM. The absence of GOM in skin biopsies has also been recently described in another CADASIL cohort. 27 The absence of GOM, therefore, does not exclude a CADASIL diagnosis, which is relevant for clinical practice, as to date GOM are deemed to have very high sensitivity and specificity for CADASIL. 7 , 27 , 28 , 29
The strengths of this study are the prospective design, the quantitative analysis of both the ultrastructural GOM and the NOTCH3ECD staining, as well as assessing NOTCH3ECD aggregation in both skin and brain vasculature. A limitation is the relatively small sample size, especially of brain tissue.
In conclusion, this study shows that CADASIL patients with an EGFr 7–34 variant have less vascular NOTCH3ECD aggregation than CADASIL patients with an EGFr 1–6 variant. This finding sheds the first light on the relation between NOTCH3 cys variant position, mutant NOTCH3 protein aggregation and CADASIL disease severity.
CONFLICT OF INTEREST
RJH is funded by Netherlands Organisation for Health Research and Development (ZonMW 91717325). JWR is funded by Netherlands Organisation for Health Research and Development (ZonMW 91717325) and the Netherlands Brain Foundation (HA2016‐02‐03). SAJLO receives financial support from Netherlands Organisation for Health Research and Development (ZonMW 91717325) and the Netherlands Brain Foundation (HA2016‐02‐03), receives institutional support from Leiden University Medical Center, and is co‐inventor on an LUMC owned patent, not related to this work. GG, AAM, MNC, RvD, IMH and CRJ have no conflicts of interest to declare that are relevant to the content of this article.
ETHICS STATEMENT
All procedures performed in studies involving human participants were in accordance with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This study was approved by the medical ethics committee of the Leiden University Medical Center (P18.164). Written informed consent was given by each participant. Patients gave written informed consent for autopsy and the post‐mortem use of material for CADASIL research.
AUTHOR CONTRIBUTIONS
Gido Gravesteijn, Remco J. Hack, Julie W. Rutten, Saskia A.J. Lesnik Oberstein contributed to the study conception and design. Data collection was performed by Gido Gravesteijn, Minne N. Cerfontaine, Aat A. Mulder, Remco J. Hack, Remco van Doorn, Ingrid Hegeman. Data analyses were performed by Gido Gravesteijn, Minne N. Cerfontaine, Remco J. Hack, Julie W. Rutten and Saskia A.J. Lesnik Oberstein. The first draft of the manuscript was written by Gido Gravesteijn, Julie W. Rutten, Saskia A.J. Lesnik Oberstein and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/nan.12751.
Supporting information
Data S1. Supporting Information 1. The correlation between NOTCH3 score of the 10 most‐affected vessels and all vessels.
Supporting Information 2. NOTCH3 aggregation measures (NOTCH3 score and GOM count) and vessel characteristics by EGFr group and disease severity.
Supporting Information 4. Correlation between NOTCH3 score and NOTCH3 cys variant position
Supporting Information 3. Association between GOM count, EGFr group and disease severity.
ACKNOWLEDGEMENTS
This work was funded by Netherlands Organisation for Health Research and Development (ZonMW 91717325) and the Netherlands Brain Foundation (Hersenstichting HA2016‐02‐03). We gratefully thank all CADASIL patients who participated in this study. Furthermore, we thank P.H. Neeskens for electron microscopy sample preparation, and I.F.A.C Fokkema, M. Kroon and M.A. Santcroos for their ImageJ scripting advise.
Gravesteijn G, Hack RJ, Mulder AA, et al. NO TCH3 variant position is associated with NOTCH3 aggregation load in CADASIL vasculature. Neuropathol Appl Neurobiol. 2022;48(1):e12751. 10.1111/nan.12751
Julie W. Rutten and Saskia A.J. Lesnik Oberstein contributed equally to this work.
[Correction added on 23 September 2021, after first online publication: Peer review history statement has been added.]
Funding information Netherlands Organisation for Health Research and Development, Grant/Award Number: ZonMW 91717325; Netherlands Brain Foundation, Grant/Award Number: HA2016‐02‐03
Data availability statement
The authors confirm that the summary data supporting the findings of this study are available within the article and its supplementary material. The raw data that support the findings of this study are available on request from the corresponding author upon reasonable request.
REFERENCES
- 1. Dichgans M, Markus HS, Salloway S, et al. Donepezil in patients with subcortical vascular cognitive impairment: a randomised double‐blind trial in CADASIL. Lancet Neurol. 2008;7(4):310‐318. [DOI] [PubMed] [Google Scholar]
- 2. Rutten JW, Haan J, Terwindt GM, van Duinen SG, Boon EMJ, Lesnik Oberstein SAJ. Interpretation of NOTCH3 mutations in the diagnosis of CADASIL. Expert Rev Mol Diagn. 2014;14(5):593‐603. [DOI] [PubMed] [Google Scholar]
- 3. Joutel A, Andreux F, Gaulis S, et al. The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. J Clin Invest. 2000;105(5):597‐605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Opherk C, Duering M, Peters N, et al. CADASIL mutations enhance spontaneous multimerization of NOTCH3. Hum Mol Genet. 2009;18(15):2761‐2767. [DOI] [PubMed] [Google Scholar]
- 5. Joutel A, Favrole P, Labauge P, et al. Skin biopsy immunostaining with a Notch3 monoclonal antibody for CADASIL diagnosis. Lancet. 2001;358(9298):2049‐2051. [DOI] [PubMed] [Google Scholar]
- 6. Lesnik Oberstein SAJ, van Duinen SG, van den Boom R, et al. Evaluation of diagnostic NOTCH3 immunostaining in CADASIL. Acta Neuropathol. 2003;106(2):107‐111. [DOI] [PubMed] [Google Scholar]
- 7. Tikka S, Mykkänen K, Ruchoux MM, et al. Congruence between NOTCH3 mutations and GOM in 131 CADASIL patients. Brain. 2009;132(4):933‐939. 10.1093/brain/awn364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brulin P, Godfraind C, Leteurtre E, Ruchoux M‐M. Morphometric analysis of ultrastructural vascular changes in CADASIL: analysis of 50 skin biopsy specimens and pathogenic implications. Acta Neuropathol. 2002;104(3):241‐248. [DOI] [PubMed] [Google Scholar]
- 9. Rutten JW, Van Eijsden BJ, Duering M, et al. The effect of NOTCH3 pathogenic variant position on CADASIL disease severity: NOTCH3 EGFr 1–6 pathogenic variant are associated with a more severe phenotype and lower survival compared with EGFr 7–34 pathogenic variant. Genet Med. 2019;21(3):676‐682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rutten JW, Dauwerse HG, Gravesteijn G, et al. Archetypal NOTCH3 mutations frequent in public exome: implications for CADASIL. Ann Clin Transl Neurol. 2016;3(11):844‐853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mukai M, Mizuta I, Watanabe‐Hosomi A, et al. Genotype–phenotype correlations and effect of mutation location in Japanese CADASIL patients. J Hum Genet. 2020;65(8):637‐646. [DOI] [PubMed] [Google Scholar]
- 12. Cho BPH, Nannoni S, Harshfield EL, et al. NOTCH3 variants are more common than expected in the general population and associated with stroke and vascular dementia: an analysis of 200 000 participants. J Neurol Neurosurg Psychiatry. 2021;92(7):694‐701. 10.1136/jnnp-2020-325838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van den Boom R, Lesnik Oberstein Saskia AJ, Ferrari MD, Haan J, van Buchem MA. Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy: MR Imaging Findings at Different Ages—3rd–6th Decades. Radiology. 2003;229(3):683‐690. 10.1148/radiol.2293021354 [DOI] [PubMed] [Google Scholar]
- 14. Gravesteijn G, Hack RJ, van Opstal AM, et al. Eighteen‐year disease progression and survival in CADASIL. J Stroke. 2021;23(1):132‐134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wardlaw JM, Smith EE, Biessels GJ, et al. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol. 2013;12(8):822‐838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fazekas F, Chawluk JB, Alavi A. MR signal abnormalities at 1.5 T in Alzheimer's dementia and normal aging. Am J Neuroradiol. 1987;8:421‐426. [DOI] [PubMed] [Google Scholar]
- 17. Griffanti L, Zamboni G, Khan A, et al. BIANCA (Brain Intensity AbNormality Classification Algorithm): a new tool for automated segmentation of white matter hyperintensities. Neuroimage. 2016;141:191‐205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ling Y, Jouvent E, Cousyn L, Chabriat H, De Guio F. Validation and optimization of BIANCA for the segmentation of extensive white matter hyperintensities. Neuroinformatics. 2018;16(2):269‐281. [DOI] [PubMed] [Google Scholar]
- 19. Gravesteijn G, Dauwerse JG, Overzier M, et al. Naturally occurring NOTCH3 exon skipping attenuates NOTCH3 protein aggregation and disease severity in CADASIL patients. Hum Mol Genet. 2020;29(11):1853‐1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rutten JW, Klever RR, Hegeman IM, et al. The NOTCH3 score: a pre‐clinical CADASIL biomarker in a novel human genomic NOTCH3 transgenic mouse model with early progressive vascular NOTCH3 accumulation. Acta Neuropathol Commun. 2015;3(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. MacGregor Sharp M, Criswell TP, Dobson H, Finucane C, Verma A, Carare RO. Solving an old dogma: is it an arteriole or a venule? Front Aging Neurosci. 2019;11:1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duering M, Karpinska A, Rosner S, et al. Co‐aggregate formation of CADASIL‐mutant NOTCH3: a single‐particle analysis. Hum Mol Genet [Internet]. 2011;20:3256‐3265. Available from. http://www.ncbi.nlm.nih.gov/pubmed/21628316 [DOI] [PubMed] [Google Scholar]
- 23. Kast J, Hanecker P, Beaufort N, et al. Sequestration of latent TGF‐β binding protein 1 into CADASIL‐related Notch3‐ECD deposits. Acta Neuropathol Commun. 2014;2:1. 10.1186/s40478-014-0096-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zellner A, Scharrer E, Arzberger T, et al. CADASIL brain vessels show a HTRA1 loss‐of‐function profile. Acta Neuropathol. 2018;136(1):111‐125. [DOI] [PubMed] [Google Scholar]
- 25. Monet‐Leprêtre M, Haddad I, Baron‐Menguy C, et al. Abnormal recruitment of extracellular matrix proteins by excess Notch3 ECD: a new pathomechanism in CADASIL. Brain. 2013;136(6):1830‐1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Young KZ, Lee SJ, Zhang X, et al. NOTCH3 is non‐enzymatically fragmented in inherited cerebral small‐vessel disease. J Biol Chem. 2020;295(7):1960‐1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shindo A, Tabei K, Taniguchi A, et al. A nationwide survey and multicenter registry‐based database of cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy in Japan. Front Aging Neurosci. 2020;12:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morroni M, Marzioni D, Ragno M, et al. Role of electron microscopy in the diagnosis of cadasil syndrome: a study of 32 patients. PLoS ONE. 2013;8:6‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Markus HS, Martin RJ, Simpson MA, et al. Diagnostic strategies in CADASIL. Neurology. 2002;59(8):1134‐1138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting Information 1. The correlation between NOTCH3 score of the 10 most‐affected vessels and all vessels.
Supporting Information 2. NOTCH3 aggregation measures (NOTCH3 score and GOM count) and vessel characteristics by EGFr group and disease severity.
Supporting Information 4. Correlation between NOTCH3 score and NOTCH3 cys variant position
Supporting Information 3. Association between GOM count, EGFr group and disease severity.
Data Availability Statement
The authors confirm that the summary data supporting the findings of this study are available within the article and its supplementary material. The raw data that support the findings of this study are available on request from the corresponding author upon reasonable request.


