Abstract
Bacteria often cooperate by secreting molecules that can be shared as public goods between cells. Because the production of public goods is subject to cheating by mutants that exploit the good without contributing to it, there has been great interest in elucidating the evolutionary forces that maintain cooperation. However, little is known about how bacterial cooperation evolves under conditions where cheating is unlikely to be of importance. Here we use experimental evolution to follow changes in the production of a model public good, the iron‐scavenging siderophore pyoverdine, of the bacterium Pseudomonas aeruginosa. After 1200 generations of evolution in nine different environments, we observed that cheaters only reached high frequency in liquid medium with low iron availability. Conversely, when adding iron to reduce the cost of producing pyoverdine, we observed selection for pyoverdine hyperproducers. Similarly, hyperproducers also spread in populations evolved in highly viscous media, where relatedness between interacting individuals is increased. Whole‐genome sequencing of evolved clones revealed that hyperproduction is associated with mutations involving genes encoding quorum‐sensing communication systems, while cheater clones had mutations in the iron‐starvation sigma factor or in pyoverdine biosynthesis genes. Our findings demonstrate that bacterial social traits can evolve rapidly in divergent directions, with particularly strong selection for increased levels of cooperation occurring in environments where individual dispersal is reduced, as predicted by social evolution theory. Moreover, we establish a regulatory link between pyoverdine production and quorum‐sensing, showing that increased cooperation with respect to one trait (pyoverdine) can be associated with the loss (quorum‐sensing) of another social trait.
Keywords: bacterial cooperation, cheating, experimental evolution, genomics, pyoverdine, quorum sensing
1. INTRODUCTION
Microbes are social organisms. Over the last three decades, a wealth of research has uncovered that microbial communities are shaped by complex networks of competitive and cooperative interactions (Figueiredo & Kramer, 2020; Ghoul & Mitri, 2016; Granato et al., 2019; Little et al., 2008; West et al., 2007a). Competitive interactions may involve the secretion of toxins against competitors, hunting or competitive exclusion (Granato et al., 2019; Hibbing et al., 2010; Pérez et al., 2016). Examples of cooperative behaviours include mutualistic cross‐feeding, communication via signalling molecules, and sharing the benefits of secreted molecules such as proteases, siderophores and biofilm components (D’Souza et al., 2018; Dragoš et al., 2018; Kramer et al., 2020; Robinson et al., 2020; Wilder et al., 2011). Microbial cooperation often underlies important biological processes, including the establishment of infections (Ackermann et al., 2008; Granato et al., 2018), nutrient fixation in the rhizosphere (Denison et al., 2003) and mutualistic interactions with hosts (Verma & Miyashiro, 2013).
Microbial cooperation has attracted the attention of evolutionary biologists not only because of its variety in form and function, but also because it incurs a cost for the actor while benefitting other individuals (West et al., 2007b). Cooperation could thus select for “cheating” variants that do not cooperate themselves, but benefit from cooperative acts performed by others (Ghoul, Griffin et al., 2014). How then can cooperative traits be maintained on evolutionary timescales? This question has spurred an enormous body of research focusing on how microbes cope with the threat of cheating (Özkaya et al., 2017; Smith & Schuster, 2019; Strassmann & Queller, 2011; Travisano & Velicer, 2004; Wechsler et al., 2019). While interesting in its own right, the focus on cheating might have diverted attention from other factors that could also influence the evolution of cooperative traits (see Zhang & Rainey, 2013 for a critique). While we currently know much about the evolution of cheating resistance mechanisms and environmental factors that maintain cooperation in the face of cheating, we know little about cooperative trait evolution in environments where cheating plays no role. For example, can higher levels of cooperation be selected for, or can cooperative traits be lost for reasons other than cheating, for instance through disuse (Velicer et al., 1998; Zhang & Rainey, 2013)?
Here, we tackle these questions by focusing on the evolution of pyoverdine production in the bacterium Pseudomonas aeruginosa, one of the most widely studied social traits in microbes (Buckling et al., 2007; Dumas & Kümmerli, 2012; Griffin et al., 2004; Harrison, 2013; Harrison et al., 2017; O’Brien et al., 2017; Ross‐Gillespie et al., 2015). Upon sensing iron scarcity, cells secrete pyoverdine, a siderophore that chelates environmental or host‐bound ferric iron. Iron‐loaded pyoverdine is then imported via a specific receptor, which is followed by iron reduction, release from the siderophore and subsequent recycling of pyoverdine (Kramer et al., 2020; Schalk et al., 2020). Pyoverdine production is a cooperative trait, because pyoverdines are costly for the individual cell to produce, but once loaded with iron they become available to other cells in the local neighbourhood (Buckling et al., 2007; Harrison, 2013).
It is well established that pyoverdine, as a so‐called “public good,” can select for cheating in severely iron‐limited and well‐mixed environments, where cheats can freely access secreted pyoverdines (Griffin et al., 2004; Kümmerli et al., 2015). It is less clear, however, how pyoverdine production would evolve in other environments where, for example, iron is less stringently limited, thereby reducing the advantage of cheating, and/or where environmental viscosity would limit cell mixing and thus cheating opportunities. Several scenarios can be envisaged. First, in iron‐rich environments pyoverdine production might be selectively lost because it is not required (Zhang & Rainey, 2013). Second, in viscous environments increased levels of pyoverdine production and thus cooperation could be favoured because viscosity reduces cell dispersal and siderophore diffusion (Julou et al., 2013; Kümmerli et al., 2009), which ensures that pyoverdine‐mediated social interactions occur more often between genetically related individuals (Dobay et al., 2014). Third, iron availability and environmental viscosity might interact and jointly affect the cost and benefit of pyoverdine production, and thereby select for an altered production level that matches the optimal cost‐to‐benefit ratio of the environment in which that bacteria evolved. Finally, the social trait might diversify without the production levels being affected. Indeed, a great diversity of pyoverdine variants exists among Pseudomonas strains (Butaite et al., 2017), but each strain produces only one type of pyoverdine, together with its cognate receptor. Mathematical models suggest that pyoverdine and receptor structure diversification could help individuals escape cheating (Lee et al., 2012) or gain an edge over competitors in the race for iron, even under iron‐rich conditions (Niehus et al., 2017).
To test for these alternative evolutionary outcomes, we experimentally evolved replicated populations of the laboratory strain P. aeruginosa PAO1 for 200 days (~1200 generations) in nine different environments. We manipulated environmental conditions along two gradients, iron availability and media viscosity, with each gradient entailing three levels in a 3 × 3 full‐factorial design. We hypothesized that evolution under increased iron availabilities should result in a reduction of pyoverdine production, because less pyoverdine is required when iron is abundant. In contrast, evolution under increased environmental viscosities should result in higher pyoverdine production, due to increased relatedness between interacting individuals. After experimental evolution, we quantified changes in pyoverdine production and investigated the extent to which pyoverdine remained shareable among cells. We then assessed how phenotypic changes affect the fitness of evolved populations and clones. Finally, we sequenced the genomes of 119 evolved clones to map evolved phenotypes to genotypic changes. We expected that reduced pyoverdine production should be associated with mutations in the pyoverdine locus (synthesis and regulation), while increased pyoverdine production should be associated with mutations in loci responsible for gene expression regulation.
2. MATERIAL AND METHODS
2.1. Strains and culturing conditions
We used the Pseudomonas aeruginosa wildtype strain PAO1 (ATCC 15692) and a fluorescent variant (PAO1‐gfp), directly derived from the wildtype strain. PAO1‐gfp constitutively expresses GFP (green fluorescent protein), due to a single copy insertion of gfp in the bacterium's chromosome (attTn7::ptac‐gfp). We used the pyoverdine‐negative mutant PAO1ΔpvdD (henceforth pyoverdine‐null) and the pyoverdine‐pyochelin double‐negative mutant PAO1ΔpvdDΔpchEF (henceforth siderophore‐null) as controls. The siderophore‐null mutant was used because pyochelin, a secondary siderophore of P. aeruginosa, can partly compensate for the lack of pyoverdine (Ross‐Gillespie et al., 2015).
We grew overnight precultures in 8 ml Lysogeny Broth (LB, 2% w/v) and incubated at 37°C, 210 rpm, for 16–18 h. We performed all experiments in liquid CAA medium (5 g/L casamino acids, 1.18 g/L K2HPO4.3H2O, 0.25 g/L MgSO4.7H2O, 25 mm HEPES buffer), to which we added 400 µm of the iron chelator 2,2′‐bipyridyl to bind the residual iron present in the medium. For the experimental evolution and follow‐up experiments, we manipulated iron availability of the CAA medium by adding no FeCl3, 2 µm FeCl3, or 20 µm of FeCl3 to achieve conditions of low, intermediate and high iron availability, respectively. We further manipulated the viscosity of the CAA medium by adding either no, 0.1% or 0.2% (w/v) agar. All media components were purchased from Sigma‐Aldrich. We conducted all experiments in 96‐well plates, in 200 µl of media, at 37°C and 170 rpm shaking conditions.
2.2. Experimental evolution
We experimentally evolved strains PAO1 and PAO1‐gfp in nine different environments, in a 3 × 3 full factorial design, combining the iron concentrations with the three environmental viscosities. Prior to experimental evolution, we grew overnight precultures of PAO1 and PAO1‐gfp. From these overnight cultures, we prepared glycerol stocks (750 µl culture in 750 µl glycerol [85% v/v]), stored at −80°C, to be used later as references for the ancestral strain states. We washed the remaining part of the precultures in 0.85% NaCl and adjusted them to an OD600 of 10−2 in NaCl (0.85% w/v), from which we inoculated 2 µl of culture into 198 µl of medium, so that experimental populations started at an OD600 of 10−4 (~20,000 cells/ per well). We evolved 24 replicates in each of our nine environments (12 populations each for PAO1 and PAO1‐gfp, respectively), resulting in a total of 216 independently evolving populations. We inoculated the 24 replicated populations in different wells on the same 96‐well plate. To reduce the risk of cross‐contamination, we kept all wells adjacent to an evolving population blank. Additionally, we inoculated PAO1 and PAO1‐gfp strains in a checkerboard pattern, so that the closest vertical and horizontal neighbours could be distinguished based on the GFP marker. We let each bacterial culture grow for 48 h and then transferred a sample to fresh medium. We repeated this procedure for 100 consecutive growth rounds, corresponding to ~1200 bacterial generations. At the end of each round, we used a multimode plate reader (Infinite M‐200, Tecan) to measure: (i) culture growth at OD600, (ii) pyoverdine fluorescence (excitation|emission = 400|460 nm) and (iii) GFP fluorescence (excitation|emission = 488|520 nm). Subsequently, we diluted 2 µl of each population in 198 µl NaCl (0.85% w/v), and then transferred 2 µl of this diluted culture to 198 µl of fresh medium to begin the next round of growth, resulting in a 1:10,000 dilution rate. After every other round, we mixed 50 µl of each population with 50 µl glycerol (85% v/v) for long‐term storage at −80°C. Furthermore, we performed three cross‐contamination tests to determine whether populations should be excluded from further analysis. A detailed protocol for these tests and a list of excluded populations (Table S1) can be found in the online Supporting Information.
2.3. Pyoverdine production assay
To quantify pyoverdine production changes during experimental evolution, we isolated and screened a total of 2880 evolved clones (i.e., 20 per population) for their pyoverdine production profiles in the environments in which they evolved. We first inoculated aliquots (from freezer stocks) of all 144 evolved populations from the last round of experimental evolution in 200 µl of LB in 96‐well plates. We also inoculated the ancestral wildtype strains and the pyoverdine‐null mutant as controls. We grew all cultures for 16–18 h, at 37°C and 170 rpm. Subsequently, we serially diluted each culture in 0.85% NaCl up to 10−6 and 10−7‐fold, and spread 50 µl of these diluted cultures onto iron‐supplemented LB‐agar plates (2% w/v LB, 1.2% w/v agar, 20 µm FeCl3). We incubated plates for 16–18 h at 37°C. Subsequently, we picked 20 random colonies from each evolved population and transferred them on 96‐well plates to 200 µl of the medium in which the selected clones had evolved. We further picked four colonies for each of the two ancestors (PAO1 and PAO1‐gfp), and the pyoverdine‐null mutant strains per 96‐well plate. We incubated the resulting plates at 37°C and 170 rpm for 48 h, and subsequently measured OD600 and pyoverdine fluorescence, using the plate reader as described above. We then quantified the evolved pyoverdine level of each clone relative to its ancestor (as a percentage of the ancestral production) and allocated evolved clones to four categories. These categories comprise: (i) nonproducers, producing less than 10% of the ancestral level; (ii) reduced producers, producing between 10% and 70%; (iii) regular producers, producing between 70% and 130%; and (iv) hyperproducers, producing more than 130% (Dumas & Kümmerli, 2012). Following our measurements, we preserved evolved clones individually by mixing 50 µl of a culture containing cells from the clone with 50 µl 85% glycerol for storage at −80°C.
2.4. Pyoverdine growth stimulation assay
One evolutionary response to the emergence of cheaters or other competing lineages could involve changes to the peptide backbone of pyoverdines, together with mutations in receptors, such that the altered pyoverdines would become more exclusively available for members of the respective producer lineage and less exploitable by cheaters or competitors (Inglis et al., 2016; Lee et al., 2012) To explore this possibility, we performed a common garden experiment where we used the siderophore‐null mutant as a biosensor (Jiricny et al., 2010), and examined how well this mutant can use the pyoverdines secreted by evolved producers. Specifically, we fed the pyoverdine‐containing supernatants of evolved producer and hyperproducer clones to the ancestral siderophore‐null mutant and measured its growth in low iron – low viscosity conditions. For this assay, we used six evolved clones from each of the 16 populations evolved in one of the nine media (i.e., 864 clones in total). Whenever clones within a population produced similar amounts of pyoverdine, we chose six random clones. In contrast, whenever clones from the same population produced different amounts of pyoverdine (i.e., regular and hyperproducers), we chose at least one clone per pyoverdine production phenotype and drew the remaining clones at random.
To generate pyoverdine‐enriched supernatants, we individually grew the selected 864 evolved clones in 200 µl of the low iron – low viscosity condition in 96‐well plates and incubated the plates for 24 h at 37°C and 170 rpm. Following incubation, we first measured the OD600 and pyoverdine fluorescence of all cultures, and then separated the supernatant from the bacterial cells by centrifugation at 698 g for 10 min. We filter‐sterilized the supernatants by transferring them to 96‐well 1‐ml filter‐plates (VWR International), followed by centrifugation at 198 g for 15 min. We then performed a supernatant cross‐feeding assay to quantify the extent to which each supernatant stimulates the growth of the ancestral siderophore‐null mutant. Specifically, we added 20 µl of each supernatant, in triplicate, to individual wells of a 96‐well plate containing 178 µl of the low iron – low viscosity medium. To this mixture, we added 2 µl of the siderophore‐null mutant from an overnight LB culture, washed and resuspended in 0.85% NaCl, and diluted to a starting OD600 of 10−3. We incubated plates at 37°C and 170 rpm, and measured the growth (OD600) of the siderophore‐null mutant after 24 h. As positive and negative controls, we also fed the siderophore‐null mutant with either supernatants of the ancestral strain or with its own supernatant. A pre‐experiment revealed that the concentration of ancestral pyoverdine in the supernatant required to fully stimulate the nonproducer is 2000 rfu (data not shown). Because some of the evolved producers (20.5%) made less than this threshold value, we excluded them from further analysis. For each evolved pyoverdine producer, we calculated the cross‐feeding effect as the growth (final OD600) of the pyoverdine‐null mutant in the evolved supernatant divided by its growth in the ancestral supernatant. Values above and below 1 indicate increased and decreased growth stimulation, respectively.
2.5. Genomic analysis of evolved clones
We sequenced the genomes of 119 evolved clones to identify the genetic changes that occurred during experimental evolution. A detailed description of clone selection and the DNA extraction methods can be found in the online Supporting Information. We assessed the quality of the raw reads with fastqc (Andrews, 2010) and multiqc (Ewels et al., 2016). We then aligned the reads to the P. aeruginosa PAO1 reference genome using the bwa "mem” algorithm with default settings (Li, 2013), followed by variant‐calling with the bcftools (Li et al., 2009) “mpileup” and “call” commands. We quality‐filtered the detected variants by excluding single nucleotide polymorphisms and indels below 50 “QUAL,” as well as indels with IDV <10. Subsequently, we excluded variants detected in the genomes of evolved clones that were present in the ancestral clones with bcftools isec. We then annotated the final list of variants with snpeff (Cingolani et al., 2012). We detected deletions and duplications using clc genomic workbench (version 11.0.1, https://digitalinsights.qiagen.com). After creating a "Mapping Graph Track", we identified graph threshold areas (window size = 5) using an upper coverage threshold of 0.5 for deletions and a lower coverage threshold of 500 for duplications. We subsequently visually confirmed these results with the integrated genomics viewer (Robinson et al., 2011). We identified 314 loci that independently mutated in two or more populations, and classified them according to their biological function using pseudocap (Winsor et al., 2005) and the primary literature.
2.6. Statistical analysis
We performed all statistical analyses with R version 3.6.3. (R Core Team, 2020) and used linear models, linear mixed models, one‐tailed t tests or Fisher's Exact tests to analyse the data. Prior to analysis, we used (i) the Shapiro–Wilk test and QQ‐plots to check whether the model residuals were normally distributed; and (ii) the Levene test to test for homogeneity of variances. If these assumptions were not met, we log‐transformed the affected response variables. Where appropriate, we used the false discovery rate (FDR) p‐value adjustment method to account for multiple testing (Benjamini & Hochberg, 1995). A detailed description of which statistical test was applied to which data set can be found in the online Supporting Information.
3. RESULTS
3.1. Interaction between iron availability and environmental viscosity determines pyoverdine production and growth in ancestral P. aeruginosa
We first evaluated how iron availability and media viscosity affect the growth and pyoverdine production of ancestral Pseudomonas aeruginosa prior to evolution (Figure 1). This analysis provides important information on the extent to which these two phenotypes differ across environments and thus the potential for selection to act differentially upon them. We found that ancestral P. aeruginosa growth was influenced by a significant interaction between iron availability and media viscosity (ANOVA: F 4,135 = 55.43; p < .0001, Figure 1a, Table S2). While growth decreased with more stringent levels of iron limitation, increased media viscosity dampened this effect. Pyoverdine production was also influenced by a significant interaction between iron availability and media viscosity (ANOVA: F 4,135 = 237.67; p < .0001, Figure 1b, Table S2). While pyoverdine production gradually decreased from low to high iron availability, the decrease was again less pronounced in more viscous environments. These findings show that P. aeruginosa adjusts its pyoverdine production profile in response both to iron availability and to environmental viscosity, and we might thus expect selection pressures on this trait to vary across environments.
FIGURE 1.
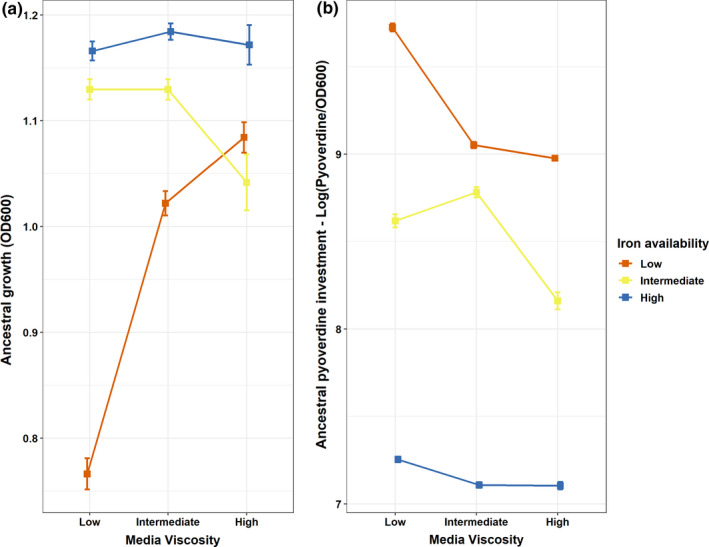
Iron availability and media viscosity jointly influence growth and pyoverdine production in the ancestral Pseudomonas aeruginosa. (a) Strain growth (OD600) in the nine different environments, which vary in medium viscosity (amount of agar added: low =0.0%; intermediate =0.1%, high =0.2%) and iron availability (low =0 µm, red line; intermediate =2 µm, yellow line; high =20 µm, blue line). (b) Pyoverdine investment, shown as log‐transformed pyoverdine fluorescence values normalized by OD, in all nine experimental conditions. Each square represents the mean of 16 independent populations, whereas the vertical bars represent the standard error of the mean
3.2. Environment‐dependent selection for decreased and increased pyoverdine production
To test whether different environments select for different pyoverdine production levels, we quantified the pyoverdine investment of evolved populations relative to the ancestor after ~1200 generations of experimental evolution. We observed that pyoverdine production levels changed significantly in five out of the nine environments (Figure 2). Consistent with previous studies, we found that populations evolved reduced pyoverdine production levels under the low iron – low viscosity condition (t 15 = −2.617, p adj = .035). Because iron limitation is most stringent and pyoverdine most needed in this environment (Figure 1), evolution towards lower pyoverdine production levels is compatible with the rise and spreading of cheating mutants. In stark contrast, populations that evolved in low‐viscosity environments but with intermediate and high iron availabilities significantly increased pyoverdine production levels compared to the ancestral wildtype (intermediate iron – low viscosity: t 15 = 7.134, p adj < .0001; high iron – low viscosity: t 15 = 5.830, p adj < .0001).
FIGURE 2.
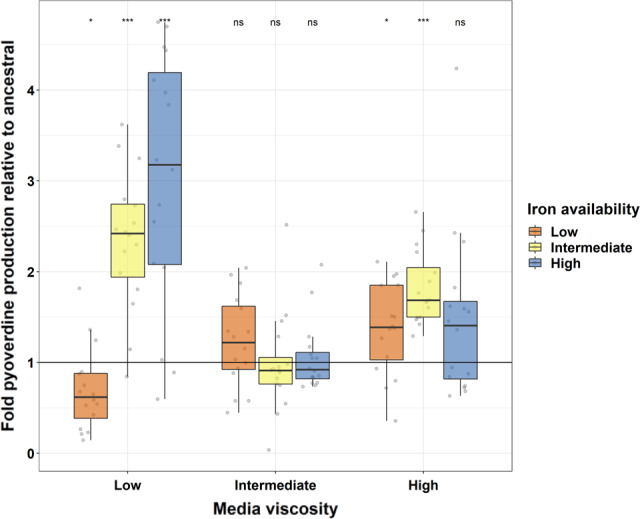
Evolution causes a media‐dependent divergence in pyoverdine production. Population‐level pyoverdine production after 100 rounds of experimental evolution, relative to the ancestral phenotype (black horizontal line) in nine different environments. We measured pyoverdine production via its natural fluorescence, scaled to the optical density of the population (OD600). Box plots show the median and the 1st and 3rd quartiles across the 16 independently evolved populations (depicted by the individual dots). Whiskers represent 1.5× interquartile range. Asterisks signify FDR‐corrected p‐values for one‐sample t tests: *p < .05, ***p < .0001, ns = not significant
Selection for altered pyoverdine production seemed to be lowest in media with intermediate viscosity, where the population‐level pyoverdine production levels did not change during experimental evolution regardless of iron concentration (intermediate viscosity with low iron: t 15 = 1.865, p adj = .105; intermediate iron: t 15 = −0.134, p adj = .895; high iron: t 15 = 0.584, p adj = .639). Conversely, in populations evolved in high‐viscosity media, we observed an increase in pyoverdine production levels in most populations. This increase was significant for two out of the three iron conditions (high viscosity ‐ with low iron: t 15 = 2.985, p adj = .021; with intermediate iron: t 15 = 8.186, p adj < .0001; with high iron: t 15 = 2.112, p adj = 0.078). Together, our results do not support the hypothesis that pyoverdine production is selected against in iron‐rich environments because of potential disuse. Instead, we found support for the hypothesis that high environmental viscosity can favour increased levels of cooperation and that pyoverdine production levels diverge across environments, possibly to match environment‐specific requirements for this siderophore.
3.3. Iron availability and viscosity both influence heterogeneity in pyoverdine production among clones from the same population
In a next step, we studied evolved populations individually and asked whether clones with distinct pyoverdine phenotypes coexist within populations. We predict highest population diversity in environments where different social strategies are likely to compete, such as under low iron – low viscosity, where cheats and pyoverdine producers can closely interact. Conversely, we expect lower levels of diversity among clones in environments that are generally favourable for cooperation (e.g., under high viscosity). Indeed, we observed that the level of within‐population heterogeneity in pyoverdine production varied both between environments and between replicates of the same condition (Figure S1). In line with our hypothesis, we found that the coefficient of variation, which quantifies phenotypic heterogeneity among clones within a population, significantly increased with more stringent iron limitation (ANOVA: F 2,139 = 4.85; p = .009; Tukey post hoc tests: low vs. intermediate t = −2.993, p = .009; low vs. high [marginally significant] t = −2.243, p = .067) and lower environmental viscosity (ANOVA: F 2,139 = 11.34; p < .0001; Tukey post hoc tests: low vs. intermediate t = −3.823, p < .001; low vs. high t = −4.37, p < .001) (Figure 3a, Table S3).
FIGURE 3.
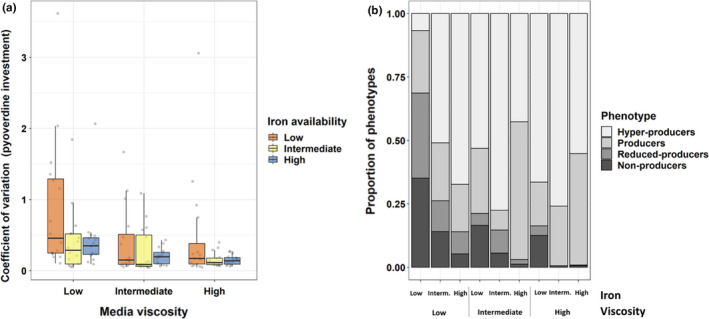
Variation and classification of pyoverdine phenotypes in clones evolved in nine different environments. (a) Coefficient of variation (SD/mean) for pyoverdine production phenotypes across clones from the same population. We measured pyoverdine production via its natural fluorescence and scaled to the optical density (OD600) of the population. Box plots show the median and the 1st and 3rd quartiles across the 16 independently evolved populations (depicted by the individual dots). (b) Distribution of discrete pyoverdine production phenotypes for all nine environments after 100 rounds of experimental evolution. We allocated clones to phenotype categories according to their pyoverdine production levels scaled relative to the ancestor (100%). (1) Nonproducers: less than 10%; (2) reduced‐producers: 10%–70%; (3) producers: 70%–130%; (4) hyperproducers: more than 130% of ancestral production. Fisher's exact test reveals significant differences in category frequencies between any two environments
We then examined whether the different environments affect the prevalence of clones with distinct pyoverdine production levels. Consequently, we allocated each isolated clone into one of four discrete pyoverdine production categories, covering the range from nonproducers to hyperproducers (Figure 3b). We found that the frequency of clones in the four categories varied significantly between any two environments (global Fisher's exact test: p < .0005; FDR‐corrected pairwise Fisher's exact tests maximum: p < .0079). Most importantly, the frequency of nonproducers decreased in environments with increased iron availability and media viscosity, whereas the prevalence of regular and hyperproducers increased in more viscous environments under all iron availabilities (Figure 3b). This analysis thus reinforces the view that there is a shift from selection for cheating to selection for increased cooperation when moving from low iron – low viscosity to higher iron – higher viscosity conditions.
3.4. Pyoverdine under‐ and hyperproduction comes at a fitness cost
The high frequency of pyoverdine non‐ and hyperproducers suggests that this trait was under selection during experimental evolution. Here, we explore whether changes in pyoverdine phenotypes have fitness consequences for evolved clones in the environments in which they had evolved. If nonproducers evolved because of cheating, then their fitness should decrease when growing in monoculture. Conversely, if over‐producers evolved because higher levels of cooperation are beneficial then their fitness should exceed the fitness of regular producers. Indeed, we observed significant growth differences between evolved clones from the four pyoverdine production categories (linear mixed model: t 373 = 51.91, p < .0001, Figure 4). As predicted by the cheating hypothesis, nonproducers grew significantly more poorly than regular pyoverdine producers (Tukey post hoc test: z = 4.024; p < .001). However, regular pyoverdine producers also grew significantly better than hyperproducers (z = −9.45, p < .001), thus refuting our hypothesis that over‐production is associated with an overall growth advantage.
FIGURE 4.
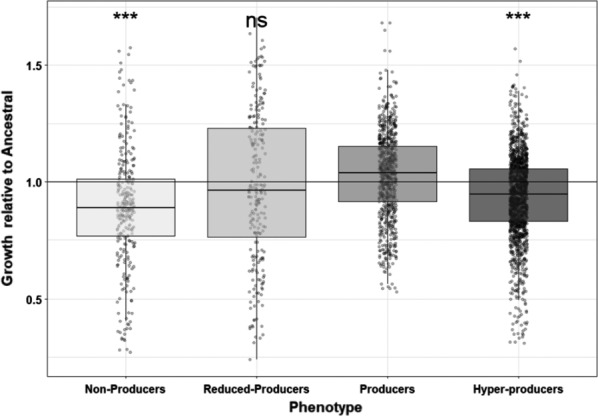
Evolution of non‐ and hyperpyoverdine production is associated with growth declines. (a) Growth of evolved clones allocated to four different pyoverdine production categories, scaled relative to the ancestor (100%): (1) nonproducers: less than 10%; (2) reduced‐producers: 10%–70%; (3) regular producers: 70%–130%; (4) hyperproducers: more than 130% of ancestral production. Significance levels (***p < .0001, ns = nonsignificant) refer to pairwise comparisons between producers and the other three phenotype categories. Horizontal black line (y = 1) indicates the ancestral phenotype
One shortcoming of the above analysis is that it combines clones across all environments, which does not allow us to test whether pyoverdine hyperproduction is associated with fitness benefits only in specific environments. To address this issue, we repeated our analysis for the nine environments separately. While we still observed that hyperproducers generally grew less well than regular producers (Figure S2; Table S4), we also found that their growth decline was most pronounced in the low iron – low viscosity condition. In contrast, growth differences between producer and hyperproducer clones became negligible in environments with increased iron or increased viscosity, suggesting that pyoverdine hyperproduction might be equally successful as regular production in these environments. Moreover, it is possible that hyperproducers compensate for reduced end‐point growth with an increase in other fitness components, such as an increased growth‐rate or shorter lag‐phase. The high‐throughput nature of our experimental design did not allow us to capture these fitness metrics.
3.5. Environment‐dependent change in the extent to which pyoverdines from evolved producers can stimulate the growth of an engineered nonproducer
Next, we explored whether pyoverdine producers and hyperproducers responded to the presence of competing lineages in their populations by changing the extent to which their pyoverdines are compatible. To test this hypothesis, we harvested pyoverdine‐containing supernatants from ancestral and evolved producers and hyperproducers, and fed these supernatants to an engineered siderophore‐null mutant. We found that the supernatants of all evolved clones still stimulated the growth of the ancestral nonproducer (Figure 5). However, the growth stimulation experienced by the siderophore‐null mutant depended on a significant interaction between the viscosity and iron availability of the environments in which the producers and hyperproducer evolved (LMER: = 26.35, p < .0001, Figure 5, Table S5). Specifically, the relative growth stimulation of the siderophore‐null mutant was significantly reduced when subjected to supernatants from producers evolved in the low iron – low viscosity environment (t 35 = −6.40, p < .0001) (Figure 5). In all other environments, the supernatant‐induced growth stimulation of the siderophore‐null mutant was either unchanged or even higher compared to the supernatant of the ancestral pyoverdine producer (Figure 5). Crucially, we included evolved pyoverdine production levels as covariates into our model and indeed found that this covariate was a significant positive predictor of growth stimulation (LMER: = 278.31, p < .0001, Table S5), in addition to the treatment effects reported above. Together, these results suggest that only pyoverdine (hyper‐)producing clones evolved under low iron – low viscosity have adapted to nonproducers and reduced the exploitability of their pyoverdine.
FIGURE 5.
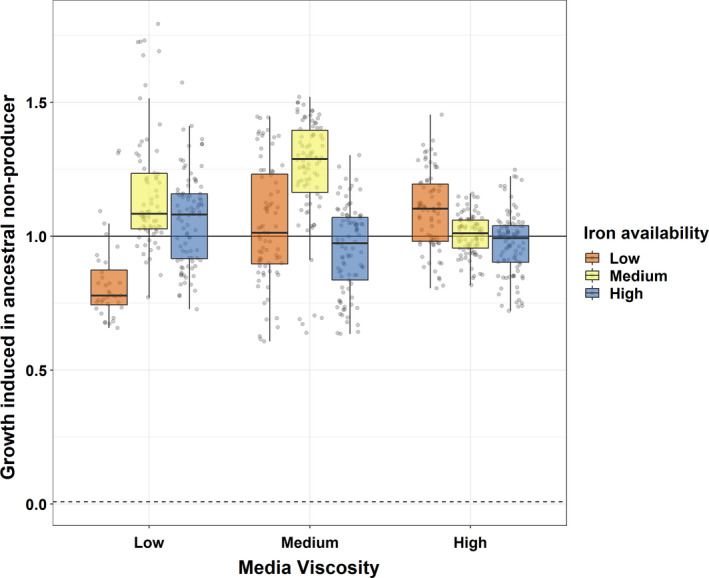
Evolutionary change in the extent to which pyoverdines from evolved clones can be used by an ancestral nonproducer. Each individual circle shows the average effect (three replicates) of supernatant collected from an evolved pyoverdine (hyper‐)producer on the growth of the siderophore‐null mutant. All growth values are scaled relative to the growth stimulation this mutant experienced in the supernatant of the ancestral wildtype (solid horizontal line at y = 1). The dashed line represents the growth of the siderophore‐null mutant when fed its own supernatant. Box plots show the median and the 1st and 3rd quartiles, whereas whiskers represent 1.5× interquartile range
3.6. Genomic features of evolved clones
To map the evolved phenotypes to genetic changes, we sequenced the genomes of 119 evolved clones from 68 independent populations to an average sequencing depth of 230× (min = 177×, max = 334×), and compared them to their respective ancestors (PAO1 and PAO1‐gfp). We found a total of 3472 mutations (SNPs, duplications and deletions), distributed over 1897 loci (coding and intergenic regions). Individual clones had between two and 191 mutations (median = 7, Figure 6a). The distribution of mutations per clone followed a bimodal pattern: most clones (78.99%) had few mutations (<20), while a minority of clones (21.01%) had many mutations. This discrepancy in mutations per clone can be explained by the occurrence of nonsynonymous mutations in the coding regions of the mutS and mutL genes in almost all clones (92%, 23 out of 25) possessing more than 20 mutations (Figure 6a). These genes code for parts of the DNA mismatch repair system of P. aeruginosa, and mutations in their coding regions result in substantially elevated mutation rates (Oliver et al., 2002; Wiegand et al., 2008). We henceforth call clones harbouring such mutations hypermutators.
FIGURE 6.
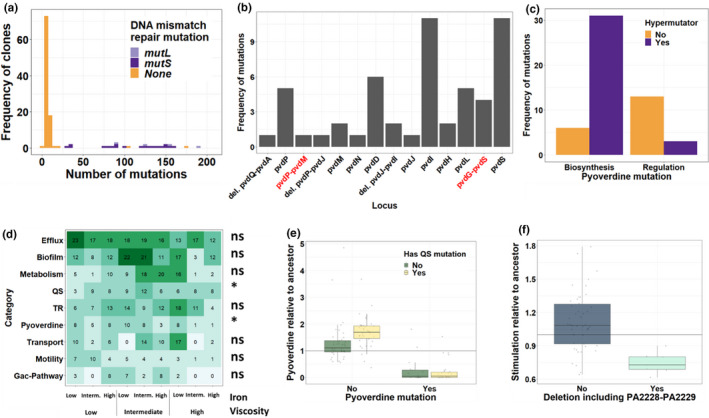
Mutations in the pyoverdine locus and quorum sensing genes respectively result in pyoverdine under‐ and hyperproduction. (a) Distribution of the number of mutations per clone. All but two clones with more than 20 mutations have mutations in the DNA mismatch repair machinery (hypermutators). (b) Distribution of mutations in genes (black labels) and intergenic regions (red labels) of the pyoverdine locus across clones. Genes are ordered according to the physical map of the locus. (c) Nonhypermutator clones have more mutations in pyoverdine regulatory genes, whereas mutations in pyoverdine biosynthesis genes are over‐represented among hypermutator clones. (d) Heatmap of the most common mutational targets sorted by functional categories, across our nine different environments. TR, transcriptional regulation; QS, quorum sensing. Labels to the right of the heatmap indicate whether mutations in the respective category had a significant (*p < .05) or nonsignificant (ns, p > .05) effect on evolved pyoverdine production level. (e) Box plots showing the evolved level of pyoverdine production for clones with mutations in the pyoverdine locus, QS genes, both sets of genes or none of these genes. (f) Box plots showing the evolved level of growth stimulation of the ancestral nonproducer, for clones that have, or not, a deletion that includes the upstream region of the qsrO‐vqsM‐PA2228 operon. Box plots in (e) and (f) depict the median, the 1st and 3rd quartiles and 1.5× interquartile range (whiskers). The horizontal black lines (y = 1) indicate ancestral phenotypes
3.7. Mutations in the pyoverdine locus are associated with reduced pyoverdine production
We then focused on nonsynonymous mutations in the pyoverdine locus, that is in the regulatory and biosynthetic genes for pyoverdine production, together with their intergenic regions. We asked whether mutations in these genes affect the pyoverdine production of evolved clones. A total of 38 clones had mutations in the pyoverdine locus, and these mutants produced significantly less pyoverdine (mean ± SE: 0.27 ± 0.07) than clones without such mutations (1.50 ± 0.08) (ANOVA: F 1,117 = 88.86; p < .0001, Figure S3). Only one clone produced no pyoverdine without having a mutation in the pyoverdine locus. We could attribute its phenotype to a nonsynonymous mutation in tatC. Mutations in this gene are known to impair pyoverdine maturation (Lee et al., 2016; Voulhoux et al., 2006), and we thus included this mutant in the class of pyoverdine mutants.
Almost all genes of the pyoverdine locus harboured mutations (Figure 6b). The most frequent mutations occurred in or upstream of pvdS, the sigma factor that positively regulates the expression of pyoverdine, and in pvdI, the largest gene in the cluster, encoding a nonribosomal peptide synthetase (Figure 6b). While pvdS mutations have frequently surfaced in previous more short‐term evolution experiments (Granato et al., 2018; Kümmerli et al., 2015), mutations in pyoverdine biosynthesis genes are much less common (O’Brien et al., 2019a). We suspected that this difference is linked to the evolution of hypermutators in our experiment. Indeed, hypermutators incurred significantly fewer mutations in regulatory genes, but more mutations in pyoverdine biosynthesis genes compared to nonhypermutators (Fisher's exact test: p < .0001, Figure 6c). For all those populations in which we sequenced more than one clone, we examined the order in which mutations appeared (Figure S4). We found that mutations in the pyoverdine locus arose after mutS/mutL mutations in eight populations, but never the other way round. In six populations, we were not able to resolve the order in which mutations occurred. Together, we observed a high prevalence of mutations in the pyoverdine locus, a shift in the mutational landscape between non‐ and hypermutators, and that most mutations in this locus are associated with decreased pyoverdine production.
3.8. Mutations in quorum‐sensing systems are associated with pyoverdine hyperproduction
Next, we investigated possible links between mutational patterns and pyoverdine hyperproduction. For this purpose, we classified mutations into functional categories. Figure 6d shows the nine most frequent categories (in which we detected at least 25 independent mutations per category), and the prevalence of mutations in them across our nine environments. One of these categories (“pyoverdine”) includes mutations in the pyoverdine locus itself, which we have shown to be associated with under‐production. We thus asked whether mutations in any of the other eight categories are associated with pyoverdine hyperproduction, while statistically controlling for the presence of mutations in the pyoverdine locus. We found that mutations in quorum‐sensing (QS) genes were significantly associated with increased pyoverdine production in clones with no mutations in the pyoverdine locus (ANOVA: F 1:115 = 8.211, p adj = .039) (Figure 6e; Table S6). Mutations occurred in all three known P. aeruginosa QS systems (Las, Rhl and PQS) and included nonsynonymous SNPs, small indels, and large duplications and deletions (Table S7). In addition, the mutational patterns of Figure 6d also suggest that some mutations are associated with adaptation to the growth medium. The most obvious candidates are mutations in efflux pump‐related genes, particularly those responsible for the regulation of the mexAB‐oprM pump (Table S8), which occurred in 108 out of the 119 clones. Their high prevalence could result from the use of bipyridyl, a compound that not only chelates iron outside the cell, but has also toxic effects when entering cells. Efflux pump upregulation has been reported to improve growth in the presence of bipyridyl (Liu et al., 2010).
3.9. Deletions in the qsrO‐vqsM‐PA2228 operon and its upstream region are associated with a reduction in pyoverdine‐mediated growth stimulation
Finally, we investigated whether the reduction in pyoverdine‐mediated growth stimulation observed for some evolved producer clones (Figure 5) is associated with specific genetic changes. We followed a similar strategy as above (Figure 6d), testing whether mutations in genes from specific functional categories are associated with a reduction in pyoverdine‐mediated growth stimulation. This approach yielded no significant association for any category (Table S9). We therefore explored whether mutations in specific loci were associated with changes in this phenotype. We found a single hit: supernatants from clones with a deletion (130 bp to 21 kb) comprising the qsrO‐vqsM‐PA2228 operon and its upstream region (seven cases) stimulated the growth of the ancestral nonproducer significantly less than clones without such deletions (Figure 6f, ANOVA: F 1:51 = 13.979, p adj = .007, Table S10).
4. DISCUSSION
Because bacterial cooperative traits affect virulence, ecosystem functioning and microbiome assembly, enormous interest has focused on their evolution (Davis & Isberg, 2019; Ebrahimi et al., 2019; Magnúsdóttir et al., 2015). Particular attention has been paid to cheating (Harrison et al., 2017; Strassmann & Queller, 2011; Tarnita, 2017)—the loss of cooperation through mutants that exploit the cooperative efforts of others. In contrast, cooperative trait evolution remains poorly examined in environments where cheating plays no role (but see Velicer et al., 1998). Here we explored how different environments shape the evolutionary trajectory of a model bacterial cooperative trait, the production of the iron‐scavenging siderophore pyoverdine by the opportunistic pathogen Pseudomonas aeruginosa. We experimentally evolved this species for ~1200 generations in nine different environments that varied in iron availability and environmental viscosity. We found that selection for reduced pyoverdine production due to cheating occurred only in one of the nine environments (low iron – low viscosity), where relatedness among individuals is low and the benefit of exploitation is high. In stark contrast, pyoverdine production either did not change or even increased over time in four environments each (Figure 2). Selection for increased pyoverdine production occurred predominantly in environments with either high iron availability, where the social trait is expressed at a relatively low level and thus incurs reduced costs, or with high viscosity, where interacting individuals are more closely related. Whole‐genome sequencing revealed that point mutations in genes of the pyoverdine locus, particularly the iron‐starvation sigma factor pvdS, are significantly associated with decreased pyoverdine production. Conversely, SNPs and large‐scale deletions in QS genes are associated with pyoverdine hyperproduction. Together, our results suggest that bacterial social traits can undergo rapid evolutionary change and allow bacteria to adapt to the prevailing environmental and social conditions.
The evolution of pyoverdine nonproducers or reduced‐producers only consistently occurred in one environment (low iron, low viscosity). Because pyoverdine is most needed under low iron availability, our findings indicate that non‐ and reduced‐producers have a selective advantage due to cheating (Figure 2; Figure S1). This pattern is consistent with previous studies, where nonproducing cheats spread because they used pyoverdine produced by others (Ghoul, West et al., 2014; Harrison et al., 2017; Kümmerli et al., 2015). The mutational patterns discovered are also consistent with previous work showing that pyoverdine non‐ or reduced‐production predominantly arose by mutations in pvdS and its promotor region in nonhypermutator clones (Granato & Kümmerli, 2017; Kümmerli et al., 2015; O’Brien et al., 2019; Tostado‐Islas et al., 2021). The sigma factor pvdS regulates the expression of the entire pyoverdine biosynthesis machinery (Cunliffe et al., 1995; Ringel & Brüser, 2018). Mutations in it are thus often associated with (i) reduced production of this public good, and (ii) major cost saving when the entire pyoverdine locus is shut down. In contrast to nonhypermutators, we found that mutations in pyoverdine biosynthesis genes were overrepresented in hypermutators. We hypothesize that mutations in these genes that reduce pyoverdine production costs are rare, and thus only surface in clones with elevated mutation rates.
Our data further suggest that pyoverdine producers can adapt to cheats by reducing the compatibility of their pyoverdines (Figure 5). This phenomenon predominantly surfaced in the low‐iron/low‐viscosity environment, where nonproducer prevalence was highest. Specifically, we observed that pyoverdines of evolved producers stimulated the growth of an engineered nonproducer less than the pyoverdine of the ancestral wildtype. However, our genomic analysis revealed that this change in compatibility is not due to modification of the pyoverdine backbone structure, a commonly proposed (yet unproven) adaptative response to cheating (Inglis et al., 2016; Lee et al., 2012; Smith et al., 2005; Stilwell et al., 2018). Instead, we observed that reduced pyoverdine‐mediated growth benefits for nonproducers were associated with mutations in the qsrO‐vqsM‐PA2228 operon and its upstream region (Figure 6f). The exact function of this operon is unknown, but deletions in it can lead to a complete shutdown of all three QS systems of P. aeruginosa (Köhler et al., 2014; Liang et al., 2014). Given that vqsM is a putative transcriptional regulator (Huang et al., 2019), we believe that the change in pyoverdine compatibility is not a direct consequence of QS silencing, but rather a pleiotropic effect of an as yet to be described regulatory link of this operon and pyoverdine. We speculate that this operon is involved in post‐synthetic pyoverdine modifications, affecting the compatibility of this molecule among different members of the community.
The main finding of our experiments is the evolution of pyoverdine hyperproduction in multiple environments. It highlights that increased levels of cooperation can evolve across a range of conditions. In line with our hypothesis, we found that pyoverdine production increased in high‐viscosity environments regardless of iron availability. High viscosity reduces individual dispersal and molecule diffusion (Kümmerli et al., 2009). Both effects increase the relatedness among interacting individuals, as clonemates are more likely to stay together and social interactions take place across a smaller physical range (i.e., in smaller groups) (Julou et al., 2013; Weigert & Kümmerli, 2017). Limited dispersal and small group size are known to favour cooperation (Dobay et al., 2014; El Mouden & Gardner, 2008; Hamilton, 1964; Queller, 1994). While previous work showed that high relatedness can maintain cooperation when cooperators and genetically engineered cheaters are mixed in short‐term experiments (Bastiaans et al., 2016; Gilbert et al., 2007; Griffin et al., 2004; Kümmerli et al., 2009), we here demonstrate that it can actually favour increased levels of cooperation during more long‐term experiments.
We further observed that hyperproducers also emerged and spread in low‐viscosity environments with intermediate and high levels of iron. This finding opposes our original hypothesis that increased iron availability should select for reduced pyoverdine production, because fewer molecules are necessary to scavenge iron when it is abundant. An alternative explanation for our findings is that increased pyoverdine production could be sustainable in these environments, because the cost‐to‐benefit ratio of this trait is altered. Costs might be relatively low because the pyoverdine production level is still considerably lower compared to what strains produce in the low‐iron/low‐viscosity environment (Figure 1). Benefits might be relatively high because pyoverdine is still required to scavenge iron bound to the bipyridyl chelator. Overall, our results suggest that increased resource availability may favour the evolution of higher cooperative efforts because cooperation becomes cheaper while its benefits persist (Brockhurst et al., 2008).
We found that mutations in QS regulatory genes (lasR, rhlR and pqsR) and full deletions of the Las‐regulon were associated with an increase in pyoverdine production, suggesting a link between pyoverdine and QS regulons. We propose two non‐mutually exclusive reasons to explain why these mutations were favoured in our experiment. First, mutations in QS loci may be advantageous per se, and pyoverdine hyperproduction may simply be a by‐product of these mutations. In our growth medium, QS‐controlled traits such as proteases or biosurfactants are not needed, so abolishing QS could save substantial metabolic costs (Özkaya et al., 2018). In support of this by‐product hypothesis, we found that QS‐mutants occurred in all environments (Figure 6d), suggesting that it is generally favourable to lose QS. Second, mutations in QS genes may be advantageous because they cause pyoverdine hyperproduction. Our results also support this hypothesis, because pyoverdine hyperproducers did not spread in the low‐iron/low‐viscosity environment, where cheaters dominated. Instead, they reached high frequencies in high‐viscosity environments, where cooperation is predicted to be favourable. Thus, mutations in the QS‐regulon may accomplish two goals at once: silencing of a superfluous regulon, and the possibility to increase pyoverdine cooperation.
The evolutionary trajectories and associated genetic changes we describe are remarkably similar to patterns of P. aeruginosa evolution during chronic infections in humans (Marvig et al., 2015; Winstanley et al., 2016). For example, longitudinal studies on cystic fibrosis patients with P. aeruginosa lung infections revealed that social traits are often under selection (Andersen et al., 2015; Jiricny et al., 2014). For instance, both pyoverdine hyper‐ and nonproducers evolve in human patients in sequential steps (Andersen et al., 2018). Furthermore, the widespread accumulation of QS mutants that we observed in the laboratory has parallels in human lungs (Bjarnsholt et al., 2010; Wilder et al., 2009) and in animal infection models (Granato et al., 2018; Jansen et al., 2015). Moreover, hypermutators and isolates that over‐express efflux pumps are also commonly observed in clinical isolates (Mena et al., 2008; Rees et al., 2019; Sobel et al., 2005), where they seem to contribute to antibiotic resistance. Consistently, we found hypermutators in almost all our experimental treatments, and the widespread spreading of efflux pump mutants, probably in response to bipyridyl toxicity. Together, these parallels suggest that in vitro studies, despite their obvious limitations, may help understand general evolutionary trajectories taken by pathogens.
In conclusion, we found that P. aeruginosa can quickly evolve alternative social phenotypes to match prevailing abiotic (iron availability) and biotic (relatedness) conditions. Cheating, which was the focus of many previous studies, seems to be only favoured in one of the environments studied here. Instead, our data suggest that P. aeruginosa adapts its pyoverdine production profile to match environmental requirements, often by up‐regulating pyoverdine production, but never by losing the trait altogether. We further show that social traits should not be studied in isolation, as they are connected through an intricate regulatory network (Balasubramanian et al., 2013). Selection for changes in one trait, such as the loss of QS, affect other traits, such as increased pyoverdine production. An integrative approach that considers the multifaceted social profiles of bacteria is needed to fully understand the evolution of sociality in these microbes.
AUTHOR CONTRIBUTIONS
A.R.T.F., A.W. and R.K. designed the research; A.R.T.F. performed the research and analysed the data; A.R.T.F., A.W. and R.K. interpreted the data and wrote the paper.
CONFLICTS OF INTERESTS
The authors declare no conflict of interest.
Supporting information
Supplementary Material
Table S1, S6‐S10
ACKNOWLEDGMENTS
The authors would like to thank Carla Bello‐Cabrera and Michael Baumgartner for bioinformatics assistance, Jos Kramer for statistical advice and two anonymous reviewers for valuable comments. This work was supported by the University Research Priority Program “Evolution in Action” of the University of Zurich; the European Research Council (Grant Agreement No. 739874 to A.W.; Grant Agreement No. 681295 to R.K.), and by Swiss National Science Foundation (grant 31003A_172887 to A.W.). Open Access Funding provided by Universitat Zurich.
Figueiredo, A. R. T. , Wagner, A. , & Kümmerli, R. (2021). Ecology drives the evolution of diverse social strategies in Pseudomonas aeruginosa . Molecular Ecology, 30, 5214–5228. 10.1111/mec.16119
DATA AVAILABILITY STATEMENT
The phenotypic data underlying this article are available in the Dryad Digital Repository (https://doi.org/10.5061/dryad.34tmpg4kw). The sequencing data can be found in the European Nucleotide Database (https://www.ebi.ac.uk/ena/), under accession no. PRJEB45376.
REFERENCES
- Ackermann, M. , Stecher, B. , Freed, N. E. , Songhet, P. , Hardt, W. D. , & Doebeli, M. (2008). Self‐destructive cooperation mediated by phenotypic noise. Nature, 454, 987–990. 10.1038/nature07067 [DOI] [PubMed] [Google Scholar]
- Andersen, S. B. , Ghoul, M. , Marvig, R. L. , Lee, Z. B. , Molin, S. , Johansen, H. K. , & Griffin, A. S. (2018). Privatisation rescues function following loss of cooperation. Elife, 7:e38594. 10.7554/eLife.38594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, S. B. , Marvig, R. L. , Molin, S. , Johansen, H. K. , & Griffin, A. S. (2015). Long‐term social dynamics drive loss of function in pathogenic bacteria. Proceedings of the National Academy of Sciences of the United States of America, 112(34), 10756–10761. 10.1073/pnas.1508324112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S. (2010). FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc [Google Scholar]
- Balasubramanian, D. , Schneper, L. , Kumari, H. , & Mathee, K. (2013). A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Research, 41(1), 1–20. 10.1093/nar/gks1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiaans, E. , Debets, A. J. M. , & Aanen, D. K. (2016). Experimental evolution reveals that high relatedness protects multicellular cooperation from cheaters. Nature Communications, 7:11435. 10.1038/ncomms11435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. , & Hochberg, Y. (1995). Controlling the False Discovery Rate : A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological), 57(1), 289–300. [Google Scholar]
- Bjarnsholt, T. , Jensen, P. Ø. , Jakobsen, T. H. , Phipps, R. , Nielsen, A. K. , Rybtke, M. T. , Tolker‐Nielsen, T. , Givskov, M. , Høiby, N. , Ciofu, O. , & the Scandinavian Cystic Fibrosis Study Consortium (2010). Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS One, 5(4), e10115–. 10.1371/journal.pone.0010115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst, M. A. , Buckling, A. , Racey, D. , & Gardner, A. (2008). Resource supply and the evolution of public‐goods cooperation in bacteria. BMC Biology, 6:20. 10.1186/1741-7007-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling, A. , Harrison, F. , Vos, M. , Brockhurst, M. A. , Gardner, A. , West, S. A. , & Griffin, A. (2007). Siderophore‐mediated cooperation and virulence in Pseudomonas aeruginosa. FEMS Microbiology Ecology, 62(2), 135–141. 10.1111/j.1574-6941.2007.00388.x [DOI] [PubMed] [Google Scholar]
- Butaite, E. , Baumgartner, M. , Wyder, S. , & Kümmerli, R. (2017). Siderophore cheating and cheating resistance shape competition for iron in soil and freshwater Pseudomonas communities. Nature Communications, 8: 414. 10.1038/s41467-017-00509-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani, P. , Platts, A. , Wang, L. L. , Coon, M. , Nguyen, T. , Wang, L. , Land, S. J. , Lu, X. , & Ruden, D. M. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly, 6(2), 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe, H. E. , Merriman, T. R. , & Lamont, I. L. (1995). Cloning and characterization of pvdS, a gene required for pyoverdine synthesis in Pseudomonas aeruginosa: PvdS is probably an alternative sigma factor. Journal of Bacteriology, 177(10), 2744–2750. 10.1128/jb.177.10.2744-2750.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza, G. , Shitut, S. , Preussger, D. , Yousif, G. , Waschina, S. , & Kost, C. (2018). Ecology and evolution of metabolic cross‐feeding interactions in bacteria. Natural Product Reports, 35, 455–488. 10.1039/c8np00009c [DOI] [PubMed] [Google Scholar]
- Davis, K. M. , & Isberg, R. R. (2019). One for All, but Not All for One: Social Behavior during Bacterial Diseases. Trends in Microbiology, 27(1), 64–74. 10.1016/j.tim.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison, R. F. , Bledsoe, C. , Kahn, M. , O’Gara, F. , Simms, E. L. , & Thomashow, L. S. (2003). Cooperation in the rhizosphere and the “free rider” problem. Ecology, 84(4), 838–845. 10.1890/0012‐9658(2003)084[0838:CITRAT]2.0.CO;2 [Google Scholar]
- Dobay, A. , Bagheri, H. C. , Messina, A. , Kümmerli, R. , & Rankin, D. J. (2014). Interaction effects of cell diffusion, cell density and public goods properties on the evolution of cooperation in digital microbes. Journal of Evolutionary Biology, 27(9), 1869–1877. 10.1111/jeb.12437 [DOI] [PubMed] [Google Scholar]
- Dragoš, A. , Kiesewalter, H. , Martin, M. , Hsu, C.‐Y. , Hartmann, R. , Wechsler, T. , Eriksen, C. , Brix, S. , Drescher, K. , Stanley‐Wall, N. , Kümmerli, R. , & Kovács, Á. T. (2018). Division of Labor during Biofilm Matrix Production. Current Biology, 28(12), 1903–1913.e5. 10.1016/j.cub.2018.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas, Z. , & Kümmerli, R. (2012). Cost of cooperation rules selection for cheats in bacterial metapopulations. Journal of Evolutionary Biology, 25(3), 473–484. 10.1111/j.1420-9101.2011.02437.x [DOI] [PubMed] [Google Scholar]
- Ebrahimi, A. , Schwartzman, J. , & Cordero, O. X. (2019). Cooperation and spatial self‐organization determine rate and efficiency of particulate organic matter degradation in marine bacteria. Proceedings of the National Academy of Sciences of the United States of America, 116(46), 23309–23316. 10.1073/pnas.1908512116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mouden, C. , & Gardner, A. (2008). Nice natives and mean migrants: The evolution of dispersal‐dependent social behaviour in viscous populations. Journal of Evolutionary Biology, 21(6), 1480–1491. 10.1111/j.1420-9101.2008.01614.x [DOI] [PubMed] [Google Scholar]
- Ewels, P. , Magnusson, M. , Lundin, S. , & Käller, M. (2016). MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics, 32(19), 3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo, A. R. T. , & Kramer, J. (2020). Cooperation and Conflict Within the Microbiota and Their Effects On Animal Hosts. Frontiers in Ecology and Evolution, 8: 132. 10.3389/fevo.2020.00132 [DOI] [Google Scholar]
- Ghoul, M. , Griffin, A. S. , & West, S. A. (2014. a). Toward an evolutionary definition of cheating. Evolution, 68(2), 318–331. 10.1111/evo.12266 [DOI] [PubMed] [Google Scholar]
- Ghoul, M. , & Mitri, S. (2016). The Ecology and Evolution of Microbial Competition. Trends in Microbiology, 24(10), 833–845. 10.1016/j.tim.2016.06.011 [DOI] [PubMed] [Google Scholar]
- Ghoul, M. , West, S. A. , Diggle, S. P. , & Griffin, A. S. (2014. b). An experimental test of whether cheating is context dependent. Journal of Evolutionary Biology, 27(3), 551–556. 10.1111/jeb.12319 [DOI] [PubMed] [Google Scholar]
- Gilbert, O. M. , Foster, K. R. , Mehdiabadi, N. J. , Strassmann, J. E. , & Queller, D. C. (2007). High relatedness maintains multicellular cooperation in a social amoeba by controlling cheater mutants. Proceedings of the National Academy of Sciences of the United States of America, 104(21), 8913–8917. 10.1073/pnas.0702723104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato, E. T. , & Kümmerli, R. (2017). The path to re‐evolve cooperation is constrained in Pseudomonas aeruginosa. BMC Evolutionary Biology, 17, 214. 10.1186/s12862-017-1060-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato, E. T. , Meiller‐Legrand, T. A. , & Foster, K. R. (2019). The Evolution and Ecology of Bacterial Warfare. Current Biology, 29(11), R521–R537. 10.1016/j.cub.2019.04.024 [DOI] [PubMed] [Google Scholar]
- Granato, E. T. , Ziegenhain, C. , Marvig, R. L. , & Kümmerli, R. (2018). Low spatial structure and selection against secreted virulence factors attenuates pathogenicity in Pseudomonas aeruginosa. ISME Journal, 12(12), 2907–2918. 10.1038/s41396-018-0231-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin, A. S. , West, S. A. , & Buckling, A. (2004). Cooperation and competition in pathogenic bacteria. Nature, 430, 1024–1027. 10.1038/nature02802.1 [DOI] [PubMed] [Google Scholar]
- Hamilton, W. D. (1964). The genetical evolution of social behaviour. I. Journal of Theoretical Biology, 7(1), 1–16. 10.1016/0022-5193(64)90038-4 [DOI] [PubMed] [Google Scholar]
- Harrison, F. (2013). Dynamic social behaviour in a bacterium: Pseudomonas aeruginosa partially compensates for siderophore loss to cheats. Journal of Evolutionary Biology, 26(6), 1370–1378. 10.1111/jeb.12126 [DOI] [PubMed] [Google Scholar]
- Harrison, F. , Mcnally, A. , Silva, A. C. , Heeb, S. , & Diggle, S. P. (2017). Optimised chronic infection models demonstrate that siderophore ‘cheating’ in Pseudomonas aeruginosa is context specific. ISME Journal, 11, 2492–2509. 10.1038/ismej.2017.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbing, M. E. , Fuqua, C. , Parsek, M. R. , & Peterson, S. B. (2010). Bacterial competition: Surviving and thriving in the microbial jungle. Nature Reviews Microbiology, 8, 15–25. 10.1038/nrmicro2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. , Shao, X. , Xie, Y. , Wang, T. , Zhang, Y. , Wang, X. , & Deng, X. (2019). An integrated genomic regulatory network of virulence‐related transcriptional factors in Pseudomonas aeruginosa. Nature Communications, 10: 2931. 10.1038/s41467-019-10778-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis, R. F. , Biernaskie, J. M. , Gardner, A. , & Kümmerli, R. (2016). Presence of a loner strain maintains cooperation and diversity in well‐mixed bacterial communities. Proceedings of the Royal Society B: Biological Sciences, 283: 20152682, 10.1098/rspb.2015.2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, G. , Crummenerl, L. L. , Gilbert, F. , Mohr, T. , Pfefferkorn, R. , Thänert, R. , Rosenstiel, P. , & Schulenburg, H. (2015). Evolutionary transition from pathogenicity to commensalism: Global regulator mutations mediate fitness gains through virulence attenuation. Molecular Biology and Evolution, 32(11), 2883–2896. 10.1093/molbev/msv160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny, N. , Diggle, S. P. , West, S. A. , Evans, B. A. , Ballantyne, G. , Ross‐Gillespie, A. , & Griffin, A. S. (2010). Fitness correlates with the extent of cheating in a bacterium. Journal of Evolutionary Biology, 23(4), 738–747. 10.1111/j.1420-9101.2010.01939.x [DOI] [PubMed] [Google Scholar]
- Julou, T. , Mora, T. , Guillon, L. , Croquette, V. , Schalk, I. J. , Bensimon, D. , & Desprat, N. (2013). Cell‐cell contacts confine public goods diffusion inside Pseudomonas aeruginosa clonal microcolonies. Proceedings of the National Academy of Sciences, 110(31), 12577–12582. 10.1073/pnas.1301428110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, T. , Ouertatani‐Sakouhi, H. , Cosson, P. , & Van Delden, C. (2014). QsrO a novel regulator of quorum‐sensing and virulence in Pseudomonas aeruginosa. PLoS One, 9(2), e87814. 10.1371/journal.pone.0087814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, J. , Özkaya, Ö. , & Kümmerli, R. (2020). Bacterial siderophores in community and host interactions. Nature Reviews Microbiology, 18, 152–163. 10.1038/s41579-019-0284-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmerli, R. , Griffin, A. S. , West, S. A. , Buckling, A. , & Harrison, F. (2009). Viscous medium promotes cooperation in the pathogenic bacterium Pseudomonas aeruginosa . Proceedings of the Royal Society B: Biological Sciences, 276, 3531–3538. 10.1098/rspb.2009.0861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmerli, R. , Santorelli, L. A. , Granato, E. T. , Dumas, Z. , Dobay, A. , Griffin, A. S. , & West, S. A. (2015a). Co‐evolutionary dynamics between public good producers and cheats in the bacterium Pseudomonas aeruginosa. Journal of Evolutionary Biology, 28(12), 2264–2274. 10.1111/jeb.12751 [DOI] [PubMed] [Google Scholar]
- Lee, W. , van Baalen, M. , & Jansen, V. A. A. (2012). An evolutionary mechanism for diversity in siderophore‐producing bacteria. Ecology Letters, 15(2), 119–125. 10.1111/j.1461-0248.2011.01717.x [DOI] [PubMed] [Google Scholar]
- Lee, Y. , Kim, Y. J. , Lee, J. H. , Yu, H. E. , Lee, K. , Jin, S. , & Ha, U. H. (2016). TatC‐dependent translocation of pyoverdine is responsible for the microbial growth suppression. Journal of Microbiology, 54(2), 122–130. 10.1007/s12275-016-5542-9 [DOI] [PubMed] [Google Scholar]
- Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA‐MEM. arXiv (pp. 1–3). Retrieved from http://arxiv.org/abs/1303.3997 [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , Marth, G. , Abecasis, G. , Durbin, R. , & 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25(16), 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, H. , Deng, X. , Li, X. , Ye, Y. , & Wu, M. (2014). Molecular mechanisms of master regulator VqsM mediating quorum‐sensing and antibiotic resistance in Pseudomonas aeruginosa. Nucleic Acids Research, 42(16), 10307–10320. 10.1093/nar/gku586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, A. E. F. , Robinson, C. J. , Peterson, S. B. , Raffa, K. F. , & Handelsman, J. (2008). Rules of engagement: Interspecies interactions that regulate microbial communities. Annual Review of Microbiology, 62, 375–401. 10.1146/annurev.micro.030608.101423 [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Yang, L. , & Molin, S. (2010). Synergistic activities of an efflux pump inhibitor and iron chelators against Pseudomonas aeruginosa growth and biofilm formation. Antimicrobial Agents and Chemotherapy, 54(9), 3960–3963. 10.1128/AAC.00463-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnúsdóttir, S. , Ravcheev, D. , Crécy‐Lagard, V. , & Thiele, I. (2015). Systematic genome assessment of B‐vitamin biosynthesis suggests cooperation among gut microbes. Frontiers in Genetics, 6: 148. 10.3389/fgene.2015.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvig, R. L. , Sommer, L. M. , Molin, S. , & Johansen, H. K. (2015). Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nature Genetics, 47, 57–64. 10.1038/ng.3148 [DOI] [PubMed] [Google Scholar]
- Mena, A. , Smith, E. E. , Burns, J. L. , Speert, D. P. , Moskowitz, S. M. , Perez, J. L. , & Oliver, A. (2008). Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. Journal of Bacteriology, 190(24), 7910–7917. 10.1128/JB.01147-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehus, R. , Picot, A. , Oliveira, N. M. , Mitri, S. , & Foster, K. R. (2017). The evolution of siderophore production as a competitive trait. Evolution, 71(6), 1443–1455. 10.1111/evo.13230 [DOI] [PubMed] [Google Scholar]
- O’Brien, S. , Kümmerli, R. , Paterson, S. , Winstanley, C. , & Brockhurst, M. A. (2019). Transposable temperate phages promote the evolution of divergent social strategies in Pseudomonas aeruginosa populations. Proceedings of the Royal Society B‐Biological Sciences, 286, 28620191794. 10.1101/593798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien, S. , Luján, A. M. , Paterson, S. , Cant, M. A. , & Buckling, A. (2017). Adaptation to public goods cheats in Pseudomonas aeruginosa. Proceedings of the Royal Society B: Biological Sciences, 284: 20171089. 10.1098/rspb.2017.1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, A. , Baquero, F. , & Blázquez, J. (2002). The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: Molecular characterization of naturally occurring mutants. Molecular Microbiology, 43(6), 1641–1650. 10.1046/j.1365-2958.2002.02855.x [DOI] [PubMed] [Google Scholar]
- Özkaya, Ö. , Balbontín, R. , Gordo, I. , & Xavier, K. B. (2018). Cheating on Cheaters Stabilizes Cooperation in Pseudomonas aeruginosa. Current Biology, 28(13), 2070–2080. 10.1016/j.cub.2018.04.093 [DOI] [PubMed] [Google Scholar]
- Özkaya, Ö. , Xavier, K. B. , Dionisio, F. , & Balbontín, R. (2017). Maintenance of microbial cooperation mediated by public goods in single‐ and multiple‐trait scenarios. Journal of Bacteriology, 199:e00297‐17. 10.1128/JB.00297-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez, J. , Moraleda‐Muñoz, A. , Marcos‐Torres, F. J. , & Muñoz‐Dorado, J. (2016). Bacterial predation: 75 years and counting! Environmental Microbiology, 18(3), 766–779. 10.1111/1462-2920.13171 [DOI] [PubMed] [Google Scholar]
- Queller, D. C. (1994). Genetic relatedness in viscous populations. Evolutionary Ecology, 8, 70–73. [Google Scholar]
- R Core Team (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing. [Google Scholar]
- Rees, V. E. , Deveson Lucas, D. S. , López‐Causapé, C. , Huang, Y. , Kotsimbos, T. , Bulitta, J. B. , Rees, M. C. , Barugahare, A. , Peleg, A. Y. , Nation, R. L. , Oliver, A. , Boyce, J. D. , & Landersdorfer, C. B. (2019). Characterization of Hypermutator Pseudomonas aeruginosa Isolates from Patients with Cystic Fibrosis in Australia. Antimicrobial Agents and Chemotherapy, 63(4), e02538–‐8. 10.1128/AAC.02538-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringel, M. T. , & Brüser, T. (2018). The Biosynthesis of Pyoverdines. Microbial Cell, 5(10), 424–437. 10.15698/mic2018.10.649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J. T. , Thorvaldsdóttir, H. , Winckler, W. , Guttman, M. , Lander, E. S. , Getz, G. , & Mesirov, J. P. (2011). Integrative Genome Viewer. Nature Biotechnology, 29, 24–26. 10.1038/nbt.1754.Integrative [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, T. , Smith, P. , Alberts, E. R. , Colussi‐Pelaez, M. , & Schuster, M. (2020). Cooperation and cheating through a secreted aminopeptidase in the pseudomonas aeruginosa RpoS response. MBio, 11(2), e03090–19. 10.1128/mBio.03090-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross‐Gillespie, A. , Dumas, Z. , & Kümmerli, R. (2015). Evolutionary dynamics of interlinked public goods traits: An experimental study of siderophore production in Pseudomonas aeruginosa. Journal of Evolutionary Biology, 28(1), 29–39. 10.1111/jeb.12559 [DOI] [PubMed] [Google Scholar]
- Schalk, I. J. , Rigouin, C. , & Godet, J. (2020). An overview of siderophore biosynthesis among fluorescent Pseudomonads and new insights into their complex cellular organization. Environmental Microbiology, 22(4), 1447–1466. 10.1111/1462-2920.14937 [DOI] [PubMed] [Google Scholar]
- Smith, E. E. , Sims, E. H. , Spencer, D. H. , Kaul, R. , & Olson, M. V. (2005). Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. Journal of Bacteriology, 187(6), 2138–2147. 10.1128/JB.187.6.2138-2147.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, P. , & Schuster, M. (2019). Public goods and cheating in microbes. Current Biology, 29(11), R442–R447. 10.1016/j.cub.2019.03.001 [DOI] [PubMed] [Google Scholar]
- Sobel, M. L. , Hocquet, D. , Cao, L. , Plesiat, P. , & Poole, K. (2005). Mutations in PA3574 (nalD) lead to increased MexAB‐OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy, 49(5), 1782–1786. 10.1128/AAC.49.5.1782-1786.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilwell, P. , Lowe, C. , & Buckling, A. (2018). The effect of cheats on siderophore diversity in Pseudomonas aeruginosa. Journal of Evolutionary Biology, 31(9), 1330–1339. 10.1111/jeb.13307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassmann, J. E. , & Queller, D. C. (2011). Evolution of cooperation and control of cheating in a social microbe. Proceedings of the National Academy of Sciences of the United States of America, 108(Suppl 2), 10855–10862. 10.1073/pnas.1102451108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnita, C. E. (2017). The ecology and evolution of social behavior in microbes. Journal of Experimental Biology, 220(1), 18–24. 10.1242/jeb.145631 [DOI] [PubMed] [Google Scholar]
- Tostado‐Islas, O. , Mendoza‐Ortiz, A. , Ramírez‐García, G. , Cabrera‐Takane, I. D. , Loarca, D. , Pérez‐González, C. , Jasso‐Chávez, R. , Jiménez‐Cortés, J. G. , Hoshiko, Y. , Maeda, T. , Cazares, A. , & García‐Contreras, R. (2021). Iron limitation by transferrin promotes simultaneous cheating of pyoverdine and exoprotease in Pseudomonas aeruginosa. ISME Journal, 15, 2379–2389. 10.1038/s41396‐021‐00938‐6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travisano, M. , & Velicer, G. J. (2004). Strategies of microbial cheater control. Trends in Microbiology, 12(2), 72–78. 10.1016/j.tim.2003.12.009 [DOI] [PubMed] [Google Scholar]
- Velicer, G. J. , Kroos, L. , & Lenski, R. E. (1998). Loss of social behaviors by Myxococcus xanthus during evolution in an unstructured habitat. Proceedings of the National Academy of Sciences of the United States of America, 95(21), 12376–12380. 10.1073/pnas.95.21.12376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, S. C. , & Miyashiro, T. (2013). Quorum sensing in the squid‐Vibrio symbiosis. International Journal of Molecular Sciences, 14(8), 16386–16401. 10.3390/ijms140816386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux, R. , Filloux, A. , & Schalk, I. J. (2006). Pyoverdine‐mediated iron uptake in Pseudomonas aeruginosa: The Tat system is required for PvdN but not for FpvA transport. Journal of Bacteriology, 188(9), 3317–3323. 10.1128/JB.188.9.3317-3323.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler, T. , Kümmerli, R. , & Dobay, A. (2019). Understanding policing as a mechanism of cheater control in cooperating bacteria. Journal of Evolutionary Biology, 32(5), 412–424. 10.1111/jeb.13423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigert, M. , & Kümmerli, R. (2017). The physical boundaries of public goods cooperation between surface‐attached bacterial cells. Proceedings of the Royal Society B: Biological Sciences, 284: :20170631. 10.1098/rspb.2017.0631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, S. A. , Diggle, S. P. , Buckling, A. , Gardner, A. , & Griffin, A. S. (2007a). The Social Lives of Microbes. Annual Review of Ecology Evolution and Systematics, 38, 53–77. 10.1146/annurev.ecolsys.38.091206.095740 [DOI] [Google Scholar]
- West, S. A. , Griffin, A. S. , & Gardner, A. (2007b). Social semantics: Altruism, cooperation, mutualism, strong reciprocity and group selection. Journal of Evolutionary Biology, 20(2), 415–432. 10.1111/j.1420-9101.2006.01258.x [DOI] [PubMed] [Google Scholar]
- Wiegand, I. , Marr, A. K. , Breidenstein, E. B. M. , Schurek, K. N. , Taylor, P. , & Hancock, R. E. W. (2008). Mutator genes giving rise to decreased antibiotic susceptibility in Pseudomonas Aeruginosa . Antimicrobial Agents and Chemotherapy, 52(10), 3810–3813. 10.1128/AAC.00233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder, C. N. , Allada, G. , & Schuster, M. (2009). Instantaneous within‐patient diversity of Pseudomonas aeruginosa quorum‐sensing populations from cystic fibrosis lung infections. Infection and Immunity, 77(12), 5631–5639. 10.1128/IAI.00755-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder, C. N. , Diggle, S. P. , & Schuster, M. (2011). Cooperation and cheating in Pseudomonas aeruginosa: the roles of the las, rhl and pqs quorum‐sensing systems. ISME Journal, 5, 1332–1343. 10.1038/ismej.2011.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor, G. L. , Lo, R. , Ho Sui, S. J. , Ung, K. S. E. , Huang, S. , Cheng, D. , & Brinkman, F. S. L. (2005). Pseudomonas aeruginosa Genome Database and PseudoCAP: Facilitating community‐based, continually updated, genome annotation. Nucleic Acids Research, 33(suppl_1), D338–D343. 10.1093/nar/gki047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstanley, C. , O’Brien, S. , & Brockhurst, M. A. (2016). Pseudomonas aeruginosa Evolutionary Adaptation and Diversification in Cystic Fibrosis Chronic Lung Infections. Trends in Microbiology, 24(5), 327–337. 10.1016/j.tim.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , & Rainey, P. B. (2013). Exploring the sociobiology of pyoverdin‐producing pseudomonas. Evolution, 67(11), 3161–3174. 10.1111/evo.12183 [DOI] [PubMed] [Google Scholar]
- Jiricny N., Molin S., Foster K., Diggle S., Scanlan P., Ghoul M., Johansen H. K., Santorelli L. A., Popat R., West S. A., Griffin A. S. (2014). Loss of Social Behaviours in Populations of Pseudomonas aeruginosa Infecting Lungs of Patients with Cystic Fibrosis. PLoS ONE, 9(1), e83124. 10.1371/journal.pone.0083124 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Table S1, S6‐S10
Data Availability Statement
The phenotypic data underlying this article are available in the Dryad Digital Repository (https://doi.org/10.5061/dryad.34tmpg4kw). The sequencing data can be found in the European Nucleotide Database (https://www.ebi.ac.uk/ena/), under accession no. PRJEB45376.


