Abstract
Acral melanoma is the major subtype of melanoma in Chinese patients. However, a majority of current studies focused on non‐acral melanoma. Most immortalized melanoma cell lines and primary cells were not from acral melanoma. Besides, there are rarely reports about methods for establishing primary acral melanoma cell cultures and related animal models. Here, we present four new human primary acral melanoma cell lines. To determine the mutational profile of the established primary melanoma cells for future targeted use, we performed exome sequencing. We next examined cell proliferation of the primary acral melanoma cells by colony‐formation assays and CCK8 assay. We also evaluated the proliferative and metastatic potential of XYAM‐4 in vivo. We report a detailed protocol for establishing cultured primary acral melanoma cells for Chinese patients and related animal models. We also summarize the features in our acral melanoma cell lines and the existing acral melanoma cell lines. This will provide an effective research tool for research on drug responses and individualized treatment for Chinese patients and comparative studies of melanomas between western and Chinese populations.
Keywords: acral melanoma, Chinese patients, melanoma mutation, primary cell, primary cell culture
1. INTRODUCTION
Malignant melanoma has a poor survival rate and is the main subtype of melanoma in Asian populations, accounting for more than 60% of all melanomas (Haugh et al., 2018). However, a majority of current studies focus on non‐acral melanoma instead and immortalized melanoma cell lines are widely used—that is, the primary cells are not acral melanoma cells. Acral melanoma has unique clinical, morphological, and genetic characteristics, as well as aggressive histopathologic features and a poorer survival rate than other melanomas. Most cutaneous melanomas have gene mutations, including BRAF, NRAS, or NF1 mutations. In contrast, most acral melanomas lack these mutations, and rates of KIT and KRAS mutations were found to be higher in acral melanomas (Moon et al., 2018 ). Additionally, acral melanoma has a wider range of structural variants and a lower mutational burden than cutaneous melanoma (Zhang et al., 2019). Therefore, neither immortalized melanoma cell lines nor other primary melanoma cells are reliable models for acral melanoma‐related research. It thus becomes necessary to culture primary acral melanoma cells. In the present study, we established a simple method for culturing primary acral melanoma cells and established animal models for Chinese patients.
2. MATERIALS AND METHODS
2.1. Sources of patient samples
Samples were removed from patients in the Department of Dermatology, at Xiangya Hospital, who were diagnosed with malignant melanoma between August 2018 and June 2019. All enrolled patients provided written informed consent, and the study was approved by the Institutional Review Board.
2.2. Reagent preparation
The wash buffer consisted of phosphate‐buffered solution (PBS) supplemented with 500 U/ml penicillin, 500 U/ml streptomycin and 6.25 U/ml nystatin (GENVIEW, Cat# GA3503). Our collection medium was Opti‐MEM® I reduced‐serum medium supplemented with 5% foetal bovine serum (GIBCO, Cat# 31985070) and antibiotics (200 U/ml penicillin, 200 U/ml streptomycin and 2.5 U/ml nystatin). The complete Opti‐MEM medium included Opti‐MEM® I Reduced Serum Medium supplemented with 5% foetal bovine serum (GIBCO, Cat# 10099‐141) and antibiotics (100 U/ml penicillin, 100 U/ml streptomycin and 1.25 U/ml nystatin).
2.3. Sample transport and processing
Tumour tissues were placed into 50 ml centrifuge tubes, each containing 10 ml of collection medium, after removal from the patients. The samples, kept at a temperature of 4°C, were immediately transported to the laboratory. As all samples were excised from the heels or the thumbs of the patients, the prevention of fungal contamination was crucial. Therefore, portions of tumour tissue in the epidermis were removed, with the aim of preserving as much material as possible due to its rarity. The tissues were then rinsed in wash buffer five times, and once more in PBS.
2.4. Preparation of single‐cell suspensions from melanoma tumour tissues
To obtain single melanoma cell suspensions using sterile scissors and tweezers, we first removed adipose or and necrotic tissue before cutting melanoma tumour tissues into fragments (with volumes less than 1 mm3) on sterile cell culture dishes. Tissue digestion was required for our experiment, but acral melanoma tumour tissues are rigid and not easily digested. By exploring different procedures, we found that a combination of 0.2% collagenase IV (Sigma, Cat# V900893) and 0.1% Dispase II enzymes (Sigma, Cat# D4693) was optimal for the digestion of acral melanoma tumour tissues. The tissues were incubated in a solution containing collagenase IV and dispase II enzymes for approximately 30 min at 37°C. Then, the cell suspensions were filtered through a sterile cell strainer and centrifuged at 1000 rpm for 10 min at room temperature. Then, the resulting pellets were collected and resuspended in 2 ml of complete Opti‐MEM each, along with 100 U/ml penicillin, 100 U/ml streptomycin and 1.25 U/ml nystatin, in a 3.5 cm dish. These dishes were then incubated at 37°C in 5% CO2. After 2 days, the medium was removed and the cells were washed with PBS. The medium was refreshed every 2 days until the primary cells reached confluence.
2.5. Cell culture and cell purification
Cells obtained from the abovementioned steps may contain fibroblasts. However, fibroblasts adhere less tightly to the dish than malignant cells and can be detached by short treatments with EDTA‐free trypsin digestion solutions. Cells suspended after the first 1 min of centrifugation were discarded, and the same treatment was repeated several times to obtain a more pure collection of primary melanoma cells.
2.6. DNA Extraction and Exome Sequencing
DNA was extracted using a QIAamp DNA Mini Kit (QIAGEN GmbH, Cat# 51304), and exome capture was performed using a Nimble Gen SeqCap Med Exome 44M Kit. This was followed by paired‐end sequencing on the Illumina HiSeq platform. The calling of variants, including single‐nucleotide variants (SNVs) and indels, was accomplished by GATK. The called variants were filtered by Hard filtering and Variant Quality Score Recalibration and further annotated using ANNOVAR.
2.7. Colony‐formation assays
Primary acral melanoma cells were plated in 6‐well (800 cells per well) or in 12‐well plates (400 cells per well). Fresh medium was added every 3 days. Two weeks after planting, cells in 6‐well plates were fixed with methanol (Servicebio, Cat#G1101) and stained with crystal violet (Beyotime Biotechnology, Cat#0121) for 20 min. The same process was applied to cells in 12‐well plates 4 weeks after planting.
2.8. Cell proliferation
Primary acral melanoma cells were plated in 96‐well plates (800 cells per well). At various time points, 90 μl fresh medium and 10 μl CCK8 (Bimake, Cat#B34302) reagent were added to each well. Absorbance values were measured at 450 nm after 2.5 hr of incubation.
2.9. Animal study
Female NSG (NOD scid gamma) mice (6–8 weeks old) were obtained from the Department of Laboratory Animals at Central South University. All studies were conducted according to experimental protocols approved by the Ethical Review of Experimental Animals at Central South University. 2 × 106 XYAM‐4 cells in 100 μl PBS were injected subcutaneously into the right flank of each NSG mouse. Tumour size and body weight were recorded at the indicated time points, with the former determined via vernier calliper measurements and calculated using [length × width2]/2. The mice were euthanized at the end point of the experiment, or when they became moribund with rapid weight loss, hunched postures and laboured breathing. All mice were euthanized via cervical dislocation after intraperitoneal injection of 0.1% pentobarbital sodium (30 mg/kg). All efforts were made to minimize animal suffering and reduce the number of animals involved in this experiment.
3. RESULTS
3.1. Establishment of cultured primary acral melanoma cell cultures
In this study, we report methods for establishing primary acral melanoma cell cultures (Figure 1). Clinical data from each patient are summarized in Table 1. All patients were diagnosed with malignant acral melanoma at the Department of Dermatology at Xiangya Hospital. We also summarize the clinical data of the existing acral melanoma cell lines in Table 1, which can used by researchers when choosing cell line models.
FIGURE 1.
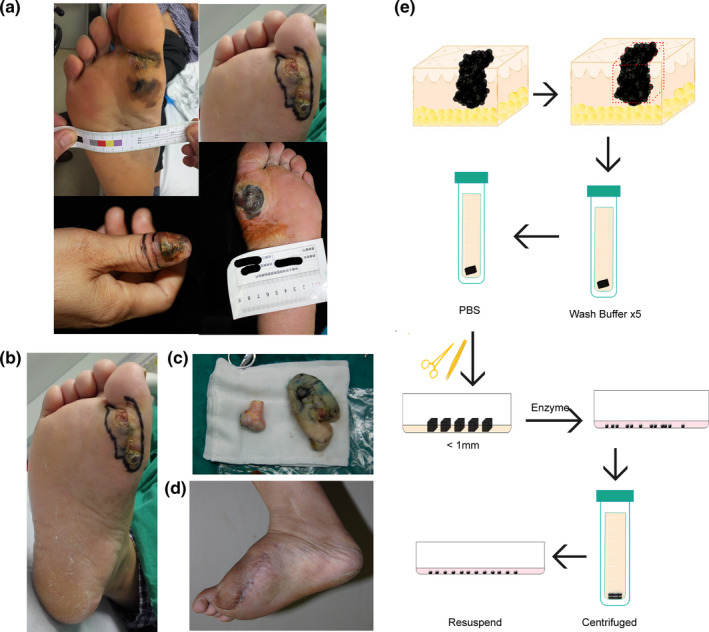
Representative images of acral melanoma in patients and the abridged general view of methods used to establish primary acral melanoma cell cultures. (a) Representative images of acral melanoma in patients; (b‐d) Preoperative photographs of an acral melanoma patient; (e) The abridged general view of the methods used to acquire primary acral melanoma cells
TABLE 1.
Clinical characteristics of the patients
| Cell | Patient ethnicity | Age (year) | Sex | Chief complaint | Source | Primary site | Primary histology |
|---|---|---|---|---|---|---|---|
| XYAM‐1 | Chinese | 55 | M | Mass increased for 2 months | Primary tumour | Right heel | ALM |
| XYAM‐2 | Chinese | 84 | M | Black spot increased and thicken for 3 months | Primary tumour | Right thumb | ALM |
| XYAM‐3 | Chinese | 50 | M | Papule increased for 7 months, extravasated for 1 month | Primary tumour | Right heel | ALM |
| XYAM‐4 | Chinese | 71 | M | Mass increased with ulcer for 6 months | Primary tumour | Left heel | ALM |
| SMYM‐PRGP | Japanese | 69 | M | Unknown | Primary tumour | Sole | ALM (Radial Growth Phase) |
| MMG1 | Japanese | Unknown | Unknown | Unknown | Primary tumour | Sole | ALM (Vertical Growth Phase) |
| WM3211 | Unknown | Unknown | Unknown | Unknown | Primary tumour | Ankle | ALM (Radial Growth Phase) |
| SM3 | Japanese | Unknown | Unknown | Unknown | Primary tumour | Sole | ALM (Radial Growth Phase) |
| Mel‐2 | Japanese | Unknown | Unknown | Unknown | In‐transit subcutaneous metastasis | Sole | ALM |
| SM2‐1 | Japanese | Unknown | Unknown | Unknown | In‐transit subcutaneous metastasis | Sole | Regressed |
| Mel18 | Japanese | Unknown | Unknown | Unknown | Lymph node metastasis | Nailbed | ALM |
3.2. Morphology and melanoma marker expression in primary acral melanoma cell cultures
The primary melanoma cells obtained are shown in Figure 2a. We used a fluorescence assay to confirm whether the cells we obtained were melanoma cells, labelling them with S100 and vimentin, which are relatively specific diagnostic markers for melanoma. Encouragingly, our results showed positive protein expression of S100 and vimentin in our cells, which can be seen in Figure 2b,c.
FIGURE 2.
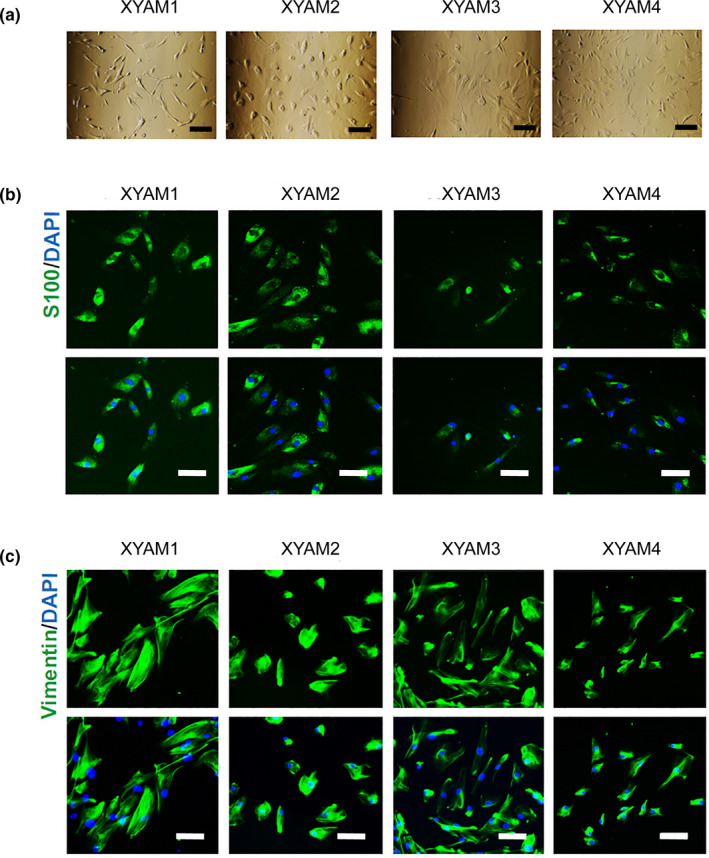
Morphology and melanoma marker expression in primary acral melanoma cell cultures after seeding. (a) Images of primary acral melanoma cells captured at X200 magnification. Scale bars: 200 μm; (b‐c) Detection of S100 and vimentin‐labelled primary melanoma cells via fluorescence assay. Representative images of primary melanoma cells captured at X200 magnification; (b) Detection of S100‐labelled primary melanoma cells; (c) Detection of vimentin‐labelled primary melanoma cells
3.3. Mutation in acral melanoma cells
To examine the mutational profiles of acral melanoma cells, we extracted DNA from primary acral melanoma cells and performed exome sequencing. To summarize mutational alterations observed in our acral melanoma cell lines and the existing acral melanoma cell lines, we assembled a tabular resource, which may be useful for researchers to decide which one to choose in their studies (Figure 3). Detail mutation‐related information was summarized in Table S1. We found only one of our primary cell lines harboured a KRAS mutation. However, three of them had TP53 mutations (Figure 3 and Table S1).
FIGURE 3.

Exon mutation summary of acral melanoma cells. The selected genetic events include 25 possible mutations (BRAF, NRAS, KRAS, NF1, TP53, IDH1, GNA11, GNAQ, CDK4, CDKN2A, DDX3X, KIT, MAP2K1, PPP6C, PTEN, RAC1, RB1, ARID2, TERT, CTNNB1, ITGA4, GPR179, CBL, GRK7 and APC) determined to be significantly altered in the landmark melanoma study by The Cancer Genome Atlas
3.4. Proliferation and metastasis of primary acral melanoma cells
We then examined the population dependence and cell proliferation of primary acral melanoma cells using Cell Counting Kit 8 and colony‐formation assays. XYAM‐1 and XYAM‐4 cells had faster proliferation rates than XYAM‐2 and XYAM‐3 cells in vitro (Figure 4a,b). XYAM‐4 cells formed 100 mm3 tumours approximately 3 weeks after subcutaneous injection into NSG mice (Figure 4c‐f). To evaluate the metastatic potential of XYAM‐4 in vivo, NSG mice were injected subcutaneously with XYAM‐4 cells. Debulking surgery was conducted on the mice when XYAM‐4 cells formed tumours approximated 1000 mm3 in size, at around 5 weeks. The mice were euthanized after a further 3 weeks, when new 1000 mm3 tumours formed (Figure 4g). Ultimately, micrometastasis of XYAM‐4 cells was discovered through haematoxylin and eosin staining (Figure 4h). We examined the expressions of S100 and vimentin and found that both were expressed in the lung tissues of XYAM‐4 tumour‐bearing NSG mice (Figure 4i,j).
FIGURE 4.
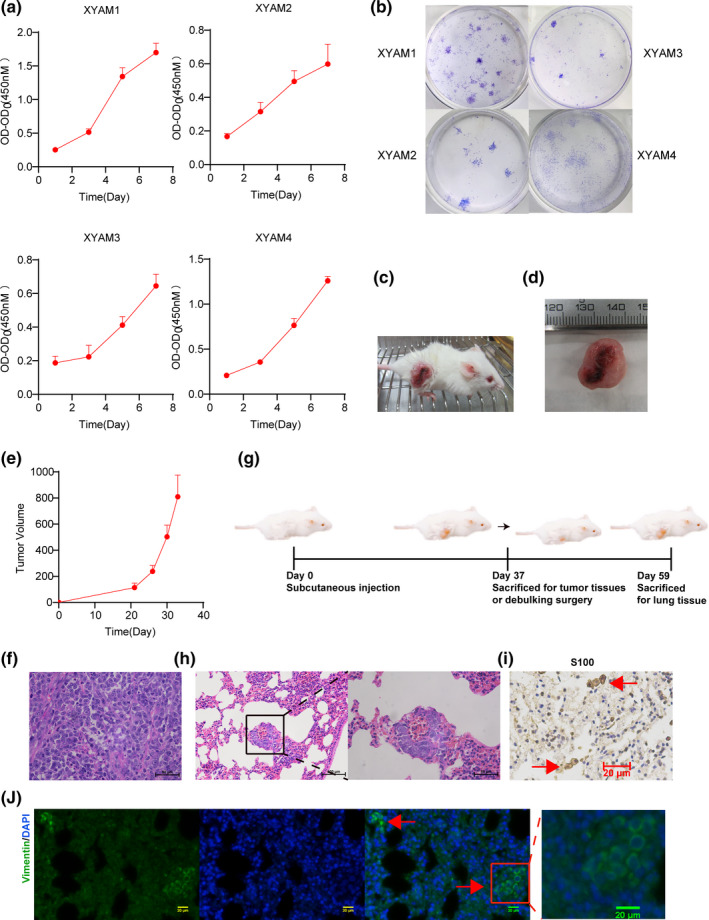
Growth characteristics of XYAM cells. (a) The proliferative ability of XYAM‐1/2/3/4 cells was investigated using CCK8; (b) Colony formation assays of XYAM‐1/2/3/4 cells; (c‐e) NSG mice were injected subcutaneously with XYAM‐4 cells. Tumour volume was calculated at the indicated time points. Representative images (c) and tumour grow curves (e) of an XYAM‐4 tumour‐bearing NSG mouse; (d) Representative XYAM‐4 tumour image; (f) Representative H&E stain of XYAM‐4 tumour tissues; (g) Experimental protocol of XYAM‐4 metastatic potential in animal model; (h) Representative H&E staining of lung tissue in XYAM‐4 tumour‐bearing NSG mice; (i) Immunohistochemistry (IHC) for S100 in the lung tissues of XYAM‐4 tumour‐bearing NSG mice; (j) Immunofluorescence stain for vimentin in the lung tissues of XYAM‐4 tumour‐bearing NSG mice
4. DISCUSSION
Acral melanoma is the major subtype of melanoma in Chinese patients, and its treatment remains a challenge to current methods. There is currently a growing awareness of the necessity of using primary cells for research on tumour biology and therapeutics. However, acral melanoma cell lines are more difficult to establish and there are few reports on methods for establishing primary acral melanoma cell cultures (Furney et al., 2012; Murata et al., 2007). Here, we describe our simple and effective culturing methods and present four new human primary acral melanoma cell lines, gathered from a Chinese population. We also summarize the features in our acral melanoma cell lines and the existing acral melanoma cell lines and it will be valuable tools for researchers investigating this melanoma subtype.
S100 and vimentin are relatively specific diagnostic markers of melanoma (Romano et al., 2015), and the cell lines tested positive for S100 and vimentin. We performed exome sequencing to determine the mutational profiles of the established primary melanoma cells for future targeted use, finding that most of the acral melanoma primary cells had mutations in TP53. The TP53 mutation is the most frequent mutation in human tumours and is suggested to be related to both tumour evasion and tumour progression. Notably, previous works indicated that acral melanomas do not have mutations in TP53 or only 9.1% of metastatic melanoma patients had previously been noted to harbour TP53 mutations (Xiao et al., 2018; Hayward et al., 2017), whereas three‐quarters of the acral melanoma primary cells that we obtained had a TP53 mutation. One primary acral melanoma cell line also harboured a KRAS mutation. It has been reported that melanoma patients with high KRAS expression have reduced overall survival, and previous research showed that 3%–13.3% of acral melanomas had KRAS mutations (Sheen et al., 2020). Our results may be due to our small sample size, but they may also reflect the different genetic backgrounds of different races.
Acral melanoma is a major subtype of melanoma in Chinese patients. We published a detailed protocol for establishing cultured primary acral melanoma cells from Chinese patients and evaluated their proliferative and metastatic potential using related animal models. This protocol will provide an effective tool for research on drug responses and for individualized treatments for Chinese patients, as well as for comparative studies of melanomas between Western and Chinese populations.
However, this study has several limitations. There are no therapeutic responses to evaluate cell lines and animal models. It may also be necessary to evaluate whether human tumours have changed in murine stroma during serial passaging. These limitations will be addressed in future studies.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interests.
AUTHOR CONTRIBUTION
R.H., X. C. and M. Y. contributed to conception and design, development of methodology and writing, review and/or revision of the manuscript; R.H. and S. Z. contributed to acquisition of data; S. Z. and J.S. contributed to analysis and interpretation of data; X. C. and M. Y. contributed to study supervision; All authors read and approved the final manuscript.
Supporting information
Table S1
ACKNOWLEDGEMENTS
This study was supported by grants No. 81874138 from General Program, The National Natural Science Foundation of China, Major Projects of International Cooperation and Exchanges NSFC Grand No.81620108024.
Hu R, Zhao S, Su J, Chen X, & Yin M. (2021). Establishment of cultured primary acral melanoma cells and animal models for Chinese patients. Pigment Cell & Melanoma Research, 34, 1131–1137. 10.1111/pcmr.12996
Contributor Information
Xiang Chen, Email: chenxiangck@126.com.
Mingzhu Yin, Email: yinmingzhu2008@126.com.
DATA AVAILABILITY STATEMENT
The exome sequencing data referenced in this study are available under PRJNA725194 in the SRA database. All the data supporting this study are available within the article and from the corresponding authors upon reasonable request.
REFERENCES
- Furney, S. J. , Turajlic, S. , Fenwick, K. , Lambros, M. B. , MacKay, A. , Ricken, G. , Mitsopoulos, C. , Kozarewa, I. , Hakas, J. , Zvelebil, M. , Lord, C. J. , Ashworth, A. , Reis‐Filho, J. S. , Herlyn, M. , Murata, H. , & Marais, R. (2012). Genomic characterisation of acral melanoma cell lines [J]. Pigment Cell & Melanoma Research, 25(4), 488–492. [DOI] [PubMed] [Google Scholar]
- Haugh, A. M. , Zhang, B. , Quan, V. L. , Garfield, E. M. , Bubley, J. A. , Kudalkar, E. , Verzi, A. E. , Walton, K. , VandenBoom, T. , Merkel, E. A. , Lee, C. Y. , Tan, T. , Isales, M. C. , Kong, B. Y. , Wenzel, A. T. , Bunick, C. G. , Choi, J. , Sosman, J. , & Gerami, P. (2018). Distinct patterns of acral melanoma based on site and relative sun exposure [J]. The Journal of Investigative Dermatology, 138(2), 384–393. [DOI] [PubMed] [Google Scholar]
- Hayward, N. K. , Wilmott, J. S. , Waddell, N. , Johansson, P. A. , Field, M. A. , Nones, K. , Patch, A. M. , Kakavand, H. , Alexandrov, L. B. , Burke, H. , Jakrot, V. , Kazakoff, S. , Holmes, O. , Leonard, C. , Sabarinathan, R. , Mularoni, L. , Wood, S. , Qinying, X. , Waddell, N. , … Mann, G. J. (2017). Whole‐genome landscapes of major melanoma subtypes [J]. Nature, 545(7653), 175–180. [DOI] [PubMed] [Google Scholar]
- Moon, K. R. , Choi, Y. D. , Kim, J. M. , Jin, S. , Shin, M. H. , Shim, H. J. , Lee, J. B. , & Yun, S. J. (2018). Genetic alterations in primary acral melanoma and acral melanocytic nevus in Korea: Common mutated genes show distinct cytomorphological features [J]. The Journal of Investigative Dermatology, 138(4), 933–945. [DOI] [PubMed] [Google Scholar]
- Murata, H. , Ashida, A. , Takata, M. , Yamaura, M. , Bastian, B. C. , & Saida, T. (2007). Establishment of a novel melanoma cell line SMYM‐PRGP showing cytogenetic and biological characteristics of the radial growth phase of acral melanomas [J]. Cancer Science, 98(7), 958–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, R. C. , Carter, J. M. , & Folpe, A. L. (2015). Aberrant intermediate filament and synaptophysin expression is a frequent event in malignant melanoma: An immunohistochemical study of 73 cases [J]. Modern Pathology, 28(8), 1033–1042. [DOI] [PubMed] [Google Scholar]
- Sheen, Y. S. , Tan, K. T. , Tse, K. P. , Liao, Y. H. , Lin, M. H. , Chen, J. S. , Liau, J. Y. , Tseng, Y. J. , Lee, C. H. , Hong, C. H. , Liao, J. B. , Chang, H. T. , & Chu, C. Y. (2020). Genetic alterations in primary melanoma in Taiwan [J]. British Journal of Dermatology, 182(5), 1205–1213. [DOI] [PubMed] [Google Scholar]
- Xiao, W. , Du, N. , Huang, T. , Guo, J. , Mo, X. , Yuan, T. , Chen, Y. , Ye, T. , Xu, C. , Wang, W. , Wang, G. , Cai, S. , & Chen, J. (2018). TP53 mutation as potential negative predictor for response of anti‐CTLA‐4 therapy in metastatic melanoma [J]. EBioMedicine, 32, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Peng, Y. , Li, C. , Li, Q. , Yu, Z. , Pang, Y. , Wu, A. R. , Huang, Y. , & Li, H. (2019). Genomic heterogeneity and branched evolution of early stage primary acral melanoma shown by multiregional microdissection sequencing [J]. The Journal of Investigative Dermatology, 139(7), 1526–1534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Data Availability Statement
The exome sequencing data referenced in this study are available under PRJNA725194 in the SRA database. All the data supporting this study are available within the article and from the corresponding authors upon reasonable request.


