Summary
The biological importance of lanthanides has only recently been identified, initially as the active site metal of the alternative methanol dehydrogenase (MDH) Xox‐MDH. So far, the effect of lanthanide (Ln) has only been studied in relatively few organisms. This work investigated the effects of Ln on gene transcription and protein expression in the facultative methanotroph Methylocella silvestris BL2, a widely distributed methane‐oxidizing bacterium with the unique ability to grow not just on methane but also on other typical components of natural gas, ethane and propane. Expression of calcium‐ or Ln‐dependent MDH was controlled by Ln (the lanthanide switch) during growth on one‐, two‐ or three‐carbon substrates, and Ln imparted a considerable advantage during growth on propane, a novel result extending the importance of Ln to consumers of this component of natural gas. Two Xox‐MDHs were expressed and regulated by Ln in M. silvestris, but interestingly Ln repressed rather than induced expression of the second Xox‐MDH. Despite the metabolic versatility of M. silvestris, no other alcohol dehydrogenases were expressed, and in double‐mutant strains lacking genes encoding both Ca‐ and Ln‐dependent MDHs (mxaF and xoxF5 or xoxF1), growth on methanol and ethanol appeared to be enabled by expression of the soluble methane monooxygenase.
Introduction
Methane, the most abundant hydrocarbon in the atmosphere and a potent greenhouse gas, is one of the most significant contributors to climate change (Saunois et al., 2016). Biogenic methane results from the metabolism of methanogenic archaea, but in addition, thermogenic methane‐rich natural gas is produced as a result of heat‐ and pressure‐mediated breakdown of buried organic material. Much natural gas is released to the atmosphere as a result of fossil fuel extraction, but naturally released thermogenic methane, from diffuse seepage or point releases such as macro‐seeps, mud volcanoes and volcanoes, accounts for 14% of all methane from natural sources and usually contains other short‐chain alkanes, mostly ethane and propane. Approximately 3 and 2 Tg y−1 of ethane and propane, respectively, are released to the atmosphere in natural gas (Etiope and Ciccioli, 2009; Dalsøren et al., 2018), which sometimes contains up to 35 vol% ethane and propane (Etiope and Klusman, 2010; Schimmelmann et al., 2018). Release of methane to the atmosphere is reduced by methane‐oxidizing microbes (methanotrophs), which consume over half of the methane produced in soils, sediments and the water column (Reeburgh, 2007) and emissions of ethane and propane are similarly mediated by microbes, but whose relative contribution is unknown (Beilen and Funhoff, 2005; Shennan, 2006; Kinnaman et al., 2007; Redmond et al., 2010).
The genus Methylocella comprises several species of aerobic facultative methanotrophs from the Alphaproteobacteria (type II), which are active in many environments and are sometimes the most abundant methanotrophs (Farhan Ul Haque et al., 2020). Methylocella is unique among methanotrophs as it grows robustly on a range of multi‐carbon compounds, including, for several of the Methylocella strains, not only methane but also ethane and propane (Dedysh et al., 2005; Crombie and Murrell, 2014; Farhan Ul Haque et al., 2019). These short‐chain alkanes are oxidized by soluble diiron centre monooxygenases to the corresponding alcohols: methanol, ethanol and 1‐ and 2‐propanol (Crombie and Murrell, 2014).
Methanol dehydrogenase (MDH) is the second enzyme of the methane metabolic pathway, catalyzing the conversion of methanol to formaldehyde. For many years, this was thought to be exclusively carried out, in methanotrophs, by a soluble pyrroloquinoline quinone (PQQ)‐containing periplasmic enzyme (Mxa‐MDH), made up of large and small subunits (MxaF and MxaI), in an α2β2 structure, with a calcium ion at the active site (Anthony, 2004). A specific cytochrome c L (MxaG) accepts electrons from MDH, and another protein, MxaJ, annotated as a solute‐binding protein but of unknown function, is required for functional MDH (Amaratunga et al., 1997), and several genes at the same locus are involved in metal insertion and enzyme assembly and maturation. Recently, it was shown that a gene often present in methylotrophs (xoxF) encodes an homologous MDH (Xox‐MDH, approximately 50% amino acid identity between XoxF and MxaF), which uses a rare earth lanthanide (Ln), rather than calcium as cofactor (Fitriyanto et al., 2011; Hibi et al., 2011; Nakagawa et al., 2012; Pol et al., 2014), reviewed by Picone and Op den Camp (2018). The xox gene clusters also typically encode MxaG and MxaJ homologues (XoxG and XoxJ), but Xox‐MDH does not usually include a small subunit (although the enzyme from Candidatus Methylomirabilis oxyfera was purified as an α2β2 hybrid of XoxF and MxaI; Wu et al., 2015). Several studies have suggested that Xox‐MDH is as environmentally relevant, if not more so, than Mxa‐MDH (Taubert et al., 2015; Chistoserdova, 2016; Huang et al., 2019; Li et al., 2019), and it is also worth noting that rare earth elements are not, in fact, particularly rare in the Earth's crust (Tyler, 2004). Knowledge of Ln‐dependent MDH has enabled identification of Ln‐dependent methanol utilization in methanotrophs, methylotrophs, and members of genera not previously known for methylotrophy (Pol et al., 2014; Lv et al., 2018; Farhan Ul Haque et al., 2019; Huang et al., 2019; Kato et al., 2020). Based on sequence data, five clades of Xox‐MDHs can be identified, XoxF1–XoxF5, with additional Ln‐dependent homologues with activity towards other substrates (Keltjens et al., 2014). Clade 5 appears to be the most abundant, although many organisms contain multiple enzymes from the same or different clades (Taubert et al., 2015). Several Xox enzymes have been purified and the structures of examples from clades 2 and 5 have been determined, in addition to Mxa‐MDH. In both Mxa‐MDH and Xox‐MDH, the large subunit forms a superbarrel made up of eight four‐stranded beta sheets, arranged radially like the blades of a propeller with, in Mxa‐MDH, the small subunit folded round the surface (Anthony, 2004; Keltjens et al., 2014; Pol et al., 2014; Deng et al., 2018; Jahn et al., 2018; Good et al., 2020).
Both Mxa‐ and Xox‐MDHs contain a conserved catalytic Asp residue (position D303 in MxaF from Methylorubrum extorquens, formerly Methylobacterium extorquens) and the Ln‐dependent Xox‐MDHs can be identified by a second Asp residue two positions downstream, replacing Ala in MxaF (A305 in MxaF from M. extorquens), required for coordination of the lanthanide atom (Anthony, 2004; Keltjens et al., 2014; Good et al., 2020).
The advantage to a methanotroph of this methanol‐oxidizing flexibility is unclear, but it may be important as a response to changing environmental conditions or for controlling the symbiotic transfer of metabolites (e.g. methanol) to other members of the microbial community (Krause et al., 2016; Yu et al., 2017).
Although several examples lack one or other form of MDH, both Mxa‐MDH and Xox‐MDH are present in many methylotrophs and methanotrophs, where they are differentially regulated by the lanthanides also required for Xox‐MDH activity (Skovran and Martinez‐Gomez, 2015; Chistoserdova, 2019). This ‘lanthanide switch’ has been studied in a few methylotrophs and methanotrophs (Keltjens et al., 2014; Skovran and Martinez‐Gomez, 2015; Chistoserdova, 2016; Semrau et al., 2018; Chistoserdova, 2019; Skovran et al., 2019), demonstrating that in the presence of Ln, transcription and expression of Mxa‐MDH is repressed and that of Xox‐MDH is enhanced. Our understanding of the mechanisms of regulation of MDH, both in response to different substrates and to metals, is incomplete, but the best‐studied system is probably from Methylorubrum extorquens. The genome of this methylotroph encodes a pair of two‐component regulators, mxcQE and mxbDM, and a response regulator mxaB, all of which are proposed to be involved in Ln‐dependent regulation of methanol oxidation, and in addition the XoxF protein itself was shown to be part of the regulatory cascade (reviewed by Skovran et al., 2019 and Chistoserdova, 2019). The lanthanide switch has been investigated in a few methanotrophs including the alphaproteobacterium Methylosinus trichosporium OB3b, and the gammaproteobacteria Methylomonas sp. strain LW13 and Methylotuvimicrobium buryatense 5GB1C (formerly Methylomicrobium) (Farhan Ul Haque et al., 2015; Chu et al., 2016; Chu and Lidstrom, 2016; Zheng et al., 2018). Clearly, there are differences in the regulation of the lanthanide switch between facultative methylotrophs (Methylorubrum) and many methanotrophs. For example, neither Methylosinus trichosporium OB3b nor Methylotuvimicrobium buryatense 5GB1C contain close homologues of mxcQE or mxbDM, although both contain sensor histidine kinases which may sense lanthanides (Semrau et al., 2018; Skovran et al., 2019).
The role of lanthanides in biology was only recently identified (Cotruvo, 2019) and much remains unknown or poorly understood regarding the importance and role of these elements in one‐ and multi‐carbon metabolism. The genus Methylocella contains facultative methanotrophs able to grow well on both one‐carbon and multi‐carbon compounds and the aims of this study were to take the first steps towards an understanding of the role of lanthanides in the best‐studied example, Methylocella silvestris BL2.
Results and discussion
MDH gene transcription
The M. silvestris genome contains five predicted PQQ‐dependent MDH genes, Msil_0471, Msil_1587, Msil_2260, Msil_3149 and Msil_3387. Based on amino acid sequence and the presence of the Ln‐coordinating aspartate residue in Xox‐MDH, these can be identified as encoding Mxa‐ and Xox‐MDHs from clades 5 (two copies), clade 1 and clade 3 (Fig. 1 and Supporting Information Fig. S1), herein referred to as MxaF, XoxF5, XoxF5(2), XoxF1 and XoxF3, respectively. The genome also contains two predicted PQQ‐ADH sequences, Msil_2867 and Msil_3733, clustering with group 9 PQQ‐ADH (Keltjens et al., 2014), both of which also contain the Ln‐coordinating Asp residue, but whose function is unknown. Despite the fact that M. silvestris grows well on ethanol, no other sequences related to characterized PQQ‐dependent ethanol dehydrogenases were found, such as ExaF from Methylorubrum extorquens or PedE/H from Pseudomonas putida (Good et al., 2016; Wehrmann et al., 2017). Examination of the genome sequence in the vicinity of each of these putative MDH genes revealed the presence of the MDH‐associated cytochrome c L (xoxG) and solute‐binding protein (xoxJ), except for the second xoxF5 copy, which lacks any other recognizable MDH‐related sequences nearby (Fig. 2). Since these Ca2+‐ and Ln3+ ‐dependent MDHs were expected to be transcriptionally regulated by the presence of Ln, cells were grown in the absence or presence of Ln and gene expression levels were evaluated by RT‐qPCR (Fig. 3). During growth on methane, the addition of Ln (10 μM) to cultures increased transcription of xoxF5 and repressed mxaF 180‐ and 80‐fold respectively. Transcription of xoxF5(2) and xoxF3 was not significantly affected by the addition of Ln, and the transcription of these two genes appeared to be at a low level based on the cycle threshold (Ct) values. Interestingly, in contrast to xoxF5, xoxF1 was downregulated 24‐fold by the addition of Ln, thus responding similarly to mxaF, despite the presence of the predicted Ln‐coordinating aspartate residue (Supporting Information Fig. S1). It is interesting to note that the XoxF (clade 1) purified from Candidatus Methylomirabilis oxyfera appeared to be a calcium‐containing enzyme (Wu et al., 2015). If this is also the case for the Methylocella protein (68% amino acid identity with the sequence from Ca. M. oxyfera), downregulation in response to Ln might be expected.
Figure 1.
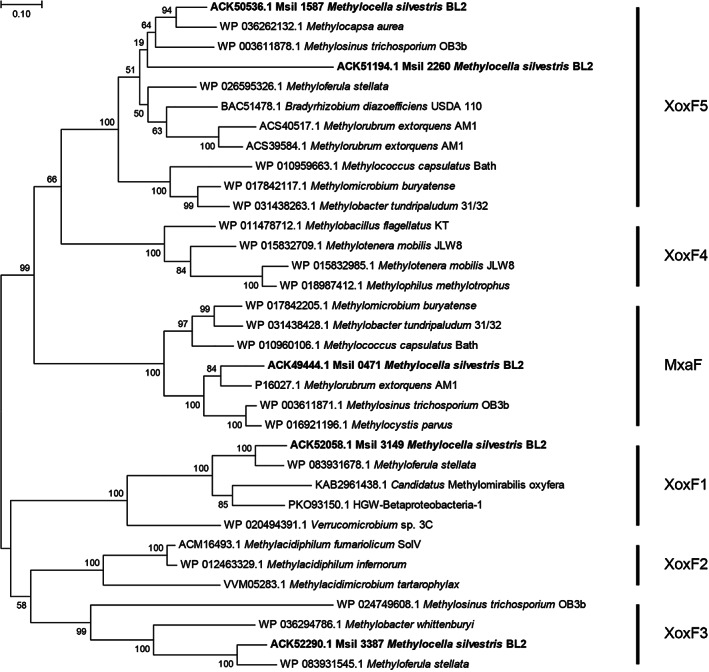
Relationship, based on amino acid sequences, of the putative M. silvestris MDHs (in bold type) with those of representative examples from the databases. The tree was constructed using the maximum likelihood method in MEGA 7 (Kumar et al., 2016). Gaps were eliminated, there were 566 positions in the final dataset and the scale bar shows substitutions per site. Bootstrap values (500 replications) are shown at the nodes.
Figure 2.
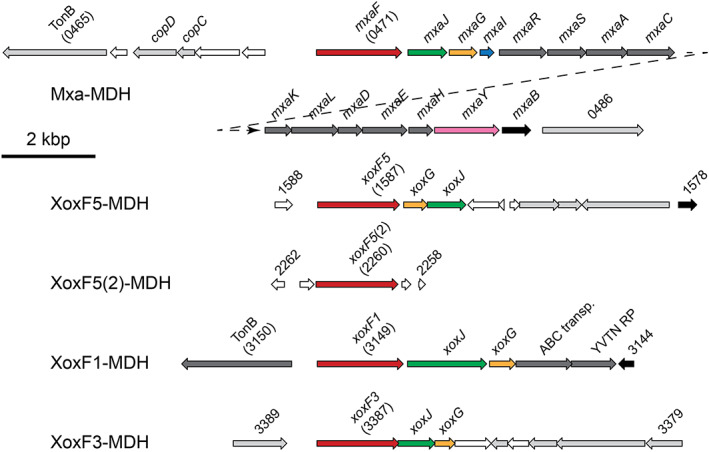
MDH gene clusters in M. silvestris. The genes are colour coded: red, MDH large subunit; blue, small subunit; yellow, cytochrome c; green, solute binding protein; pink, sensor histidine kinase; black, response regulators; dark grey, other MDH‐associated genes; light grey, no direct MDH association; white, hypothetical proteins of unknown function. Four‐digit gene identifications refer to abbreviated locus tags, e.g. 0471, Msil_0471 (mxaF). YVTN RP, 40 residue YVTN repeat protein; TonB, TonB‐dependent receptor.
Figure 3.
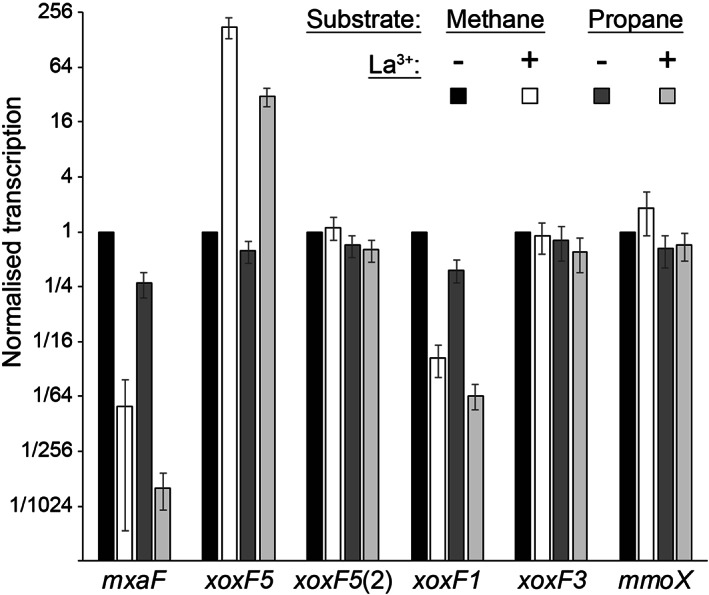
MDH gene transcription in cells grown on methane or propane in the presence or absence of Ln. Transcript levels (fold‐change, normalized to reference gene rpoB) are shown relative to growth on methane without Ln. Error bars show the standard error of the mean (n = 3)
Since M. silvestris can grow on propane, which is oxidized to both 1‐propanol and 2‐propanol (Crombie and Murrell, 2014), MDH transcription was also assessed during growth on propane, and the same pattern of MDH regulation was observed (Fig. 3). Transcription of mmoX, encoding the sMMO alpha subunit and included as an additional control, was not significantly affected, either by growth substrate (as expected, Crombie and Murrell, 2014) or by the addition of Ln. Hence, the lanthanide switch operates in Methylocella, as has been previously observed in both non‐methane‐utilizing methylotrophs and obligate methanotrophs, see Skovran et al. (2019) for a review.
Growth phenotype in the presence or absence of lanthanum
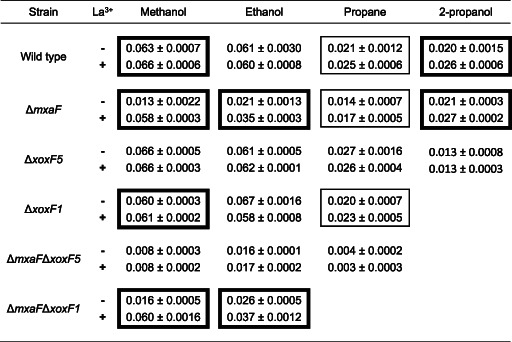
M. silvestris grew slightly better on methanol in the presence compared to the absence of Ln (5 μM) (approximately 6% improvement in growth rate, Table 1; Supporting Information Fig. S2A). Since this indicated an effect of added Ln, unmarked mutants were constructed with deletions of the MDH genes regulated by Ln: mxaF, xoxF5, and xoxF1 and double mutants in mxaF/xoxF5 and mxaF/xoxF1, and the growth phenotype of the mutants was assessed. The presence or absence of Ln made no difference to growth on pyruvate, and strain ΔmxaF grew similarly to the wild type on methanol in Ln‐containing medium, but at approximately one quarter of the rate in the absence of Ln, and without achieving exponential growth (Table 1, Fig. 4A), clearly showing the importance of the Ca‐containing enzyme in medium without Ln, but implying that another enzyme is able to substitute, to a certain extent, for Mxa‐MDH. One possible candidate might be XoxF1, transcriptionally regulated in the same sense as mxaF. However, Ln had a very similar effect on growth of mutant strain ΔxoxF1 as it did in the wild type, i.e. a 3% increase in growth rate in the presence compared to absence of Ln, implying at most a minor role for XoxF1 during growth on methanol, also confirmed by the phenotype of double‐mutant ΔmxaFΔxoxF1, which closely resembled that of strain ΔmxaF (Table 1). Alternatively, it might be that XoxF5 was sufficiently expressed and active in the absence of supplied Ln, for example, if cells were able to scavenge sufficient Ln from glassware (despite precautions), or if the enzyme had sufficient levels of expression and activity in the absence of Ln, to enable this comparatively low growth rate. However, although double‐mutant ΔmxaFΔxoxF5 did not grow discernibly for approximately 300 h, it subsequently grew at approximately 13% of the wild‐type rate (Fig. 4A), irrespective of the presence or absence of Ln. Due to the technical difficulties experienced in construction of these double mutants, construction of a triple mxaF/xoxF1/xoxF5 mutant was not attempted. However, the lack of a distinct phenotype for strain ΔxoxF1 during growth on methanol suggests that a triple mutant would behave similarly to strain ΔmxaFΔxoxF5 under the experimental conditions described here. Together, these data suggest that the combined activity of XoxF5 and XoxF1 was not solely responsible for residual methanol oxidation in strain ΔmxaF without Ln, or in strain ΔmxaFΔxoxF5, and that other non Ln‐dependent methanol‐oxidizing enzyme(s) were expressed under these conditions (discussed below).
Table 1.
Specific growth rate (h−1 ± standard error of the mean) of M. silvestris strains grown on methanol, ethanol, propane or 2‐propanol in the absence or presence of lanthanum.
|
Significant differences of each strain/substrate combination in the absence of presence of lanthanum are shown with wide borders (P ≤ 0.01) or narrow borders (P ≤ 0.05). All cultures in triplicate except: wild type, methanol, n = 9; strain ΔxoxF5, methanol, n = 6; strain ΔxoxF5, ethanol, +La3+, n = 2. For strain ΔmxaFΔxoxF5, the growth rates shown with methanol and propane are following extended lag phases of 300 and 600 h, respectively, as described in the text.
Figure 4.
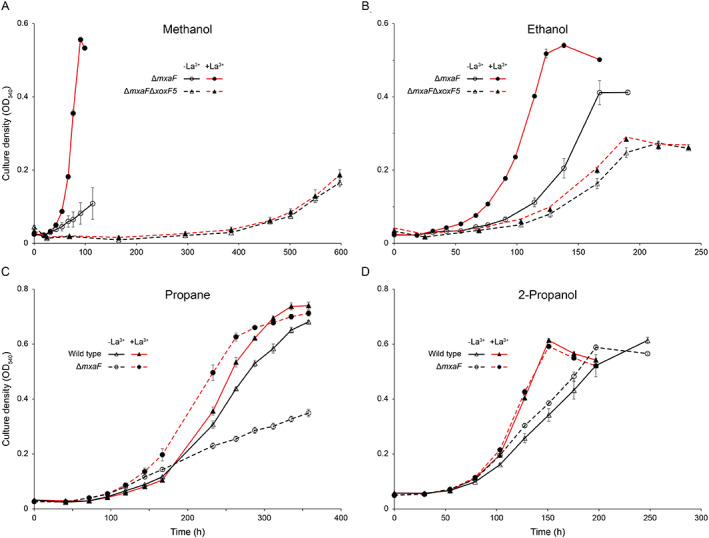
Upper panels, growth of strain ΔmxaF (solid lines) and of double mutant ΔmxaFΔxoxF5 (dashed lines) on (A) methanol (0.1% v/v) or B) ethanol in the absence (black) or presence (red) of Ln (5 μM). Lower panels, growth of the wild type (solid lines) and strain ΔmxaF (dashed lines) on (C) propane (20% v/v) or (D) 2‐propanol (0.05% v/v) in the absence (black) or presence (red) of Ln (5 μM). Error bars show the standard error of the mean (n = 3).
Finally, however, and perhaps surprisingly given the transcriptional switch from mxaF to xoxF5 induced by Ln, strain ΔxoxF5 grew at wild‐type rates irrespective of the presence or absence of Ln (Table 1).
Promoter activity
This lack of a Ln‐dependent growth phenotype in strain ΔxoxF5 suggested either, that under these conditions an alternative enzyme was responsible for methanol oxidation, or that deletion of xoxF5 interfered with the differential transcriptional regulation of mxaF and xoxF5 described above (as has been observed previously in other studies; Chu and Lidstrom, 2016; Zheng et al., 2018; Groom et al., 2019; Ochsner et al., 2019; Yanpirat et al., 2020). Therefore, following the strategy of these earlier researchers, the activity of the promoters of the MDH genes was assessed using promoter‐reporter plasmids, consisting of DNA fragments upstream of mxaF, xoxF5 and xoxF1 (952, 619 and 588 bp respectively) cloned in front of a gfp gene (encoding green fluorescent protein, GFP), which were introduced into the wild type and the single deletion strains. These GFP‐expressing reporter strains were then grown on methanol in the presence or absence of Ln and fluorescence was quantified (Fig. 5). In the wild type, promoter activity mirrored the RT‐qPCR data shown in Fig. 3, with the mxaF and xoxF1 promoter activities reduced 53‐ and 6‐fold, respectively, in the presence compared to the absence of Ln, whereas xoxF5 promoter activity was increased 10‐fold. This same Ln‐dependent promoter activity was evident in mutant strains ΔmxaF and ΔxoxF1. In contrast, in strain ΔxoxF5, the promoter activities of mxaF and xoxF5 were entirely unaffected by Ln while only a minor (not statistically significant) difference in the activity of the xoxF1 promoter was detected. While confirming the operation of the lanthanide switch in the wild type shown by the RT‐qPCR results above, these data show that this transcriptional control of mxaF and xoxF5 (and likely xoxF1) was disabled by deletion of xoxF5, suggesting either that the XoxF5 protein is itself involved in gene regulation, as shown previously (Skovran et al., 2011; Good et al., 2020), and/or that a suppressor mutation developed following gene deletion, allowing transcription of mxaF in the presence of Ln. It was previously shown in both alphaproteobacterial methylotrophs and gammaproteobacterial methanotrophs that suppressor mutations occurred in sensor kinases likely involved directly or indirectly in Ln sensing, thus allowing transcription and expression of Mxa‐MDH in the presence of Ln (Chu and Lidstrom, 2016; Zheng et al., 2018; Ochsner et al., 2019; Yanpirat et al., 2020). Genes encoding a two‐component system comprising sensor histidine kinase MxaY (Msil_0484) and response regulator MxaB (Msil_0485) are located at the end of the Mxa‐MDH gene cluster in M. silvestris. This arrangement is present in several alphaproteobacterial methanotrophs including Methylosinus and Methylocystis strains but differs from Methylorubrum extorquens AM1, which contains only an orphan response regulator mxaB at this locus (Springer et al., 1998). Despite the low sequence identity between M. silvestris MxaY and homologues in the databases (max 43% amino acid identity with examples outside the family Beijerinckiaceae), the location of the mxaY gene, adjacent to Mxa‐MDH, suggested that a mutation might have occurred in this gene. Sequencing of the PCR products, using primers targeting the entire mxaY gene in the wild type and strain ΔxoxF5, revealed that compared with the wild type, all mutant sequences (6/6) had a deletion after nucleotide 402, resulting in a predicted truncated protein of 172 amino acids (compared with 473 in the wild type). While these data do not prove a connection between mxaY mutation and mxaF expression in the presence of Ln, they suggest a role for MxaY in the lanthanide switch in M. silvestris. Interestingly, strain ΔxoxF5 never exhibited a Ln‐dependent phenotype, did not appear to adapt over time to growth in the presence of Ln and grew in Ln‐containing medium without an extended lag phase, suggesting that the suppressor mutation was generated immediately and illustrating the strong selective pressure for efficient methanol oxidation in this strain.
Figure 5.
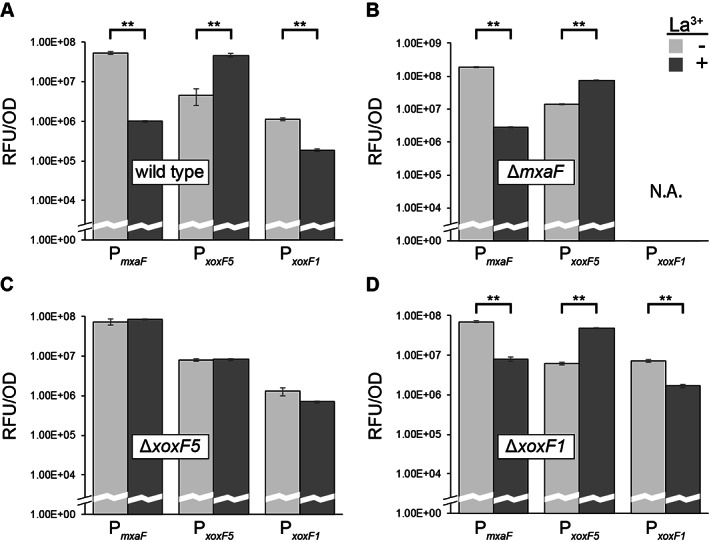
MDH promoter activity of wild‐type and mutant strains during growth on methanol. The bar chart shows background‐subtracted relative fluorescent units (RFU), normalized to cell density, of (A), wild‐type strain; (B), strain ΔmxaF; (C), strain ΔxoxF5 and (D), strain ΔxoxF1, containing reporter constructs consisting of a green fluorescent protein (gfp) gene under the control of the mxaF, xoxF5, or xoxF1 promoters, grown in the presence or absence of Ln (5 μM). Error bars show the standard error of the mean (n = 3) and significant differences between strains grown with or without Ln are shown as ** (P ≤ 0.01). N.A., not assayed.
Growth on ethanol
The effect of Ln on M. silvestris while growing on ethanol resembled its effect during growth on methanol, except that the addition of Ln made no difference to the growth rate of the wild type (Table 1; Supporting Information Fig. S2B). As before, strain ΔmxaF exhibited a severe growth defect without Ln, which was partially alleviated by addition of Ln. Double‐mutant ΔmxaFΔxoxF5 was capable of relatively poor growth irrespective of the presence or absence of Ln, albeit at a low growth rate and to a comparatively low final cell density, but without the extremely long lag phase noticeable during growth on methanol (Fig. 4B). Furthermore, unlike on methanol, strain ΔmxaF achieved exponential rather than linear growth on ethanol in the absence of Ln. As was the case for methanol, the presence or absence of Ln had no effect on the growth of strain ΔxoxF5 on ethanol, which grew at similar rates to the wild type under both conditions, and double‐mutant ΔmxaFΔxoxF1 behaved similarly to strain ΔmxaF (Table 1). These data demonstrate that the lanthanide switch also operates during growth on ethanol and confirm that not only methanol‐oxidizing activity but also ethanol‐oxidizing activity exists in cells lacking both principal MDHs, irrespective of the presence of Ln.
Growth on propane and 2‐propanol
Since M. silvestris can also grow on propane, which is oxidized by M. silvestris to both 1‐propanol and 2‐propanol (although the strain does not grow on 1‐propanol; Crombie and Murrell, 2014), and MDH transcription was regulated by Ln during growth on propane, the response of the wild type and mutant strains to Ln was tested during growth on propane. Notably, the wild type, strain ΔmxaF and strain ΔxoxF1 all grew significantly better (16%–22% increase in growth rate) on propane in the presence compared to the absence of Ln (Table 1, Fig. 4C), demonstrating a Ln‐dependent phenotype. As before, deletion of XoxF5 removed the effect of Ln on growth on propane, confirming that a similar mode of regulation of these enzymes operates during growth on propane as during growth on 1‐ and 2‐carbon substrates. Double‐mutant ΔmxaFΔxoxF5 eventually grew at approximately 16% of the wild‐type rate, but only after an extremely long lag phase of approximately 600 h, during which there was no growth whatsoever, again suggesting the eventual expression of an alternative enzyme in the absence of MDH. Growth on 2‐propanol, one of the products of propane oxidation, was also evaluated with and without Ln. The wild type and strain ΔmxaF behaved in an almost identical manner and addition of Ln stimulated growth almost identically in both strains (27%– 34% increase in growth rate, Fig. 4D), whereas strain ΔxoxF5, which was not affected by Ln, grew at half the rate of the Ln‐supplied wild type (Table 1). These data suggest a minor role for Mxa‐MDH in 2‐propanol oxidation and that Xox‐MDH was more efficient in oxidation of this secondary alcohol. There was a clear Ln‐mediated growth advantage to cells growing on both propane and 2‐propanol and the results suggest that the benefit of Ln during growth on propane is at least partly due to enhanced oxidation of 2‐propanol. While Ln‐dependent enzymes capable of oxidation of secondary alcohols have been described, in general Ln‐dependent enzymes for which methanol is not the primary substrate are distinct from Xox‐MDH and have comparatively poor catalytic properties with methanol (Keltjens et al., 2014; Good et al., 2016; Wehrmann et al., 2017; Huang et al., 2018; Picone and Op den Camp, 2018; Wang et al., 2019). Hence, the finding that M. silvestris uses the same pair of versatile enzymes, one dependent on Ca and one on Ln, for oxidation of C1–C3 primary and secondary alcohols is both novel and surprising.
Protein expression
To observe how gene transcription was correlated with protein expression and to examine the wider impact of the lanthanide switch, protein was extracted from cells grown on methanol or ethanol with or without Ln and the proteomes were determined by liquid chromatography/mass spectrometry (LC/MS). Since double‐mutant strains ΔmxaFΔxoxF5 and ΔmxaFΔxoxF1 were able to grow to a certain extent on all substrates tested, both with and without Ln, albeit, in some cases, with a long lag phase, these strains were included in the proteomic analysis. Strain ΔmxaFΔxoxF5 was grown on methanol or ethanol with or without Ln, and strain ΔmxaFΔxoxF1 was grown on methanol or ethanol, without Ln only, resulting in 10 strain/condition combinations. A total of 2472 proteins were identified by two or more unique peptides. As predicted from transcriptional data, of the five predicted PQQ‐dependent MDHs, only MxaF, XoxF5 and XoxF1 were expressed at appreciable levels. XoxF5(2) (Msil_2260) was not detected in any sample, and XoxF3 (Msil_3387) was expressed at very low levels ranging from 0.00003% to 0.00017% of total protein, Supporting Information Table S1. In comparison, MxaF and XoxF5 were among the most abundant proteins in the wild type, comprising 9.7% and 4.5% of the total during growth on methanol in the absence or presence of Ln respectively (Fig. 6). XoxF1 was expressed at comparatively low levels, three orders of magnitude less than MxaF. To investigate the effect of Ln on protein expression, proteins expressed by cells grown on methanol in the presence of Ln were compared with proteins expressed in its absence, and the same comparison was made with cells grown on ethanol. In total, 44 proteins were differentially expressed (at least fourfold, P ≤ 0.01) during wild‐type growth on methanol with/without Ln, of which 9 were upregulated and 35 downregulated in the presence of Ln (Supporting Information Table S1). During growth on ethanol, 49 proteins were similarly differentially expressed, of which 23 were upregulated. The Ln‐induced regulation of MDH proteins, similar for growth on both methanol and ethanol (Table 2), reflected the RT‐qPCR transcriptional data. During growth on methanol, in comparison with the Ln‐free condition, the presence of Ln induced 54‐fold higher expression of XoxF5 (Fig. 6) and the XoxF5‐associated cytochrome c L and SBP (XoxG and XoxJ) were also up‐regulated (ninefold and sevenfold, Table 2). In contrast, MxaF was downregulated ninefold in Ln‐containing medium, together with the associated proteins of the Mxa‐MDH operon (Msil_0472–0485), downregulated between sixfold and 28‐fold. Interestingly, and as predicted by RT‐qPCR data, XoxF1 (Msil_3149) was also downregulated (sevenfold) by Ln, together with its associated cytochrome c L, XoxG (Msil_3147) and solute‐binding protein XoxJ (Msil_3148) (sevenfold and 62‐fold, respectively). Of the Xox‐MDH enzymes, clades 5, 4 and 2 have been most well characterized (Picone and Op den Camp, 2018), with the exception of the XoxF clade 1 enzyme purified from the anaerobe Ca. Methylomirabilis oxyfera (Wu et al., 2015). This unusual methanotroph contains a single cluster of genes predicted to encode three MDH homologues, Mxa‐MDH and two Xox enzymes from clades 1 and 2. The XoxF1‐MDH from Ca. M. oxyfera was purified with high activity and surprisingly seemed to contain Ca rather than Ln (as mentioned above), despite the presence of the diagnostic Ln‐coordinating aspartate residue (D305 in the XoxF5 from Methylorubrum extorquens), also present in M. silvestris XoxF1 (Supporting Information Fig. S1) (Keltjens et al., 2014; Wu et al., 2015; Good et al., 2020). In M. silvestris, the mutant phenotypes suggested that XoxF1 plays a minor role under the experimental conditions used here, but XoxF1 was clearly down‐regulated, at both RNA and protein level, by Ln in the wild type, perhaps suggesting that this enzyme might also contain Ca rather than Ln. Therefore, more work is needed to determine the role and properties of this enzyme in M. silvestris.
Figure 6.
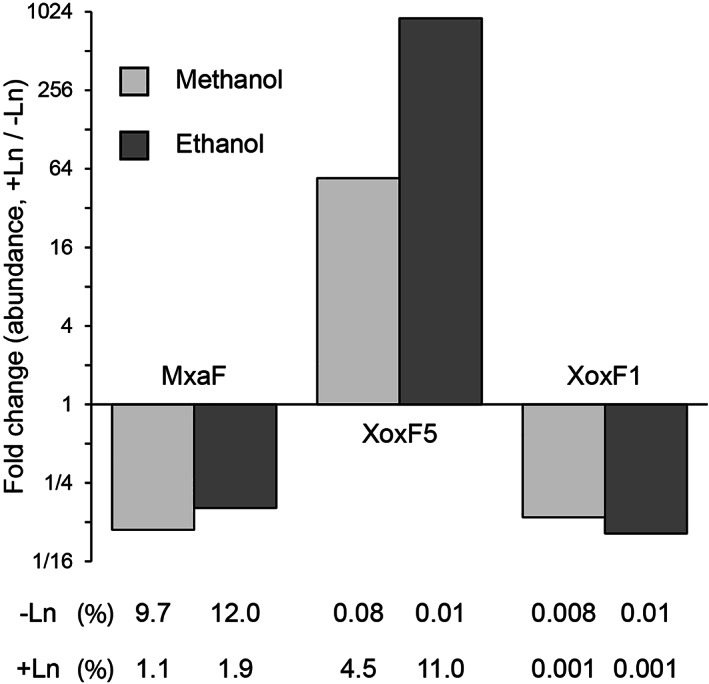
Change in abundance of MDH proteins in wild type M. silvestris cells grown on methanol or ethanol in the presence Ln, compared with growth without Ln. The figures below the bars show the abundance of each protein in the presence or absence of Ln as percentage of total protein.
Table 2.
Relative abundance of MDH proteins and proteins thought to be involved in the lanthanide switch in wild‐type M. silvestris grown on methanol or ethanol in the presence compared with the absence of lanthanum.
| Methanol | Ethanol | ||||
|---|---|---|---|---|---|
| log2 (FC) | P value | log2 (FC) | P value | ||
| Mxa‐MDH | |||||
| Msil_0471 | MxaF (large s/u) | −3.2 | 2.8E‐12 | −2.6 | 1.2E‐03 |
| Msil_0472 | MxaJ (SBP) | −4.8 | 2.3E‐15 | −4.2 | 8.6E‐10 |
| Msil_0473 | MxaG (Cyt c) | −3.9 | 2.3E‐15 | −2.9 | 2.6E‐04 |
| Msil_0475 | MxaR | −3.1 | 5.2E‐12 | −2.5 | 2.2E‐03 |
| Msil_0476 | MxaS | −3.4 | 4.8E‐14 | −2.4 | 4.1E‐03 |
| Msil_0482 | MxaE | −2.4 | 7.6E‐07 | −2.9 | 2.8E‐04 |
| Msil_0483 | MxaH | −3.2 | 4.3E‐12 | −2.7 | 7.6E‐04 |
| Msil_0485 | MxaB (RR) | −4.1 | 2.3E‐15 | −3.4 | 6.2E‐06 |
| XoxF5‐MDH | |||||
| Msil_1585 | XoxJ5 (SBP) | 2.5 | 5.6E‐07 | 3.4 | 1.5E‐06 |
| Msil_1586 | XoxG5 (Cyt c) | 3.1 | 5.0E‐11 | 4.2 | 3.1E‐10 |
| Msil_1587 | XoxF5 (large s/u) | 5.8 | 2.3E‐15 | 9.8 | 7.0E‐15 |
| XoxF1‐MDH | |||||
| Msil_3147 | XoxG1 (Cyt c) | −2.7 | 4.3E‐09 | −2.5 | 3.0E‐03 |
| Msil_3148 | XoxJ1 (SBP) | −6.0 | 2.3E‐15 | −4.5 | 4.8E‐11 |
| Msil_3149 | XoxF1 (large s/u) | −2.9 | 5.1E‐10 | −3.3 | 1.1E‐05 |
| Others | |||||
| Msil_0271 | LanM | 5.3 | 2.3E‐15 | 4.7 | 4.1E‐13 |
| Msil_3146 | ABC‐transporter | −2.2 | 4.0E‐06 | −2.8 | 5.0E‐04 |
The data show the log2 (fold‐change, FC) of proteins upregulated or downregulated ≥ 4‐fold (|log2| ≥ 2 and P ≤ 0.01), in both the methanol and ethanol growth condition, together with adjusted P values. Cells are shaded to show upregulation and downregulation (red and blue, respectively).
As well as the MDHs, also highly regulated was lanmodulin (LanM, Msil_0271), 39‐ and 26‐fold upregulated by Ln in methanol‐ and ethanol‐grown cells respectively (Table 2). Lanmodulins, which bind lanthanides, are recently discovered EF‐hand domain‐containing homologues of the well‐studied calcium‐binding calmodulin (Featherston and Cotruvo, 2021). LanM was proposed to be a periplasmic receptor for Ln transported through the outer membrane by a TonB transporter (Cotruvo et al., 2018). Upregulation of LanM in the presence of Ln is consistent with its proposed role as a periplasmic shuttle of Ln to Xox‐MDH (Cotruvo et al., 2018) and these data support similar findings of Ochsner et al. (2019), although, interestingly, the lanM gene was not differentially expressed when the transcriptome of Methylorubrum extorquens AM1 was examined in response to Ln (Good et al., 2019), and close homologues of this gene do not appear to be present in several gammaproteobacterial methanotrophs. In Methylorubrum, this gene is located in a 10‐gene cluster encoding proteins including a TonB receptor, components of an ABC transport system and other periplasmic proteins of unknown function, predicted to be responsible for Ln transport (Ochsner et al., 2019; Roszczenko‐Jasińska et al., 2020). Although M. silvestris has homologues of several of these genes, they are not co‐located with lanM. Three of them, encoding a YVTN family beta‐propeller repeat protein, an ABC‐transporter substrate‐binding protein and a TonB receptor (homologues of LutBAH as defined in the study by Roszczenko‐Jasińska et al., 2020) are adjacent to the xoxF1 genes (Fig. 2) and were moderately downregulated (threefold to fivefold) by Ln (Supporting Information Table S1). The TonB receptor shares 40% amino acid identity with a Methylosinus trichosporium OB3b gene product (WP_003609830.1), which was similarly transcriptionally downregulated in response to cerium (Gu and Semrau, 2017). Hence, in M. silvestris, under these conditions, expression of these proteins was regulated differently than expression of LanM. The Ln concentration (5 μM) used here is orders of magnitude higher than the bio‐available concentrations commonly encountered in the environment (Glass et al., 2020) suggesting that toxicity (due to mis‐metalation) might be important. Under these circumstances, it might be an appropriate strategy to reduce membrane transport and increase abundance of LanM to minimize availability of free Ln3+.
Apart from MDH, MDH‐associated proteins and Ln trafficking proteins, only two other proteins were differentially expressed during growth on both methanol and ethanol: the RNA chaperone Hfq and a subunit of nitrate reductase (Supporting Information Table S1), although several other enzymes associated with nitrogen metabolism were regulated during growth on methanol (but not ethanol). Hfq is an RNA‐binding protein with a role in global regulation of gene expression (Vogel and Luisi, 2011). Although a direct role for Ln in nitrogen metabolism cannot be ruled out, and expression of nitrogen metabolism genes in Methylosinus trichosporium varied depending on metal availability (Gu and Semrau, 2017), it seems more likely that these changes are indirect results of the lanthanide switch, possibly due to Ln‐mediated changes in growth rate or carbon utilization efficiency, or perhaps due to differences in the product distribution (formaldehyde or formate) of the Ca‐ and Ln‐containing MDH enzymes and consequent metabolic re‐programming (Keltjens et al., 2014).
As described above, the expression pattern of MDHs was similar during growth on both methanol and ethanol (Fig. 6; Supporting Information Tables S1 and S2). Methylorubrum extorquens expresses a Ln‐dependent ethanol dehydrogenase, ExaF, that contributes to ethanol oxidation and is also able to sustain growth on methanol in MDH knockouts (Good et al., 2016). On the other hand, comparison of gene transcription in the facultative methanotroph Methylocystis sp. SB2, grown on methane or ethanol, showed that the switch to ethanol repressed mxaF transcription 49‐fold and induced transcription of an NAD(P)‐dependent short‐chain dehydrogenase (Vorobev et al., 2014), suggesting that Mxa‐MDH may not be the primary enzyme responsible for ethanol oxidation in this Methylocystis strain. However, in M. silvestris, no other predicted alcohol dehydrogenases were highly expressed and the abundance of MxaF or XoxF5 increased when cells were grown on ethanol in comparison to on methanol (Fig. 6). Together with growth data presented above, these data show that M. silvestris uses the same Mxa/Xox enzymes to oxidize both methanol and ethanol and that the lanthanide switch operates with both substrates.
To investigate which alcohol‐oxidizing enzyme(s) enabled the comparatively slow growth of double‐mutants ΔmxaFΔxoxF5 and ΔmxaFΔxoxF1, proteins upregulated in these strains in comparison with the wild type under the same growth conditions were identified. Several proteins (19–73) were regulated (≥4‐fold, P ≤ 0.01) in mutant ΔmxaFΔxoxF5 during growth on both methanol and ethanol, either without or with Ln, and in mutant ΔmxaFΔxoxF1 during growth on these substrates without Ln, in comparison with the wild type under the same conditions, indicative of disrupted core metabolic processes (Supporting Information Table S4). None of these are annotated with a possible short‐chain alcohol‐metabolizing ability or are homologues of short‐chain alcohol dehydrogenases, suggesting that no alternative alcohol dehydrogenases took the place of PQQ‐MDH. In particular, XoxF1 was not upregulated in strain ΔmxaFΔxoxF5. Instead, all the mutant strain/growth conditions expressed the soluble methane monooxygenase (sMMO) polypeptides. The wild type does not express the sMMO at high levels during growth on methanol or ethanol, but in these mutants, the structural polypeptides were upregulated 22‐ to over 1000‐fold over the corresponding wild‐type expression levels (Fig. 7), approximating the levels of the wild type during growth on methane (Patel et al., 2012), with the sMMO comprising 14%–27% of total protein in mutants cells. The sMMO has a well‐documented wide substrate range, including methanol and ethanol (Colby et al., 1977; Patel et al., 1982; Tinberg and Lippard, 2010), suggesting that suppressor mutations may have arisen in the sMMO regulatory network (of which little is known in Methylocella), to allow sMMO‐mediated alcohol oxidation. To verify that the sMMO was responsible for the eventual growth of double‐mutant strain ΔmxaFΔxoxF5 on methanol, cells were subcultured into methanol‐containing medium during active growth and incubated in the presence or absence of the sMMO inhibitor, acetylene. Cells incubated with acetylene did not grow during the 640 h of the experiment, whereas without acetylene cells grew at the low growth rate previously observed with this strain. In contrast, acetylene had little or no effect on growth of the wild type on methanol (Supporting Information Fig. S2) demonstrating that the sMMO could enable weak growth on methanol in the absence of both MxaF and XoxF5.
Figure 7.
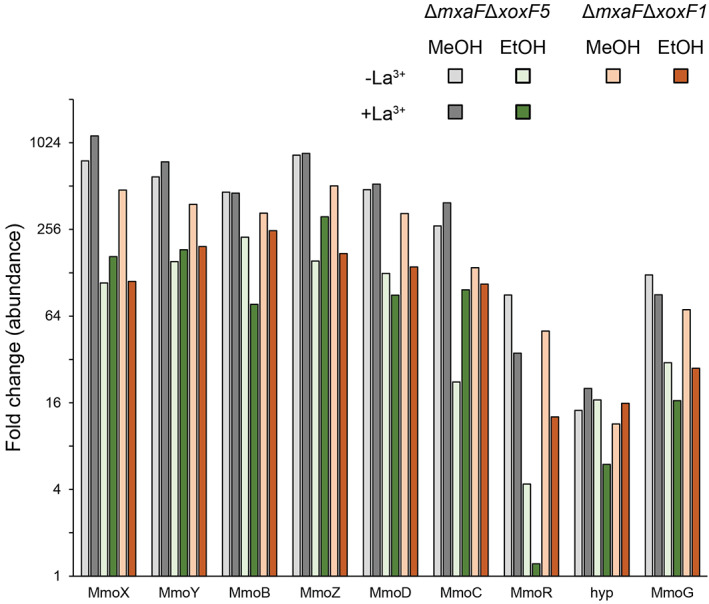
Relative abundance of the sMMO in mutant strains compared with the wild type. The sMMO structural polypeptides (MmoXYBZDC) were upregulated 22‐ to over 1000‐fold, with more modest levels for the regulatory protein and chaperone (MmoRG) in mutant strains ΔmxaFΔxoxF5 and ΔmxaFΔxoxF1 during growth on methanol or ethanol, irrespective of the presence of Ln, compared with the wild type. All P‐values ≤ 0.01, see Supporting Information Table S4 for full details. hyp, hypothetical protein of unknown function.
Conclusions
The participation of lanthanides in biological processes was only discovered recently, and there is still much to learn. The data presented here expand the breadth of our knowledge of Xox‐MDHs in the family Beijerinckiaceae, not previously studied in the context of Ln regulation, and show that the lanthanide switch operates in the facultative methanotroph M. silvestris. Lanthanides were first implicated in methanol metabolism and subsequently in ethanol metabolism (Hibi et al., 2011; Good et al., 2016) and the data presented here extend the role of Ln‐containing enzymes to growth on propane. Methylocella are environmentally important bacteria, which are extremely abundant in some environments where methane and other short‐chain alkanes co‐exist (Farhan Ul Haque et al., 2018; Farhan Ul Haque et al., 2019), suggesting that the ability to benefit from these multiple carbon sources may be important for its competitive success (Crombie and Murrell, 2014). In contrast to other ethanol‐oxidizing methylotrophs such as Methylorubrum extorquens and Methylocystis sp. SB2, however, M. silvestris expresses only the same pair of enzymes, depending on Ln availability, to oxidize not only C1 and C2 but also C3 alcohols, despite the fact that most Ca‐containing MDHs do not efficiently oxidize secondary alcohols such as 2‐propanol (Goodwin and Anthony, 1998). The data suggest that in M. silvestris, Xox‐MDH is more efficient than Mxa‐MDH at oxidizing 2‐propanol, explaining the increased benefit of Ln during growth on propane and 2‐propanol (21% and 34% increase in growth rate due to Ln, respectively) compared to during growth on methanol (6% increase) (Table 1).
Most work has focussed so far on only a few examples of Xox‐MDH, mainly from clades 4 and 5 (Huang et al., 2019). The finding here that the XoxF1 of M. silvestris is also regulated by Ln, but in the same sense as Mxa‐MDH rather than as XoxF5‐MDH, is significant. Enzymes from this clade are distributed among a relatively small number of diverse taxa, including alpha‐ and gammaproteobacteria, verrucomicrobia and members of the candidate phylum NC10 (Keltjens et al., 2014). However, under the experimental conditions used here, there was no prominent phenotype of strain ΔxoxF1, and therefore the role of this enzyme is not clear. The location of xoxF1 in M. silvestris adjacent to putative Ln‐trafficking genes suggests a possible regulatory role, as has already been shown for XoxF clade 5 enzymes in Methylorubrum (Skovran et al., 2011; Vu et al., 2016; Ochsner et al., 2019), and this warrants further investigation.
Deletion of xoxF5 resulted in a suppressor mutation in a sensor kinase‐encoding gene associated with the Mxa‐MDH gene cluster, mxaY, in line with several previous studies which have identified mutations in homologous genes in other organisms (Chu and Lidstrom, 2016; Zheng et al., 2018; Ochsner et al., 2019; Yanpirat et al., 2020). So far, this is the only gene predicted to be involved in MDH regulation in M. silvestris, but the limited data available suggest that regulation is enabled by the two component regulator comprising MxaY and MxaB, with plasma‐membrane‐located sensor kinase MxaY responsive to Ln (directly or more likely indirectly), in the periplasm, and the cognate response regulator MxaB controlling transcription of mxaF and xox genes, with the effect of repressing mxa and xoxF1, and activating xoxF5 in the presence of Ln. This is broadly in line with the model proposed for the gammaproteobacterial methanotroph Methylotuvimicrobium buryatense 5GB1C (Groom et al., 2019), although we should note that there are several differences between Ln‐trafficking and the lanthanide switch in M. buryatense and in alphaproteobacteria.
Generation of a strain constitutively expressing the sMMO in double Mxa‐Xox‐MDH mutants was surprising and fortuitous and suggests that repression of sMMO expression by alternative carbon sources (such as methanol), while presumably distinct from the lanthanide switch, was disabled in these strains. Little is known about sMMO regulation in Methylocella, and analysis of the genetic basis of constitutive sMMO expression may provide insights into MMO regulation, the subject of further study in this laboratory.
In conclusion, this work has expanded our knowledge of Ln‐dependent microbiology and established base data for Methylocella, which now enable more detailed examination of the properties and regulation of Mxa‐ and Xox‐MDHs in this uniquely versatile and environmentally important organism.
Methods
Growth of Methylocella
M. silvestris BL2 and mutant strains were grown in dilute mineral salts (DNMS) medium in 120 ml glass vials as previously described (Crombie and Murrell, 2014). All media were made using ultrapure water (18 MΩ cm) produced using an Elga Purelab Classic or Elga Purelab Chorus 1 Life Scence water purification system (Veolia Water Technologies, High Wycombe, UK). For evaluation of growth, gene transcription and protein expression, special precautions were taken to minimize contamination with lanthanides. At the completion of each experiment, cultures were killed by autoclaving. The vials were extensively rinsed in water, filled with de‐ionized water and re‐autoclaved and then acid washed by soaking overnight in 10% v/v nitric acid, followed by repeated rinsing in ultrapure water. For growth in the presence of Ln, lanthanum chloride was added to 5 μM just before autoclaving. Vials were segregated into those that had been used with Ln and those without Ln, for the duration of the study. Gases were added to the headspace by injection through the septum and other substrates were added from sterile stock solutions to the concentrations indicated. For acetylene inhibition experiments, acetylene (4 ml) was injected through the septum to give a headspace concentration of approximately 4% v/v.
RT‐qPCR analysis of gene transcription
Cells were grown in 25 ml volume on methane or propane (20% v/v), with or without lanthanum (10 μM), in triplicate and harvested at mid‐exponential phase (OD540 = 0.21–0.36). Aliquots of the culture (4 ml) were immediately treated with RNA Protect Bacteria reagent (Qiagen, Hilden, Germany) following the manufacturer's instructions and the pellet was stored at −80°C, prior to RNA extraction using a hot acid phenol method as previously described (Gilbert et al., 2000). Following two treatments with RNase‐free DNase and purification using an RNeasy kit (Qiagen), RNA was eluted in 40 μl water and quantified using a Qubit fluorometer (Invitrogen, Carlsbad, USA). Synthesis of cDNA used Superscript III reverse transcriptase (RT) (Invitrogen) with 100 ng RNA and random hexamers in 20 μl reactions, following the manufacturer's instructions, and including controls in which RT was omitted. Standard‐curve‐based quantitative PCR was conducted in 25 μl reactions in 96‐well plates with SensiFAST SYBR Hi‐ROX master mix (Bioline, London, UK), 400–800 nM of each primer and 2 μl of 1/10 dilution of cDNA template, using an Applied Biosystems StepOnePlus instrument (Applied Biosystems, Waltham, USA). Standards for each target gene and no‐RT controls were included on each plate and transcription was quantified with respect to rpoB as reference gene. Primer sequences are given in Supporting Information Table S3. Two technical replicates were analysed for each of three biological replicates for both substrates, with and without Ln. Amplification efficiency was between 85% and 105% for all targets, except mmoX (75%). Target gene transcription, normalized to rpoB, is shown relative to levels in cultures grown on methane, without Ln.
Construction of mutant strains
Unmarked deletions of mxaF, xoxF5 and xoxF1 were generated as described previously (Crombie and Murrell, 2011). Since MDH genes were the deletion targets, M. silvestris cells were grown on pyruvate (8 mM) or succinate (5 mM). Briefly, removal of the gene of interest was accomplished by homologous recombination between arms A and B (DNA sequences identical to approximately 500 bp either side of the gene of interest), which were cloned either side of a kanamycin resistance cassette flanked by loxP sites. Arms A and B were generated by PCR and cloned into pCM184 (Marx and Lidstrom, 2002) using the primers and restriction sites shown in Supporting Information Table S3, followed by excision of the linear fragment by restriction digest, purification and transfer into the wild‐type or single‐deletion background strains by electroporation and selection on kanamycin‐containing plates as described (Crombie and Murrell, 2011). Removal of the kanamycin antibiotic cassette was accomplished by Cre recombinase, expressed from plasmid pCM157 (Marx and Lidstrom, 2002). Successful removal of the gene of interest was confirmed by PCR and sequencing using primers located outside the regions of homology (Supporting Information Table S3).
Promoter reporter strains
Activity of the mxaF, xoxF5 and xoxF1 promoters was assayed by cloning the promoter fragments in front of a gfp gene in plasmid pMHA200 (Ali and Murrell, 2009). Upstream DNA (971 bp, 639 bp and 608 bp for mxaF, xoxF5, and xoxF1 respectively) was amplified by PCR using the primers shown in Supporting Information Table S3. The resultant amplicons were cloned into pJet1.2 (Thermo Fisher Scientific, UK), digested with XbaI, blunted using T4 polymerase (Fermentas, Thermo Fisher Scientific), excised with KpnI, purified from a gel and ligated with pMHA200 treated similarly, except cut initially with EcoRI, resulting in plasmids pMHA_0471, pMHA_1587 and pMHA_3149 for mxaF, xoxF5 and xoxF1, respectively. Integrity of these plasmids was checked by PCR, using primers pMHA_TF/pMHA_TR (Supporting Information Table S3) annealing to the pMHA200 backbone, and restriction digest. These constructs, together with the parental vector pMHA200, were transferred into all strains by electroporation, as previously described (Crombie and Murrell, 2011), and selected on kanamycin‐containing plates. Colonies were transferred to liquid medium containing kanamycin and grown to late exponential phase without Ln and then transferred to plastic universal tubes (5 ml culture volume) and grown with or without lanthanum (5 μM), in triplicate. Cells were harvested by centrifugation, resuspended in 400 μl phosphate buffer (pH 5.5, 20 mM) and 200 μl of the cells suspension was assayed for fluorescence in 96‐well black microplates using a Spectramax iD5 plate reader (Molecular Devices, San Jose, CA, USA) with excitation and emission at 485 and 535 nm respectively. The density of a 1/4 dilution was determined at 540 nm and fluorescence expressed as relative fluorescence units per unit culture density, subtracting the background values obtained with the promoter‐less vector.
Sequencing of mxaY
DNA was extracted from the wild type and strain ΔxoxF5 using standard methods (Sambrook et al., 2001). The entire mxaY gene (Msil_0484) was amplified by PCR using primers 484_F/484_R (Supporting Information Table S3) and sequenced using the same primers.
Statistical analysis
Growth rate of cultures was determined during the exponential growth phase using a minimum of three data points. Since minor differences in culture medium or the condition (e.g. growth stage) of inoculating cultures can affect growth rates, statistical differences were only computed based on the growth of the same strain, in the presence or absence of Ln, that is, between cultures set up using the same parental culture, and not between strains. Comparisons between experimental conditions were carried out using Student's t‐test.
Proteomic analysis
Wild‐type cells were grown in 25 ml of DNMS medium in 120 ml serum vials supplied with methanol (0.1% v/v) or ethanol (0.05% v/v) with or without lanthanum (5 μM) and harvested at late exponential phase. For mutant strains ΔmxaFΔxoxF5 and ΔmxaFΔxoxF1, cells were harvested by pooling three replicates of each growth condition at the conclusion of the growth experiments described above. Cells were harvested by centrifugation (3500g, 20 min, 20°C), washed in lysis buffer (25 mM Tris (pH 7.5), 25 mM NaCl, protease inhibitor #2 (Merck 539132) (reconstituted as recommended and used at 1% v/v) and resuspended in 0.5 ml of the same buffer. Cells were broken by sonication (5 cycles of 15 s on and 20 s off) and centrifuged at 16 000g, 15 min, 4°C to remove cell debris. The extract was submitted to the Proteomics Facility, John Innes Centre (Norwich, UK) for analysis. The proteins were precipitated from the extract with five volumes of acetone (Nickerson and Doucette, 2020). The resulting pellets were dissolved in 100 μl of 2.5% sodium deoxycholate (SDC; Merck). Protein was quantified using the bicinchoninic acid assay (Thermo Fisher Scientific) and 100 μg of protein were reduced, alkylated, and digested with trypsin in the presence of 0.2 M EPPS buffer (Merck) and 2.5% SDC according to standard procedures. After the digest, the SDC was precipitated by adjusting to 0.2% trifluoroacetic acid (TFA), and the clear supernatant subjected to C18 solid‐phase extraction (SPE) (OMIX tips; Agilent). Tandem mass tag (TMT) labelling was performed using a TMT10plex™ kit (Lot TL274393, Thermo) according to the manufacturer's instructions with slight modification; the dried peptides were dissolved in 90 μl of 0.2 M EPPS buffer/10% acetonitrile, and 200 μg TMT reagent dissolved in 22 μl of acetonitrile was added. Aliquots of 2 μl from each sample were combined and analysed on the mass spectrometer (see below) to check labelling efficiency and estimate total sample abundances. The main sample aliquots were combined correspondingly and desalted using a C18 Sep‐pak cartridge (Waters). The eluted peptides were dissolved in 500 μl of 10 mM NH4HCO3 and fractionated by high pH reversed‐phase HPLC. Using an ACQUITY Arc Bio System (Waters), the samples were loaded to a Kinetex® 5 μm EVO C18 100 Å LC Column 250 × 4.6 mm (Phenomenex). Fractionation was performed with the following gradient of solvents A (water), B (acetonitrile), and C (25 mM NH4HCO3) at a flow rate of 1 ml min−1. Solvent C was kept at 10% throughout the gradient; solvent B: 0–5 min: 5%, 5–80 min: 5%–60%, 80–90 min: 60%–80%, followed by 5 min at 80% and re‐equilibration to 5%. Fractions of 1 ml were collected, dried down, and concatenated to produce 25 final fractions for MS analysis.
Aliquots were analysed by nanoLC‐MS/MS on an Orbitrap Eclipse™ Tribrid™ mass spectrometer coupled to an UltiMate® 3000 RSLCnano LC system (Thermo Fisher Scientific, Hemel Hempstead, UK). The samples were loaded onto a trap column (nanoEase M/Z Symmetry C18 Trap Column, Waters, Wilmslow, UK) with 0.1% TFA at 15 μl min−1 for 3 min. The trap column was then switched in‐line with the analytical column (nanoEase M/Z column, HSS C18 T3, 1.8 μm, 100 Å, 250 mm × 0.75 μm, Waters) for separation using the following gradient of solvents A (water, 0.1% formic acid) and B (80% acetonitrile, 0.1% formic acid) at a flow rate of 0.2 μl min−1: 0–3 min 3% B (parallel to trapping); 3–10 min linear increase B to 12%; 10–105 min increase B to 50%; followed by a ramp to 99% B and re‐equilibration to 3% B. Data were acquired with the following parameters in positive ion mode: MS1/OT: resolution 120 K, profile mode, mass range m/z 400–1800, automatic gain control (AGC) target 100%, max inject time 50 ms; MS2/IT: data dependent analysis with the following parameters: top10 in IT turbo mode, centroid mode, quadrupole isolation window 1.0 Da, charge states 2–5, threshold 1.9e4, collision energy (CE) = 33, AGC target 200%, max inject time 70 ms, dynamic exclusion 1 count/7 s/±7 ppm; MS3 synchronous precursor selection (SPS): 10 SPS precursors, isolation window 1 Da, higher‐energy C‐trap dissociation (HCD) fragmentation with CE = 65, Orbitrap Turbo TMT and TMTpro resolution 15 k, AGC target 200%, max inject time 105 ms, real‐time search: protein database Methylocella sylvestris (NCBI, GCA_000021745.1_ASM2174v1_translated_cds.faa, number of entries: 3821), 1 missed cleavage, oxidation (M) as variable, carbamidomethyl (C) and TMT6plex as fixed modifications, Xcorr = 1, dCn = 0.05.
The raw data were processed and quantified in Proteome Discoverer 2.4.1.15 (Thermo) using the incorporated search engine Sequest HT and the Mascot search engine (Matrix Science, London, UK; Mascot version 2.7.0). The processing workflow included recalibration of MS1 spectra (RC), precursor ion quantification by most confident centroid (20 ppm), fasta database M. sylvestris and common contaminants, precursor/fragment tolerance 6 ppm/0.6 Da, variable/fixed modifications were oxidation (M)/carbamidomethyl (C) and TMT6plex. The consensus workflow included the following parameters: unique peptides (protein groups), intensity‐based abundance, TMT channel correction values applied (TL274393), co‐isolation/SPS matches thresholds 50%/80%, normalization on total peptide abundances, protein abundance‐based ratio calculation, missing values imputation by low abundance resampling, hypothesis testing by t‐test (background based), adjusted P‐value calculation by BH method.
Supporting information
Table S1. Abundance (% of total protein for each sample) of PQQ‐ADH in cell extract of Methylocella silvestris grown on methanol or ethanol in the absence or presence of lanthanum.
Table S2. Proteins differentially regulated in M. silvestris BL2 cells grown on A) methanol or B) ethanol, in response to lanthanum. The fold‐change (FC) of proteins upregulated in lanthanum‐containing medium is shown with a positive log2 (FC) value and downregulated proteins are shown with a negative value. Only proteins differentially expressed at |log2 (FC)| ≥ 2 and adjusted p value ≤0.01 are shown. Proteins regulated in both methanol‐ and ethanol‐grown cells are shown in bold type.
Table S3. Primer sequences.
Table S4. Table S4 is included as a separate Excel spreadsheet file.
Fig. S1. Detail of alignment of M. silvestris PQQ‐ADH sequences with other representative examples. The catalytic aspartate residue and calcium or lanthanide‐coordinating residue (A or D) (at positions 303 and 305 respectively in MxaF from Methylorubrum extorquens) are indicated with arrows.
Fig. S2. Growth curves of the wild type during growth on methanol (A) or ethanol (B). Error bars show the standard error of the mean (n = 3). Panel A shows a representative of three independent experiments.
Fig. S3. Double mutant strain ΔmxaFΔxoxF5 (A) or the wild type (B) were grown on methanol (0.1% v/v) in the presence (red) or absence (black) of 4% v/v acetylene in the vial headspace. Error bars show the standard error of the mean (n = 3).
Acknowledgements
This work was funded by a Leverhulme Trust Early Career Fellowship (ECF‐2016‐626) to ATC. The author thanks Gerhard Saalbach and colleagues at the John Innes Centre Proteomics Facility for proteomic analysis, and Colin Murrell for generous help and support.
References
- Ali, H. , and Murrell, J.C. (2009) Development and validation of promoter‐probe vectors for the study of methane monooxygenase gene expression in Methylococcus capsulatus Bath. Microbiology 155: 761–771. [DOI] [PubMed] [Google Scholar]
- Amaratunga, K. , Goodwin, P.M. , O'Connor, C.D. , and Anthony, C. (1997) The methanol oxidation genes mxaFJGIR(S)ACKLD in Methylobacterium extorquens . FEMS Microbiol Lett 146: 31–38. [DOI] [PubMed] [Google Scholar]
- Anthony, C. (2004) The quinoprotein dehydrogenases for methanol and glucose. Arch Biochem Biophys 428: 2–9. [DOI] [PubMed] [Google Scholar]
- Beilen, J.B.V. , and Funhoff, E.G. (2005) Expanding the alkane oxygenase toolbox: new enzymes and applications. Curr Opin Biotechnol 16: 308–314. [DOI] [PubMed] [Google Scholar]
- Chistoserdova, L. (2016) Lanthanides: new life metals? World J Microb Biot 32: 1–7. [DOI] [PubMed] [Google Scholar]
- Chistoserdova, L. (2019) New pieces to the lanthanide puzzle. Mol Microbiol 111: 1127–1131. [DOI] [PubMed] [Google Scholar]
- Chu, F. , and Lidstrom, M.E. (2016) XoxF acts as the predominant methanol dehydrogenase in the type I methanotroph Methylomicrobium buryatense . J Bacteriol, 8: 1317–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, F. , Beck, D.A.C. , and Lidstrom, M.E. (2016) MxaY regulates the lanthanide‐mediated methanol dehydrogenase switch in Methylomicrobium buryatense . PeerJ 4: e2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby, J. , Stirling, D.I. , and Dalton, H. (1977) The soluble methane mono‐oxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenate n‐alkanes, n‐alkenes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem J 165: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotruvo, J.A. (2019) The chemistry of lanthanides in biology: recent discoveries, emerging principles, and technological applications. ACS Cent Sci, 5: 1496–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotruvo, J.J.A. , Featherston, E.R. , Mattocks, J.A. , Ho, J.V. , and Laremore, T.N. (2018) Lanmodulin: a highly selective lanthanide‐binding protein from a lanthanide‐utilizing bacterium. J Am Chem Soc, 140: 15056–15061. [DOI] [PubMed] [Google Scholar]
- Crombie, A. , and Murrell, J.C. (2011) Development of a system for genetic manipulation of the facultative methanotroph Methylocella silvestris BL2. Methods Enzymol 495: 119–133. [DOI] [PubMed] [Google Scholar]
- Crombie, A.T. , and Murrell, J.C. (2014) Trace‐gas metabolic versatility of the facultative methanotroph Methylocella silvestris . Nature 510: 148–151. [DOI] [PubMed] [Google Scholar]
- Dalsøren, S.B. , Myhre, G. , Hodnebrog, Ø. , Myhre, C.L. , Stohl, A. , Pisso, I. , et al. (2018) Discrepancy between simulated and observed ethane and propane levels explained by underestimated fossil emissions. Nat Geosci 11: 178–184. [Google Scholar]
- Dedysh, S.N. , Knief, C. , and Dunfield, P.F. (2005) Methylocella species are facultatively methanotrophic. J Bacteriol 187: 4665–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y.W. , Ro, S.Y. , and Rosenzweig, A.C. (2018) Structure and function of the lanthanide‐dependent methanol dehydrogenase XoxF from the methanotroph Methylomicrobium buryatense 5GB1C. J Biol Inorg Chem 23: 1037–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etiope, G. , and Ciccioli, P. (2009) Earth's degassing: a missing ethane and propane source. Science 323: 478. [DOI] [PubMed] [Google Scholar]
- Etiope, G. , and Klusman, R.W. (2010) Microseepage in drylands: flux and implications in the global atmospheric source/sink budget of methane. Glob Planet Change 72: 265–274. [Google Scholar]
- Farhan Ul Haque, M. , Crombie, A.T. , and Murrell, J.C. (2019) Novel facultative Methylocella strains are active methane consumers at terrestrial natural gas seeps. Microbiome 7: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan Ul Haque, M. , Xu, H.‐J. , Murrell, J.C. , and Crombie, A. (2020) Facultative methanotrophs – diversity, genetics, molecular ecology and biotechnological potential: a mini‐review. Microbiology 166: 894–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan Ul Haque, M. , Crombie, A.T. , Ensminger, S.A. , Baciu, C. , and Murrell, J.C. (2018) Facultative methanotrophs are abundant at terrestrial natural gas seeps. Microbiome 6: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan Ul Haque, M. , Kalidass, B. , Bandow, N. , Turpin, E.A. , DiSpirito, A.A. , and Semrau, J.D. (2015) Cerium regulates expression of alternative methanol dehydrogenases in Methylosinus trichosporium OB3b. Appl Environ Microb 81: 7546–7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherston, E.R. , and Cotruvo, J.A. (2021) The biochemistry of lanthanide acquisition, trafficking, and utilization. Biochim Biophys Acta Mol Cell Res 1868: 118864. [DOI] [PubMed] [Google Scholar]
- Fitriyanto, N.A. , Fushimi, M. , Matsunaga, M. , Pertiwiningrum, A. , Iwama, T. , and Kawai, K. (2011) Molecular structure and gene analysis of Ce3+‐induced methanol dehydrogenase of Bradyrhizobium sp. MAFF211645. J Biosci Bioeng 111: 613–617. [DOI] [PubMed] [Google Scholar]
- Gilbert, B. , McDonald, I.R. , Finch, R. , Stafford, G.P. , Nielsen, A.K. , and Murrell, J.C. (2000) Molecular analysis of the pmo (particulate methane monooxygenase) operons from two type II methanotrophs. Appl Environ Microbiol 66: 966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass, J.B. , Cowan, E.T. , and Johannesson, K.H. (2020) Lanthanide rarity in natural waters: implications for microbial C1 metabolism. FEMS Microbiol Lett 367: fnaa165. [DOI] [PubMed] [Google Scholar]
- Good, N.M. , Moore, R.S. , Suriano, C.J. , and Martinez‐Gomez, N.C. (2019) Contrasting in vitro and in vivo methanol oxidation activities of lanthanide‐dependent alcohol dehydrogenases XoxF1 and ExaF from Methylobacterium extorquens AM1. Sci Rep 9: 4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good, N.M. , Vu, H.N. , Suriano, C.J. , Subuyuj, G.A. , Skovran, E. , and Martinez‐Gomez, N.C. (2016) Pyrroloquinoline quinone‐containing ethanol dehydrogenase in Methylobacterium extorquens AM1 extends lanthanide‐dependent metabolism to multi‐carbon substrates. J Bacteriol 198: 3109–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good, N.M. , Fellner, M. , Demirer, K. , Hu, J. , Hausinger, R.P. , and Martinez‐Gomez, N.C. (2020) Lanthanide‐dependent alcohol dehydrogenases require an essential aspartate residue for metal coordination and enzymatic function. J Biol Chem 295: 8272–8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, P.M. , and Anthony, C. (1998) The biochemistry, physiology and genetics of PQQ and PQQ‐containing enzymes. Adv Microb Physiol 40: 1–80. [DOI] [PubMed] [Google Scholar]
- Groom, J.D. , Ford, S.M. , Pesesky, M.W. , and Lidstrom, M.E. (2019) A mutagenic screen identifies a TonB‐dependent receptor required for the lanthanide metal switch in the type I methanotroph Methylotuvimicrobium buryatense 5GB1C. J Bacteriol 201: e00120–e00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, W. , and Semrau, J.D. (2017) Copper and cerium‐regulated gene expression in Methylosinus trichosporium OB3b. Appl Microbiol Biotechnol, 101: 8499–8516. [DOI] [PubMed] [Google Scholar]
- Hibi, Y. , Asai, K. , Arafuka, H. , Hamajima, M. , Iwama, T. , and Kawai, K. (2011) Molecular structure of La3+‐induced methanol dehydrogenase‐like protein in Methylobacterium radiotolerans . J Biosci Bioeng 111: 547–549. [DOI] [PubMed] [Google Scholar]
- Huang, J. , Yu, Z. , and Chistoserdova, L. (2018) Lanthanide‐dependent methanol dehydrogenases of XoxF4 and XoxF5 clades are differentially distributed among methylotrophic bacteria and they reveal different biochemical properties. Front Microbiol 9: 1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Yu, Z. , Groom, J. , Cheng, J.‐F. , Tarver, A. , Yoshikuni, Y. , and Chistoserdova, L. (2019) Rare earth element alcohol dehydrogenases widely occur among globally distributed, numerically abundant and environmentally important microbes. ISME J 13: 2005–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn, B. , Pol, A. , Lumpe, H. , Barends, T. , Dietl, A. , Hogendoorn, C. , et al. (2018) Similar but not the same: first kinetic and structural analyses of a methanol dehydrogenase containing a europium ion in the active site. Chembiochem 19: 1147–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, S. , Takashino, M. , Igarashi, K. , and Kitagawa, W. (2020) Isolation and genomic characterization of a proteobacterial methanotroph requiring lanthanides. Microbes Environ 35: ME19128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keltjens, J. , Pol, A. , Reimann, J. , and Op den Camp, H.M. (2014) PQQ‐dependent methanol dehydrogenases: rare‐earth elements make a difference. Appl Microbiol Biotechnol 98: 6163–6183. [DOI] [PubMed] [Google Scholar]
- Kinnaman, F.S. , Valentine, D.L. , and Tyler, S.C. (2007) Carbon and hydrogen isotope fractionation associated with the aerobic microbial oxidation of methane, ethane, propane and butane. Geochim Cosmochim Acta 71: 271–283. [Google Scholar]
- Krause, S.M.B. , Johnson, T. , Samadhi Karunaratne, Y. , Fu, Y. , Beck, D.A.C. , Chistoserdova, L. , and Lidstrom, M.E. (2016) Lanthanide‐dependent cross‐feeding of methane‐derived carbon is linked by microbial community interactions. Proc Natl Acad Sci 114: 358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , and Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Yao, Q. , Guo, X. , Crits‐Christoph, A. , Mayes, M.A., IV , et al. (2019) Genome‐resolved proteomic stable isotope probing of soil microbial communities using 13CO2 and 13C‐methanol. Front Microbiol 10: 2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, H. , Sahin, N. , and Tani, A. (2018) Isolation and genomic characterization of Novimethylophilus kurashikiensis gen. Nov. sp. nov., a new lanthanide‐dependent methylotrophic species of Methylophilaceae . Environ Microbiol 20: 1204–1223. [DOI] [PubMed] [Google Scholar]
- Marx, C.J. , and Lidstrom, M.E. (2002) Broad‐host‐range cre‐lox system for antibiotic marker recycling in gram‐negative bacteria. Biotechniques 33: 1062–1067. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T. , Mitsui, R. , Tani, A. , Sasa, K. , Tashiro, S. , Iwama, T. , et al. (2012) A catalytic role of XoxF1 as La3+‐dependent methanol dehydrogenase in Methylobacterium extorquens strain AM1. PLoS One 7: e50480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson, J.L. , and Doucette, A.A. (2020) Rapid and quantitative protein precipitation for proteome analysis by mass spectrometry. J Proteome Res 19: 2035–2042. [DOI] [PubMed] [Google Scholar]
- Ochsner, A.M. , Hemmerle, L. , Vonderach, T. , Nussli, R. , Bortfeld‐Miller, M. , Hattendorf, B. , and Vorholt, J.A. (2019) Use of rare‐earth elements in the phyllosphere colonizer Methylobacterium extorquens PA1. Mol Microbiol, 111: 1152–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, N.A. , Crombie, A. , Slade, S.E. , Thalassinos, K. , Hughes, C. , Connolly, J.B. , et al. (2012) Comparison of one‐ and two‐dimensional liquid chromatography approaches in the label‐free quantitative analysis of Methylocella silvestris . J Proteome Res 11: 4755–4763. [DOI] [PubMed] [Google Scholar]
- Patel, R.N. , Hou, C.T. , Laskin, A.I. , and Felix, A. (1982) Microbial oxidation of hydrocarbons: properties of a soluble methane monooxygenase from a facultative methane‐utilizing organism, Methylobacterium sp. strain CRL‐26. Appl Environ Microb 44: 1130–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picone, N. , and Op den Camp, H.J. (2018) Role of rare earth elements in methanol oxidation. Curr Opin Chem Biol 49: 39–44. [DOI] [PubMed] [Google Scholar]
- Pol, A. , Barends, T.R.M. , Dietl, A. , Khadem, A.F. , Eygensteyn, J. , Jetten, M.S.M. , and Op den Camp, H.J.M. (2014) Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ Microbiol 16: 255–264. [DOI] [PubMed] [Google Scholar]
- Redmond, M.C. , Valentine, D.L. , and Sessions, A.L. (2010) Identification of novel methane‐, ethane‐, and propane‐oxidizing bacteria at marine hydrocarbon seeps by stable isotope probing. Appl Environ Microb 76: 6412–6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeburgh, W.S. (2007) Global methane biogeochemistry. In Treatise on Geochemistry, Holland, H.D. , and Turekian, K.K. (eds). Amsterdam: Elsevier, pp. 1–32. [Google Scholar]
- Roszczenko‐Jasińska, P. , Vu, H.N. , Subuyuj, G.A. , Crisostomo, R.V. , Cai, J. , Lien, N.F. , et al. (2020) Gene products and processes contributing to lanthanide homeostasis and methanol metabolism in Methylorubrum extorquens AM1. Sci Rep 10: 12663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. , and Maniatis, T. (2001) Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Saunois, M. , Bousquet, P. , Poulter, B. , Peregon, A. , Ciais, P. , Canadell, J.G. , et al. (2016) The global methane budget 2000–2012. Earth Syst Sci Data 8: 697–751. [Google Scholar]
- Schimmelmann, A. , Ensminger, S.A. , Drobniak, A. , Mastalerz, M. , Etiope, G. , Jacobi, R.D. , and Frankenberg, C. (2018) Natural geological seepage of hydrocarbon gas in the Appalachian Basin and Midwest USA in relation to shale tectonic fracturing and past industrial hydrocarbon production. Sci Total Environ 644: 982–993. [DOI] [PubMed] [Google Scholar]
- Semrau, J.D. , DiSpirito, A.A. , Gu, W. , and Yoon, S. (2018) Metals and methanotrophy. Appl Environ Microb 84: e02289–e02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shennan, J.L. (2006) Utilisation of C2–C4 gaseous hydrocarbons and isoprene by microorganisms. J Chem Technol Biotechnol 81: 237–256. [Google Scholar]
- Skovran, E. , and Martinez‐Gomez, N.C. (2015) Just add lanthanides. Science 348: 862–863. [DOI] [PubMed] [Google Scholar]
- Skovran, E. , Raghuraman, C. , and Martinez‐Gomez, N. (2019) Lanthanides in methylotrophy. In Methylotrophs and Methylotroph Communities, Chistoserdova, L.V. (ed). Poole, UK: Caister Academic Press. [Google Scholar]
- Skovran, E. , Palmer, A.D. , Rountree, A.M. , Good, N.M. , and Lidstrom, M.E. (2011) XoxF is required for expression of methanol dehydrogenase in Methylobacterium extorquens AM1. J Bacteriol 193: 6032–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, A.L. , Auman, A.J. , and Lidstrom, M.E. (1998) Sequence and characterization of mxaB, a response regulator involved in regulation of methanol oxidation, and of mxaW, a methanol‐regulated gene in Methylobacterium extorquens AM1. FEMS Microbiol Lett 160: 119–124. [DOI] [PubMed] [Google Scholar]
- Taubert, M. , Grob, C. , Howat, A.M. , Burns, O.J. , Dixon, J.L. , Chen, Y. , and Murrell, J.C. (2015) xoxF encoding an alternative methanol dehydrogenase is widespread in coastal marine environments. Environ Microbiol 17: 3937–3948. [DOI] [PubMed] [Google Scholar]
- Tinberg, C.E. , and Lippard, S.J. (2010) Oxidation reactions performed by soluble methane monooxygenase hydroxylase intermediates Hperoxo and Q proceed by distinct mechanisms. Biochemistry 49: 7902–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, G. (2004) Rare earth elements in soil and plant systems ‐ a review. Plant Soil 267: 191–206. [Google Scholar]
- Vogel, J. , and Luisi, B.F. (2011) Hfq and its constellation of RNA. Nat Rev Microbiol 9: 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobev, A. , Jagadevan, S. , Jain, S. , Anantharaman, K. , Dick, G.J. , Vuilleumier, S. , and Semrau, J.D. (2014) Genomic and transcriptomic analyses of the facultative methanotroph Methylocystis sp. strain SB2 grown on methane or ethanol. Appl Environ Microb 80: 3044–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu, H.N. , Subuyuj, G.A. , Vijayakumar, S. , Good, N.M. , Martinez‐Gomez, N.C. , and Skovran, E. (2016) Lanthanide‐dependent regulation of methanol oxidation systems in Methylobacterium extorquens AM1 and their contribution to methanol growth. J Bacteriol, 198: 1250–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Suganuma, S. , Hibino, A. , Mitsui, R. , Tani, A. , Matsumoto, T. , et al. (2019) Lanthanide‐dependent methanol dehydrogenase from the legume symbiotic nitrogen‐fixing bacterium Bradyrhizobium diazoefficiens strain USDA110. Enzyme Microb Technol 130: 109371. [DOI] [PubMed] [Google Scholar]
- Wehrmann, M. , Billard, P. , Martin‐Meriadec, A. , Zegeye, A. , and Klebensberger, J. (2017) Functional role of lanthanides in enzymatic activity and transcriptional regulation of pyrroloquinoline quinone‐dependent alcohol dehydrogenases in pseudomonas putida KT2440. mBio 8: e00570–e00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M.L. , Wessels, J.C. , Pol, A. , Op den Camp, H.J. , Jetten, M.S. , and van Niftrik, L. (2015) XoxF‐type methanol dehydrogenase from the anaerobic methanotroph "Candidatus Methylomirabilis oxyfera". Appl Environ Microbiol 81: 1442–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanpirat, P. , Nakatsuji, Y. , Hiraga, S. , Fujitani, Y. , Izumi, T. , Masuda, S. , et al. (2020) Lanthanide‐dependent methanol and formaldehyde oxidation in Methylobacterium aquaticum strain 22A. Microorganisms 8: 822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Z. , Beck, D.A.C. , and Chistoserdova, L. (2017) Natural selection in synthetic communities highlights the roles of Methylococcaceae and Methylophilaceae and suggests differential roles for alternative methanol dehydrogenases in methane consumption. Front Microbiol 8: 2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y. , Huang, J. , Zhao, F. , and Chistoserdova, L. (2018) Physiological effect of XoxG(4) on lanthanide‐dependent methanotrophy. mBio 9: e02430‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Abundance (% of total protein for each sample) of PQQ‐ADH in cell extract of Methylocella silvestris grown on methanol or ethanol in the absence or presence of lanthanum.
Table S2. Proteins differentially regulated in M. silvestris BL2 cells grown on A) methanol or B) ethanol, in response to lanthanum. The fold‐change (FC) of proteins upregulated in lanthanum‐containing medium is shown with a positive log2 (FC) value and downregulated proteins are shown with a negative value. Only proteins differentially expressed at |log2 (FC)| ≥ 2 and adjusted p value ≤0.01 are shown. Proteins regulated in both methanol‐ and ethanol‐grown cells are shown in bold type.
Table S3. Primer sequences.
Table S4. Table S4 is included as a separate Excel spreadsheet file.
Fig. S1. Detail of alignment of M. silvestris PQQ‐ADH sequences with other representative examples. The catalytic aspartate residue and calcium or lanthanide‐coordinating residue (A or D) (at positions 303 and 305 respectively in MxaF from Methylorubrum extorquens) are indicated with arrows.
Fig. S2. Growth curves of the wild type during growth on methanol (A) or ethanol (B). Error bars show the standard error of the mean (n = 3). Panel A shows a representative of three independent experiments.
Fig. S3. Double mutant strain ΔmxaFΔxoxF5 (A) or the wild type (B) were grown on methanol (0.1% v/v) in the presence (red) or absence (black) of 4% v/v acetylene in the vial headspace. Error bars show the standard error of the mean (n = 3).


