Abstract
Our immune system is critical for preventing and treating SARS-CoV-2 infections, but aberrant immune responses can have deleterious effects. While antibodies to glycans could recognize the virus and influence the clinical outcome, little is known about their roles. Using a carbohydrate antigen microarray, we profiled serum antibodies in healthy control subjects and COVID-19 patients from two separate cohorts. COVID-19 patients had numerous autoantibodies to self-glycans, including antiganglioside antibodies that can cause neurological disorders. Additionally, nearly all antiglycan IgM signals were lower in COVID-19 patients, indicating a global dysregulation of this class of antibodies. Autoantibodies to certain N-linked glycans correlated with more severe disease, as did low levels of antibodies to the Forssman antigen and ovalbumin. Collectively, this study indicates that expanded testing for antiglycan antibodies could be beneficial for clinical analysis of COVID-19 patients and illustrates the importance of including host and viral carbohydrate antigens when studying immune responses to viruses.
Significance Statement.
Antibody responses induced by SARS-CoV-2 infection can provide protection, but aberrant responses can be harmful. Since the surface of the virus is heavily glycosylated, glycan-binding antibodies can be induced and may affect disease severity and clinical outcome. To evaluate their roles, we used a glycan microarray to profile serum antiglycan antibodies in COVID-19 patients and healthy control subjects. Overall, we uncovered striking differences, including global dysregulation of antiglycan IgM and induction of numerous autoantibodies to glycans in COVID-19 patients, including some that cause neurological problems. The results provide a deeper understanding of antibody responses, illustrate the importance of evaluating antibody responses to glycans, and indicate that assessment of antiglycan antibodies could aid the clinical analysis of COVID-19 patients.
Introduction
COVID-19 is a respiratory disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Since emerging, this virus has caused millions of deaths worldwide (1). An especially troubling issue is that some symptoms can last for months beyond the primary infection, even in the absence of detectable virus (2–4). It is unclear why some patients, often referred to as “long haulers,” have prolonged effects. Beyond the severe impact on human health, SARS-CoV-2 has caused major disruptions to many aspects of life, including the economy, education, travel, and personal life. As a result, an unprecedented global effort is underway to develop effective methods to prevent and treat COVID-19. Because this is a new infectious virus, our understanding of the factors that influence clinical outcomes are limited. Therefore, there is an urgent need to improve our basic understanding of how the virus works, why it causes severe disease outcomes, and how we can intervene to protect human life.
Our immune system plays a critical role in preventing, clearing, and treating SARS-CoV-2. Therefore, understanding host immune responses to SARS-CoV-2 is essential for developing effective therapies and vaccines to control this pandemic. While the immune response can involve many elements of the innate and adaptive arms of the immune system, antibody responses are one of the most important features. Most patients develop a robust antibody response to the virus (5–7), and neutralizing antibodies in recovering patients may help prevent new infections through the administration of convalescent serum (8–11). While often beneficial, overly aggressive and/or aberrant antibody responses can also be harmful in COVID-19 patients (12–14). For example, several studies have shown that SARS-CoV-2 induces autoantibodies (15–22). Alternatively, certain antibody responses can actually enhance infection (23). For these reasons, a thorough understanding of antibody responses to SARS-CoV-2 is vital for prevention and treatment of COVID-19.
Numerous groups have been studying antibody responses to SARS-CoV-2 (12–15,24–28), however, the vast majority of these studies focus on antibodies that bind proteins. Several studies have demonstrated that the SARS-CoV-2 spike protein is heavily glycosylated, raising the possibility that pre-existing and/or induced antibodies to carbohydrates may recognize the virus (29–34). A few studies have begun to address the roles of antiglycan antibodies. For example, a recent study reported an inverse relationship between COVID-19 disease severity and serum anti-α-Gal antibodies (35). Low levels of antibodies to the Tn antigen (GalNAcα-Ser/Thr) have also been observed in COVID-19 patients (36). A longitudinal study of four patients found antibodies to several N-linked glycans in COVID-19 patients (37). Autoantibodies to certain gangliosides have also been observed in several case studies of COVID-19 patients with Guillain–Barre Syndrome (GBS) related symptoms (16, 17). Lastly, recent reports have shown a small correlation with ABO blood type and susceptibility to COVID-19, and this effect may involve pre-existing serum antibodies to the blood group A (BG-A) and/or blood group B (BG-B) carbohydrates (38–42). Collectively, these studies suggest that glycans and antiglycan antibodies may play an important role in the prevention, severity, and treatment of COVID-19.
To better understand the roles of glycans in the immune response to SARS-CoV-2, we compared serum antiglycan IgG and IgM antibody repertoires of uninfected control subjects with two different cohorts of COVID-19 patients. To monitor a large and diverse assortment of antibody populations, we profiled each serum sample using a carbohydrate antigen microarray with over 800 components. These studies revealed that COVID-19 patients had substantial differences in antiglycan antibodies, including unusual antibodies to a variety of self-glycans.
Results
Study design
Serum from 40 SARS-CoV-2-infected patients from Raybiotech, 70 SARS-CoV-2-infected patients from the National Institutes of Health (NIH), and 38 uninfected, healthy individuals (hereafter, referred to as “controls”) were used in the study (Table 1). All control serum samples were collected before the outbreak of SARS-CoV-2 began. All COVID-19 patients had a positive antibody test for IgG, IgM, or both to the spike protein receptor binding domain. Information about patient symptoms and outcomes were not available for the Raybiotech cohort. Blood type was not available for any of the COVID-19 patients. The differences in age between the control group and the COVID-19-positive groups were statistically significant and may have some influence on the results (see below). Samples from the two COVID-19 cohorts were collected at different times, in different countries, and as part of different studies, helping to ensure that any observed antibody responses or differences were consistent in COVID-19 patients.
Table 1.
Demographics of cohorts.
| Control group | Raybiotech COVID-19 | NIH COVID-19 | |
|---|---|---|---|
| Total | 38 | 40 | 70 |
| Sex (M/F) | 22/16 | 20/20 | 33/37 |
| Average age (range) | 38 (18–65) | 64 (41–92)* | 53 (0–83)* |
| Country of sample collection | United States | United States | Italy |
| Disease severity—no. (%) | |||
| Asymptomatic or mild | na | 35 (50%) | |
| Severe or critical | na | 35 (50%) | |
| Comorbidities—no. (%) | |||
| Any | na | na | 37 (59%) |
| Diabetes | na | na | 5 (8%) |
| Obesity | na | na | 5 (8%) |
| Cardiovascular disease | na | na | 14 (22%) |
| Treatment—no. (%) | |||
| Hydroxychloroquine only | na | na | 13 (19%) |
| Dexamethasone only | na | na | 2 (3%) |
| Hydroxychloroquine and dexamethasone | na | na | 12 (17%) |
| Median days from onset of symptoms to sample collection (range) | na | 33 (1–34) | 18 (2–80); P = 0.19 |
na = not available; *P < 0.05.
To assess the antiglycan repertoires of patients with COVID-19, we profiled IgG and IgM from serum samples on a carbohydrate antigen microarray containing 816 components. The microarray included a diverse collection of N- and O-linked glycans, glycans from glycolipids, glycopeptides, bacterial and fungal glycans, and some natural glycoproteins. This set of glycans allows for rapid profiling of a broad range of antiglycan antibody populations in serum including those to both foreign and self-antigens (for designations of self/nonself, see Supporting Information and Supporting Excel file). Antibody signals from each Raybiotech COVID-19 patient were compared to the control set to identify signals with statistically different averages. These initial hits were further evaluated using the NIH COVID-19 patients as a validation set.
Global dysregulation of serum antiglycan IgM in SARS-CoV-2-positive patients
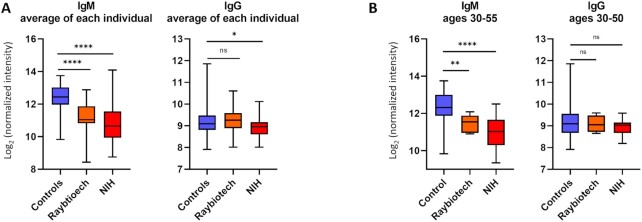
We started by evaluating mean IgG and IgM signals of all the array components for each patient to assess global differences in antiglycan antibody levels in control and COVID-19 patient samples and to provide context for individual differences (Fig. 1A). For nearly every glycan, the mean IgM signals to glycans were 2- to 4-fold lower in both cohorts of SARS-CoV-2-positive patients compared to controls, while the total mean IgG signals were similar. Across the entire array, the average IgM signals in the control group were 2.3-fold higher than COVID-19 patients. To determine if this effect was specific to carbohydrate-binding IgM or due to differences in total serum IgM levels, we measured the total IgM in the Raybiotech cohort and a subset of control samples. The average total IgM levels in the COVID-19 patient samples were 30% lower than the average total IgM in the control samples (Figure S1, Supplementary Material). Thus, differences in total IgM only partially explain the substantially lower IgM signals observed on the array in SARS-CoV-2-positive patients. In addition, when comparing signals from a subset of patients of similar ages [30 to 50; average = 40.5 for controls, 43.7 for Raybiotech (P = 0.32 versus controls), and 41.3 for NIH (P = 0.66 versus controls) cohorts], there was still a considerable difference in IgM signals (Fig. 1B). Thus, age and total IgM only partially explain the lower overall IgM signals in COVID-19 patients. Since we observed this difference in cohorts from different countries, the effect does not appear to be due to variations in location. Also, COVID-19 patients and controls were randomized, so it does not appear to be due to technical issues with the arrays or assay. Average IgM signal in the NIH cohort did not correlate with titers to nucleocapsid, titers to spike protein, or comorbidities, including diabetes, obesity, or cardiovascular disease. Many (39%) of the COVID-19 patients in the NIH cohort were treated with hydroxychloroquine and/or dexamethasone, both of which are immunosuppressive; however, we did not observe a correlation between the average IgM signal on the array and treatment with either drug. Average IgM signal in the NIH cohort was not correlated with the presence of a secondary infection. For IgG, there was a small but statistically significant different in average signals between the controls and the NIH cohort. This difference was not significant when comparing the subset of patients ages 30 to 50.
Figure 1.
Average IgG and IgM antibody signals to all glycans. (A) Box and whisker plots of the average array signals (log-transformed base 2) to all array components for IgG and IgM antibodies from control and COVID-19 serum samples. Mean IgG of control samples was 9.21 raw fluorescence units (RFU) on a log2 scale. Mean IgG signal for the Raybiotech COVID-19 cohort was 9.30, and the mean of NIH COVID-19 cohort was 8.90. An unpaired t test with Welch's correction showed no significant difference between mean IgG values of the controls and Raybiotech cohort and a small difference between the controls and NIH COVID-19 cohort IgG values (*, P = 0.0250). Mean IgM of controls was 12.41. Mean IgM of Raybtioech COVID-19 cohort was 11.21. Mean IgM of NIH COVID-19 cohort was 10.83. Unpaired t test with Welch's correction for mean IgM values was significant for both COVID-19 cohorts compared to controls (****, P < 0.0001). (B) Box and whisker plots of the average array signals for IgM and IgG of a select age range of patients. ns, not significant. Boxes depict quartiles and whiskers depict the min and max.
Differences in antiglycan antibodies based on disease status
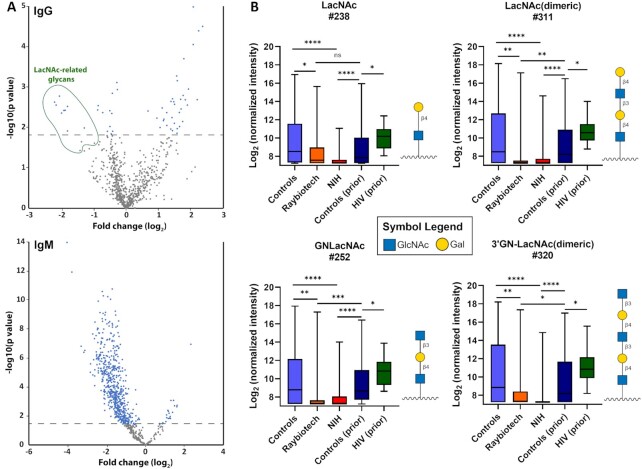
Next, we evaluated potential differences in the mean signals between the COVID-19 and control cohorts for each of the carbohydrate antigens on the array. We used the Raybiotech cohort as a discovery set and the NIH cohort as an independent confirmation set. Overall comparisons of the Raybiotech cohort and controls are illustrated in Fig. 2(A). Given the large, systematic differences in IgM signals for COVID-19 patients and control subjects, we decided to focus on individual array components for IgG. Signals for a subset of 19 array components displayed statistically significant differences with consistent directionality in both cohorts relative to the control group (see Materials and Methods for details). A total of nine of these glycans were LacNAc derivatives; six were glycoproteins, glycopeptides, or peptides; three were single glycans; and one was a nonhuman xylose derivative (Fig. 2B; Supplementary Excel file; Figure S2, Supplementary Material).
Figure 2.
Differences between COVID-19 patients and controls. (A). Volcano plots with the fold change (log2) for the average Raybiotech COVID-19 patient signal relative to the controls on the x-axis and the negative P-value (log10) on the y-axis. Dots above the dashed line were statistically significant using a false discovery rate of 20%. (B) Box and whisker plots of the average array signals (log-transformed base 2) to LacNAc glycan array components for IgG antibodies from control and Raybiotech and NIH COVID-19 serum samples. Also, plotted are the data from a previous studies analyzing healthy control samples and patients with HIV infection. See Materials and Methods for the method of discovery and statistical analysis. Each row was analyzed individually, without assuming a consistent SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001; ns, not significant. Glycan structures were created using GlycoGlyph (45). Boxes depict quartiles and whiskers depict the min and max.
To further evaluate these results, we next compared the 19 hits to antibody signals measured previously by our group in separate studies. One study included 220 healthy subjects (43), and the other study included 27 HIV-positive patients approximately 1 year after diagnosis (44). In both cases, the version of the microarray used in those studies had about half as many array components as the current microarray, so only some of the signals could be compared. Nevertheless, the analysis provides additional context in those cases where it is possible. For example, LacNAc related components, such as LacNAc (array component #238) and LacNAc (dimeric; array component #311; also known as the i antigen) were significantly lower in our COVID-19 patients than in our prior/historical healthy control set and the HIV-positive patients. These results indicate that low IgG to these components is consistent when compared to a larger cohort of healthy controls and that low IgG to LacNAc-related glycans is not a general feature of viral infection.
We also evaluated several potential covariates. Signals for the 19 hits did not correlate with age or sex in any of the individual cohorts. We have previously found no differences in antiglycan IgG profiles based on age or sex in healthy subjects (43), so the data reported in this study are consistent with the prior results. We did not have sufficient information to analyze potential correlations with race. IgG signals to LacNAc-related glycans did not correlate with anti-SARS-CoV2 spike or nucleocapsid titers. Within the NIH cohort, these IgG did not correlate with any comorbidities, including diabetes, obesity, or cardiovascular disease, nor did they correlate with treatment with hydroxychloroquine or dexamethasone.
Potential correlations of antibodies to ABH blood group antigens, alpha-Gal, and the Tn antigen
Prior studies have reported altered levels of antibodies to blood group antigens, the alpha-Gal antigen, and the Tn antigen in COVID-19 patients. Therefore, we evaluated each of these in more detail. For each of these antigens, we have many structural variants on our array, including different carrier glycan chains, different linkers, and different densities on the array surface. For the Tn antigen, GalNAc alpha-linked to serine or threonine of a peptide chain, we have many different peptide sequences as well as sequences with multiple Tn residues. These variations allow us to evaluate different antibody subpopulations that may have unique recognition requirements for capture on the array surface.
A previous study by Urra et al. reported an inverse correlation for IgG and IgM antibodies to alpha-Gal [Galα1–3Galβ1–3(4)GlcNAc] and COVID-19 disease severity; those with the most severe outcomes had the lowest levels of alpha-Gal antibodies (35). In their study, COVID-19 patients as a group had lower antibody levels than healthy subjects. For our study, the alpha-Gal antigen displayed inconsistent directionality. For the Raybiotech COVID-19 cohort, COVID-19 patients had higher IgG to alpha-Gal than controls, whereas the NIH COVID-19 cohort had lower IgG to alpha-Gal relative to the controls. Given the inconsistent directionality of the differences, antibodies to the alpha-Gal antigen were not considered further in this study. For IgM, we observed strong correlations with disease status when evaluating both the discovery set and the NIH set. In both cohorts, COVID-19 patients had substantially lower IgM signals than controls. The effect was quite large, with the median value of the NIH cohort being as much as 60-fold lower than that of the controls. While consistent, lower IgM signals were also observed for numerous other glycan antigens and appear to be a general, overall effect rather than a specific difference for certain antigens.
There have been several studies that have shown a correlation between blood type and SARS-CoV-2 infection rate (38–41). In particular, individuals with blood type A have a slightly higher infection rate than those with blood type O. Since serum antibodies to blood group antigens are highly correlated with blood type, we next examined this family of antibodies. Thus, we might have expected to see lower antibody signals to blood group A antigens. Instead, none of the blood group A or B antigens on our array displayed a significant correlation with disease status in our initial discovery phase for IgG. Similar to the alpha-Gal antigen and many other glycans, we observed significantly lower IgM to blood group A and B antigens in COVID-19 patients versus controls.
For the Tn antigen, a prior study reported lower antibody levels in COVID-19 patients relative to controls (36). This study measured combined signals from IgG, IgM, and IgA. In our study, none of the various Tn peptides displayed a significant correlation with disease status for IgG. For IgM, many Tn peptides showed very strong correlations with disease status in both the discovery cohort and the NIH cohort. In some cases, the median value for COVID-19 patients was as much as 8-fold lower than the control subjects. While large in magnitude and consistent, this appears to be a general effect for IgM.
Autoantibodies to self-carbohydrates in SARS-CoV-2-positive patients
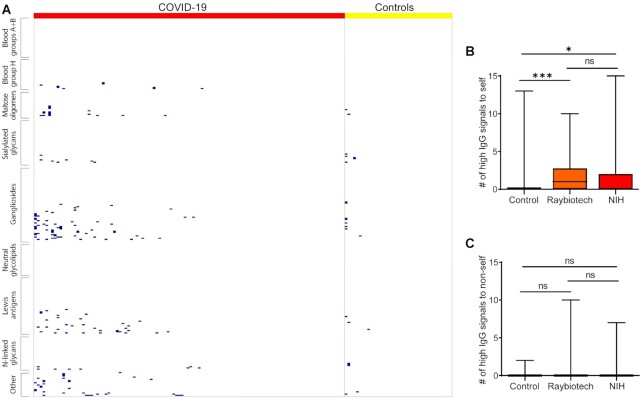
One striking difference between serum samples from COVID-19 patients and healthy controls were unusually high antibody signals to self-carbohydrates (Fig. 3). “Unusually high” was defined as a signal that was greater than 5 SD above the mean of the control group and a signal of at least 1,500 RFU, which is 10-fold above the floor value for our assay. In those cases where data were available, we also required that the signals be at least 3 SD above the mean for our prior study of 220 healthy subjects (43). For individual instances, unusually high signals were typically observed in 1 to 5 patients, with the most frequent instance occurring in 11/110 patients. For IgG, the instances were distributed over many different patients, with 56% of the patients having at least one unusually high signal to a self-glycan and 32% having at least two. For IgM, most of the unusually high antibody signals to self-glycans occurred in three patients, accounting for 71% of the instances. About 17% of patients had at least one unusually high IgM signal and 7% had at least two.
Figure 3.
Abnormally high IgG signals for selected glycan families. (A) Heat map depicting the abnormally high IgG signals for selected self-glycans. Glycans in rows, and patients in columns. Blue boxes represented high signals. (B) Bar graphs comparing the number of abnormally high IgG signals for self-glycans among the three cohorts. (C) Bar graphs comparing the number of abnormally high IgG signals for nonself glycans among the three cohorts. *, P < 0.05; **, P < 0.01; and ***, P < 0.001; ns, not significant. Boxes depict quartiles and whiskers depict the min and max.
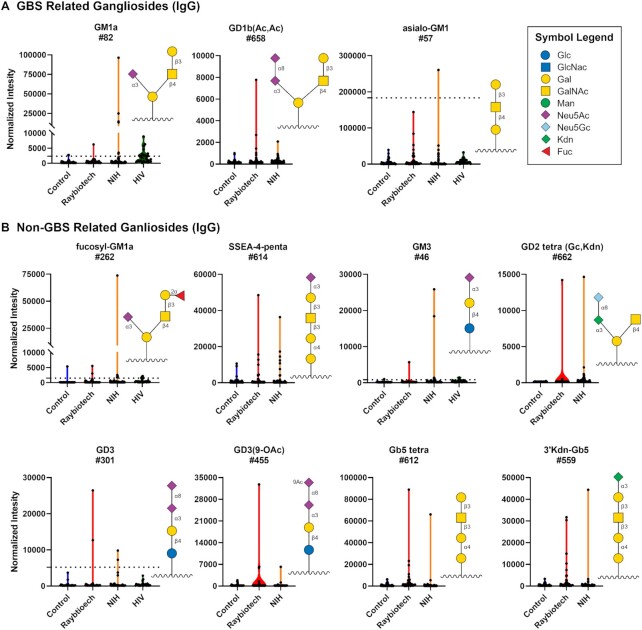
The glycan family most frequently targeted by abnormally high antibodies were gangliosides and other glycolipids (Fig. 3; Figure S3, Supplementary Material). While very uncommon in healthy individuals, antiganglioside antibodies are often found in patients with autoimmune diseases and other nervous system dysfunctions (46). For example, antibodies to asialo-GM1, GM1a, GD1a, and GD1b are frequently observed in patients with GBS. We observed unusually high IgG antibodies to several GBS glycans in certain patients (Fig. 4A). Large IgG signals were also observed to several other gangliosides/glycolipids not associated with GBS, such as GD3, fucosyl-GM1, Gb5, stage-specific embryonic antigen 4 (SSEA-4), and GM3 (Fig. 4B). In many cases, such as for GM1a, GM3, fucoysl-GM1, and 9-O-acetylated GD3, the largest signal in the COVID-19 patients was > 10-fold higher than the highest signal in the controls and > 10-fold higher the average signal from the prior/historical cohort of 220 patients plus 3 SD. High signals to gangliosides were not observed in HIV-infected patients, with the exception of anti-GT2 antibodies (Figure S4, Supplementary Material). No abnormally high signals were observed for neutral glycolipids and relatively few were observed for other sialylated glycans, indicating a selective response to gangliosides.
Figure 4.
High antibody signals to select ganglioside glycans in COVID-19 patient serum. Violin plots showing high IgG signals to various gangliosides/glycolipids for COVID-19 patients from Raybiotech and NIH versus controls, with each point representing data from an individual patient. Where available, graphs also include data from a prior study analyzing antiglycan antibodies of HIV-infected patients (44). To compare high signals to a larger subset of healthy controls, the dashed lines represent 3 SD above the mean of 220 control samples from a previous study (43). (A) GBS-associated ganglioside and (B) other gangliosides/glycolipids. Glycan structures were created using GlycoGlyph (45).
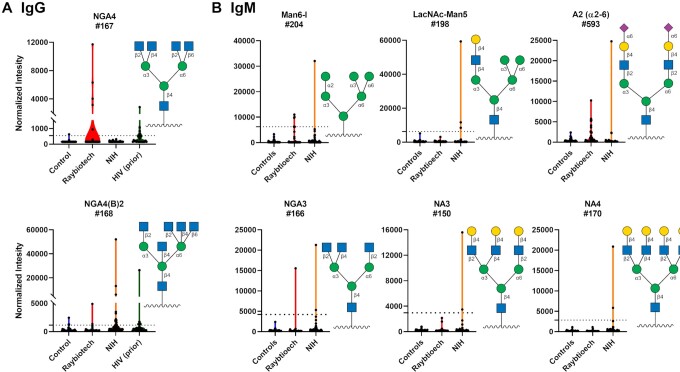
In addition to the antibodies to gangliosides/glycolipids, we also observed unusually large IgG and IgM signals to various N-linked glycans and oligomannose substructures found in N-linked glycans (Fig. 5; Figure S5, Supplementary Material). N-linked glycans are abundant in the human body, and they also cover the spike proteins of SARS-CoV-2. The most remarkable examples were four patients with abnormally high IgG antibodies to NGA4 a complex, tetraantennary N-glycan with the following sequence: GlcNAcβ1–2(GlcNAcβ1–6)Manα1–6[GlcNAcβ1–2(GlcNAcβ1–4)Manα1–3]Manβ1–4GlcNAcβ. The highest COVID-19 patient signal was > 20-fold higher than the controls from this study as well as 20-fold higher than the mean of our prior/historical 220 healthy subjects plus 3 SD (and 10-fold higher than the highest signal for all 220 subjects). For three of the four patients, the anti-NGA4 antibodies were highly selective for NGA4; they did not cross-react with any other related N-glycans structures on our array, including NGA3 (GlcNAcβ1–2Manα1–6[GlcNAcβ1–2(GlcNAcβ1–4)Manα1–3]Manβ1–4GlcNAc), a closely related triantennary N-glycan, NGA3B (GlcNAcβ1–2Manα1–6[GlcNAcβ1–2(GlcNAcβ1–4)Manα1–3](GlcNAcβ1–4)Manβ1–4GlcNAc), and NA4 (Galβ1–4GlcNAcβ1–2(Galβ1–4GlcNAcβ1–6)Manα1–6[Galβ1–4GlcNAcβ1–2(Galβ1–4GlcNAcβ1–4)Manα1–3]Manβ1–4GlcNAcβ. These results indicate a specific response to NGA4, rather than a nonspecific or polyreactive response. The fourth patient's antibodies reacted with NGA3 and NGA4, but no other N-linked glycans. Monoclonal antibodies to N-linked glycans (PGT121, PGT126, and PGT128) bound to the spike protein but did not neutralize the virus (Supplementary Figures S6 and S7, Supplementary Material).
Figure 5.
High IgG and IgM signals to select N-linked glycans in COVID-19 patient serum. Violin plots show high antibody signals to select N-linked glycan array components for serum from COVID-19 patients compared to baseline signals seen in serum from controls, with each point representing data from an individual patient. Where available, graphs also include data from a prior study analyzing antiglycan antibodies of HIV-infected patients (44). To compare high signals to a larger subset of healthy controls, the dashed lines represent 3 SD above the mean of 220 control samples from a previous study (43). (A) IgG. (B) IgM. See symbol legend in Fig. 4. Glycan structures were created using GlycoGlyph (45).
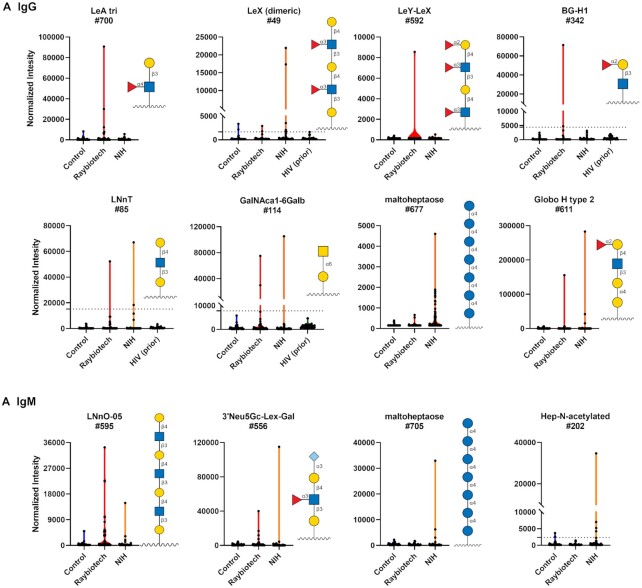
Beyond gangliosides/glycolipids and N-linked glycans, we also observed abnormally high antibody signals to a variety of other self-glycans (Fig. 6; Figures S8 and S9, Supplementary Material). Several of these antibodies targeted Lewis/blood group antigens such as Lewis A (LeA), Lewis X (LeX), Sialyl Lewis X (SLeX), and blood group H (BG-H1 and Globo H-related glycans). Several patients had unusually high IgG and IgM to malto-oligosaccharides such as maltoheptaose; these glycans are substructures found in glycogen. We also observed several patients with unusually high antibodies to hyaluronic acid oligosaccharides, although this does not appear to be specific for COVID-19 as antibodies to hyaluronic acid were also observed in HIV-infected patients (Figure S4, Supplementary Material).
Figure 6.
High antibody signals to self-glycans in COVID-19 patient serum. Violin plots show high antibody signals to select self glycans for serum from COVID-19 patients compared to baseline signals seen in serum from control donors, with each point representing data from an individual patient. Where available, graphs also include data from a prior study analyzing antiglycan antibodies of HIV-infected patients (44). To compare high signals to a larger subset of healthy controls, the dashed lines represent 3 SD above the mean of 220 control samples from a previous study (43). (A) IgG. (B) IgM. See symbol legend in Fig. 4. Glycan structures were created using GlycoGlyph (45).
Antiglycan antibodies correlate with disease severity in SARS-CoV-2-positive patients
We next evaluated potential differences in antiglycan antibodies based on the disease severity. This information was available for the NIH COVID-19 cohort but not Raybiotech. The average age of patients with severe/critical disease was 57 (range = 23 to 83), versus 49 (range = 0 to 82) for patients with asymptomatic/mild/moderate disease. The difference in age was statistically significant (P = 0.0323). There were 17 females and 18 males in the severe/critical group, and there were 19 females and 16 males in the asymptomatic/mild/moderate cohort. There was no statistically significant difference in the sexes.
To identify potential relationships between antiglycan antibodies and disease severity, we looked for significant differences between groups that also displayed consistent trends with the level of severity (e.g. asymptomatic < mild < moderate < severe < critical). After identifying potential correlates, we also compared signals for healthy controls from this study, as well as from our prior/historical healthy controls, to provide additional perspective for the trends.
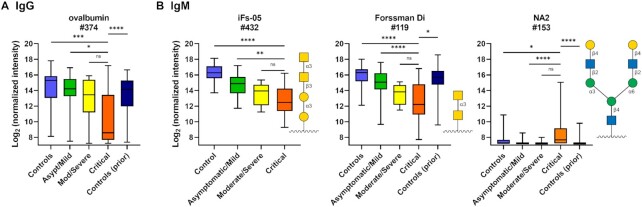
For IgG, antibody signals to the glycoprotein ovalbumin and its periodate-treated variant were the only components that correlated with disease severity, with lower signals associated with increasing disease severity (see Fig. 7A). The differences were largely driven by especially low signals in critical patients. Since sodium periodate treatment degrades the glycans on glycoproteins, the relationship does not appear to be glycan-dependent. Since no other antiglycan IgG antibodies displayed this type of relationship, low signals do not appear to be due to a general effect such as a weakened immune system of critical patients. Antiovalbumin IgG did not correlate with sex, age, titers to nucleocapsid, titers to spike protein, or any comorbidities, such as diabetes, obesity, or cardiovascular disease. They also did not correlate with treatment with hydroxychloroquine or dexamethasone, or with the presence of secondary infection.
Figure 7.
Antiglycan IgG and IgM antibody signals that correlate with disease severity. (A) Box and whisker plots of the average IgG signals (log-transformed base 2) to ovalbumin for controls, NIH COVID-19 patients designated as either asymptomatic/mild, moderate/severe, or critical, and controls from a prior study. (B) Box and whisker plots of the average array IgM signals (log-transformed base 2) to iso-Forssman, the Forssman disaccharide, and NA2 for controls, NIH COVID-19 patients designated as either asymptomatic/mild, moderate/severe, or critical, and controls from a prior study. Method of discovery is ANOVA using the step-up method of Benjamini and Hochberg with a false discovery rate (FDR) = 0.05 to account for multiple comparisons, and demonstration of consistent trends with increasing disease severity. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. See symbol legend in Fig. 4. Glycan structures were created using GlycoGlyph (45). Boxes depict quartiles and whiskers depict the min and max.
For IgM signals, several antiglycan antibody populations displayed significant correlations with disease severity (Fig. 7B; Figure S10, Supplementary Material). For maltotriose and the N-linked glycan NA2, higher signals were associated with more severe disease, especially in critical patients. The relationship was especially notable for NA2, where signals were significantly higher than the controls from this study and our prior/historical controls, despite the fact that the most IgM signals on the array were decreased in COVID-19 patients relative to controls. A total of three patients had IgM signals for NA2 that met our strict criteria for “abnormally high,” and several others had high signals that were high but did not quite meet the criteria. Strong correlations were also observed for antiglycan antibodies that target the Forssman disaccharide (Forssman Di), iso-Forssman (iFs), and a core 5 glycopeptide [Ac-Ser-(GalNAcα1–3GalNAcα1-)Ser-Ser-Gly], glycans sharing the same terminal disaccharide motif, GalNAcα1–3GalNAc. In these cases, however, lower signals were associated with more severe disease. (Fig. 7B). IgM signals to core 5, Forssman antigen, iso-Forssman antigen, NA2, and maltotriose did not correlate significantly with sex, age, titers to nucleocapsid, titers to spike protein, or comorbidities, nor did they correlate with the presence of a secondary infection. IgM to iFs and maltotriose displayed a significant correlation (P < 0.05) to treatment with hydroxychloroquine. However, this association was largely driven by the difference in treatment between mild patients (only 1/35 received hydroxychloroquine; 0/35 received dexamethasone) and severe/critical patients (24/35 received hydroxychloroquine and 9/35 received dexamethasone). If one only includes severe/critical patients, IgM signals to these glycans no longer correlate with hydroxychloroquine treatment, indicating that antibodies to these glycans were not caused by treatment. In addition, the average IgM signals did not decrease with the use of hydroxychloroquine (or dexamethasone), so there does not appear to be a general effect on IgM levels. The average IgM signals across the microarray did not correlate with disease severity.
Long term follow up was available for 22 patients from the NIH cohort. A total of four of 22 patients with 6 months follow up information experienced paresthesia, a common symptom of GBS. In total, one of these patients had IgG autoantibodies to several gangliosides, including Fuc-GM1, GD1b, GM1, and GM2. A second patient had high IgG to Fuc-GM1. In total, one patient with numerous IgM autoantibodies experienced prolonged skin peeling and hair loss. Additional studies will be needed to evaluate these relationships in more detail.
Discussion
Understanding immune responses to SARS-CoV-2 infection is critical for preventing and treating the disease. For example, SARS-CoV-2 can trigger an overly aggressive immune response leading to excessive damage to the patient, and uncovering this problem has led to the use of the anti-inflammatory agent dexamethasone as an effective treatment for COVID-19 (47). While there is considerable information being reported on various aspects of the response (5, 12–14, 28, 48, 49), very little is known about immune responses to carbohydrates. Since the SARS-CoV-2 spike protein is heavily glycosylated and enveloped viruses can also incorporate host glycolipids and glycoproteins into their envelope (29–34), pre-existing or induced antiglycan antibodies could potentially recognize the virus and influence disease progression. To address these possibilities, we used a large carbohydrate antigen microarray to profile serum antiglycan IgG and IgM antibody repertoires in two separate COVID-19 patient cohorts versus controls.
COVID-19 patients displayed unique antiglycan antibody profiles relative to healthy controls. One of the most prominent differences was a global dysregulation of antiglycan IgM in COVID-19 patients. The vast majority of antiglycan IgM signals were substantially lower than in healthy controls, while no general alterations were observed for antiglycan IgG. Many antiglycan IgM antibodies are produced by B1 cells, and this subset has been reported to be decreased in COVID-19 patients (50). Antibodies produced by B1 cells are critical for protection from infections and for clearing debris derived from dead cells in the body (51). The low levels of antiglycan IgM could render COVID-19 patients more susceptible to secondary infections and/or disrupt the proper removal of cell debris.
A second striking difference in COVID-19 patients relative to controls were autoantibodies to numerous self-carbohydrates, including gangliosides, N-linked glycans, and Lewis antigens, in certain patients. In some cases, the antibody signals observed in COVID-19 patients were > 20 times higher than the largest signal in the control group. Antibodies to a small subset of gangliosides have been reported previously in several COVID-19 patients (16, 52, 53). Our study provides further support of those observations and uncovers antibodies to a much larger assortment of gangliosides/glycolipids than previously reported. These antibodies could arise via recognition of gangliosides/glycolipids incorporated into the SARS-CoV-2 envelope. In addition, the detection of abnormally high antibodies to N-linked glycans, Lewis antigens, and other self-carbohydrates has not been previously reported in COVID-19 patients. These antibodies could be induced by glycans on the spike protein or host glycans incorporated into the envelope. Taken together, our results demonstrate much more extensive development of autoantibodies to glycans in COVID-19 patients than previously known.
Several lines of evidence indicate that the autoantibodies observed in COVID-19 patients are unique and specific to SARS-CoV-2 infection. We have investigated antiglycan antibody repertoires in numerous human serum samples previously, including over 200 healthy subjects and over 100 cancer patients before and after treatment with various cancer vaccines (44,54–58). Based on our prior work, abnormally high antibodies to human gangliosides, N-linked glycans, and other self-glycans are uncommon. Some instances where we have observed high antibodies to some of the glycans are HIV-infected patients (antibodies to Man9, GT2, and GT3) (44) and cancer patients immunized with a whole cell cancer vaccine (antibodies to GM2, GM3, Gb5, and sialyl Lewis X) (57). In these cases, antibodies to self-glycans were present in fewer patients and for fewer glycans than what we observed in COVID-19 patients. In prior studies, we found that serum IgG and IgM levels to nearly all glycans on our array are stable over time frames of up to 3 years (44, 58), indicating that high signals in certain patients are not simply due to high variability or random fluctuations over time. Lastly, our prior studies on healthy subjects of varying age indicate that these high antibody populations are not merely due to increasing age.
Antibodies to self-glycans could be clinically relevant for a variety of reasons. Autoantibodies to self-glycans are associated with a variety of autoimmune disorders (46, 59–61). For example, antibodies to gangliosides are often linked to neurological disorders such as GBS and Miller Fisher Syndrome. Gangliosides are expressed at high levels on nerve cells, and antibodies to these glycans can have a variety of effects, including destruction of the neuromuscular junction of nerve cells and disruption of the blood–nerve barrier and/or blood–brain barrier (62, 63). Gangliosides also play roles in immune tolerance, signal transduction, and cell adhesion, and antibodies to gangliosides can disrupt these processes as well (64). From a clinical perspective, antibodies to GM1, GD1a, GM1b, and GalNAc-GD1a are linked to acute motor axonal neuropathy, and antibodies to GQ1b, GT1a, GD1b, and GD3 are associated with cranial, bulbar, and sensory variants of GBS (60, 61,65). Much less is known about clinical effects of antibodies to N-linked glycans and other self-glycans, but these glycans are present on numerous cells in the human body.
A third difference between COVID-19 patients and healthy subjects was significantly lower than normal IgG antibodies to the i antigen (Galβ1–4GlcNAcβ1–3Galβ1–4GlcNAc; also known as dimeric LacNAc) and other LacNAc-containing structures. The i antigen is expressed at high levels on fetal red blood cells and tissues as well as in B cells, T cells, monocytes, and macrophages. Most adults have weak autoantibodies to these antigens that bind in a temperature dependent manner (66). They are often referred to as “cold agglutinins” because they will agglutinate red blood cells at lower temperatures, but not at 37°C. In most people, they do not cause any clinical symptoms, but they can cause anemia in some patients, especially elderly individuals susceptible to cold temperatures in their extremities.
The effects of low levels of IgG to the i antigen and other LacNAc glycans have not been well-studied. In the context of COVID-19, several possibilities are conceivable. LacNAc-terminal glycans are expressed on the SARS-CoV-2 spike protein on both N-linked and O-linked glycans (29–34, 67), so antibodies that target LacNAc could bind to the virus helping to provide immune protection. People with lower levels of these antibodies would have reduced binding and, therefore, reduced protection from infection. Alternatively, low levels may be observed because SARS-CoV-2 causes considerable cell death. Antibodies that recognize cell debris may be depleted and/or complexed with cellular materials rendering them unable to bind on our array.
Several serum antiglycan antibody populations were correlated with disease severity. Lower than normal IgM levels to the Forssman antigen, iso-Forssman antigen, and core 5 were all correlated with more severe disease. Anti-Forssman antibodies are typically very abundant in human serum, and they are thought to provide protection against infections from a variety of viruses and bacteria (68). The presence of autoantibodies to the N-linked glycan NA2 were correlated with more severe disease as well. This glycan is found on numerous human glycoproteins, and antibodies to it could contribute to autoimmune-like features of COVID-19. Lastly, although not a carbohydrate, we also found that low levels of IgG to ovalbumin correlated with more severe disease. Ovalbumin is an abundant protein in chickens, and 90% of adults have serum IgG to ovalbumin (69). Exposure to ovalbumin can occur via diet or via immunization with common flu vaccines. Flu vaccination is associated with a lower risk of severe disease in COVID-19 patients (70), so low IgG to ovalbumin in our study may be related to whether patients have received a flu vaccine.
Our study has several important implications for understanding and treating COVID-19. A substantial proportion of COVID-19 patients experience neurological symptoms, such as reduced sense of smell, headaches, muscle pain and spasms, and paresthesia (71, 72). Antibodies to gangliosides are known to cause similar neurological symptoms in individuals with GBS. While several case reports have identified antiganglioside antibodies in COVID-19 patients, testing for these autoantibodies is not routinely carried out. Our results indicate that these antibodies are more widespread than previously known and that testing for antiganglioside antibodies should be expanded in COVID-19 patients, especially those with acute or prolonged neurological symptoms. If detected, treatments commonly used for GBS like IVIG may be beneficial. Second, convalescent plasma therapy has been granted emergency use authorization (10, 73), and anticoronavirus hyperimmune intravenous immunoglobulin (hIVIG) has entered Phase III clinical trials (74). Our results indicate that convalescent sera may contain autoantibodies to glycans, and that screening for potential autoantibodies may be important to minimize potential complications.
Several limitations of this study should be mentioned. First, our glycan microarray only contains a small portion of the glycans found in the human glycome. Thus, there may be other important antiglycan antibody populations that were not detected. Second, the overall number of patients and healthy subjects in the study is relatively small, and we only had information about disease severity for one of the COVID-19 cohorts. We attempted to minimize false positive correlations by including two separate COVID-19 cohorts when comparing COVID-19 patients to healthy subjects, using a conservative statistical threshold for evaluating relationships with disease severity, and comparing antibody signals to our prior, historical data where possible. Nevertheless, follow up studies will be useful to further evaluate the results of this study. Also, longitudinal samples were not evaluated in this study. Therefore, additional studies will be needed to determine how long the autoantibodies and other alterations persist. Third, the screening was carried out at a single dilution which precludes determination of titers. From our prior studies, we know that > 95% of the signals are not saturated (in terms of the capacity of the spot to capture antibody). However, some of the abnormally high signals may be saturated and could be even higher than what we measured at a dilution of 1:50. Fourth, our cohorts contained samples collected at a variety of times after the initial onset of symptoms. As a result, we are likely to only identify differences that are relatively persistent and may have missed important changes that occurred early after infection. Lastly, we only measured IgG and IgM isotypes. Since IgA are often produced in response to mucosal infections, analysis of antiglycan IgA could reveal additional autoantibodies and relevant responses in the future.
Overall, our study highlights the importance of studying immune responses to carbohydrates. Glycans are one of the major families of antigens found on SARS-CoV-2 and other viruses, but responses to these antigens are often difficult to study. By profiling serum antibodies with a large and diverse carbohydrate antigen microarray, we were able to rapidly identify substantial differences in antiglycan antibodies between COVID-19 patients and healthy subjects as well as antibodies that correlate with disease severity. These results provide new insight into the immune response to SARS-CoV-2 and illustrate the importance of studying antibodies to host antigens in addition to viral antigens. The results also provide a more complete understanding of the risks associated with SARS-CoV-2 infection, which is critical for making informed health decisions.
Materials and Methods
Serum samples
Publicly available, deidentified serum samples from 40 individuals with SARS-CoV-2 infections and 10 healthy controls were purchased from Raybiotech, Inc. (Peachtree Corners, GA). All individuals with samples collected by Raybiotech and designated to be infected with SARS-CoV-2 were symptomatic. These samples were collected on-site at multiple Raybiotech locations within the United States approximately 3 to 4 weeks after initial symptoms. Among the Raybiotech COVID-19-positive samples, 10 were IgM positive, 10 were IgG positive, and 20 were not specified as either IgM or IgG positive. Deidentified serum samples from 70 individuals with SARS-CoV-2 infections were obtained from the NIH COVID-19 repository. These samples were collected in Brescia, Italy, approximately 1 to 10 weeks after initial symptoms. Information about patient enrollment, sample collection, and ethical approval (Comitato Etico Provinciale, Brescia, protocol NP 4000, CORONAlab) have been published previously (75). Of these samples, three were characterized as asymptomatic, 30 were mild, one was moderate, nine severe, and 26 critical. A total of 28 additional healthy control samples were obtained from Valley Biomedical Products and Services (Winchester, VA). All non-COVID-19 samples were collected prior to the SARS-CoV-2 pandemic. The “reference serum” used to normalize data was pooled from 10 samples purchased from Valley Biomedical Products and Services. Samples were stored at −70°C prior to use. All COVID-19 samples were collected in March through May of 2020. None of the patients or controls in this study received a COVID-19 vaccine. Given the early stage of the pandemic, we have assumed it was the first infection for each patient.
Microarray fabrication and assay
The glycan microarrays were fabricated as previously described (76, 77). The microarray contained 816 array components and included a variety of human glycans (N-linked glycans, O-linked glycans, and glycan portions of glycolipids), nonhuman glycans, glycopeptides, and glycoproteins. Each array component was printed in duplicate to produce a full array, and eight copies of the full array were printed on each slide. Prior to each experiment, each microarray slide was scanned in an InnoScan 1100 AL fluorescence scanner to check for any defects and missing print spots. The slides were fitted with an 8-well module (Sigma-Aldrich) to allow eight independent assays on each slide. In the assay, arrays were blocked with 3% BSA in PBS buffer (400 µL/well) overnight at 4°C, then washed six times with PBST buffer (PBS with 0.05% v/v Tween 20). Serum samples diluted at 1:50 in 3% BSA and 1% HSA in PBST were added onto each slide (100 µL/well). To minimize technical variations, all samples were assayed in duplicate on separate slides. A mix of COVID-19 and control samples were run on each slide to minimize effects of day-to-day or slide-to-slide variability. Raybiotech and control samples were evaluated in a randomized order, and the NIH samples were assayed in a randomized order with all information blinded to the operator. After agitation at 100 rpm for 4 hours at 37°C, slides were washed six times with PBST (200 µL/well). The bound serum antibodies were detected by incubating with Cy3 antiHuman IgG (Cy3 AffiniPure Goat AntiHuman IgG, Fcγ fragment specific; Jackson ImmunoResearch; catalog #109–165–098) and DyLight 647 antihuman IgM (Alexa Fluor 647 AffiniPure Goat AntiHuman IgM, Fc5μ fragment specific; Jackson ImmunoResearch; catalog #109–605–129) at 3 µg/mL in PBS buffer with 3% BSA and 1% HSA (100 µL/well) under agitation at 37°C for 2 hours in the dark. Based on our prior analysis and data for our IgG, IgA, and IgM control spots, the secondary reagents provide selective recognition of the appropriate isotype (78). After washing with PBST eight times (200 µL/well), the slides were removed from the modules and soaked in PBST for 5 minutes prior to being dried by centrifugation at 1,000 rpm (112 × g) for 10 minutes. Slides were then scanned with an InnoScan 1100 AL (Innopsys) at 5-µm resolution. The photomultiplier tube (PMT) settings were the same for all experiments to limit unintentional signal variation. Slides were scanned at “high” and “low” PMT settings (for the 532 nm laser, high PMT = 5 and low = 1; for the 635 nm laser, high = 25 and low = 9) to increase the dynamic range and appropriately scale-saturated components. Signal intensities ≤ 50,000 RFU were used directly from the high PMT setting; intensities above 50,000 RFU were corrected using the low PMT scans and the algorithm of Lyng et al. (79, 80). The fluorescence intensity of each array spot was quantified with GenePix Pro 7 software (Molecular Devices). Any features marked as missing or defective in the prescan were excluded from further analysis. The local background corrected median was used for data analysis, and spots with intensity lower than 150 RFU (a value that is ∼50% of the typical IgM background) were set to 150. The signals for replicate spots on duplicate wells were averaged and log-transformed (base 2) for future analysis. See Figure S11 (Supplementary Material) for representative preassay and postassay array images.
Biostatistical analyses were carried out using Partek Genomics Suite (St. Louis, MO). Controls were not included in the analysis. Also, if no patient signals were > 10 (log2) RFU for a particular glycan, then that glycan was removed from statistical analyses so as not to overinterpret variations of weak signals. After removing these components, 752 remained. To identify differences between COVID-19 patients and controls, we used a two-phase approach. In the discovery phase, we identified 57 IgG signals that were statistically different using ANOVA and the false discovery rate (FDR) method of Benjamini and Hochberg with an FDR of 20% (P-value < 0.0144; Supporting Excel file). This subset of components was then evaluated within the NIH cohort to assess consistency of P-values and directionality. In the second phase, 19 out of 57 IgG signals were found to have statistically significant P-values and consistent directionality in both cohorts relative to the controls. For analysis of potential correlations with disease severity, we used ANOVA using the step-up method of Benjamini and Hochberg with a FDR = 0.05 to account for multiple comparisons, and demonstration of consistent trends with increasing disease severity.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Consortium for Functional Glycomics (GM62116; The Scripps Research Institute), Xuefei Huang (Michigan State University), Thomas Tolbert (University of Kansas), Lai-Xi Wang (University of Maryland), Joseph Barchi, Jr. (National Cancer Institute), Todd Lowary (University of Alberta), Beat Ernst (University of Basel), Omicron Biochemicals Inc., GlycoHub, and Glycan Therapeutics for generously contributing glycans for the array. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: anti-HIV-1 gp120 monoclonal antibody (PGT121) from IAVI (cat# 12343); anti-HIV-1 gp120 monoclonal antibody (PGT126) from IAVI (cat# 12344); and anti-HIV-1 gp120 monoclonal antibody (PGT128) from IAVI (81, 82). We thank the following NIAID COVID-19 Consortium members for logistical, infrastructure, and analysis support: Jason Barnett, Samuel Chauvin, Xi Cheng, Jeffrey Danielson, Kerry Dobbs, Elizabeth Garabedian, Vasu Kuram, William Lau, Zhiwen Li, Mary Magliocco, Helen Matthews, Marshall Nambiar, Smilee Samuel, Elana Shaw, Michael Stack, Sarah Weber, Sandhya Xirasagar, and Yu Zhang. The authors thank all the patients who consented to participate in our study and their health care providers during their hospitalization. We would like to thank Charles Rainwater for his assistance with the material transfer agreements between the Italian hospital and the NIAID/NIH. We also thank the staff of the ASST Spedali Civili of Brescia, Italy, in which clinical laboratory tests were performed. We thank the NIAID Office of Cyber Infrastructure and Computational Biology, Bioinformatics and Computational Biosciences Branch (contract HHSN316201300006W/HHSN27200002 to MSC, Inc.) and Operations and Engineering Branch for developing the HGRepo system to enable streamlined access to the data.
Notes
Competing Interest: The authors declare no competing interest.
Contributor Information
Dorothy L Butler, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, USA.
Luisa Imberti, Centro di Ricerca Emato-oncologica AIL (CREA) and Diagnostic Department, ASST Spedali Civili di Brescia, Brescia, Italy.
Virginia Quaresima, Centro di Ricerca Emato-oncologica AIL (CREA) and Diagnostic Department, ASST Spedali Civili di Brescia, Brescia, Italy.
Chiara Fiorini, Centro di Ricerca Emato-oncologica AIL (CREA) and Diagnostic Department, ASST Spedali Civili di Brescia, Brescia, Italy.
Jeffrey C Gildersleeve, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, USA.
NIAID COVID-19 Consortium:
Jason Barnett, Samuel Chauvin, Xi Cheng, Jeffrey Danielson, Kerry Dobbs, Elizabeth Garabedian, Vasu Kuram, William Lau, Zhiwen Li, Mary Magliocco, Helen Matthews, Marshall Nambiar, Smilee Samuel, Elana Shaw, Michael Stack, Sarah Weber, Sandhya Xirasagar, and Yu Zhang
Funding
This work was supported by the Intramural Research Program of the National Cancer Institute, NIH, and by Regione Lombardia, Italy (project “Risposta immune in pazienti con COVID-19 e co-morbidita’”).
Authors' Contributions
D.L.B carried out all the glycan array assays, and D.L.B and J.C.G performed the analysis of glycan array data. L.I., V.Q., and C.F. cared for patients, collected samples, and collected the clinical data. L.I., V.Q, and C.F. clinically characterized patients with help from the NIAID COVID Consortium and the COVID Clinicians. NIAID COVID Consortium provided logistical, infrastructure, and analysis support. D.L.B. and J.C.G. wrote the initial draft of the manuscript, and L.I., V.Q, and C.F. provided additional edits and analysis.
Data Availability
Full glycan microarray data and patient information can be found in the Supporting Excel file.
References
- 1. Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. 2020. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 324(8):782–793. [DOI] [PubMed] [Google Scholar]
- 2. Davido B, Seang S, Tubiana R, de Truchis P. Post-COVID-19 chronic symptoms: a postinfectious entity?. Clin Microbiol Infect. 26:1448–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carfì A, Bernabei R, Landi F, Gemelli Against COVID-19 Post-Acute Care Study Group . 2020. Persistent symptoms in patients after acute COVID-19. JAMA. 324(6):603–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weerahandi H, et al. 2021. Post-discharge health status and symptoms in patients with severe COVID-19. J Gen Intern Med. 36:738–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xiang F, et al. 2020. Antibody detection and dynamic characteristics in patients with COVID-19. Clin Infect Dis. 71:1930–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xie J, et al. 2020. Characteristics of patients with coronavirus disease (COVID-19) confirmed using an IgM-IgG antibody test. J Med Virol. 92:2004–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Theel ES, et al. 2020. The role of antibody testing for SARS-CoV-2: is there one?. J Clin Microbiol. 58(8):e00797–e00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shen C, et al. 2020. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 323(16):1582–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duan K, et al. 2020. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci USA. 117(17):9490–9496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Joyner MJ, et al. 2020. Effect of convalescent plasma on mortality among hospitalized patients with COVID-19: initial three-month experience. medRxiv. doi: 10.1101/2020.08.12.20169359. [Google Scholar]
- 11. Joyner MJ, et al. 2020. Early safety indicators of COVID-19 convalescent plasma in 5000 patients. J Clin Invest. 130(9):4791–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shah VK, Firmal P, Alam A, Ganguly D, Chattopadhyay S. 2020. Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front Immunol. 11:1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi Y, et al. 2020. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 27(5):1451–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mathew D, et al. 2020. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 369(6508):eabc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gruber C, et al. 2020. Mapping systemic inflammation and antibody responses in Multisystem Inflammatory Syndrome in Children (MIS-C. Cell. 183:982–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Civardi C, Collini A, Geda DJ, Geda C. 2020. Antiganglioside antibodies in Guillain-Barré syndrome associated with SARS-CoV-2 infection. J Neurol Neurosurg Psychiatry. 91:1361–1362. [DOI] [PubMed] [Google Scholar]
- 17. Abu-Rumeileh S, Abdelhak A, Foschi M, Tumani H, Otto M. 2020. Guillain-Barré syndrome spectrum associated with COVID-19: an up-to-date systematic review of 73 cases. J Neurol. 268:1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Casciola-Rosen L, et al. 2022. IgM autoantibodies recognizing ACE2 are associated with severe COVID-19. JCI Insight. 7(9):e158362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Franke C, et al. 2020. High frequency of cerebrospinal fluid autoantibodies in COVID-19 patients with neurological symptoms. Brain Behav Immun. 93:415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zuo Y, et al. 2020. Prothrombotic autoantibodies in serum from patients hospitalized with COVID-19. Sci Transl Med. 12(570):eabd3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wijst MGPvd, et al. 2021. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci Transl Med. 13(612):eabh2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bastard P, et al. 2021. Autoantibodies neutralizing type I IFNs are present in ∼4% of uninfected individuals over 70 years old and account for ∼20% of COVID-19 deaths. Sci Immunol. 6(62):eabl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kulkarni R. 2019. Antibody-sependent enhancement of viral infections. In: Dynamics of immune activation in viral diseases. Berlin: Springer. p. 9–41. [Google Scholar]
- 24. Kuri-Cervantes L, et al. 2020. Immunologic perturbations in severe COVID-19/SARS-CoV-2 infection. bioRxiv. doi: 10.1101/2020.05.18.101717. [Google Scholar]
- 25. Bastard P, et al. 2020. Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Science. 370:eabd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gudbjartsson DF, et al. 2020. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med. 383:1724–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. 2020. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 20(6):363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hueston L, et al. 2020. The antibody response to SARS-CoV-2 infection. Open Forum Infect Dis. 7(9):ofaa387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shajahan A, Supekar NT, Gleinich AS, Azadi P. 2020. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 30:981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. 2020. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 369(6501):330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y, et al. 2020. Site-specific N-glycosylation characterization of recombinant SARS-CoV-2 spike proteins. Mol Cell Proteomics. 20:100058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao P, et al. 2020. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 Receptor. Cell Host Microbe. 28(4):586–601.e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Casalino L, et al. 2020. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent Sci. 6:1722–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Woo H, et al. 2020. Developing a fully glycosylated full-length SARS-CoV-2 spike protein model in a viral membrane. J Phys Chem B. 124(33):7128–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Urra JM, et al. 2020. The antibody response to the glycan α-Gal correlates with COVID-19 disease symptoms. J Med Virol. 93:2065–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Breiman A, et al. 2021. Low levels of natural anti-α-N-acetylgalactosamine (Tn) antibodies are associated with COVID-19. Front Microbiol.12:641460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heidepriem J, et al. 2021. Longitudinal development of antibody responses in COVID-19 patients of different severity with ELISA, peptide, and glycan arrays: an immunological case series. Pathogens. 10(4):438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Latz CA, et al. 2020. Blood type and outcomes in patients with COVID-19. Ann Hematol. 99(9):2113–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao J, et al. 2020. Relationship between the ABO blood group and the COVID-19 susceptibility. Clin Infect Dis. 73:328–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zietz M, Tatonetti NP. 2020. Testing the association between blood type and COVID-19 infection, intubation, and death. Nat Comm. 11:5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dzik S, Eliason K, Morris EB, Kaufman RM, North CM. 2020. COVID-19 and ABO blood groups. Transfusion. 60(8):1883–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ellinghaus D, et al. 2020. The ABO blood group locus and a chromosome 3 gene cluster associate with SARS-CoV-2 respiratory failure in an Italian-Spanish genome-wide association analysis. medRxiv. doi: 10.1101/2020.05.31.20114991. [Google Scholar]
- 43. Muthana SM, Gildersleeve JC. 2016. Factors affecting anti-glycan IgG and IgM repertoires in human serum. Sci Rep. 6:19509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Scheepers C, et al. 2017. Serum glycan-binding IgG antibodies in HIV-1 infection and during the development of broadly neutralizing responses. AIDS. 31(16):2199–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mehta AY, Cummings RD. 2020. GlycoGlyph: a glycan visualizing, drawing and naming application. Bioinformatics. 36(11):3613–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lardone RD, Irazoqui FJ, Nores GA. 2019. Most of anti-glycolipid IgG-antibodies associated to neurological disorders occur without their IgM counterpart. J Biomed Sci. 26(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Horby P, et al. 2021. Effect of dexamethasone in hospitalized patients with COVID-19: preliminary report. New Engl J Med. 384:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tang Y, et al. 2020. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front Immunol. 11:1708–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Del Valle DM, et al. 2020. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 26:1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wen W, et al. 2020. Immune cell profiling of COVID-19 patients in the recovery stageby single-cell sequencing. Cell Discov. 6(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aziz M, Brenner M, Wang P. 2020. Therapeutic potential of B-1a cells in COVID-19. Shock. 54(5):586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gutiérrez-Ortiz C, et al. 2020. Miller Fisher syndrome and polyneuritis cranialis in COVID-19. Neurology. 95(5):e601. [DOI] [PubMed] [Google Scholar]
- 53. Guilmot A, et al. 2020. Immune-mediated neurological syndromes in SARS-CoV-2-infected patients. J Neurol. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Muthana SM, Gildersleeve JC. 2016. Factors affecting anti-glycan IgG and IgM repertoires in human serum. Sci Rep. 6(1):19509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Campbell CT, et al. 2014. Humoral response to a viral glycan correlates with survival on PROSTVAC-VF. Proc Natl Acad Sci USA. 111(17):E1749–E1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Campbell CT, et al. 2013. Serum antibodies to blood group A predict survival on PROSTVAC-VF. Clin Cancer Res. 19(5):1290–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xia L, Schrump DS, Gildersleeve JC. 2016. Whole-cell cancer vaccines induce large antibody responses to carbohydrates and glycoproteins. Cell Chem Biol. 23(12):1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Durbin SV, Wright WS, Gildersleeve JC. 2018. Development of a multiplex glycan microarray assay and comparative analysis of human serum anti-glycan IgA, IgG, and IgM repertoires. ACS Omega. 3(12):16882–16891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gleeson PA. 1994. Glycoconjugates in autoimmunity. Biochim Biophys Acta Gene Struct Exp. 1197(3):237–255. [DOI] [PubMed] [Google Scholar]
- 60. Nores GA, et al. 2008. Anti-GM1 antibodies as a model of the immune response to self-glycans. Biochim Biophys Acta Chem Subj. 1780(3):538–545. [DOI] [PubMed] [Google Scholar]
- 61. Cutillo G, Saariaho A-H, Meri S. 2020. Physiology of gangliosides and the role of antiganglioside antibodies in human diseases. Cell Mol Immunol. 17(4):313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Plomp JJ, Willison HJ. 2009. Pathophysiological actions of neuropathy-related anti-ganglioside antibodies at the neuromuscular junction. J Physiol. 587(Pt 16):3979–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kaida K, Ariga T, Yu RK. 2009. Antiganglioside antibodies and their pathophysiological effects on Guillain–Barré syndrome and related disorders—a review. Glycobiology. 19(7):676–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yu RK, Tsai Y-T, Ariga T, Yanagisawa M. 2011. Structures, biosynthesis, and functions of gangliosides—an overview. J Oleo Sci. 60(10):537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Willison HJ, Yuki N. 2002. Peripheral neuropathies and anti-glycolipid antibodies. Brain. 125(Pt 12):2591–2625. [DOI] [PubMed] [Google Scholar]
- 66. Daniels G. 2013. I and i antigens, and cold agglutination. In: Human blood groups. 3rd ed. New York (NY): Wiley. [Google Scholar]
- 67. Sanda M, Morrison L, Goldman R. 2021. N- and O-glycosylation of the SARS-CoV-2 spike protein. Anal Chem. 93(4):2003–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Galili U. 2020. Human natural antibodies to mammalian carbohydrate antigens as unsung heroes protecting against past, present, and future viral infections. Antibodies. 9(2):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kilshaw PJ, McEwan FJ, Baker KC, Cant AJ. 1986. Studies on the specificity of antibodies to ovalbumin in normal human serum: technical considerations in the use of ELISA methods. Clin Exp Immunol. 66(2):481–489. [PMC free article] [PubMed] [Google Scholar]
- 70. Huang K, Lin S-W, Sheng W-H, Wang C-C. 2021. Influenza vaccination and the risk of COVID-19 infection and severe illness in older adults in the United States. Sci Rep. 11(1):11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yavarpour-Bali H, Ghasemi-Kasman M. 2020. Update on neurological manifestations of COVID-19. Life Sci. 257:118063–118063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Orsucci D, Ienco EC, Nocita G, Napolitano A, Vista M. 2020. Neurological features of COVID-19 and their treatment: a review. Drugs Context. 9:2020–2025-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liu STH, et al. 2020. Convalescent plasma treatment of severe COVID-19: a propensity score–matched control study. Nat Med. 26:1708–1713. [DOI] [PubMed] [Google Scholar]
- 74. Sakamoto J, et al. 1986. Expression of Lewisa, Lewisb, X, and Y blood group antigens in human colonic tumors and normal tissue and in human tumor-derived cell Llnes. Cancer Res. 46:1553–1561. [PubMed] [Google Scholar]
- 75. Abers MS, et al. 2021. An immune-based biomarker signature is associated with mortality in COVID-19 patients. JCI Insight. 6(1):e144455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Campbell CT, Zhang Y, Gildersleeve JC. 2010. Construction and use of glycan microarrays. Curr Protoc Chem Biol. 2(1):37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Xia L, Gildersleeve JC. 2015. The glycan array platform as a tool to identify carbohydrate antigens. In: Lepenies B, editor. Carbohydrate-based vaccines: methods and protocols. New York (NY): Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Muthana S, Xia L, Campbell CT, Zhang Y, Gildersleeve JC. 2015. Competition between serum IgG, IgM, and IgA anti-glycan antibodies. Plos ONE. 10(3):e0119298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lyng H, et al. 2004. Profound influence of microarray scanner characteristics on gene expression ratios: analysis and procedure for correction. BMC Genomics. 5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Oyelaran O, McShane LM, Dodd L, Gildersleeve JC. 2009. Profiling human serum antibodies with a carbohydrate antigen microarray. J Proteome Res. 8:4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Walker LM, et al. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 477(7365):466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pejchal R, et al. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 334(6059):1097–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Full glycan microarray data and patient information can be found in the Supporting Excel file.