Abstract
Background
Prostate magnetic resonance imaging (MRI) is technically demanding, requiring high image quality to reach its full diagnostic potential. An automated method to identify diagnostically inadequate images could help optimize image quality.
Purpose
To develop a convolutional neural networks (CNNs) based analysis pipeline for the classification of prostate MRI image quality.
Study Type
Retrospective.
Subjects
Three hundred sixteen prostate mpMRI scans and 312 men (median age 67).
Field Strength/Sequence
A 3 T; fast spin echo T2WI, echo planar imaging DWI, ADC, gradient‐echo dynamic contrast enhanced (DCE).
Assessment
MRI scans were reviewed by three genitourinary radiologists (V.P., M.D.M., S.C.) with 21, 12, and 5 years of experience, respectively. Sequences were labeled as high quality (Q1) or low quality (Q0) and used as the reference standard for all analyses.
Statistical Tests
Sequences were split into training, validation, and testing sets (869, 250, and 120 sequences, respectively). Inter‐reader agreement was assessed with the Fleiss kappa. Following preprocessing and data augmentation, 28 CNNs were trained on MRI slices for each sequence. Model performance was assessed on both a per‐slice and a per‐sequence basis. A pairwise t‐test was performed to compare performances of the classifiers.
Results
The number of sequences labeled as Q0 or Q1 was 38 vs. 278 for T2WI, 43 vs. 273 for DWI, 41 vs. 275 for ADC, and 38 vs. 253 for DCE. Inter‐reader agreement was almost perfect for T2WI and DCE and substantial for DWI and ADC. On the per‐slice analysis, accuracy was 89.95% ± 0.02% for T2WI, 79.83% ± 0.04% for DWI, 76.64% ± 0.04% for ADC, 96.62% ± 0.01% for DCE. On the per‐sequence analysis, accuracy was 100% ± 0.00% for T2WI, DWI, and DCE, and 92.31% ± 0.00% for ADC. The three best algorithms performed significantly better than the remaining ones on every sequence (P‐value < 0.05).
Data Conclusion
CNNs achieved high accuracy in classifying prostate MRI image quality on an individual‐slice basis and almost perfect accuracy when classifying the entire sequences.
Evidence Level
4
Technical Efficacy
Stage 1
Keywords: multiparametric MRI, prostate cancer, deep learning, artificial intelligence, quality control
Technical refinements, standardization and widespread availability of prostate multiparametric MRI (mpMRI) have led to a paradigm shift in the early detection of clinically significant prostate cancer (csPCa). This shift has been from the traditional approach based on prostate‐specific antigen (PSA) and systematic biopsy to the so‐called “MRI pathway,” based on mpMRI and MRI‐targeted biopsy. 1 The central role of mpMRI in the diagnostic workup of PCa is confirmed by the European Association of Urology (EAU) guidelines that recommend MRI as the first diagnostic study to perform in patients with suspicion of PCa. 2 These milestones have been achieved thanks to the high diagnostic performance of mpMRI reported by multiple studies. 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 However, the evidence shows a high variability in the diagnostic performance among centers, likely due to factors that affect overall image quality and interpretation, such as the equipment and the acquisition protocol used, and the radiologist's expertise. 11
The Prostate Imaging Reporting and Data System (PI‐RADS) establishes minimum technical standards and includes guidelines for standardizing the acquisition parameters, aiming at optimizing image quality and reducing variability. 12 However, many specifications of the technical details of the mpMRI acquisition are not specified, leading to inconsistencies in image quality regardless of adherence to the recommendations, mostly in centers with little expertise. 13 Additionally, several patient‐related factors can undermine image quality, including patient movement and the presence of air in the rectum. 14 Therefore, even if the appropriate equipment is used and the optimal acquisition protocol is implemented, quality control of mpMRI images is important. In the recent European Society of Urogenital Radiology (ESUR)/EAU Section of Urological Imaging (ESUI) consensus statements on quality requirements the authors recommended that image quality be checked and reported regularly, for determining diagnostic appropriateness. 15 Although visual assessment of mpMRI image quality by the radiologist is appropriate and can be standardized in its methodology, prompt identification of suboptimal scans while it is possible to intervene to optimize the acquisition might not always be feasible in real‐world clinical practice. 15 , 16
A fully automated technology capable of performing real‐time quality control of MRI scans, identifying those sequences that are of sub‐optimal diagnostic quality would be helpful to both the radiologist and the technologist. The use of artificial intelligence (AI), specifically deep neural networks, for image quality assessment is a field which has been already explored in nonmedical applications. 17 , 18 Briefly, deep neural networks use multiple layers to process input data and extract features of interest that are then used to provide an output. By processing the input data during the training phase, the network automatically determines, for each layer, what the kernels should look like to detect specific image characteristics, optimized in the direction of the specific task of interest. Once the network is trained, it is ready to have the kernels perform convolutions, extracting features to conclude with the desired class label. 19 Despite the high performance of this technology in image analysis, only a few studies have investigated the use of deep neural networks to classify images based on their quality, none of them focusing on prostate MRI. 20 , 21
The aim of this study was to develop an ad hoc computational image analysis pipeline based on the use of deep neural networks for the automated assessment of prostate mpMRI image quality.
Materials and Methods
Patient Population and MRI Assessment
In this retrospective IRB‐approved study with waiver of informed consent all scans of men who underwent prostate mpMRI between January 2020 and July 2020 for suspicious PCa, active surveillance or staging of known PCa was included. There were no exclusion criteria. A total of 316 prostate mpMRI scans from 312 men with a median age of 67 (IQ range 62–74) were retrospectively reviewed. These included 316 T2‐weighted imaging (T2WI) sequences, 316 diffusion‐weighted imaging (DWI) sequences, 316 apparent diffusion coefficient (ADC) sequences, and 291 dynamic contrast‐enhanced (DCE) sequences.
All exams were performed on a 3 T MR scanner (Discovery 750, GE Healthcare, USA) using a 32 multichannel surface phased‐array body coil (TORSOPA). A list of the acquisition parameters used is detailed in Table 1.
TABLE 1.
Summary of the MR Acquisition Parameters
| T2WI | DWI | DCE | |
|---|---|---|---|
| Sequence type | Fast recovery fast spin echo (FRFSE) | Echo planar imaging (EPI) | LAVA gradient echo |
| TE (msec) | 134 | 75 | 1 |
| TR (msec) | 6000 | 4300 | 3 |
| Acquisition plane | Axial and Coronal | Axial | Axial |
| Number of averages | 6 | 2 (b 50); 6 (b 800); 12 (b 1500); 14 (b 2000) | 1 |
| Slice thickness (mm) | 3 | 3 | 4 |
| Matrix size | 320 × 224 | 90 × 90 | 160 × 140 |
| Field of view (cm) | 18 × 18 | 20 × 20 | 18 × 18 |
| b‐values (s/mm2) | N/A | 50–800–1500–2000 | N/A |
| Temporal resolution (s) | N/A | N/A | 6 |
| Contrast media | N/A | N/A | Gadobutrol 0.1 mmol/Kg (injection rate 3.0 mL/sec) |
DCE = Dynamic Contrast‐enhanced; DWI = Diffusion Weighted Imaging; T2WI = T2 Weighted Imaging.
Scans were evaluated by three radiologists (V.P., M.D.M., S.C.) with 21, 12, and 5 years of experience in genitourinary imaging, respectively. Individual sequences, including axial T2WI, DWI at a b value of 1500, ADC and DCE, were assessed based on several technical and visual parameters related to image quality (as specified in the Pi‐RADS v2.1 guidelines). These included the adequacy of the field of view (FOV), spatial resolution, signal‐to‐noise (S/N) ratio, motion artifacts, magnetic susceptibility artifacts, presence of significant amount of gas in the rectum, and the appropriateness of enhancement on DCE images. A binary classification label was independently assigned by each reader to every sequence. The two classes were defined as: Q0 (denoting low quality or insufficient diagnostic quality) or Q1 (denoting high quality or sufficient diagnostic quality). In case of disagreement between the readers, the label assigned by the majority of the three radiologists was considered as the definitive label, and used both for training and as the standard of reference for the evaluation of model performance.
Computational Analysis
For the generation of the prediction models, 28 convolutional neural networks (CNNs) from the following families were tested 22 , 23 : AlexNet, 24 VGG (VGG11, VGG11‐BN, VGG13, VGG13‐BN, VGG16, VGG16‐BN,VGG19, VGG19‐BN), 25 ResNet (ResNet18, ResNet34, ResNet50, ResNet101,ResNet152), 26 SqueezeNet (SqueezeNet1‐0, SqueezeNet1‐1), 27 DenseNet (DenseNet121, DenseNet169, DenseNet161, DenseNet201), 28 GoogLeNet, 29 ShuffleNet v2 (ShuffleNet v2‐x0‐5, ShuffleNet v2‐x1‐0), 30 MobileNet v2, 31 ResNeXt (ResNeXt50‐32x4d), 32 Wide ResNet (Wide ResNet50‐2), 33 and MNASNet (MNASNet0‐5, MNASNet1‐0). 34 AlexNet and VGG are spatial exploitation‐based CNNs. GoogLeNet is the first architecture that uses the block concept, the split transform, and the merge idea. ResNet is a depth and multipath‐based CNN, which uses residual learning and has identity mapping based skip connections. WideResNet and ResNeXt are width‐based multiconnection CNNs. SqeezeNet is a feature‐map exploitation‐based CNN modeling interdependencies between feature‐maps. DenseNet is a multipath‐based CNN exploiting cross‐layer information flow. MobileNet has an inverted residual structure, and it utilizes lightweight depth‐wise convolutions.
The network weights were initialized via transfer learning. The models were pretrained on the ImageNet dataset, a large database of over 14 million pictures in different categories. 35 The networks were trained with a maximum of 300 epochs with an early stopping fixed to 25 and with a batch size equal to 32. The loss function to be optimized was the binary cross‐entropy, with an SGDoptimizer (learning rate = 0.001, momentum = 0.9).
For the classification of prostate MR images, axial T2WI, DWI at a b value of 1500, ADC, and perfusion sequences were used. Before entering the network, MRI images underwent a preprocessing phase: voxel values of the DICOM slices were normalized (using the mean and standard deviation of the voxel intensities) and image size was resampled to 224 × 224 × 3. During the training phase, data augmentation was applied. Data augmentation is a technique used to increase the amount of data by adding slightly modified copies of already existing data or newly created synthetic data from existing data. It helps reduce overfitting when training a machine learning model. With a probability of 0.3, the following transformations were applied: random rotation (e.g., ±100°), flip along the vertical axis, random shift (e.g., ±7 pixels) and elastic deformation (α = (20,40), σ = 7). Each of these technique was applied to each image with a probability of 0.3. In accordance with the radiologists, the transformations and their parameters were chosen in a way to generate images coherent with the prostate MRI scenario. The networks were trained at a slice level, and all the slices within a sequence were labeled as the same class. All architectures were modified only at the last layer, by setting the number of neurons equal to the number of image labels. All networks were trained with 10‐fold cross‐validation. The division between the training, validation, and testing sets was 70%, 20%, and 10%, respectively. A random stratified cross‐validation was performed to maintain the original proportion of labels in all the data divisions. To prevent overfitting, the slices of an individual patient's sequence were all included in the same set, meaning that the entire sequence of a patient was inside the training, validation, or testing set.
All the computations were done using Python 3.7 with PyTorch on a NVIDIA Tesla V100 SXM2 32 GB. The analysis pipeline implemented for each model and for each type of sequence is shown in Fig. 1.
FIGURE 1.
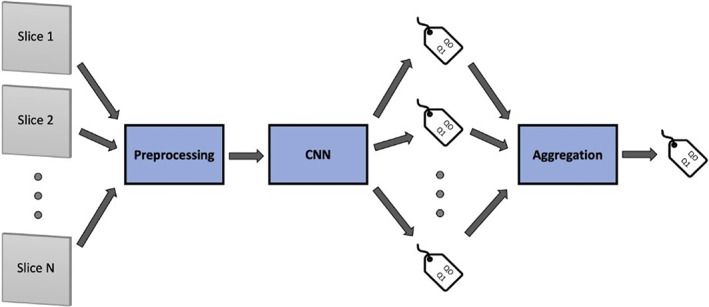
Graphical representation of the analysis pipeline. Individual slices from a given sequence are preprocessed (including normalization and voxel resampling, and data augmentation) and subsequently fed to the CNN algorithm that assigns a classification label to every slice. Classification results for all slices from the same sequence are then aggregated by means of a majority vote aggregation function, so that a classification label is assigned to the entire acquired sequence.
Statistical Analysis
To evaluate the performance of the networks, global and class‐specific accuracy were calculated on the test set slices for each individual model. Accuracy values are reported as both the global and class‐specific mean accuracies ±95% confidence intervals across the cross‐validation folds. In addition, the classification results of the individual slices were combined by using a majority vote aggregation function, meaning that the most frequent label within the sequence was assigned. 36 To assess the performances of the different models in comparison with each other, a pairwise t‐test was independently performed among the best three models and between them and all the remaining. Inter‐reader agreement was assessed using the Fleiss kappa. Agreement was considered slight for kappa values of 0.00–0.20, fair for values of 0.21–0.40 fair agreement, moderate for values of 0.41–0.60, substantial for values of 0.61–0.80, almost perfect for values of 0.81–1.00. 37 A P value of <0.05 was used for statistical significance. All statistical analysis was performed on R statistical software version 4.0.5 (http://www.R-project.org).
Results
The quality assessment resulted in the labeling of sequences as Q0 (low quality) and Q1 (high quality), respectively, as follows: 35 vs. 281 for T2WI, 46 vs. 270 for DWI, 43 vs. 273 for ADC, 37 vs. 254 for DCE (Table 2). Figures 2, 3, 4 show representative examples of high‐ and low‐quality MR images for T2WI, DWI, and DCE, respectively. Analysis of the inter‐reader agreement revealed almost perfect agreement for T2WI and DCE (kappa of 0.83 and 0.80, respectively, P < 0.05), and substantial agreement for DWI and ADC (0.77 and 0.75, respectively, P < 0.05). In total, 144,901 slices were available: 8387 slices from 316 sequences for T2WI, 8387 from 316 sequences for DWI, 8387 from 316 sequences for ADC, 119740 from 291 sequences for DCE (Table 2). The division of cases among the training, validation, and testing sets is reported in Table 3.
TABLE 2.
Summary of the MRI Dataset
| Number of Sequences (slices) | |||
|---|---|---|---|
| Sequence | All Classes | Q0 | Q1 |
| T2WI | 316 (8387) | 35 (923) | 281 (7464) |
| DWI | 316 (8387) | 46 (1235) | 270 (7152) |
| ADC | 316 (8387) | 43 (1133) | 273 (7254) |
| DCE | 291 (119,740) | 37 (15,210) | 254 (104,530) |
| Total | 1239 (144,901) | 161 (18,501) | 1078 (126,400) |
Q0 = low‐quality image; Q1 = high‐quality image.
FIGURE 2.
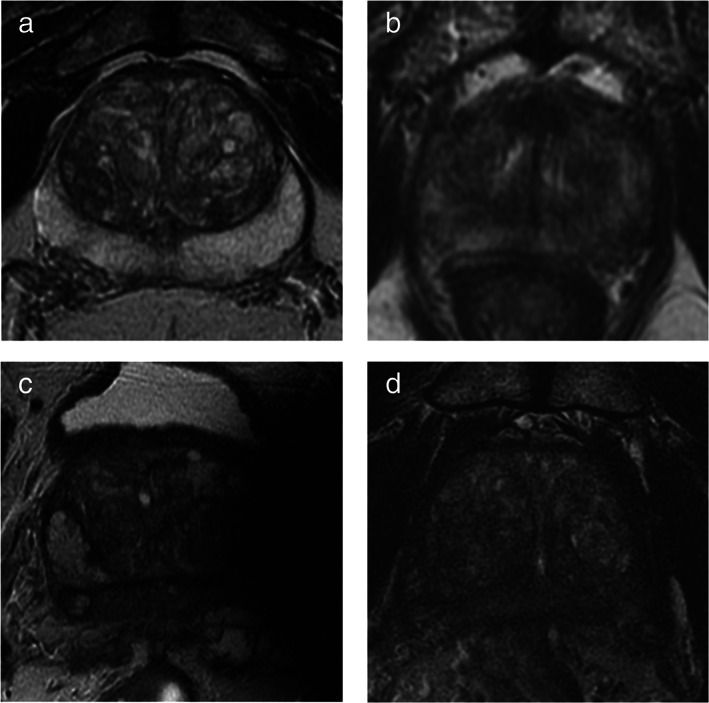
Case examples of high‐ and low‐quality scans on T2WI images. It shows examples of high‐ and low‐quality T2 images: (a) high‐quality axial T2WI image (Q1), with good spatial resolution a tissue contrast; (b) low‐quality image (Q0) with poor spatial resolution and blurred details due to patient movement during acquisition, the sequence should be repeated in order to be able to accurately interpret the study; (c) very poor‐quality acquisition (Q0) due to evident magnetic susceptibility artifacts caused by a femoral prosthesis; (d) low‐quality image (Q0) because of inadequate S/N ratio making diagnostic accuracy suboptimal, the sequence needs to be repeated following optimization of the acquisition parameters.
FIGURE 3.
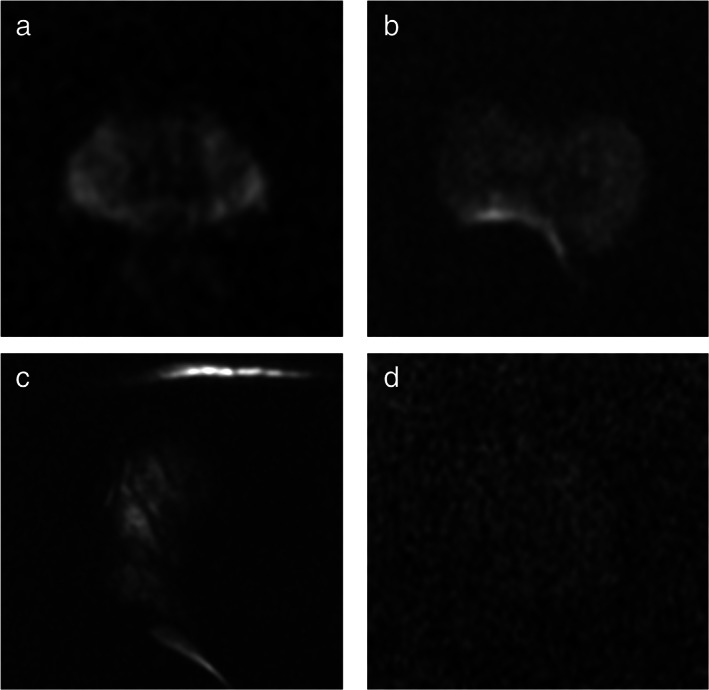
Case examples of high‐ and low‐quality scans on DWI images. It shows examples of high‐ and low‐quality DWI images: (a) high‐quality DWI image (Q1), with good S/N ratio and no evident artifacts; (b) low‐quality image (Q0) with susceptibility artifacts caused by the presence of air in the rectum—the ability to detect foci in the right posterior peripheral zone is significantly impaired—the sequence could be repeated following attempts to expel the air from the rectum; (c) inadequate acquisition (Q0) with marked distortion and signal void due to magnetic susceptibility artifacts caused by a femoral prosthesis; (d) low‐quality image (Q0) because of inadequate S/N ratio that lower significantly the diagnostic power—the sequence needs to be repeated following optimization of the acquisition parameters.
FIGURE 4.
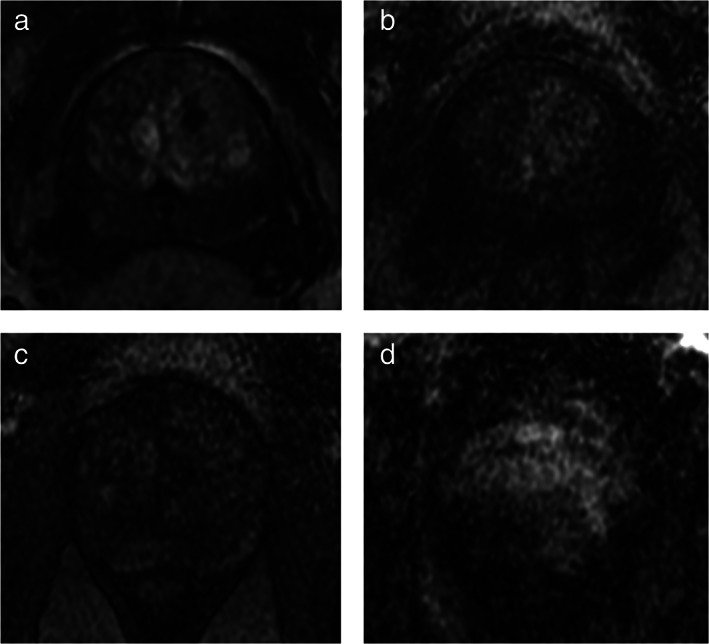
Case examples of high‐ and low‐quality scans on DCE images. It shows examples of high‐ and low‐quality perfusion images: (a) high‐quality DCE image, with good contrast enhancement of the prostate gland; (b) low‐quality image (Q0) with low contrast enhancement of the prostate gland and high noise significantly impairing the sensitivity to detect suspicious foci; (c) low‐quality acquisition (Q0) due to both low contrast enhancement of the prostate gland and to low S/N ratio, the diagnostic sensitivity of this sequence is limited; (d) poor‐quality image (Q0) because of marked motion artifacts—the ability to correctly identify areas of pathologic enhancement is compromised.
TABLE 3.
Train, validation, Test Split
| Number of Sequences | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Train | Validation | Test | |||||||
| Sequence | All | Q0 | Q1 | All | Q0 | Q1 | All | Q0 | Q1 |
| T2WI | 222 | 25 | 197 | 63 | 6 | 57 | 31 | 4 | 27 |
| DWI | 221 | 30 | 191 | 64 | 9 | 55 | 31 | 4 | 27 |
| ADC | 221 | 29 | 193 | 64 | 8 | 55 | 31 | 4 | 27 |
| DCE | 205 | 26 | 179 | 59 | 8 | 51 | 27 | 3 | 24 |
Q0 = low‐quality image; Q1 = high‐quality image.
On the slice‐based analysis, evaluation of the best‐performing models on the test set showed a global accuracy of 89.95% ± 0.02% for T2WI, 79.83% ± 0.04% for DWI, 76.64% ± 0.04% for ADC, 96.62% ± 0.01% for DCE. Q0 class‐specific accuracy values of the best performing models were 84.16% ± 0.02% for T2WI, 62.13% ± 0.05% for DWI, 64.11% ± 0.06% for ADC and 100.00% ± 0.00% for DCE, while the worst performing models achieved accuracies of 43.86% ± 0.54% for T2WI, 55.12% ± 0.37% for DWI, 24.51% ± 0.73% for ADC and 85.81% ± 0.83% for DCE.
On the sequence‐based analysis, where classification outcomes of slices were combined by using the majority vote aggregation function, the accuracy values showed optimal classification performance (100% ± 0.00% accuracy) for T2WI, DWI and DCE sequences, and a global accuracy of 92.31% ± 0.00% and a Q0‐specific accuracy of 83.33% ± 0.00% for ADC.
Overall, the different architectures performed similarly on the sequences; however, the top‐performing architectures among all the tested ones vary along the different sequences and were VGG11 for T2WI, ResNet152 for DWI, DenseNet161 for ADC and ShuffleNet(v2‐x1‐0) for DCE.
The pairwise t‐test between the different models showed that for every sequence, the best three models did not perform significantly different from each other (P value > 0.05). However, all the comparisons with the remaining models were statistically significant (P value ≤ 0.05).
Global and class‐specific accuracies of the three best performing and the three worse‐performing models over the 10‐fold cross validation for each of the four sequences are summarized in Tables 4 and 5, respectively.
TABLE 4.
Mean Accuracy Values and 95% Confidence intervals of the Three Top‐Performing Models Along the 10‐Fold Cross‐Validation for Each Sequence
| Accuracy (mean ± 95% CI) Individual Slice | Accuracy (mean ± 95% CI) Entire Sequence | ||||||
|---|---|---|---|---|---|---|---|
| Sequence | CNN Architecture | Global | Q0 Class | Q1 Class | Global | Q0 Class | Q1 Class |
| T2WI | VGG11 | 89.95 ± 0.02 | 84.16 ± 0.02 | 96.11 ± 0.04 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| ResNet101 | 87.28 ± 0.04 | 80.37 ± 0.03 | 94.34 ± 0.05 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | |
| ResNet50 | 84.93 ± 0.02 | 76.42 ± 0.04 | 93.53 ± 0.01 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | |
| DWI | ResNet152 | 79.83 ± 0.04 | 62.13 ± 0.05 | 97.46 ± 0.03 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| VGG19‐BN | 78.82 ± 0.05 | 60.08 ± 0.07 | 97.45 ± 0.03 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | |
| DenseNet121 | 75.51 ± 0.05 | 55.16 ± 0.07 | 95.78 ± 0.03 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | |
| ADC | DenseNet161 | 76.64 ± 0.04 | 64.11 ± 0.06 | 89.06 ± 0.03 | 92.31 ± 0.00 | 83.33 ± 0.00 | 100.00 ± 0.00 |
| ResNet50 | 76.09 ± 0.03 | 64.11 ± 0.05 | 87.94 ± 0.02 | 92.31 ± 0.00 | 83.33 ± 0.00 | 100.00 ± 0.00 | |
| VGG13‐BN | 73.14 ± 0.05 | 58.73 ± 0.04 | 87.51 ± 0.07 | 92.31 ± 0.00 | 83.33 ± 0.00 | 100.00 ± 0.00 | |
| DCE | ShuffleNet(v2‐x1‐0) | 96.62 ± 0.01 | 100.00 ± 0.00 | 93.96 ± 0.01 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| ShuffleNet(v2‐x0‐5) | 95.66 ± 0.04 | 99.79 ± 0.07 | 92.15 ± 0.02 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | |
| MNASNet1‐0 | 93.73 ± 0.03 | 98.90 ± 0.01 | 93.71 ± 0.05 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | |
CNN = convolutional neural networks; Q0 = low‐quality image; Q1 = high‐quality image.
TABLE 5.
Mean Accuracy Values and 95% Confidence Intervals of the Three Worst‐Performing Models Along the 10‐Fold Cross‐Validation for Each Sequence
| Accuracy (mean ± 95% CI) Individual Slice | Accuracy (mean ± 95% CI) Entire Sequence | ||||||
|---|---|---|---|---|---|---|---|
| Sequence | CNN Architecture | Global | Q0 Class | Q1 Class | Global | Q0 Class | Q1 Class |
| T2WI | SqueezeNet1‐1 | 71.07 ± 0.41 | 43.86 ± 0.54 | 88.68 ± 0.47 | 62.50 ± 0.00 | 0.00 ± 0.00 | 100.00 ± 0.00 |
| VGG16‐BN | 72.11 ± 0.74 | 46.48 ± 0.82 | 88.66 ± 0.63 | 87.50 ± 0.29 | 66.67 ± 0.32 | 100.00 ± .00 | |
| SqueezeNet1‐0 | 72.65 ± 0.61 | 57.03 ± 0.44 | 82.75 ± 0.75 | 87.50 ± 0.26 | 66.67 ± 0.28 | 100.00 ± 0.00 | |
| DWI | MobileNet‐V2 | 52.73 ± 0.45 | 55.12 ± 0.37 | 50.74 ± 0.51 | 55.57 ± 0.21 | 25.00 ± 0.17 | 80.00 ± 0.24 |
| MNASNet0‐5 | 59.12 ± 0.63 | 19.18 ± 0.61 | 91.68 ± 0.66 | 55.57 ± 0.18 | 0.00 ± .00 | 100.00 ± 0.00 | |
| AlexNet | 59.95 ± 0.66 | 14.09 ± 0.75 | 97.50 ± 0.62 | 66.67 ± 0.37 | 25.00 ± 0.00 | 100.00 ± 0.00 | |
| ADC | MNASNet0‐5 | 52.46 ± 0.76 | 24.51 ± 0.73 | 85.36 ± 0.74 | 53.86 ± 0.00 | 0.00 ± 0.00 | 100.00 ± 0.00 |
| ResNet28 | 54.25 ± 0.63 | 30.78 ± 0.65 | 81.90 ± 0.63 | 76.92 ± 0.39 | 50.00 ± 0.46 | 100.00 ± 0.00 | |
| AlexNet | 56.67 ± 0.69 | 40.43 ± 0.61 | 75.66 ± 0.76 | 84.63 ± 0.35 | 66.67 ± 0.32 | 100.00 ± 0.00 | |
| DCE | ResNet50 | 71.92 ± 0.74 | 85.81 ± 0.83 | 60.50 ± 0.72 | 89.00 ± 0.00 | 0.00 ± 0.00 | 100.00 ± 0.00 |
| VGG16‐BN | 77.31 ± 0.85 | 79.71 ± 0.77 | 75.32 ± 0.85 | 92.85 ± 0.24 | 33.33 ± 0.25 | 100.00 ± 0.00 | |
| GoogLeNet | 78.55 ± 0.71 | 89.54 ± 0.66 | 69.45 ± .70 | 96.39 ± 0.32 | 66.67 ± 0.33 | 100.00 ± 000 | |
Discussion
Prostate MRI is an accurate modality for the early detection of csPCa. 3 , 10 Its diagnostic performance is reliant on the image quality of the scans, which depends on the equipment used, the appropriateness of the protocol, and proper patient positioning and preparation. 38 In addition, several technical problems can reduce image quality of prostate MRI and need to be addressed in order not to limit diagnostic performance. 13 , 14
The CNN architectures implemented in this study were able to classify images into the low and high quality with a high degree of accuracy. Different architectures performed similarly on the sequences, meaning that the choice of the structure of the model is secondary; however, the best architectures varied among the different sequences, supporting the fact that different models interpret the same data in unique ways. Accuracy in the classification of individual MR slices was highest for DCE and T2WI, followed by DWI and ADC. Interestingly, classification accuracy was slightly different between DWI and ADC. This can be explained by the fact that for DWI classification, the b‐value of 1500, the most clinically important, was used, whereas ADC was calculated on the b 50 and b 800 acquisitions. 12
By combining the classification of individual slices with a majority vote aggregation function, almost perfect accuracy was achieved for the classification of entire sequences. Since quality‐limiting artifacts tend to be present on most slices of a given sequence, the high performance on the per‐slice analysis likely results in a much higher accuracy when classifying the entire sequence. Of note, an independent test set was used for the evaluation of model performance; therefore, the very high accuracy values are unlikely caused by overfitting.
The potential clinical implications of an automated quality control of MRI scans are linked to its integration in the MR workstation, which would make it possible for the technologist to be immediately notified of a low‐quality scan. Ideally, each MR scanner should have dedicated algorithms capable of assessing image quality of acquired sequences on‐the‐fly. This would require the MR manufacturer to develop, train, and validate specific models for each individual MR protocol, since such algorithms are likely to be application and vendor specific. This approach to quality control would be particularly helpful since the technologist may not be able to correctly interpret the clinical implications of artifacts and appropriately determine when a repeat acquisition is needed. In a recent study, Giganti et al have proposed a scoring system for visual assessment of mpMRI image quality by the radiologist. 16 However, direct visual assessment of acquired images by the radiologist is not realistically feasible in many clinical scenarios. An automated real‐time quality control of MR images can suggest the technologist to adjust the acquisition parameters and/or instruct the patient, as appropriate, in order to acquire a repeat scan of diagnostic quality. For instance, an inadequate FOV can be easily corrected, and a low S/N ratio improved by adjusting acquisition parameters. Motion artifacts can sometimes be avoided on repeated scans by asking the patient to remain still and insisting on the importance of image quality on the accuracy of the exam. The diagnostic impact of low‐quality images differs among sequences. DWI is the most important sequence for the assessment of peripheral zone of the prostate, but also the most technically demanding and the most susceptible to artifacts. 12 , 39 , 40 Artifacts on DWI can be hard to interpret, as suggested by the higher inter‐reader variability in the quality assessment of this sequence compared to T2WI and DCE revealed in this study and might therefore be misjudged by the less experienced technologist.
Conversely, image quality is less critical for the correct interpretation of perfusion images. Low‐quality DCE images are often caused by poor global enhancement of the prostate gland. However, a repeat acquisition is usually not performed because perfusion images must be acquired shortly after injection of the contrast media and in most cases the administration of an additional bolus of contrast. Fortunately, the role of DCE in the detection of suspicious PCa foci at mpMRI is marginal, being limited to the upgrading of PI‐RADS score 3 lesions to PI‐RADS score 4 lesions in the peripheral zone, and increasing evidence is demonstrating wide applicability of noncontrast prostate MRI in the clinical practice, linked to comparable accuracy in csPCa detection, provided that the radiologist's expertise and image quality are adequate enough. 12
Limitations
This study has a relatively small sample size for training. However, the models were trained on individual slices, meaning that a minimum of 8387 images were available for training and testing each model. In addition, the data augmentation techniques implemented significantly increase the number of available images. Another limitation lies in the binary nature of our classifiers: while our models are able to identify sequences of suboptimal diagnostic quality, they were not designed to grade image quality on a semiquantitative scale, nor to pinpoint to the specific underlying cause. While this would be helpful, the first and the clinically more relevant step is to promptly identify the sequences that need to be optimized. An additional limitation is the subjective nature of sequence labeling performed by the radiologists.
Lastly, the CNNs were trained on images acquired on a single MR scanner and protocol. Further studies are needed to clarify whether the models can be applied to different MR scanners and/or acquisition protocols, or if training of specific classifiers is necessary for each MR scanner/protocol.
Conclusion
This study had developed, trained, and validated a fully automated classifier based on convolutional neural networks that is capable of accurately identifying low‐quality prostate MRI images on T2WI, DWI/ADC, and DCE sequences.
Acknowledgments
Open Access Funding provided by Universita degli Studi di Roma La Sapienza within the CRUI‐CARE Agreement. [Correction added on 23 May 2022, after first online publication: CRUI‐CARE funding statement has been added.]
REFERENCES
- 1. Padhani AR, Barentsz J, Villeirs G, et al. PI‐RADS steering committee: The PI‐RADS multiparametric MRI and MRI‐directed biopsy pathway. Radiology 2019;292:464‐474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mottet N, van den Bergh RCN, Briers E, et al. EAU‐EANM‐ESTRO‐ESUR‐SIOG guidelines on prostate cancer—2020 update. Part 1: Screening, diagnosis, and local treatment with curative intent. Eur Urol 2021;79:243‐262. [DOI] [PubMed] [Google Scholar]
- 3. Drost FH, Osses DF, Nieboer D, et al. Prostate MRI, with or without MRI‐targeted biopsy, and systematic biopsy for detecting prostate cancer. Cochrane Database Syst Rev 2019;4:CD012663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kasivisvanathan V, Rannikko AS, Borghi M, et al. MRI‐targeted or standard biopsy for prostate‐cancer diagnosis. N Engl J Med 2018;378:1767‐1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rouvière O, Puech P, Renard‐Penna R, et al. Use of prostate systematic and targeted biopsy on the basis of multiparametric MRI in biopsy‐naive patients (MRI‐FIRST): A prospective, multicentre, paired diagnostic study. Lancet Oncol 2019;20:100‐109. [DOI] [PubMed] [Google Scholar]
- 6. van der Leest M, Cornel E, Israël B, et al. Head‐to‐head comparison of Transrectal ultrasound‐guided prostate biopsy versus multiparametric prostate resonance imaging with subsequent magnetic resonance‐guided biopsy in biopsy‐naïve men with elevated prostate‐specific antigen: A large prospective multicenter clinical study. Eur Urol 2019;75:570‐578. [DOI] [PubMed] [Google Scholar]
- 7. Woo S, Suh CH, Eastham JA, et al. Comparison of magnetic resonance imaging‐stratified clinical pathways and systematic transrectal ultrasound‐guided biopsy pathway for the detection of clinically significant prostate cancer: A systematic review and meta‐analysis of randomized controlled trials. Eur Urol Oncol 2019;2:605‐616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kasivisvanathan V, Stabile A, Neves JB, et al. Magnetic resonance imaging‐targeted biopsy versus systematic biopsy in the detection of prostate cancer: A systematic review and meta‐analysis. Eur Urol 2019;76:284‐303. [DOI] [PubMed] [Google Scholar]
- 9. Panebianco V, Barchetti F, Sciarra A, et al. Multiparametric magnetic resonance imaging vs. standard care in men being evaluated for prostate cancer: A randomized study. Urol Oncol Semin Orig Investig 2015;33:17.e1‐17.e7. [DOI] [PubMed] [Google Scholar]
- 10. de Rooij M, Hamoen EHJ, Fütterer JJ, Barentsz JO, Rovers MM. Accuracy of multiparametric MRI for prostate cancer detection: A meta‐analysis. Am J Roentgenol 2014;202:343‐351. [DOI] [PubMed] [Google Scholar]
- 11. Stabile A, Giganti F, Kasivisvanathan V, et al. Factors influencing variability in the performance of multiparametric magnetic resonance imaging in detecting clinically significant prostate cancer: A systematic literature review. Eur Urol Oncol 2020;3:145‐167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turkbey B, Rosenkrantz AB, Haider MA, et al. Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. Eur Urol 2019;76:340‐351. [DOI] [PubMed] [Google Scholar]
- 13. Sackett J, Shih JH, Reese SE, et al. Quality of prostate MRI: Is the PI‐RADS standard sufficient? Acad Radiol 2021;28:199‐207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Caglic I, Hansen NL, Slough RA, Patterson AJ, Barrett T. Evaluating the effect of rectal distension on prostate multiparametric MRI image quality. Eur J Radiol 2017;90:174‐180. [DOI] [PubMed] [Google Scholar]
- 15. de Rooij M, Israël B, Tummers M, et al. ESUR/ESUI consensus statements on multi‐parametric MRI for the detection of clinically significant prostate cancer: Quality requirements for image acquisition, interpretation and radiologists' training. Eur Radiol 2020;30:5404‐5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giganti F, Allen C, Emberton M, Moore CM, Kasivisvanathan V. Prostate imaging quality (PI‐QUAL): A new quality control scoring system for multiparametric magnetic resonance imaging of the prostate from the PRECISION trial. Eur Urol Oncol 2020;3:615‐619. [DOI] [PubMed] [Google Scholar]
- 17. Bosse S, Maniry D, Muller K‐R, Wiegand T, Samek W. Deep neural networks for no‐reference and full‐reference image quality assessment. IEEE Trans Image Process 2018;27:206‐219. [DOI] [PubMed] [Google Scholar]
- 18. Amirshahi SA, Pedersen M, Yu SX. Image quality assessment by comparing CNN features between images. J Imaging Sci Technol 2016;60:604101‐6041010. [Google Scholar]
- 19. Rawat W, Wang Z. Deep convolutional neural networks for image classification: A comprehensive review. Neural Comput 2017;29:2352‐2449. [DOI] [PubMed] [Google Scholar]
- 20. Mahapatra D, Roy PK, Sedai S, Garnavi R. Etinal image quality classification using saliency maps and CNNs. In: Wang L, Adeli E, Wang Q, Shi Y, Suk H‐I, editors. Machine Learning and Medical Imaging, Vol 10019. Cham: Springer International Publishing; 2016. p 172‐179. [Google Scholar]
- 21. Zhang L, Gooya A, Dong B, et al. Automated quality assessment of cardiac MR images using convolutional neural networks. In: Tsaftaris SA, Gooya A, Frangi AF, Prince JL, editors. Simulation synthesis medical imaging, Vol 9968. Cham: Springer International Publishing; 2016. p 138‐145. [Google Scholar]
- 22. Esteva A, Robicquet A, Ramsundar B, et al. A guide to deep learning in healthcare. Nat Med 2019;25:24‐29. [DOI] [PubMed] [Google Scholar]
- 23. Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer 2018;18:500‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krizhevsky A. One weird trick for parallelizing convolutional neural networks. ArXiv Prepr ArXiv 2014;14045997. [Google Scholar]
- 25. Simonyan K, Zisserman A. Very deep convolutional networks for large‐scale image recognition. ArXiv Prepr ArXiv 2014;14091556. [Google Scholar]
- 26. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. 2016 IEEE Conf Comput Vis Pattern Recognit CVPR. Las Vegas, NV, USA: IEEE; 2016. p 770‐778. [Google Scholar]
- 27. Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K. SqueezeNet: AlexNet‐level accuracy with 50x fewer parameters and< 0.5 MB model size. ArXiv Prepr ArXiv 2016;160207360. [Google Scholar]
- 28. Huang G, Liu Z, van der Maaten L, Weinberger KQ. Densely connected convolutional networks. Proc IEEE Conf Comput Vis Pattern Recognit CVPR. Silver Spring, MD, USA: IEEE Computer Society; 2017. [Google Scholar]
- 29. Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. 2015:1–9.
- 30. Ma N, Zhang X, Zheng H‐T, Sun J: Shufflenet v2: Practical guidelines for efficient CNN architecture design. 2018:116–131.
- 31. Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L‐C: Mobilenetv2: Inverted residuals and linear bottlenecks. 2018:4510–4520.
- 32. Xie S, Girshick R, Dollár P, Tu Z, He K: Aggregated residual transformations for deep neural networks 2017:1492–1500.
- 33. Zagoruyko S, Komodakis N. Wide residual networks. ArXiv Prepr ArXiv 2016;160507146. [Google Scholar]
- 34. Tan M, Chen B, Pang R, et al. Mnasnet: Platform‐aware neural architecture search for mobile. 2019:2820–2828.
- 35. Deng J, Dong W, Socher R, Li L, Li K, Fei‐Fei L. ImageNet: A large‐scale hierarchical image database. IEEE Conf Comput Vis Pattern Recognit 2009;2009:248‐255. [Google Scholar]
- 36. Kittler J. Combining classifiers: A theoretical framework. Pattern Anal Appl 1998;1:18‐27. [Google Scholar]
- 37. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977;33:159‐174. [PubMed] [Google Scholar]
- 38. Schoots IG, Barentsz JO, Bittencourt LK, et al. PI‐RADS Committee position on MRI without contrast medium in biopsy‐naive men with suspected prostate cancer: Narrative review. Am J Roentgenol 2021;216:3‐19. [DOI] [PubMed] [Google Scholar]
- 39. Valerio M, Zini C, Fierro D, et al. 3T multiparametric MRI of the prostate: Does intravoxel incoherent motion diffusion imaging have a role in the detection and stratification of prostate cancer in the peripheral zone? Eur J Radiol 2016;85:790‐794. [DOI] [PubMed] [Google Scholar]
- 40. Plodeck V, Radosa CG, Hübner H‐M, et al. Rectal gas‐induced susceptibility artefacts on prostate diffusion‐weighted MRI with epi read‐out at 3.0 T: Does a preparatory micro‐enema improve image quality? Abdom Radiol 2020;45:4244‐4251. [DOI] [PMC free article] [PubMed] [Google Scholar]


