Abstract
Animal studies in pharmaceutical drug discovery are common in preclinical research for compound evaluation before progression into human clinical trials. However, high rates of drug development attrition have prompted concerns regarding animal models and their predictive translatability to the clinic. To improve the characterization and evaluation of animal models for their translational relevance, the authors developed a tool to transparently reflect key features of a model that may be considered in both the application of the model but also the likelihood of successful translation of the outcomes to human patients. In this publication, we describe the rationale for the development of the Animal Model Quality Assessment tool, the questions used for the animal model assessment, and a high-level scoring system for the purpose of defining predictive translatability. Finally, we provide an example of a completed Animal Model Quality Assessment for the adoptive T-cell transfer model of colitis as a mouse model to mimic inflammatory bowel disease in humans.
Keywords: Animal model, Animal Model Quality Assessment (AMQA), attrition, colitis, drug discovery
INTRODUCTION
The study of animals as surrogates for humans has significantly contributed to our understanding of human health and disease for nearly 2 thousand years.1 We have recognized consistencies and conservation in mammalian biology across animal species and relied on those consistencies to make reasonable, though imperfect, extrapolations of what we learned in animals to what we understand about the human species. Those consistencies are reflected across all levels of biological hierarchy from genetic homology to cellular and systems physiology to morphology and even response to injury. The study of animals has in turn enabled progression in our understanding of basic human biology, the causes and pathogenesis of human disease, and approaches to treating disease and even preventing it. Our confidence in animal studies has progressed to the point that they are a regulatory expectation and a requirement for demonstrating the safety of a novel agent before administration to humans.2 Accordingly, a significant infrastructure has developed to produce and maintain very controlled and standardized species and strains of animals as reproducible “models” of the human condition.3
In translational biomedical research, individual animal “studies” are routinely conducted in animal “models” to either gain novel insights into human biology and disease or to test an intervention that might eventually be applied to humans. The intent of these studies has been broadly considered as either exploratory or confirmatory, representing differences in the size and procedural rigor of the studies as well as the proximate application to humans.4 In drug development, these distinctions are likely not binomial but rather represent a continuum or spectrum from early exploratory studies to validate targets and optimize promising candidate drugs to more rigorous tests of efficacy and safety that will support decisions to move into more expensive clinical trials. Accordingly, the “models” are generally intended to provide clear insights into human biology and pathobiology. The human relevance of the outcomes is dependent on both the model and the analytical rigor of the study design. This work focuses on evaluating the human relevance of a model.
High rates of drug development attrition and challenges in reproducing specific animal studies have fueled concerns that current animal studies may be a weak link in our approach to translating basic biomedical research to beneficial human applications.5 Sources of those weaknesses have been varied and include failure to adequately report the design of a study, poor alignment of the study design to the clinical intent, lack of statistical power, mitigation of bias, and even biological differences in the animal model relative to the human interest. A number of strategies6–8 have been proposed to minimize these weaknesses and ensure that the animal studies are optimized for their human relevance and translation.9
Animals of varying species, though predominated by mice, are used as models of either normal biology, as spontaneous models of a disease, or through experimentally induced disease to provide useful insights into the relevance of a pharmacologic target (ie, target identification and validation) for modulating a disease, guiding drug design, characterizing biodistribution and metabolism of a drug candidate, and identifying potential safety liabilities. Historically, animal models have inconsistently reproduced the full spectrum of etiology, mechanisms, pathogenesis, and morphology of human disease. The most translationally successful models have been those that reasonably recapitulated disease across all of those levels. Models where this might be expected include diseases that are simpler in their etiology, for example, due to single gene mutations or in infectious diseases with pathogeneses similar to that in humans.10, 12
Alhough there has been a shift to more molecular pharmacological targets for chronic and complicated diseases, approaches to animal modeling have not similarly shifted and are likely still too reliant on recapitulation of the morphology of a disease rather than the mechanism of that disease. This is further complicated by our lack of understanding of the primary molecular mediators of many complex human diseases. Recognizing inherent weaknesses in current approaches, multi-model strategies are often applied with the potential for a biased focus on those studies that provide the outcomes of most interest.
Working in a large and global pharmaceutical company, we are committed to judicious and effective use of animal studies in our efforts to develop safe and effective new medicines. At GlaxoSmithKline (GSK), we are particularly interested in the challenges of optimizing the biological and pharmacological translation of animal models and studies to patient outcomes. Others have shared this interest and suggested a disease-specific functional deficit approach,13 incorporation of a scoring system,14 and, more recently, development of the Framework to Identify Models of Disease (FIMD), which includes factors to help interpret model similarity and evidence uncertainty.15 For our purposes, we developed an Animal Model Quality Assessment (AMQA) tool to provide a consistent framework for evaluating a particular animal model that could be used to support its selection for a specific application or question across the spectrum of uses in drug development (ie, the evaluation outcomes would rationalize the usefulness of the model for a particular context of use). The aim of the evaluation was to develop a discrete line of sight to the clinical intent of the model that would optimize the likelihood of clinical translation of the outcomes.
Aside from supporting animal model selection, the AMQA has other possible uses (Table 1). In drug discovery, every novel pharmacological target presents a different biological challenge that may require a better understanding of the contextual biology or role in disease not previously characterized in a particular animal model. A critical and transparent assessment of the model provides insights into those knowledge gaps that could prompt additional characterization. The decision to progress a novel drug candidate involves an integrated evaluation of all the evidence supporting its potential benefit to patients, its safety, and acceptance by healthcare payers. A critical assessment of the translational relevance of the models used to produce that evidence enables more informed decision-making and insights into the likelihood of clinical success. Lastly, recognized gaps or weaknesses in our ability to model human-relevant biology in an animal model highlight areas for focused development of non-animal approaches.
Table 1.
Value summary for assessing the translational relevance of an animal model in a structured framework.
| Potential values for a structured assessment of the human translational relevance of an animal model |
|
Use of the tool could additionally support harm–benefit analysis (HBA). The HBA assesses whether the harm to the animals is outweighed by the potential benefit of the experiment.16 For example, a scientific justification is considered in an ethical review process but not always rigorously challenged. Although the expected harms to the animals are anticipated and mitigated, committees often base scientific justification on the scientist’s citations of published data or previous work. The AMQA tool extends this and provides, in the context of translational research, the opportunity to better understand the model quality and therefore a consideration of the “likelihood of success” of a specific experiment in a given model.
THE DEVELOPMENT OF THE AMQA
As part of its commitment to the 3Rs, GSK has a component of its corporate animal welfare and ethics group focused on design and delivery of an animal research strategy. Broadly, the strategy consists of a focus on improving animal research practices, application of iterative learning to understand preclinical-clinical concordance, and defining a future state with decreased dependency on animals. While establishing the strategy, a critical review across several therapeutic areas was conducted to gain insights into the types of animal models being used, how the models were selected, how those models were justified at ethical review (eg, in Institutional Animal Care and Use Committee reviews), the design of the studies in which the models were being used, and the ultimate clinical outcomes.
As is widely recognized, the outcomes of clinical testing for novel drug candidates is often unsuccessful with insufficient or lack of efficacy or benefit often contributing to drug development attrition17,18. Accordingly, we conducted an internal after-action review on a number of key assets with recent successful and unsuccessful clinical outcomes to identify key points of misalignment between the preclinical animal pharmacology studies and the clinical trials those studies supported. That exercise was instructive and quickly identified potential sources of translational weakness. We particularly considered features of an animal model that might contribute to differences in response between the animal and human patients. Several key features were identified and represented in our assessment, including our fundamental understanding of the human disease of interest, the biological or physiological context of the organ systems affected, historical experiences with pharmacologic responses in the model relative to human, how well the model disease reflected the human disease etiology and pathogenesis or progression, and the model’s replicability or consistency.
From that review, it was clear that a more consistent approach to establishing and recording the relevance of an animal model for a particular human disease or condition could be helpful. It was expected that such an assessment would involve multidisciplinary collaboration (eg, investigator with laboratory animal veterinarian and/or pathologist), transparently reflect the translational quality of the model and thus the data derived from the model, and provide insights into the weaknesses of a model that could be filled through either additional knowledge/investigation or mitigated by using an alternative platform (eg, complex in vitro modeling system). Notably, once a model is justified for its intended translational usefulness, the individual study design should also receive appropriate peer review with the biostatistician central to the design to ensure bias is minimized.
The AMQA tool evolved through consultation with various disciplines, including in vivo scientists/investigators, pathologists, comparative medicine experts, and non-animal modelers, and completed 3 rounds of pilots and iterative design. The challenge was to ensure applicability across a broad portfolio of models and appropriateness for both well-characterized and novel models. Once designed, feedback from investigators using the tool prompted refinement and addition of further questions. Mohanan et al., 201919 described a workshop where the principles and tool were formally shared with discovery-focused pathologists and comparative scientists. Such discussions offered opportunity for further improvements.
The assessment itself is a questions-based template that guides an investigator through key questions and considerations related to evaluating and justifying an animal model for a specific human disease interest. The questions in the template define a holistic set of information that often requires the multidisciplinary collaboration that was used to develop the tool. This question-based approach has been described broadly by de Visser.20 The question-based approach is useful in drug development and is designed to make the input explicit rather than implicit, focusing on the relevant questions being asked. We aimed to provide a simple and practical output that clearly identifies strengths and weaknesses of a particular animal model being evaluated for a particular drug development interest. Different approaches were considered for reflecting the translational quality of the features evaluated, including quantitative and qualitative methods. An absolute quantification (score of 1, 2, 3, 4, 5) was considered difficult due to the subjectivity and variability in expertise of internal expert (from preclinical to clinical) opinions in scoring specific features and the recognition that clinical experience may not exist in novel areas. Secondly, it became difficult to objectively distinguish between scores that deflected from the intent to focus on an overall assessment of model quality and to identify key areas of weakness.
Accordingly, a semi-quantitative scoring system was developed using a stoplight or red–amber–green (RAG) assessment, which is admittedly subjective but provides some measure of the strength of the evidence. The users of this tool (project team, early discovery group, academic investigator) evaluate what is known regarding the specific questions and then reflect the response in the appropriate box. In general, red = major weakness and/or differences; amber = some similarities or fair concordance, where unknowns or known weaknesses should be noted; and green = good concordance/similarities. Peer review of the outcomes of the assessment by an independent investigator would increase confidence in the assessments, and the RAG scoring can be incorporated into the larger candidate selection/drug progression considerations of pharmaceutical development.
This scoring system was not designed to rank one animal model vs another but to represent the strengths and weaknesses of a particular model in the context of its intended use and to provide an opportunity to focus effort on filling in knowledge gaps. It could be used, if required, to comparatively support the decision to use one animal model over another in a context of the intended use, for which an AMQA of each model of interest would be required.
The primary aim of the AMQA tool is to help investigators in selection of their animal models and to aid decision makers by increasing the transparency of the translational quality of animal-derived evidence.
FRAMEWORK OF THE AMQA
The AMQA is considered at 2 levels of detail represented as primary and secondary assessments (Figure 1). The primary assessment serves as an overarching framework of important considerations for an animal model intended to support pharmacological research. The questions are purposefully broad and probe fundamental principles related to the concordance of the animal model to the human condition, the underlying disease pathobiology, and previous experience with pharmacologic responses. The secondary assessment questions are more specific to the general themes probed by the primary assessment questions. The purpose of these questions is to obtain a more granular understanding of the disease in humans and the alignment with the characteristics of the animal model. Information developed for the tool helps to evaluate the translational quality of the animal model, where quality relates to human relevance, preclinical/clinical continuity, and pathobiology.
Figure 1.
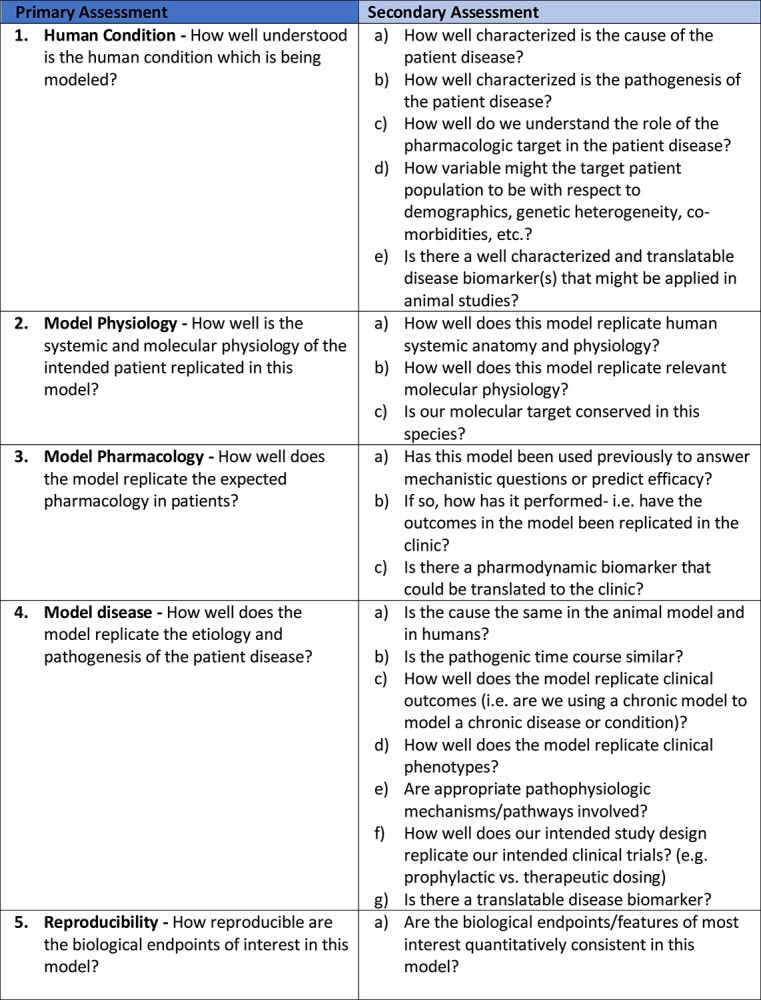
The Animal Model Quality Assessment (AMQA) framework. The primary assessment is divided into 5 sections: Human Condition, Model Physiology, Model Pharmacology, Model Disease and Reproducibility, each with a broad overarching question. The secondary assessment consists of more granular questions relating specifically to the primary areas.
ASSESSMENT QUESTIONS
Human Condition
Understanding the biological system of interest is fundamental to developing, selecting, or applying an effective “model” (ie, you cannot model what you do not know). This is a bit of a challenge when one considers that we are still trying to fully understand many of the more common and chronic progressive diseases in human society today.
Clinical human disease is the culmination of several progressive events beginning with an initiating event (an etiology), an adaptive or maladaptive host response, and progression to injury and organ dysfunction (pathogenesis). Modern medicine mostly attempts to either prevent disease onset or mitigate disease that has already initiated. Drug therapy is generally aimed at altering or reversing the progression or pathogenesis of a disease that has already initiated. Because many of the pharmacologic targets that are the focus of drug development today are mediators of pathogenesis, it is important to replicate as much as possible key events in the initiation and progression of disease when modeling the potential effectiveness of a novel therapeutic.
Though we often attempt to simplify our understanding of disease initiation and progression to facilitate our modeling, individual human responses can be quite variable as can clinical phenotypes for the same disease. That variability should be considered and may restrict the context of use of a particular model. In fact, deliberate heterogenization, including different housing/environmental conditions, ages, sex, test times, and alternate test systems, has been proposed as an approach to improving the translation of animal studies to real human conditions.21 Additionally, key measures of disease biology (ie, a “biomarker”) can be useful bridges from the human condition to an animal model to provide qualification and confidence in the relevance of that model.
Model Physiology
Mammalian biology is complex, integrated, and adaptive, enabling survival in the presence of perpetual environmental change and challenge. Much of biomedical research today is increasingly reductionist and quantitative in nature, focusing on increasing levels of molecular resolution to build understanding of the very roots of health and disease. That has the inherent danger of losing sight of a bigger “physiological picture” that significantly influences individual molecular events. Determining that a human biological target is conserved in an animal model is fairly straightforward. Ensuring that engagement and modulation of that target is quantitatively translatable from one species to another is more challenging. For example, fatty acids as a substrate for energy production in the mammalian heart are conserved across species. The quantitative dynamics of that process is likely very different in a human with a heart rate of 70 beats/min and a rat with a heart rate of 350 beats/min. That does not make the rat an irrelevant model of cardiac physiology, but it does affect the human extrapolation of rat cardiac biology.
Model Pharmacology
Although science based and regulated, drug development is still a relatively new endeavor of 100 years or so even though humans have taken natural or synthetic substances to modify health for hundreds if not thousands of years. That experience provides an unprecedented opportunity to understand how humans respond to pharmacologic agents against which animal responses can be compared and qualified. Most modern medicines were supported by preclinical animal studies. Successful medicines with concordant efficacy in the supporting animal model may provide confidence in the potential for translation of the respective animal model. Paradoxically, even failures in translation can be useful because they provide insights into potential modeling weaknesses that should be explored further. Alternatively, and also important to consider, an animal model may have a limited “domain of validity” exceeded by a novel candidate therapeutic engaging a novel pharmacologic target, thus the need to consider the full spectrum of primary and secondary assessments of animal model quality.
Model Human Disease
As noted above, understanding what we do and do not know about the human disease or condition of interest is fundamental to a strategy for modeling that disease. Other than very simple etiologies, like an infectious disease or a disease originating from a heritable single gene mutation, the complexity of a human disease is exceedingly difficult to reproduce in a modeling system. Accordingly, modeling of that disease may need to be targeted to key biological events in the initiation and progression of that disease. For example, though a diabetic state can be induced in a rodent model with streptozotocin producing a human-like clinical phenotype, human diabetes is not initiated by streptozotocin and likely contributes to significantly different pathogeneses in the 2 species. A model of streptozotocin-induced diabetes may be useful for modeling the consequences of chronic hyperglycemia but might not be useful for modeling mitigation of pancreatic islet cell loss that drives the progression of diabetes or peripheral tissue insulin resistance that potentiates it.
Reproducibility
The ultimate aim of an animal study, particularly in drug development, is to enable a conclusion about the outcomes and a decision. In the case of drug development and pharmacology modeling, the conclusion likely relates to the likelihood that an intervention will modulate a disease and the decision to progress a drug candidate. The ability to reproduce the outcomes of that experiment or study enables confidence in the decision. Much of the current interest in animal study reproducibility has focused on how well a study or experiment is reported, how well the analytical outcomes are supported by appropriate statistical power, and control of bias. Another “reproducibility” of interest is the ability to reliably replicate the normal or perturbed biology of interest. Though biological variability is inherent to the human condition and arguably useful for a modeling system, that variability can significantly challenge the ability to make a defensible conclusion from an animal study (Voelkl, et al.21). Large studies with significant biological variability interrogating broad questions have to be considered against more discrete studies interrogating discrete hypotheses in models with clear lines of sight to the human clinical condition.
SCORING USING SEMI-QUANTITATIVE INDICATORS
Responses to questions in the framework can be represented as colors depending on the strength of the evidence supporting that key feature, for example, RAG. Assigning these colors should reflect the best scientific judgement of the team developing the assessment. Red should be used to indicate major weakness and/or differences, that is, those differences likely to contribute to lack of meaningful translation to clinical outcomes. Amber should be used when there are some similarities/or fair concordance though not complete replication of the human features (it is useful to note known weaknesses). And green should be used to indicate good concordance/similarities. The RAG system provides a high-level, rapid visualization of the strengths and weaknesses in the animal model and its concordance to the human condition. Assignment of a specific color to a primary assessment should represent the integration of the secondary assessments, and it is recommended that if there is any red in the secondary assessment that a primary assessment for that section cannot be green.
There is no predefined weighting of the individual elements of the AMQA, leaving the influence of the various elements to the reviewer and the intended use of the model. We are not aware that anyone has reported a systematic review of individual models in the context of their relative translational alignment for the various features evaluated to know how to weight them. The “perfect” model would reproducibly reflect the morphologic, molecular, and pathogenetic features of the human disease in organ systems that are physiologically homologous and respond similarly to pharmacologic intervention. That model probably does not exist. Accordingly, professional judgement will influence model choice and response to the outcomes. The value of the AMQA is the transparency that informs those decisions and insights provided into model weaknesses that could be mitigated through additional characterization, alternative model choice, or varying the study design (eg, longer duration, younger or older animals, etc).
SOURCES OF INFORMATION UTILIZED TO COMPLETE AN AMQA
Information to complete the AMQA may be obtained from a variety of sources, including open access literature–searching services, for example, Medline, Embase, PubMed, or similar; search engines; animal model suppliers; and open access model databases. To follow preclinical models and outcome of models subsequently tested in the clinic, Trialtrove (PharmaIntelligence, London, UK), Pharmapendium (Elsevier, Inc.), and Cortellis (Clarivate, Inc.) may be useful. GSK also has internal systems, including patent searches, target dossiers, and, for some models, extensive historical data that are suitable for contextualizing models utilized previously. Notably, the data sources are varied and are mainly unstructured. Future evolution of the tool could consider use of developing technologies eg, natural language processing, to considerably reduce the manual requirement to initially retrieve information. Additionally, individuals considered subject matter experts within a field are also consulted as required. When publications are utilized, the study quality of the paper is important; statisticians may review the methodology and analysis presented, or utilization of published risk of bias tools may be useful.7
The benefit of a second peer reviewer following completion of the AMQA has proved valuable, particularly to assess the RAG indicators that have been assigned. The peer reviewer should be independent from the initial assessor but with relevant biological knowledge.
EXAMPLE APPLICATION: AMQA OF THE T-CELL TRANSFER MODEL OF COLITIS
The completed AMQA for the T-cell transfer model of colitis as a mouse model to mimic inflammatory bowel disease (IBD) is shown in Figure 2.
Figure 2.
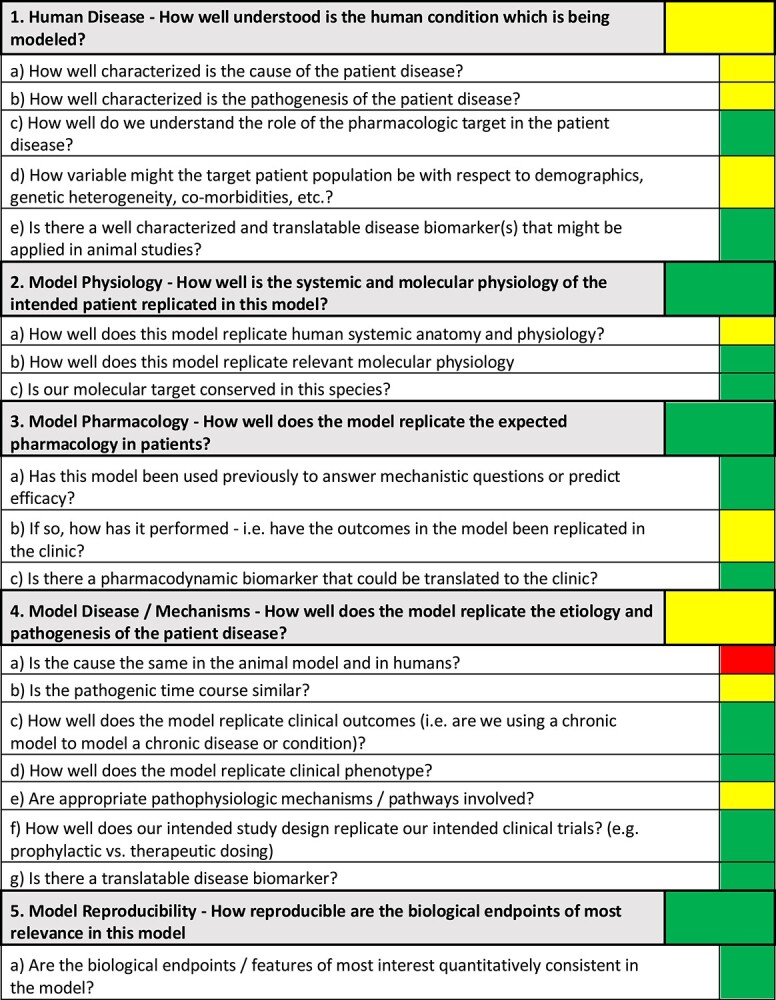
Completed AMQA for the adoptive T-cell transfer model of colitis in mice. The 5 areas of primary assessment are shown with an overall Red-Amber-Green score; the secondary assessment questions are also shown with the relative indicator.
Completing the AMQA secondary assessment questions helps guide the overall RAG indicator for the primary assessment questions. At a glance, the model’s strengths and weaknesses are apparent, as discussed in more detail below. Supporting evidence and information that resulted in the scoring color can be found in the Supplementary Data. Because no model is a perfect recapitulation of the human condition, the choice of an animal model should also depend on the specific scientific questions and the biological target being considered. Thus, the output of an AMQA for a given animal model may differ when in context for different biological pathways.
Specifically, we report here the AMQA generated for representing the translational relevance of the mouse T-cell (CD4 + CD45RBhigh) adoptive transfer model of colitis for immunological therapeutic targets. This model is the most widely used and best-characterized model of chronic colitis induced by disruption of T-cell homeostasis. We do not report specific information on the pathways of interest for GSK research, but we recognize that the T-cell transfer model of colitis is suitable to investigate the immunological mechanisms and the related biological pathways responsible for the induction, perpetuation, and/or regulation of chronic inflammation. Because the target is not declared, note that some questions, for example, 1C, 2C, 3C, in Figure 2 and are not detailed in the text below.
1. Human Disease: How Well Understood Is the Human Condition Being Modeled?
IBD is a multifactorial disorder characterized by chronic relapsing intestinal inflammation. The 2 major subtypes of IBD are ulcerative colitis (UC) and Crohn’s disease (CD). Although the cause and unpredictable recurrence of IBD are still unclear, its pathogenesis is partially known. The induction and the progression of IBDs arise from the complex interaction of 4 different factors: genetic susceptibility, environment, gut microflora, and adaptive/innate immune dysfunction.22–24 In the secondary assessment, the relevant questions “How well characterized is the cause of the patient disease?” and “How well characterized is the pathogenesis of the patient disease?” were therefore shown as amber (Fig. 2). There is also extensive variability relating to demographics and comorbidity. It is estimated that more than 1 million US and 2.5 million European residents are affected by these disorders. IBDs have also emerged in those countries where industrialization is relatively new (such as Asia, South America, and Middle East); thus, IBDs should be now considered as a rising global disease. Extra-intestinal manifestations of IBDs affecting the joints, eyes, and/or the skin can occur in up to 25% of patients.25 More than 160 genetic variants or polymorphisms are associated with IBD susceptibility. The intrinsic heterogeneity of the affected individuals is a cardinal feature of IBD. This information is captured with the amber score in the corresponding question: How variable might the target patient population be with respect to demographics, genetic heterogeneity, co-morbidities, etc? Translatable biomarkers currently used in the clinic and applicable to animal models can be disease specific (ie, body weight, diarrhea, colon histopathology, endoscopic/MRI assessments) or reflect generalized inflammation (ie, systemic cytokines).
2. Model Physiology: How Well Is the Systemic and Molecular Physiology of the Intended Patient Replicated in This Model?
One of the key matters in the comparison between our animal model and human disease is to clarify how well the mouse T-cell adoptive transfer replicates the human-relevant anatomy and physiology at the gut level. Mammalian digestive tract biology is strongly conserved. Given their shared omnivorous nature, mice and humans are quite similar. This partly explains why mouse models have been widely used in biomedical research.26 However, the anatomy, physiology, gut microflora, and immune system of the mouse and the human intestinal tract exhibit some differences that must be considered. The cells and pathways involved in the evolution of intestinal inflammation are not conserved across these species’ boundaries.27,28 Close attention must be paid to the detail in translating these mechanisms from mouse to human. This information is represented by an amber score within the model physiology section of the AMQA and specifically in relation to the question “How well does this model replicate human systemic anatomy and physiology?”
3. Model Pharmacology: How Well Does the Model Replicate the Expected Pharmacology in Patients?
There is no pharmacological cure for IBD. Currently, medications such as conventional anti-inflammatory drugs, immunomodulatory agents, biological therapies, broad spectrum antibiotics, and metronidazole for some subgroups of IBD patients are used to control symptoms or to create a period of remission that improves long-term disease outcome. This model has been used previously to answer both mechanistic questions (ie, transferring genetically naïve T cells from genetically modified donors into recipient) or to predict efficacy of several IBD drug candidates. When considering how well the mouse T-cell adoptive transfer model of colitis has predicted clinical outcomes in the past, we scored the model amber. Robust evidence supports that pivotal drivers of inflammation are shared between this model and the human disease. The CD45RBhigh adoptive transfer model has been a very useful tool to predict the clinical outcome of medications, mostly biological such as anti-TNF mAb and IL-12/IL-23 (p40) mAb (see Supplementary Information). However, data also show some limitations of the adoptive transfer, because there is not always a direct correlation between the drug efficacy in humans and mice such as for CTLA4-Ig,29,30 cyclosporine,29–31 and anti-IL-1732–36 approaches.
4. Model Disease/Mechanisms: How Well Does the Model Replicate the Etiology and Pathogenesis of the Patient Disease?
Cause and pathogenic time course are undoubtedly among the most important key drivers when assessing how the T-cell adoptive transfer model of colitis mimics the human disease. IBD is a chronic inflammatory condition, and mice with this model develop chronic colitis due to T cells homing to the intestinal mucosa and lack of functional regulatory T cells in the host that allows the development of a TH1/TH17 adaptive immune response to antigens derived from intestinal bacteria. This model is generated by the transfer of a small number of mouse naïve CD4+CD45RBhigh T cells in immunodeficient mice.37 Obviously, such an approach does not mimic the cause of IBD and justifies the red score shown in Figure 2 for the secondary assessment question: “Is the cause the same in the animal model and in humans?” The disruption of T-cell homeostasis is at the center of the adoptive transfer model and is recognized as one of the most important pathogenic mechanisms in human disease. However, it is impossible to replicate in a model the identical pathogenic time course of a very heterogenous human disease such as IBD. Therefore, the secondary assessment question “Is the pathogenic time course similar?” scored amber. Nevertheless, the mouse model replicates well the chronicity of the clinical outcome seen in humans and scored green in the corresponding question.
For the question “How well does the model replicate clinical phenotypes?” a green score was given. This was chosen because patients affected by IBD suffer from bloody diarrhea, abdominal pain, nausea, fever, and weight loss. T-cell–transferred mice, depending on the gene susceptibility of the recipient, develop a chronic progressive disease that shares several symptoms with both human CD and UC, such as weight loss, reduced physical activity, diarrhea, and loose stools. Differently from humans, there is no substantial amount of occult blood in the feces of the affected mice. The location of the lesions (large and small intestine), the histopathological profile (transmural inflammation), and the mixed Th1/Th17 inflammatory response approximate the adoptive transfer model more to CD than to UC. In addition, the chronic temporal progression of the model allows the therapeutic administration of drug in efficacy studies, mimicking settings in clinical trials.
Although IBD disease biomarkers are currently used in diagnosis for the differentiation between CD and UC, most biomarkers currently used are not disease specific but reflect generalized inflammation. With respect to the AMQA, the question in the secondary assessment relating to translatable biomarkers of disease scored green. Most IBD biomarkers, such as body weight loss, diarrhea, histopathology, serum cytokine production, calprotectin measurements, and endoscopic assessment, can be applied to this model.
5. Model Reproducibility: How Reproducible Are the Biological Endpoints of Most Relevance in This Model?
Finally, our structured framework considers the reproducibility of the endpoints in the model. Among all genetically engineered, spontaneous, and chemical models of chronic colitis, the T-cell transfer is the most widely used model. A standardized protocol can induce a highly predictable and synchronized colitis. However, the severity of the symptoms can vary according to strain, sex, and microbiota, and each protocol needs to be carefully validated in different facilities. Once done, the endpoints are usually robust and highly reproducible across studies. This is represented by the green score for the question: “Are the biological endpoints/features of most interest quantitatively consistent in this model?” Thinking further through the reproducibility of endpoints, consideration is also required around what is a reasonable or expected threshold of change needed to answer the question. For this example, based on pilot studies we have quantified the coefficient of variation for colon density (colon weight vs length ratio, one of the main endpoints of the model) as 25% among animals receiving the same treatment. We can reasonably conduct experiments in which colon density measurements are obtained on 10 animals per treatment group.
Conducting a one-sided significance test in which the alternative hypothesis is that the active treatment causes a reduction in colon density, setting a threshold for significance (alpha) of 5%, and aiming to achieve a power (1 − β) of 90%, we would expect to achieve statistical significance if the active treatment causes an approximately 30% reduction in colon density relative to the control.
In brief, this model is regarded as being reasonably representative of key features of human IBD (immune-pathology and chronic disease). Although different animal models offer some features of human diseases, none of them can be taken as a perfect surrogate. The choice of the appropriate animal model should depend on the specific question being proposed. This model is suitable to study the induction and perpetuation of colitis as well as the role of regulatory T cells in suppressing or limiting the onset of intestinal inflammation. There may be other models that are similarly suitable. Historically, the CD4+CD45RBhigh adoptive transfer model has been a very useful tool to predict clinical outcomes of drugs targeting cytokines, especially biologics. In addition, it may be useful to evaluate other models when targeting different pathways, that is, epithelial restitution and proliferation, mucosal wound repair in the presence of inflammatory response, or intestinal fibrosis.
SUMMARY
The AMQA was developed to assess the translational strength of an individual animal model for a particular biological target or pathway, particularly for support of drug discovery. The tool documents the considerations applied, through a consistent framework and a questions-based approach, and ultimately guides an investigator to consider key elements that rationalize the appropriateness of the model. Once completed, the output becomes a context for studies conducted in the model, allowing decision makers to consider the translational strength of the evidence produced by using the model. Similarly, the output can be used to represent the potential benefit or the likelihood of clinical success when utilizing a given model, which is important for HBA (ie, quantifying the “benefit”), and therefore can be utilized as a useful scientific summary to support ethical discussions for Institutional Animal Care and Use Committee, Animal Welfare Body, or Animal Welfare Ethical Review Body.
DISCUSSION
The AMQA is the first attempt internally at GSK at qualifying models from a drug discovery perspective. We know of some similar efforts13 in both industry and academia in establishing rating or scoring systems for animal models. One example described by Denayer et al14 evaluated 5 criteria: (1) the relevance of the species, (2) the complexity of the test system, (3) the simulation of the disease, (4) the predictivity using a quantal (yes or no) effect of a drug vs a graded or dose response comparison, and (5) the number of symptoms modeled in the test system. For this system, a score is given based on relevance (4 = high, down to 1 = low), and the intent is not to select one optimal model but to have a set of models to provide maximal validity for a target/program. Also, although not providing a specific assessment approach, Henderson, et al38 systematically reviewed animal experiment guidelines to identify the most common recommendations for assessing a model with additional study design recommendations. Many of the construct guidelines closely approximate features that are queried in the AMQA. More recent developments have been described to make more informative evaluations of animal models in the context of given diseases. Ferreira at al15 described a FIMD similar to above, but in more explicit detail allowing the investigator to record information on (1) epidemiology, (2) symptomatology of the disease, (3) genetics, (4) biochemical validation, (5) etiology, (6) histological validation, (7) pharmacologic validation, and (8) endpoint validation.
The FIMD assesses various aspects of the external validity of efficacy models, with both similarities with and differences from the AMQA. In terms of similarities, both include questions on (1) patient population disease and demographics, (2) disease symptoms or clinical phenotype, (3) the natural history or time course progression of the disease, (4) biomarkers, and (5) etiology. The main difference is that the AMQA places more focus on the mechanism and biological pathways of the model and the model pharmacology pertaining to clinical translation. Secondly, the AMQA questions query the clinical translatability of the disease, human physiology, and clinical trial design more than that of the FIMD. Lastly, the FIMD introduces a weighting system in an attempt to quantitate how well a model could simulate human disease. It is clear that the weighting system could be a point of debate due to differences in opinions of the relevance of each question, but with repeated use of the tool, an idea of predictive value could potentially be developed. As such, the intent of this publication is not to provide an in-depth comparison of the 2 tools or compare scoring systems but to have a way for researchers to have an internal discussion about models and record an assessment.
Although development of the AMQA is in overall alignment with the other attempts reported in the literature, the current framework takes not only the end disease into consideration but also the knowledge of the biological target and pathway. The AMQA uniquely considers the animal model holistically and prompts consideration of the contextual biology as an important mediator of both pathologic and pharmacologic outcomes. After all, it is considered an imperative to ensure that a biological pathway is operative and shared in the pathogenesis of the disease and the animal model.
Another aim of this tool is to remain relevant with external guidance such as that from the NIH’s working group on enhancing rigor, transparency, and translatability.39 One of the themes was to “improve relevance and use of animal models by establishing a framework for investigators to explain the rationale for their chosen model.” By developing the AMQA tool, we believe we are setting a foundation, not only for ourselves, but for others, because the tool itself could be adapted as required for different needs and circumstances.
The overall goal of the RAG scoring system is to provide a quick representation of key areas of strengths and weaknesses of a model with more detail in the secondary assessments. The scoring system prompts interdisciplinary collaboration in animal model characterization, supports a rationale for model selection, transparently represents strengths and weaknesses, supports HBA, and contextualizes the decision-making outputs of the study. The AMQA tool is of maximal impact when the outputs are documented to allow future learnings and iterative understanding of the scoring, firstly to refine the tool itself but also to serve the broader scientific endeavors, for example, interest in the same biological target for a different disease. Of course, the experiments that are then executed in a given model should ensure that methodological rigor are applied with appropriate inclusion of statistical principles to ensure adequate precision through power analysis and to minimize bias (randomization and blinding) and in turn reported using one of the recommended guidelines, for example, ARRIVE.40
It is recognized that the AMQA tool is dependent on current understanding of the human condition, the biology and pathobiology represented in the animal model, and the reproducibility of the experimental endpoints. This relies on comparative understanding of disease progression and pathologic changes across species. As one would expect, the less known about a disease, the more challenging the use of a model becomes for the investigator. However, it does ensure that the gaps are recognized such that an overreliance or emphasis is avoided on a particular model or study.
With continued use, there is an opportunity to improve the tool by developing a scoring system with weighting for particular questions. The challenge is arriving at a consensus for which primary or secondary assessments should hold a higher weight, so further work is required before the tool could be used as a standard across institutes.
Having formal recognition and documentation of the weaknesses of a model can also drive early consideration of non-animal alternatives and provide opportunity for targeted development of such alternatives. With some modification, one could imagine that this approach could also be applied to non-animal modeling systems where human biological relevance and reproducibility are just as important. An alignment of in vitro, in vivo, and human disease processes would vastly improve the translatability of the drug discovery and development process.
Potential conflicts of interest
All authors: No reported conflicts.
Supplementary Material
Contributor Information
Joanne Storey, Animal Research Strategy Group, Office of Animal Welfare, Ethics, Strategy and Risk, GlaxoSmithKline, Stevenage, UK.
Thomas Gobbetti, Experimental Quantitative Pharmacology Group (Immunology Research Unit), GlaxoSmithKline Medicines Research Centre, Stevenage, UK.
Alan Olzinski, Animal Research Strategy Group, Office of Animal Welfare, Ethics, Strategy and Risk, GlaxoSmithKline, Collegeville, Pennsylvania, USA.
Brian R Berridge, National Toxicology Program Division, NIH NIEHS Research Triangle Park, NC, USA.
References
- 1. Franco NH. Animal experiments in biomedical research: A historical perspective. Animals. 2013; 3(1):238–73. 10.3390/ani3010238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. ICH M3(R2) . Issued by U.S. Department of Health and Human Services Food and Drug Administration. Center for Drug Evaluation and Research. Center for Biologics Evaluation and Research 2010. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/m3r2-nonclinical-safety-studies-conduct-human-clinical-trials-and-marketing-authorization
- 3.https://www.alzforum.org/research-models
- 4. Kimmelman J, Mogil JS, Dirnagl U. Distinguishing between exploratory and confirmatory preclinical research will improve translation. PLoS Biol. 2014; 2(5):e1001863. doi: 10.1371/journal.pbio.1001863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McGonigle P, Ruggeri B. Animal models of human disease: challenges in enabling translation. Biochem Pharmacol. 2014; 87(1):162–71. 10.1016/j.bcp.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 6. Landis S, Amara S, Asadullah K, et al. A call for transparent reporting to optimize the predictive value of preclinical research. Nature 2012; 490, 187–191. 10.1038/nature11556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hooijmans CR, Rovers MM, de Vries RB, et al. SYRCLE’s risk of bias tool for animal studies. BMC Med Res Methodol. 2014; 14, 43. 10.1186/1471-2288-14-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol 2010; 8(6): e1000412. 10.1371/journal.pbio.1000412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Begley CG, Ellis LM. Raise standards for preclinical cancer research. 2012. Nature. 2012; 483:531–3 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 10. Favret JM, Weinstock NI, Feltri ML, et al. Pre-clinical mouse models of neurodegenerative lysosomal storage diseases. Front Mol Biosci. 2020; 15(7):57. 10.3389/fmolb.2020.00057. PMID: 32351971; PMCID: PMC7174556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cox ML, Lees GE, Kashtan CE, et al. Genetic cause of X-linked Alport syndrome in a family of domestic dogs. Mamm Genome. 2003; 14(6):396–403. 10.1007/s00335-002-2253-9 PMID: 12879362. [DOI] [PubMed] [Google Scholar]
- 12. Stundick MV, Albrecht MT, Houchens CR, et al. Animal models for Francisella tularensis and Burkholderia species: Scientific and regulatory gaps toward approval of antibiotics under the FDA animal rule. Vet Pathol. 2013; 50(5):877–92. 10.1177/0300985813486812 Epub 2013 Apr 29. PMID: 23628693. [DOI] [PubMed] [Google Scholar]
- 13. Samms-Dodd F. Strategies to optimize the validity of disease models in the drug discovery process. Drug Discov Today. 2006; 11(7–8):355–63. 10.1016/j.drudis.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 14. Denayer T, Stohr T, Van Roy M. Animal models in translational medicine: validation and prediction. New Horizons Translat Med. 2014; 2(1):5–11. [Google Scholar]
- 15. Ferreira GS, Veening-Griffioen DH, Boon WPC, et al. A standardised framework to identify optimal animal models for efficacy assessment in drug development. PLoS One. 2019; 14(7):e0218014. 10.1371/journal.pbio.1001863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bronstad A, Newcomer CE, Decelle T, et al. Current concepts of harm-benefit analysis of animal experiments – Report from the AALAS-FELASA working group on harm-benefit analysis-part 1. Lab Anim 2016;50(1 Suppl);1–20. 10.1177/0023677216642398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harrison, R. Phase II and phase III failures: 2013–2015. Nat Rev Drug Discov. 2016; 15, 817–818. 10.1038/nrd.2016.184 [DOI] [PubMed] [Google Scholar]
- 18. Wu S, Fernando K, Allerton C, et al. Reviving an R&D pipeline a step change in the phase II success rate. Drug Discov Today 2021:26(2):308–314. 10.1016/j.drudis.2020.10.019 [DOI] [PubMed] [Google Scholar]
- 19. Mohanan S, Maguire S, Klapwijk J, et al. Evolving the role of discovery-focused pathologists and comparative scientists in the pharmaceutical industry. Toxicol Pathol. 2019; 47(2):121–8. 10.1177/0192623318821333 Epub 2019 Jan 16. PMID: 30651043. [DOI] [PubMed] [Google Scholar]
- 20. de Visser SJ, Cohen AF, Kenter MJH. Integrating scientific considerations into R&D project valuation. Nat Biotechnol. 2020; 38, 14–18. 10.1038/s41587-019-0358-x [DOI] [PubMed] [Google Scholar]
- 21. Voelkl B, Altman NS, Forsman A, et al. Reproducibility of animal research in light of biological variation. Nature Reviews Nueroscience. 2020; 21:384–93. 10.1038/s41583-020-0313-3. [DOI] [PubMed] [Google Scholar]
- 22. Zhang YZ, Li YY. Inflammatory bowel disease: pathogenesis. World J Gastroenterol. 2014; 20(1):91–9. 10.3748/wjg.v20.i1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones-Hall YL, Grisham MB. Immunopathological characterization of selected mouse models of inflammatory bowel disease: comparison to human disease. Pathophysiology. 2014; 21(4):267–88. 10.1016/j.pathophys.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 24. Ye Y, Pang Z, Chen W, et al. The epidemiology and risk factors of inflammatory bowel disease. Int J Clin Exp Med. 2015; 8(12):22529–42 PMID: 26885239. [PMC free article] [PubMed] [Google Scholar]
- 25. Kaplan GG. The global burden of IBD: From 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015; 12(12):720–7. 10.1038/nrgastro.2015.150. [DOI] [PubMed] [Google Scholar]
- 26. Nguyen TL, Vieira-Silva S, Liston A, et al. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015; 8(1):1–16. 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004; 172(5):2731–8. 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 28. Gibbons DL, Spencer J. Mouse and human intestinal immunity: same ballpark, different players; different rules, same score. Mucosal Immunol. 2011; 4(2):148–57. 10.1038/mi.2010.85. [DOI] [PubMed] [Google Scholar]
- 29. Lindebo Holm T, Poulsen SS, Markholst H, Reedtz-Runge S. Pharmacological Evaluation of the SCID T Cell Transfer Model of Colitis: As a Model of Crohn's Disease. Int J Inflam. 2012; 2012:412178. 10.1155/2012/412178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Davenport CM, McAdams HA, Kou J, et al. Inhibition of pro-inflammatory cytokine generation by CTLA4-Ig in the skin and colon of mice adoptively transplanted with CD45RBhi CD4+ T cells correlates with suppression of psoriasis and colitis. Int Immunopharmacol. 2002; 2(5):653–72. [DOI] [PubMed] [Google Scholar]
- 31. Ikenoue Y, Tagami T, Murata M. Development and validation of a novel IL-10 deficient cell transfer model for colitis. Int Immunopharmacol. 2005; 5(6):993–1006. [DOI] [PubMed] [Google Scholar]
- 32. O'Connor W Jr, Kamanaka M, Booth CJ, et al. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat Immunol. 2009; 10(6):603–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leppkes M, Becker C, Ivanov II, et al. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 2009; 136(1):257–67. [DOI] [PubMed] [Google Scholar]
- 34. Wedebye Schmidt EG, Larsen HL, Kristensen NN, et al. TH17 cell induction and effects of IL-17A and IL-17F blockade in experimental colitis. Inflamm Bowel Dis. 2013; 19(8):1567–76. [DOI] [PubMed] [Google Scholar]
- 35. Yen D, Cheung J, Scheerens H, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006; 116(5):1310–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Noguchi D, Wakita D, Tajima M, et al. Blocking of IL-6 signaling pathway prevents CD4+ T cell-mediated colitis in a T(h)17-independent manner. Int Immunol. 2007; 19(12):1431–40. [DOI] [PubMed] [Google Scholar]
- 37. Powrie F, Leach MW, Mauze S, et al. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int Immunol. 1993; 5(11):1461–71. 10.1093/intimm/5.11.1461. [DOI] [PubMed] [Google Scholar]
- 38. Henderson VC, Kimmelman J, Fergusson D, et al. Threats to validity in the design and conduct of preclinical efficacy studies: a systematic review of guidelines for in vivo animal experiments. PLoS Med. 2013; 10(7):e1001489. 10.1371/journal.pmed.1001489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. NIH Advisory Committee to the Director Final Report: ACD Working Group on Enhancing Rigor , Transparency, and Translatability in Animal Research. NIH 2021. https://acd.od.nih.gov/documents/presentations/06112021_RR-AR%20Report.pdf.
- 40. Percie du Sert N, Hurst V, Ahluwalia A, et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. PLoS Biol. 2020; 18(7):e3000410. 10.1371/journal.pbio.3000410 eCollection 2020 Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


