Abstract
Mating dynamics follow from natural selection on mate choice and individuals maximizing their reproductive success. Mate discrimination reveals itself by a plethora of behaviours and morphological characteristics, each of which can be affected by pathogens. A key question is how pathogens affect mate choice and outcrossing behaviour. Here we investigated the effect of Orsay virus on the mating dynamics of the androdiecious (male and hermaphrodite) nematode Caenorhabditis elegans. We tested genetically distinct strains and found that viral susceptibility differed between sexes in a genotype‐dependent manner with males of reference strain N2 being more resistant than hermaphrodites. Males displayed a constitutively higher expression of intracellular pathogen response (IPR) genes, whereas the antiviral RNAi response did not have increased activity in males. Subsequent monitoring of sex ratios over 10 generations revealed that viral presence can change mating dynamics in isogenic populations. Sexual attraction assays showed that males preferred mating with uninfected rather than infected hermaphrodites. Together our results illustrate for the first time that viral infection can significantly affect male mating choice and suggest altered mating dynamics as a novel cause benefitting outcrossing under pathogenic stress conditions in C. elegans.
Keywords: Caenorhabditis elegans, host–virus interactions, mating dynamics, transcriptomics
1. INTRODUCTION
Sexual reproduction is the dominant reproductive strategy in the animal kingdom allowing for rapid adaptation to both abiotic and biotic selective pressures (Butlin, 2002; Lehtonen et al., 2012). Pathogens are the main biotic drivers of evolution by forcing hosts to constantly adapt to the peril of infection and can benefit sexual over asexual reproduction (Bell, 1982; Hamilton, 1980; Jaenike, 1978; Morran et al., 2011). In addition to selection as a result of direct effects (changed survival and/or reproductive output), pathogens may interfere with mating systems in many other ways, for instance by influencing sexual characteristics such as chemical cues, behaviour and courtship. Such interferences have been recorded for a range of species showing that frogs, mice, flies and humans are less likely to mate with infected conspecifics (Kavaliers & Choleris, 2018; Kiesecker et al., 1999). Furthermore, physiological, molecular and genetic differences between the sexes can underlie differences in pathogen susceptibility, thereby potentially favouring one sex over another in an infected population (Gipson et al., 2019; Klein & Flanagan, 2016; Scully et al., 2020). Although theory shows that sexual reproduction is evolutionary stable when both sexes are equally present (Fisher, 1930), skewed sex ratios are often observed in nature. Pathogen‐induced interference in populations can help explain unbalanced sex ratios (Dyson, 2012; Engelstädter & Hurst, 2009; Klein & Flanagan, 2016; Lynch et al., 2018; Masri et al., 2013; Morran et al., 2011).
The androdiecious (male and hermaphrodite) nematode Caenorhabditis elegans and its natural associated pathogens provide a versatile model to study sex‐dependent host–pathogen interactions (Cutter et al., 2019). C. elegans can reproduce both by outcrossing and self‐fertilization leading to variable male–hermaphrodite ratios. Males (XO sex determination) arise only rarely after spontaneous X chromosome nondisjunction with an estimated frequency of 0.1%–0.4% in the laboratory (Teotónio et al., 2006). Although successful outcrossing raises male frequencies as mated hermaphrodites produce 50% male and 50% hermaphrodite (XX sex determination) offspring, ineffective mating behaviour typically dwindles male frequencies (Borne et al., 2017; Chasnov et al., 2007; Garcia et al., 2007; Kleemann & Basolo, 2007; Palopoli et al., 2008; Teotónio et al., 2006). Yet, elevated outcrossing levels benefit bacteriallly infected C. elegans populations among others by having genetically more resistant offspring (Lynch et al., 2018; Masri et al., 2013; Morran et al., 2011).
Associated intracellular pathogens of C. elegans include microsporidia, oomycetes and a virus (Félix et al., 2011; Osman et al., 2018; Zhang et al., 2016). These pathogens induce a molecular defence mechanism, called the intracellular pathogen response (IPR), that is distinct from the response to bacterial infections (Chen et al., 2017; Panek et al., 2020; Reddy et al., 2017, 2019; Sowa et al., 2019). The IPR involves different expression of 80 IPR genes and is controlled by the IPR inhibitor pals‐22 and IPR activator pals‐25 (Reddy et al., 2019). Additionally, viral infection is counteracted by RNA interference (RNAi) and uridylation leading to a degradation of viral RNA (Ashe et al., 2013; Coffman et al., 2017; Félix et al., 2011; Le Pen et al., 2018; Sterken et al., 2014; Tanguy et al., 2017). Males differ largely from hermaphrodites in various molecular pathways (Cook et al., 2019; Gerstein et al., 2014; White et al., 2007), but it is currently unknown if nematodes show sex‐dependent resistance against intracellular pathogens such as viruses and if stress caused by these pathogens affects mating dynamics.
For the first time we studied infection responses for both sexes in different wild types of C. elegans to the naturally occurring intracellular pathogen Orsay virus (OrV). The nonlethal OrV is a positive‐sense, single‐stranded RNA virus (family Nodaviridae) that infects intestinal cells (Félix et al., 2011). Although relatively few individuals within most populations of hermaphrodites become infected (up to 40% for the highly susceptible wild type JU1580), infected cells show severe morphological effects leading to fitness disadvantages (Ashe et al., 2013; Félix et al., 2011; Frézal et al., 2019). We found that viral infection affects male mating choice in C. elegans and suggest mating dynamics as a novel cause benefitting outcrossing under pathogenic stress conditions in C. elegans.
2. MATERIALS AND METHODS
2.1. Caenorhabditis elegans strains and culturing
Caenorhabditis elegans wild isolate strains N2, CB4856 and JU1580 were used. Mutant strain RB859 daf‐22(ok693) was obtained from the Caenorhabditis Genetic Center. Strain ERT54 (jyIs8[pals‐5p::GFP, myo‐2p::mCherry] X) was a kind gift from Emily Troemel (Reddy et al., 2017, 2019). The strains were grown on nematode growth medium (NGM) plates seeded with Escherichia coli OP50 as a food source (Brenner, 1974). Male populations were maintained weekly by crossing adult males with L4 hermaphrodites in a 3:1 ratio. Experiments were started by transferring a starved population to a fresh NGM dish followed by bleaching after the adults were egg‐laying (3 days after transfer at 20°C, 4 days after transfer at 16°C) (Brenner, 1974). All experiments were performed at 20°C unless indicated otherwise.
2.2. Orsay virus stock and mock lysate
Standard used OrV stock was prepared as previously described (Félix et al., 2011). One modification was made, namely to use modified NGM medium with 34 g L–1 agar (mNGM) plates to grow the nematodes as this prevented the nematode from burrowing into the agar. Briefly, persistently infected JU1580 starved nematodes were washed off 100 9‐cm‐diameter mNGM plates and flash‐frozen in liquid nitrogen. The lysate was collected and filtered through a 0.2‐µm filter and stored at −80°C until used. The infectivity of the OrV stock was tested by a dose–response experiment in JU1580 (Sterken et al., 2014).
For the choice‐assay experiments additional OrV and mock stocks were created. Mock stocks were used in these experiments, because then both mock and OrV stocks contain nematode lysate that was expected to affect attraction of males. Mock and OrV stocks were made in parallel to minimize batch effects by the nematodes used. All animals were grown at 16°C. First, OrV JU1580 mock stock was prepared by bleaching a persistently infected JU1580 population to remove the OrV. After the bleached population was starved it was transferred to 100 new 9‐cm mNGM plates. Subsequently, the same protocol as for obtaining the OrV stock was followed (Félix et al., 2011; Sterken et al., 2014). Second, N2 and RB859‐based mock and OrV stocks were made by infecting and lysing N2 or RB859 nematodes instead of JU1580 nematodes. To infect the N2 or RB859 nematodes, 100 µl of previously obtained JU1580 OrV stock was added to a 9‐cm mNGM plate with a proliferating N2 population. Once this population was starved, the plate was divided over 100 fresh 9‐cm mNGM plates of which the populations were flash‐frozen upon starvation. The standard protocol was then followed (Félix et al., 2011; Sterken et al., 2014). Third, young adult mock and OrV stocks were made by infecting N2 or RB859 L1 nematodes collected from 10 9‐cm mNGM plates (24 hr post bleaching) with either 200 µl RB859 mock or OrV stock (obtained from starved populations as described above) according to the protocol described by Sterken et al., (2014). Populations were washed five times with M9 (instead of the standard three times) after exposure to the mock or OrV stock for 1 hr. Populations were collected when the first eggs appeared on the plate (90 hr post bleaching for N2 nematodes, 104 hr post bleaching for RB859 nematodes). The “young adult N2 and RB859 stocks” were used in the daf‐22 choice assay experiment.
2.3. Orsay virus infection experiments
For the single‐sex experiments, male and hermaphrodite nematodes were separated ~44 hr post bleaching by placing them on fresh plates. After 200 nematodes were handpicked per plate the populations were washed into an Eppendorf tube and infected 48 hr post bleaching as described previously (Sterken et al., 2014). Mock infections were performed by adding M9 instead of OrV unless indicated that a mock lysate (lysate of uninfected nematodes) was used. The experiment was performed in technical duplicate, and therefore every sample contained ~400 nematodes. The samples for small RNA (sRNA) sequencing contained ~600 nematodes. However, for some experiments, not enough males (fewer than 400) were present on the plates to pick males from and in these cases a total of 200 male nematodes were picked per sample (and also for the hermaphrodite samples within the same experimental batch). Infected populations were collected 30 hr post infection (78 hr post bleaching). Viral loads were obtained as described by Sterken et al. (2014).
As the viral loads for single‐sex infections did not follow normal distributions and some samples were zero‐inflated, a nonparametric bootstrap approach was used to estimate sampling distributions and perform statistical tests (Kulesa et al., 2015). Bootstrap samples were drawn 10,000,000 times and used to calculate the one‐sided bootstrap test statistic (the resampled difference in mean viral load of males and hermaphrodites). Each bootstrap sample was drawn by randomly selecting a set of n experimentally observed viral loads (n being the number of observations made for both males and hermaphrodites: 20 for N2 and CB4856, 16 for JU1580) from the full data set with replacement (meaning that the same observed viral load can be drawn multiple times). Every bootstrap sample draws half of the observations from male samples and half of the observations from hermaphrodite samples. The bootstrapped p‐value was calculated by dividing the number of bootstrapped test statistics that were greater than the experimentally observed test statistic by the total number of bootstrap test statistics of 10,000,000. Bootstrap analysis was performed in r (version 4.0.2).
For the mixed‐sex experiments, 30 L4 hermaphrodites and 90 adult males were transferred to a plate that was incubated overnight. Hermaphrodites in the absence of males were picked as a control from the same starting populations. Subsequently, 30 male‐ or self‐fertilized nematodes were transferred to a new plate where they were allowed to lay eggs for 6 hr before the hermaphrodites were removed from the plate. The resulting eggs were incubated for 20 hr after which they were infected with OrV as previously described (Sterken et al., 2014). Nematodes were collected as young adults 30 hr post infection (56 hr post start of egg‐laying). Viral loads were obtained as described by Sterken et al. (2014) and mixed and hermaphrodite‐only samples were compared with a paired t test with equal variances.
2.4. RNA isolation
RNA isolation was performed using the Maxwell 16 AS2000 instrument with the Maxwell 16 LEV simply RNA Tissue Kit (Promega). The recommended protocol was followed, except for adding 10 mg of proteinase K per sample after addition of the lysis buffer. Lysis was performed in a shaker for 10 min at 65°C (1,000 rpm) (Eppendorf). RNA sample quantity and quality were measured by using NanoDrop (Thermo Scientific).
2.5. Gene expression measurements by RT‐qPCR
cDNA was made from 1 µg of RNA using the GoScript Reverse Transcriptase kit (Promega) and following the recommended protocol with random hexamers (Thermo Scientific). Gene‐expression measurements were performed on cDNA of ~100 untreated male or hermaphrodite nematodes (collected 48 hr post bleaching). Samples were collected for five biological replicates. Gene expression was quantified by RT‐qPCR (real‐time quantitative PCR) using custom designed primers (Table S7) that overlap at least one exon–exon border to prevent unintended amplification of any remaining DNA. RT‐qPCR was performed on the MyIQ using iQ SYBR Green Supermix (Bio‐Rad) and the recommended protocol. Primer efficiencies were checked by testing dilution ranges for N2 and JU1580 male populations. Correct primer annealing to the CB4856 genome was tested in silico (Thompson et al., 2015). Gene expression in each sample was quantified for the gene of interest and two reference genes (Y37E3.8 and rpl‐6) in technical duplicate. Gene expression was determined according to Sterken et al. (2014) and the effect of sex and strain were determined per gene via a linear model:
with Y being the log2 gene expression. Y was explained over sex (S; male or hermaphrodite), genotype (G; N2, CB4856 or JU1580), the interaction between sex and strain. and the error term ε. When gene expression for a certain combination of gene and sample remained under the detection limit by RT‐qPCR (Ct > 40) that measurement was excluded from the analysis.
2.6. Whole‐genome gene expression by microarray
For whole‐genome gene‐expression measurements, microarrays were used on eight biological replicates of N2 mock‐treated or OrV‐infected males and hermaphrodites (obtained via the “single‐sex” infection protocol). Only samples where the OrV could be detected by RT‐qPCR were used for this analysis. Gene Expression Microarray 4X44K C. elegans V2 slides were used for the microarrays (Agilent Technologies). Microarrays were performed based on the “Two‐Color Microarray‐Based Gene Expression Analysis; Low Input Quick Amp Labeling” protocol, version 6.0 from Agilent Technologies, starting from step five. The microarrays were scanned (Agilent High Resolution C Scanner) and extracted using the Agilent Feature Extraction Software (version 10.7.1.1) with the recommended settings. Normalization was performed separately per sex using the limma package in r (version 4.0.2) (Ritchie et al., 2015). The Loess method was used for within‐array normalization and the Quantile method for between‐array normalization (Ritchie et al., 2015; Smyth & Speed, 2003; Zahurak et al., 2007). The obtained single channel normalized intensities were log2 transformed. The log2 intensities were used in further analysis using the package tidyverse (1.3.0) in r (version 4.0.2) (Wickham et al., 2019). Gene expression of control genes that should be either similar (rpl‐6) or differentially (tra‐1, fem‐3) expressed between the sexes were checked and confirmed the validity of the data.
Basal gene expression differences in IPR gene expression were determined by selecting transcriptional data of the mock‐treated samples (log2 intensities) and running the following linear model:
where Y is the log2 normalized intensity of spot i (1, 2,…, 45,220) that was explained over sex (S, male or hermaphrodite), and the error term ε. A significance threshold was set by the p.adjust function, using the Benjamini & Hochberg correction (FDR < 0.05) (Benjamini & Hochberg, 1995).
The effect of treatment on the IPR gene response was analysed by selecting transcriptional data (log2 intensities) for the 80 IPR genes (as described by Reddy et al., 2019) and running the following linear model per sex:
where Y is the log2 normalized intensity of spot i (1, 2,…, 109) that was explained over treatment (T, mock or infected), and the error term ε. A common significance threshold was determined after combining the data sets of both sexes and using the p.adjust function with the Benjamini & Hochberg correction (FDR < 0.05) (Benjamini & Hochberg, 1995).
2.7. Small RNA sequencing
Samples for sRNA sequencing contained at least 1 µg of RNA with a concentration >50 ng µl–1. One biological replicate per sample was investigated. Samples were sequenced by DNBseq Small RNA sequencing (BGI) using either a 5′‐dependent or 5′‐independent protocol. The protocol type refers to ligation of the 5′‐adaptor. The 5′‐dependent protocol detects only monophosphorylated sRNA strands, whereas the 5′‐independent protocol also allows detection of sRNA strands that had 5′‐triphosphate or 5′‐capped modification. The read data were aligned to the OrV genome (GenBank: HM030970.2 and HM030971.2) using bowtie2 via the public server at usegalaxy.org to map the data (Afgan et al., 2018; Langmead & Salzberg, 2013). Reads that aligned to the OrV genome were subsequently analysed in r (version 4.0.2). Reads with a mapping quality lower than 40 were excluded from analysis. Spatial clustering of 23‐nt sRNA hotspots was determined for bins of 20 nt. As hotspots follow a binomial distribution, differences were determined via the Poisson test in r (version 4.0.2) after correcting for multiple testing via the p.adjust function with the Benjamini & Hochberg correction (FDR < 0.05) (Benjamini & Hochberg, 1995).
2.8. Pharyngeal pumping rate counts
Pharyngeal pumping rates were counted for L4 (48 hr post bleaching) and young adult (72 hr post bleaching) N2, CB4856 and JU1580 nematodes using a Leitz Greenough microscope and a counter. The pumping rates were counted twice for 20 s per nematode and the average of both measurements was used in further analysis. Pumping rate counting was performed at room temperature (~20°C) and the experiment was performed over five independent biological experiments counting six nematodes of each genotype/sex per experiment. For these experiments, the OrV was not present on the plates. The data were analysed by the two‐sample Wilcoxon signed‐rank test in r (version 4.0.2).
2.9. Fluorescent bead accumulation
To estimate the volume of food intake, nematodes were either exposed to the fluorescent beads in liquid (with OrV present) or whilst feeding on the plate (without OrV present). The plate feeding assay was performed as previously described (Bakowski et al., 2014). In short, nematodes were incubated on the plate for 30 min at 25°C before feeding was halted by placing the plates on ice. The NGM plates were incubated with a mixture of 30 µl E. coli OP50 and 5 µl of Fluoresbrite Polychromatic Red Microspheres (1.00‐µm particles, 4.55 × 1010 particles ml–1) (Polysciences, Inc.). The liquid exposed nematodes were incubated in a mixture of 30 µl E. coli OP50, 5 µl of Fluoresbrite Polychromatic Red Microspheres (1.00‐µm particles, 4.55 × 1010 particles ml–1) (Polysciences, Inc.), 105 µl 0.1% NGM and 15 µl OrV stock for 1 hr at room temperature to mimic the OrV infection assay. Then nematodes were washed once with 0.25 mm levamisole in M9 to stop the feeding and remove the surplus of fluorescent beads. For both assays nematodes were fixed in 0.25 mm levamisole in M9 and nematodes were imaged using an Axio Observer Z1m inverted microscope (Zeiss). The exposure time for the red fluorescent channel (λ = 453 nm) was fixed at 440 ms. These experiments were performed independently five times. Each biological replicate contained between 10 and 50 nematodes for imaging.
Images were analysed using the imagej software (version 1.51f, National Institutes of Health). The total surface area of the nematode was selected by using the Wand Tool on the brightfield image. Images for which the surface area of the nematode could not be selected due to nearby presence of, for example, a bubble were excluded from the analysis. Subsequently, the area size and minimum, maximum and mean fluorescence were determined for the selected area. The data were tested for normality by a Kruskal–Wallis test and subsequently analysed by the two‐sample Wilcoxon signed‐rank test in r (version 4.0.2). The normalized fluorescence intensity represents the average amount of fluorescence in the nematode (controlling for the size of the nematode) and the absolute fluorescence represents all fluorescent signal measured in that nematode.
2.10. Male frequency counts
Before the start of the experiment, 30 male and 10 hermaphrodite L4 nematodes were crossed to obtain isogenic male populations for each genotype (N2, CB4856 and JU1580) and grown until starved at 20°C. From the same plate, 10 hermaphrodites were picked that were also grown to starvation. Starved plates were transferred onto 10 9‐cm mNGM plates per genotype/sex combination and incubated at 20°C. Three days later nematodes were bleached and again incubated at 20°C. When nematodes reached the L4 stage (48 hr post bleaching), 15 males (from the mixed male–hermaphrodite plates) and 135 hermaphrodites (from the hermaphrodite‐only plates) per genotype/sex combination were placed onto a new plate. For each genotype/sex combination, four plates were made of which two were mock infected by adding 100 µl mock stock lysate and two were OrV infected by adding 100 µl OrV stock lysate. The experiment was performed in biological triplicate, resulting in six replicates per genotype/treatment combination. Biological replicates were started on different days from independently grown starting populations. After preparation of the plates the experiment was blinded by removing the name tags and replacing them with a number by a colleague not involved in the experiment. The plates were incubated at 20°C throughout the experiment.
Every 3 days the number of adult males and total adult nematodes was counted using a dissecting microscope. Counting started 6 days after bleaching and continued for 10 transfers. Plates were divided into eight parts with the use of a pen and parts were counted until a total of at least 100 adult individuals were observed. After counting, the nematodes were washed off the plate in 2 ml M9 and for each population ~100 nematodes were transferred by pipetting to a fresh mNGM plate. The remaining nematodes were pelleted and flash frozen for RNA isolation after transfer number 1, 5 and 10 and these samples were checked for OrV presence as a control. Based on this analysis two technical replicates of OrV‐infected CB4856 were excluded from the analysis as they appeared to have cleared the infection.
After the counting, the samples were unblinded and analysed using a prewritten r script. Statistical analyses were performed using a linear mixed effect model for each genotype separately, explaining the observed male frequency over transfer, treatment and replicate. The analyses were performed in r (version 4.0.2) using the package nlme (Pinheiro et al., 2021) according to the following model:
where Y is the male frequency of replicate i (1, 2,…, 34) that was explained over treatment (T, mock or infected), transfer (Tr; 1, 2,…, 10), the interaction between treatment and transfer, and the error term ε based on the autocorrelation matrix of replicate i (1, 2,…, 34) given transfer T (1, 2,…, 10). The anova function was used in r (version 4.0.2) to compute the analysis of variance (Chambers & Hastie, 1992).
2.11. Choice assay
Choice assay experiments were based on chemical and bacterial choice assays described previously (Bargmann et al., 1993; Zhang et al., 2005). Two droplets of 15 µl E. coli OP50 were placed on a 6‐cm NGM dish after which the plates were incubated at room temperature for 2 days (~20°C). Before the start of the assay either 15 µl OrV stock or 15 µl mock stock lysate was pipetted on top of the droplets. After the plates had dried, 15 N2, CB4856 or JU1580 L4 males or hermaphrodites (48 hr post bleaching) were placed in the middle between the droplets. After 2 and 24 hr the number of nematodes in each droplet was noted. Data were collected for five biological replicates. Per biological replicate the behaviour of 45 nematodes for each genotype/sex combination was tested using three separate plates with 15 nematodes each. The data in this experiment were analysed by combining the total counts of nematodes in the mock or OrV droplet and performing a Chi‐square test in r (version 4.0.2).
2.12. Mating assay
N2 male and ERT54 populations were synchronized by bleaching. 16 hr post synchronization either 50 µl OrV stock or M9 was added to an ERT54 population on a 9‐cm NGM plate. The, 48 hr post bleaching (hpb) N2 males were placed on a 9‐cm NGM plate without hermaphrodites. At the same time, (un)infected ERT54 nematodes were individually transferred to 3‐cm NGM plates. Next, young adult ERT54 nematodes (62 hpb) were inspected for pals‐5::GFP expression under an Axio Observer Z1m inverted microscope (Zeiss). Animals with high intestinal pals‐5::GFP expression were selected as positively infected nematodes, whereas animals without this expression were not used in the assay. Subsequently, N2 males from the male‐only plate were added to either a mock‐treated or an OrV‐infected ERT54 individual. Males were placed at 0.9 cm distance from the middle of the bacterial food droplet. Directly after adding the male to the plate, filming for 20 min started. Movies captured one frame per second and were made at room temperature (~20°C) using four USB cameras simultaneously (Conrad, cat. no. 191341–62) using the video capturing program virtualdub (version 1.10.2) and imagej (version 1.52) (Schindelin et al., 2012; Schneider et al., 2012). Filming was performed using 62‐ to 66‐hr‐old nematodes that had not yet started egg‐laying.
Movies were inspected manually and for each of the movies three characteristics were reported: (i) the time it took before the male entered the bacterial droplet, (ii) the time it took before the male first touched the hermaphrodite and (iii) the time it took before mating started. A mating was noted when the male's tail stayed attached to the hermaphrodite's vulva for at least three frames and mating attempts stopped afterwards (based on results of LeBoeuf et al., 2014). The time the mating started was noted as the time to mating. Statistical analyses of the data were performed with a Student's t test assuming equal variances.
Hermaphrodite movements were investigated for 30 s before and 90 s after the first contact between male and hermaphrodite. A longer time frame to measure the velocity of hermaphrodites after contact with the male was chosen, because males did often not directly start mating attempts and thus it may take longer than 30 s to see an effect of male mating attempts on the hermaphrodite velocity. Tracking was performed using the plugin manual tracking in imagej (version 1.52) (Schindelin et al., 2012; Schneider et al., 2012). The movement data were subsequently analysed in r (version 4.0.2). The velocity of the nematodes was compared using Student's t test with equal variances and the correlation between (i) the time passed between first contact of the nematodes and mating and (ii) the average velocity of the hermaphrodite was calculated by a generalized linear model:
where Y is the time between the first contact of male and hermaphrodite and mating (1, 2,…, 41) that was explained over the average velocity of the hermaphrodite (V), and the error term ε.
3. RESULTS
3.1. Viral susceptibility differs between sexes in a genotype‐dependent manner
Different genotypes of Caenorhabditis elegans hermaphrodites vary in susceptibility to OrV (Ashe et al., 2013; Félix et al., 2011), but the susceptibility of C. elegans males has not been investigated. To compare the viral susceptibility of C. elegans males and hermaphrodites, both sexes were infected for three genetically distinct genotypes: N2, CB4856 and JU1580 (Figure 1a; Table S1). N2 is the common reference genotype. Compared to N2, CB4856 is genetically highly distinct and CB4856 hermaphrodites are less susceptible to OrV after short viral exposure (Sterken et al., 2021; Thompson et al., 2015). JU1580 has higher viral susceptibility than N2, because of a deficiency in the antiviral RNAi pathway of JU1580 (Ashe et al., 2013; Félix et al., 2011; Sterken et al., 2014). We found that N2 males had lower viral loads than hermaphrodites (Figure 1b) (bootstrap, p = .025). CB4856 males and hermaphrodites were often not successfully infected and had similar viral loads (Figure 1b) (bootstrap, p = .10). JU1580 males and hermaphrodites were both highly susceptible to viral infection (Figure 1b) (bootstrap, p = .24). Since the largest difference between males and hermaphrodites was seen for the N2 strain, OrV susceptibility was also tested in mixed male and hermaphrodite N2 populations. The mixed‐sex populations contained around 30%–40% of males, contrary to a hermaphrodite‐only population. As expected, male presence had a tendency to lower the viral load in the population (t test, p = .07) (Figure S1; Table S1). Notably, all mixed‐sex populations were successfully infected, contrary to the male‐only populations.
FIGURE 1.
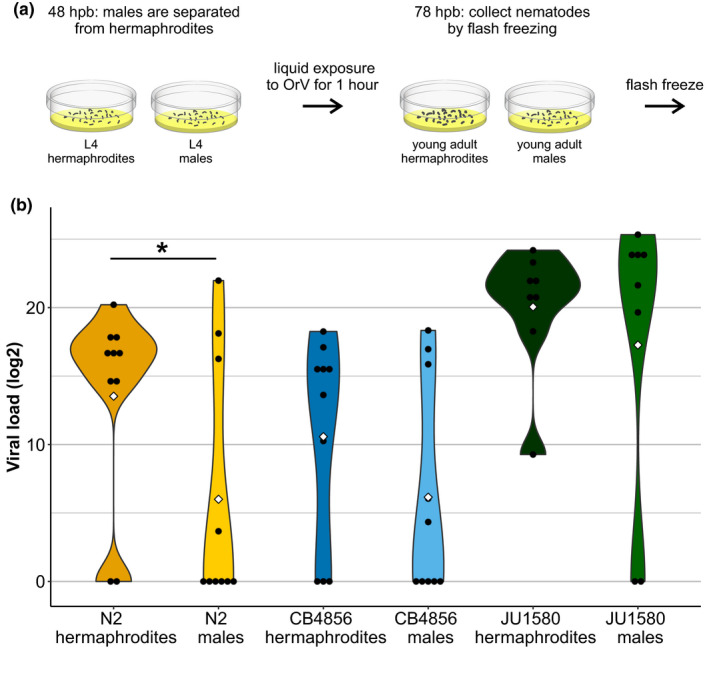
Sex‐specific viral susceptibility. (a) Male and hermaphrodite nematodes were separated 48 hr post bleaching. Subsequently, single‐sex populations were exposed to OrV in liquid for 1 hr and grown on plates until 78 hr post bleaching. Then, nematode populations were collected and viral loads were obtained by RT‐qPCR. (b) Viral loads obtained for hermaphrodites and males of the strains N2, CB4856 and JU1580. Each dot represents a biological replicate (n = 10 for N2 and CB4856, n = 8 for JU1580). The white diamond shows the mean per combination of sex and strain. Statistically significant differences are indicated by an asterisk (bootstrap, p < .05)
Since OrV is taken up whilst nematodes feed, sex‐based differences in ingestion may affect viral loads. Therefore, we quantified both the rate and volume of food intake in L4 and young adult males and hermaphrodites (Figure S2a; Table S2). The rate of food intake was measured by counting pumping rates of the pharynx, a neuromuscular feeding organ. The sex of the nematodes did not affect the food intake rate in these life stages (p > .05) (Figure S2b). To quantify the feeding volume, fluorescent beads were mixed with Escherichia coli OP50 and fed to the nematodes (Bakowski et al., 2014). In all cases the normalized fluorescent signal was at least as high in the males as in the hermaphrodites, showing that the average amount of ingestion within the nematodes was similar or males had higher ingestion (Figure S2c). The absolute amount of fluorescent signal was higher for adult hermaphrodites than for adult males, which corresponds to their larger body size (t test, p < .05) (Figure S2d). In conclusion, the ingestion rates cannot explain observed differences in viral susceptibility observed between sexes and strains.
3.2. Antiviral activity in males and hermaphrodites
Males and hermaphrodites could have a different molecular response to OrV infection. Both the RNAi activity and IPR were measured in males and hermaphrodites infected with OrV. Potent RNAi activity against OrV resulting in viral small interfering RNA (siRNA) production has been described for hermaphrodites (Ashe et al., 2013; Coffman et al., 2017), but males can differ in RNAi efficacy to other environmental triggers (Bezler et al., 2019). To explore the hypothesis that males may differ in RNA, infected N2 male and hermaphrodite populations were used for sRNA sequencing. Both sexes showed similar sRNA characteristics for the main RNAi products: primary 23‐nt long siRNAs (Figure S3a,b) and secondary 22‐nt (antisense) siRNAs with a G at the 5′‐end, because they are present in a similar ratio (Figure S3b). However, also for the samples showing a similar viral load for both sexes, the number of reads that aligned to the viral genome was lower in males (n total = 20,656,692 n viral = 57,390 [0.27%]) than in hermaphrodites (n total = 32,667,244, n viral = 420,484 [1.28%]). This contradicts that males would have a more potent RNAi response and is in line with research demonstrating that hermaphrodites produce more secondary siRNA products than males after environmental RNAi (Bezler et al., 2019). Furthermore, we compared spatial clustering of 23‐nt sRNAs (primary siRNAs) on the viral genome. Roughly 40% of the hotspots (39% for OrV RNA segment 1 and 42% for OrV RNA segment 2) were equal between males and hermaphrodites. Of the hotspots that differed between both sexes, 67% were only present in hermaphrodites (62% for OrV RNA segment 1 and 72% for OrV RNA segment 2). That, together with the lower occurrence of male siRNA reads, contradicts that higher RNAi efficacy would lower the susceptibility of males to OrV infection (Figure S3c).
As the IPR changes the nematode's gene expression, IPR activity was investigated by measuring gene expression in mock‐treated and OrV‐infected N2 adults. We selected samples with positively confirmed OrV infection by RT‐qPCR. Male samples had a lower average viral load than hermaphrodite samples (Figure S4a). Adult males had lower expression of IPR regulators pals‐22 and pals‐25 than hermaphrodites (Figure S4b). Under mock conditions, expression differed for 26 IPR genes, of which 24 were more highly expressed in males (Figure S4b; Table S3). OrV infection led to upregulation of 26 IPR genes for hermaphrodites and six IPR genes for males (Table S3). We also measured expression of the IPR regulators pals‐22 and pals‐25 and three IPR genes, pals‐6, pals‐14 and eol‐1, by RT‐qPCR in untreated L4 nematodes (for N2, CB4856 and JU1580) to explore IPR expression upon viral exposure in our assay (Figure S5; Table S3). We found that the IPR activator pals‐25 was more highly expressed in males at this stage (linear model, p = .002), suggesting other IPR genes (that are regulated by pals‐25) might also be upregulated. This would match data previously collected for the modENCODE project that showed N2 L4 males have enriched expression of multiple pals genes when compared to hermaphrodites (Gerstein et al., 2010; Leyva‐Díaz et al., 2017). Nevertheless, we did not detect increased expression for the limited set of (3/80) IPR genes we measured here (Figure S5). Together, our gene expression data collected for adults and the public modENCODE data set for L4 nematodes indicate that IPR gene expression is constitutively high for males, which may reduce viral loads throughout the course of infection or potentially protect from OrV infection.
3.3. Viral infection changes mating dynamics
The observed viral susceptibilities of males and hermaphrodites differed per strain (Figure 1). We investigated whether this genotype‐dependent viral susceptibility difference between the sexes could result in different mating dynamics per genotype. In particular, we hypothesized that N2 males, which are more resistant to OrV, could become more common in infected populations. This hypothesis was tested by monitoring the male frequency in mock‐treated and OrV‐infected populations for 10 generations. Starting populations contained 10% males of either N2, CB4856 or JU1580 nematodes in combination with hermaphrodites from the same genotype. Thus, genetic adaption of the nematodes via outcrossing to OrV was not possible in this single‐genotype set‐up.
After observing the nematodes for 10 generations, the frequency of N2 males did not increase in infected compared to mock‐treated populations, which contradicted our previously stated hypothesis (mixed linear model, p = .26) (Figure 2; Table S4). However, we found that for CB4856 more males were present in infected populations (mixed linear model, p = .046) (Figure 2), despite CB4856 males not showing a higher resistance to OrV than the hermaphrodites (Figure 1). For JU1580, viral presence increased male presence over time, contrary to male ratios in healthy populations (mixed linear model, p = .0070). After 10 generations, JU1580 males made up 25% of the infected population, compared to 14% in mock‐infected conditions and CB4856 males were 5% more prevalent in infected than in uninfected populations. Thus, we concluded that viral presence can change mating dynamics although it was not clear in which way male resistance contributes to this phenomenon.
FIGURE 2.
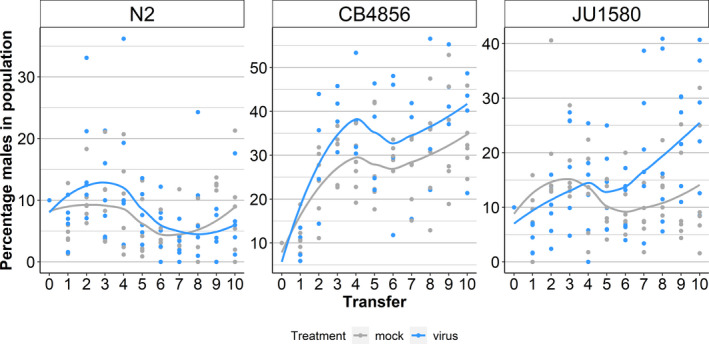
Male frequency over 10 generations. The percentage of males in isogenic N2, CB4856 and JU1580 populations over 10 generations (30 days). Counts from mock populations are shown in grey, and counts in OrV‐infected populations in blue. Each dot represents a technical replicate (plate with nematodes) and the technical replicates are equally divided over three biological replicates (having a different start date). The plotted lines represent the loess fit
3.4. Males prefer healthy over infected hermaphrodites modulating sex
To investigate if male C. elegans nematodes showed a preference for healthy hermaphrodites, a mating choice assay was designed. Healthy L4 males or hermaphrodites of the strains N2, CB4856 and JU1580 were given a choice between the lysate of mock‐treated or the lysate of OrV‐infected hermaphrodite populations (Table S5). Two types of lysate were used: lysates obtained from N2 or JU1580 nematodes. The number of nematodes on each spot was counted at 2 and 24 hr after placing them on the plate (nematode stage is L4 and young adult respectively) (Figure 3a). Our results show that in the L4 stage males and hermaphrodites did not have a preference for either the mock or virus spot on the plate (Figure 3b). However, in the young adult stage males of all three genotypes displayed a significant preference for the N2‐based mock lysate over the lysate of infected nematodes (Figure 3b). For the JU1580‐based lysate the same trend was observed but was not significant for JU1580 males (Figure 3). Hermaphrodites did not show a preference for the mock lysate (Figure 3). Together, these results suggested that adult males would be more attracted to healthy than infected hermaphrodites.
FIGURE 3.
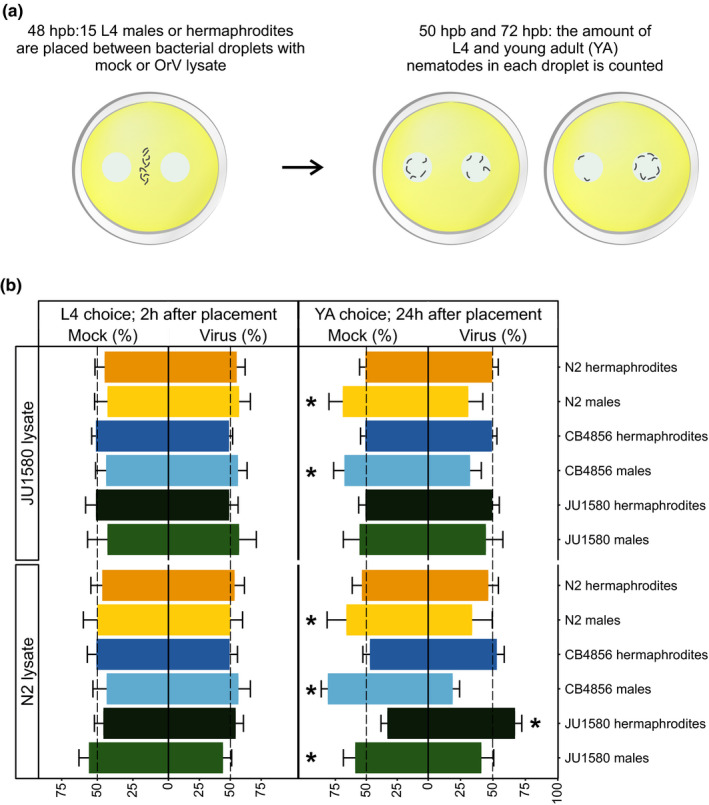
Choice assay between mock‐ and OrV‐infected nematode lysate. (a) Males and hermaphrodite of the genotypes CB4856, JU1580 and N2 were allowed to choose between a bacterial droplet containing the lysate of either a mock‐treated or infected population of mixed‐stage hermaphrodites. Fifteen male or hermaphrodite nematodes were placed in between these droplets and were observed at 2 and 24 hr after placement. (b) The percentage of nematodes that were observed in the mock or OrV droplet on the plate in the L4 and young adult (YA) stage (2 or 24 hr after placement, respectively). Only nematodes that were in either the mock or OrV droplet were counted. Mock and OrV lysates were made by lysing either N2 or JU1580 populations. The dotted line indicates where nematodes would not prefer either of the spots. Filled bars represent the percentage of nematodes in either the mock or OrV droplet and the error bars indicate standard error of the mean. In total, five biological replicates (nematodes derived from independent starting populations) containing three technical replicates (plates) with each plate containing 15 nematodes were counted. In total, 225 nematodes were observed per condition. Nematode populations that showed a significant preference for one of the spots are indicated with an asterisk (chi‐square test, p < .05)
Males may distinguish between infected‐ and mock‐treated lysates by pheromones released by the hermaphrodites. Therefore, we used RB859 daf‐22 mutants, which do not excrete male‐attracting pheromones (Von Reuss et al., 2012). We obtained the lysate of mock‐treated and OrV‐infected RB859 mutants for use in our choice assay (Figure S6; Table S5) (Von Reuss et al., 2012). Interestingly, the choice that males make between the lysates appears independent from pheromone signals, because we observed that adult males also favour uninfected over infected pheromone‐free lysate of daf‐22 nematodes (chi‐square test, p < .001) (Figure S6). Additionally, young adult hermaphrodites choose the mock lysate in this experiment (chi‐square test, p < .001), yet with a weaker preference than the males. Thus, males appear to have another way to distinguish uninfected from infected lysates.
To investigate if the preference for the lysate of healthy hermaphrodites translates into mating behaviour, we performed a mating assay. Males were placed with a single (un)infected, young adult hermaphrodite (Figure 4a). Subsequently, mating behaviour of N2 males towards mock‐treated or OrV‐infected nematodes was quantified by filming their movements for 20 min (Movie S1; Table S6). We recorded the time it took for males to (i) enter the bacterial droplet, (ii) touch the hermaphrodite and (iii) mate (Figure 4b–e). The males did not differ in the frequency of shown behaviours towards healthy or infected hermaphrodites (chi‐square test, p > .05) (Table S6). However, the videos show that when the hermaphrodite was infected, males took longer between the first contact and mating (t test, p = .028) (Figure 4d). The overall process from entering the bacterial droplet to mating took 50% longer for infected than foruninfected nematodes (t test, p = .006) (Figure 4e).
FIGURE 4.
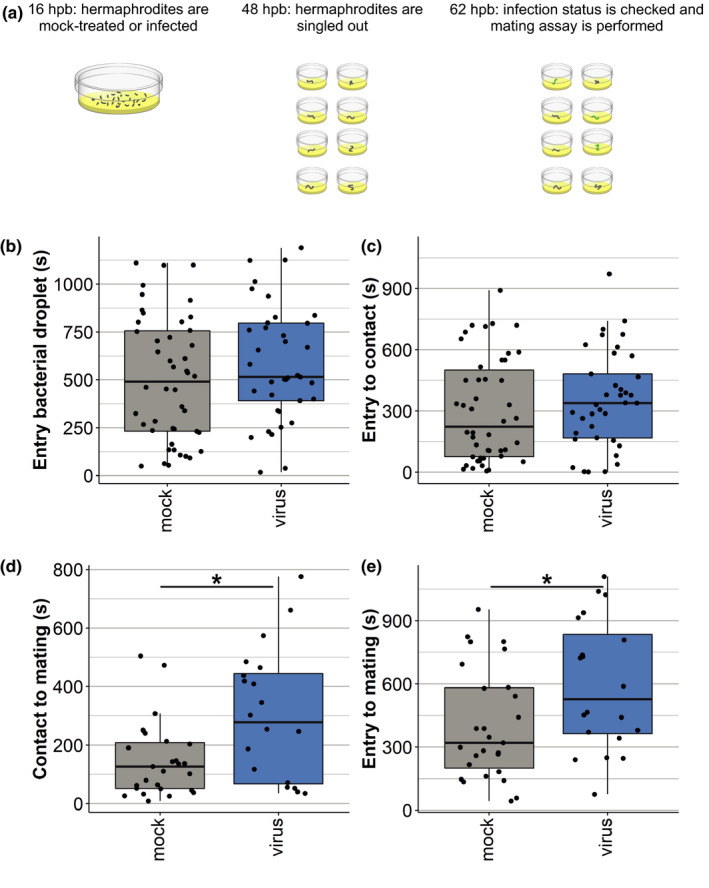
Mating assay with mock‐treated and OrV‐infected hermaphrodites. (a) For the mating assay, hermaphrodite populations of the reporter strain ERT54 (pals‐5::GFP in N2 background) were either mock‐treated or infected with OrV. Once the hermaphrodites were 48 hr old they were placed on individual plates for 14 hr. Then, nematodes were checked for expression of green fluorescent protein, indicating successful infection. Successfully infected individuals and mock‐treated individuals were placed in a camera set‐up. A young adult male was added at a set distance and mating behaviour was observed for 20 min. (b) The time recorded for the male to enter the bacterial droplet. (c) The time recorded between the male entering the bacterial droplet and the first physical contact between male and hermaphrodite. (d) The time recorded between the first contact and mating. (e) The time recorded between the male entering the bacterial droplet and mating. For (b)–(e) statistically significant samples are indicated with an asterisk (t test, p < .05). The number of technical replicates (nematodes filmed) was n = 121 for mock‐treated hermaphrodites and n = 122 for OrV‐infected hermaphrodites. The nematodes were divided over 10 biological replicates (different days). Each dot represents a nematode and when a certain behaviour was not observed in the 20‐min time frame this animal was excluded from the graph
Behavioural differences between healthy and infected hermaphrodites could determine mating efficiency. Therefore, hermaphrodite movements were measured before and after contact with the male (Table S6). OrV‐infected hermaphrodites moved at similar speeds as healthy hermaphrodites before contact with the male (t test, p = .093) (Figure S7a). Moreover, the speed of movement did not appear to determine effective mating in our assay, because (i) both fast and slower moving nematodes were mated and (ii) a correlation between velocity and time from the first contact to mating was lacking (linear model, R 2 = .02, p = .21) (Figure S7b). Hermaphrodites could also avoid male mating attempts by crawling away after contact with the male. We investigated the change in speed after male contact, but did not observe a stronger avoidance response for infected than for healthy hermaphrodites (t test, p = .21) (Figure S7c). In conclusion, both attraction assays performed here indicate that male C. elegans nematodes would sooner mate with uninfected over OrV‐infected hermaphrodites. In cases where OrV infection affects the progeny production of hermaphrodites, this altered mating behaviour could determine male percentages and outcrossing in infected populations.
4. DISCUSSION
Pathogens have various means to perturb mating system dynamics. Unravelling the mechanisms by which pathogens interfere in the mating dynamics of their hosts provides insight into the way pathogens affect the world around them. Here, we studied the differences in OrV infection of males vs. hermaphrodites of the nematode Caenorhabditis elegans. We found that males of the strain N2 were less susceptible than hermaphrodites and that males had higher IPR activity. Viral infection changed the mating dynamics in infected populations of other genotypes. Infected hermaphrodites were less attractive partners for males and viral presence can increase male presence in a population. Our findings provide a novel perspective for how pathogens shape mating dynamics in isogenic populations of C. elegans.
Caenorhabditis elegans–pathogen co‐evolution experiments illustrate that multiple factors contribute to outcrossing in C. elegans (Lopes et al., 2008; Lynch et al., 2018; Morran et al., 2009, 2011). Mated hermaphrodites produce 50% male offspring and unmated hermaphrodites only have 0.2% male offspring (Anderson et al., 2010; Cutter et al., 2019), so an increase in C. elegans males implies outcrossing takes place in that population. Presence of the bacterial pathogen Serratia marcescens increases male frequencies in C. elegans populations and their outcrossed offspring are more able to counteract infection than offspring from selfing hermaphrodites (Morran et al., 2011). On the other hand, the bacterial pathogen Bacillus thuringiensis decreased outcrossing in C. elegans populations. Interestingly, males suffer disproportionally from infection to B. thurgiensis, yet stayed stably present at a rate of about 10%. This suggests that despite the direct disadvantage of susceptible males, the overall populations still indirectly benefit from outcrossing (Masri et al., 2013). Furthermore, male frequencies can also be maintained at higher levels for certain genotypes under control conditions (Teotonio et al., 2012) or increase under stress in isogenic populations that cannot benefit from fitness‐increasing genetic recombination (Lynch et al., 2018; Morran et al., 2009). In one of the latter cases, increased male presence is linked to their higher resistance to starvation‐induced stress. Our data demonstrate that males prefer mating with healthy (instead of OrV‐infected) hermaphrodites. In highly susceptible populations OrV infection can reduce offspring numbers, but not all individuals in the population become infected (Ashe et al., 2013). Our findings suggest that males would mostly mate with healthy hermaphrodites within infected populations. These healthy mated hermaphrodites could have the highest offspring numbers and produce 50% males. On the other hand, unmated hermaphrodites with lower brood sizes produce only hermaphrodites. Over time, this would lead to an increase in male presence in infected populations. which may explain the observations we made in JU1580 and CB4856 populations.
Mating behaviour depends on more than just the presence of a pathogen. In general, C. elegans males are characterized by their inefficient mating behaviour relative to obligatory outcrossers and N2 males even belong to the poorest maters for this species (Garcia et al., 2007; Wegewitz et al., 2008). In contrast, CB4856 nematodes exhibit relatively efficient mating behaviour (Wegewitz et al., 2008). Even though our results suggest that males may become more prevalent in infected populations by selecting the best partners, they will need to mate efficiently, or male numbers will dwindle quickly. Thus, the inefficient mating behaviour of N2 males may help to explain why viral presence did not lead to an increase in males in infected N2 populations. Additionally, the mating behaviour of hermaphrodites may also change when they are infected. Infected individuals for many species behave differentially, which can lead to divergent mating frequencies (Beltran‐Bech & Richard, 2014; Burand et al., 2005; Paciência et al., 2019). Under standard conditions C. elegans hermaphrodites can avoid (costly) mating by crawling away during the mating attempts of the male (Chasnov & Chow, 2002; Garcia et al., 2007; Woodruff et al., 2014). Although we did not observe that infected hermaphrodites behaved different than healthy ones, we can also not fully reject that hermaphrodite behaviour could affect mating.
Molecular cues are commonly used to distinguish infected from noninfected potential partners (Beltran‐Bech & Richard, 2014). C. elegans males are normally attracted by hermaphrodite‐produced ascarosides (Chasnov et al., 2007; Pungaliya et al., 2009; Srinivasan et al., 2008). However, our choice experiment using an ascaroside‐lacking daf‐22 mutant (Von Reuss et al., 2012) suggests that ascaroside signals do not play an essential role for males to distinguish healthy from infected hermaphrodites. In addition, publicly available transcriptional data show that the main genes in the ascaroside‐producing pathway (acox‐1, maoc‐1, dhs‐28 and daf‐22) remain equally expressed in infected nematodes (Chen et al., 2017; Sarkies et al., 2013; van Sluijs et al., 2019). Hence, other means of (chemical) communication might contribute to the mating preference for uninfected hermaphrodites by C. elegans males. For example, C. elegans can recognize a noncoding RNA to avoid a pathogenic bacterium (Kaletsky et al., 2020), which illustrates another way by which infected nematodes may be recognized.
Our findings indicate that sex‐based susceptibility differences occur in a genotype‐dependent manner based on the viral loads of three genetically distinct strains. Of note, the viral loads in hermaphrodite populations are probably underestimations since these populations contained the first eggs at the moment of collection. Eggs cannot be infected by OrV (Félix et al., 2011), but embryos do express the reference genes that are used to normalize viral expression. Previous studies indicated IPR gene expression is enhanced in L4‐stage males (Gerstein et al., 2014; Leyva‐Díaz et al., 2017) and we found that some IPR genes are also more highly expressed in adult males. Notably, males are also more resistant to an intracellular fungus, although the connection with the IPR has not been studied (Van Den Berg et al., 2006). Not all genes in the IPR network were more active in virus‐infected males than hermaphrodites. The IPR upregulates a large network of 80 genes against a broad range of stressors (Chen et al., 2017; Reddy et al., 2017, 2019), but the biochemical reaction to counteract the stress appears more specific. For example, the IPR proteins that promote thermotolerance do not enhance pathogen resistance (Panek et al., 2020) and drh‐1 is only required for induction of the antiviral IPR (Sowa et al., 2019). Furthermore, the IPR genes in JU1580 and CB4856 hermaphrodites respond differently to OrV infection than in N2 (Sarkies et al., 2013; van Sluijs et al., 2019); this may relate to similar viral susceptibilities for JU1580 and CB4856 between males and hermaphrodites. Thus, a more thorough understanding of the IPR is necessary to draw firm conclusions about IPR involvement in sex‐specific viral susceptibility.
The importance of C. elegans males remains a topic of debate, with most studies indicating their disadvantages and only a few providing them with a potential ecological role (Cutter et al., 2019). C. elegans males are typically rare in nature, further contributing to the idea that they may be less relevant for the species (Barrière & Félix, 2005; Félix & Duveau, 2012; Richaud et al., 2018; Sivasundar & Hey, 2005). Nevertheless, males may prove beneficial under stress conditions, including viral infections, and could thereby play a supporting role in the natural history of C. elegans. Here, we have shown how the presence of an intracellular pathogen shifts mating behaviour and shapes flexible outcrossing in this species.
AUTHOR CONTRIBUTIONS
L.v.S., J.L., M.S., S.v.H. and N.Z. performed OrV infection experiments. L.v.S. and J.A.G. monitored male presence in mock‐treated or infected populations. L.v.S. performed choice assays and mating experiments. J.A.G. and N.Z. performed gene expression measurements by microarray. S.P.J.M.V., S.v.H. and M.H.S. performed gene expression measurement by RT‐qPCR. S.P.J.M.V., J.L. and L.v.S. performed food intake experiments. The data were analysed by L.v.S. and M.G.S. L.v.S., J.E.K., G.P.P. and M.G.S. wrote the manuscript. All authors read and approved the final manuscript.
Supporting information
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Figure S7
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Video S1
ACKNOWLEDGMENTS
The authors thank Emily Troemel for sending us reporter strains and advice on food intake assays. We also thank Erik Andersen, Frank Schroeder and Marie‐Anne Félix for advice regarding choice assay and mating experiments. Giel Göertz is thanked for sharing his knowledge about (processing) sRNA sequencing data. Katharina Jovic is thanked for advice about the VirtualDub script and camera set‐up to simultaneously film nematode movements. Furthermore, we thank the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), for providing strains. M.S. was supported by NWO domain Applied and Engineering Sciences VENI grant (17282). L.v.S. was funded by NWO (824.15.006).
van Sluijs, L. , Liu, J. , Schrama, M. , van Hamond, S. , Vromans, S. P. J. M. , Scholten, M. H. , Žibrat, N. , Riksen, J. A. G. , Pijlman, G. P. , Sterken, M. G. , & Kammenga, J. E. (2021). Virus infection modulates male sexual behaviour in Caenorhabditis elegans . Molecular Ecology, 30, 6776–6790. 10.1111/mec.16179
DATA AVAILABILITY STATEMENT
All custom written scripts and required data for the analyses in R can be accessed via https://git.wur.nl/published_papers/sluijs_etal_2021 Microarray and sRNA sequencing data sets are deposited at ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under accession nos. E‐MTAB‐9561 and E‐MTAB‐10642 respectively. Processed data are included in the supporting tables to support straightforward access.
REFERENCES
- Afgan, E. , Baker, D. , Batut, B. , Van Den Beek, M. , Bouvier, D. , Ech, M. , Chilton, J. , Clements, D. , Coraor, N. , Grüning, B. A. , Guerler, A. , Hillman‐Jackson, J. , Hiltemann, S. , Jalili, V. , Rasche, H. , Soranzo, N. , Goecks, J. , Taylor, J. , Nekrutenko, A. , & Blankenberg, D. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Research, 46, W537–W544. 10.1093/nar/gky379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J. L. , Morran, L. T. , & Phillips, P. C. (2010). Outcrossing and the maintenance of males within C. elegans populations. Journal of Heredity, 101(Supplement 1), S62–S74. 10.1093/jhered/esq003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe, A. , Bélicard, T. , Le Pen, J. , Sarkies, P. , Frézal, L. , Lehrbach, N. J. , Félix, M.‐A. , Miska, E. A. (2013). A deletion polymorphism in the Caenorhabditis elegans RIG‐I homolog disables viral RNA dicing and antiviral immunity. Elife, 2, e00994. 10.7554/eLife.00994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakowski, M. A. , Desjardins, C. A. , Smelkinson, M. G. , Dunbar, T. A. , Lopez‐Moyado, I. F. , Rifkin, S. A. , Cuomo, C. A. , Troemel, E. R. (2014). Ubiquitin‐mediated response to microsporidia and virus infection in C. elegans . PLoS Path, 10(6), e1004200. 10.1371/journal.ppat.1004200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann, C. I. , Hartwieg, E. , & Horvitz, H. R. (1993). Odorant‐selective genes and neurons mediate olfaction in C. elegans . Cell, 74(3), 515–527. 10.1016/0092-8674(93)80053-H [DOI] [PubMed] [Google Scholar]
- Barrière, A. , & Félix, M. A. (2005). High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Current Biology, 15(13), 1176–1184. 10.1016/j.cub.2005.06.022 [DOI] [PubMed] [Google Scholar]
- Bell, G. (1982). The masterpiece of nature: the evolution and genetics of sexuality. Berkeley: University of California Press. [Google Scholar]
- Beltran‐Bech, S. , & Richard, F. J. (2014). Impact of infection on mate choice. Animal Behaviour, 90, 159–170. 10.1016/j.anbehav.2014.01.026 [DOI] [Google Scholar]
- Benjamini, Y. , & Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, 57(1), 289–300. [Google Scholar]
- Bezler, A. , Braukmann, F. , West, S. M. , Duplan, A. , Conconi, R. , Schütz, F. , Gönczy, P. , Piano, F. , Gunsalus, K. , Miska, E. A. , & Keller, L. (2019). Tissue‐ and sex‐specific small RNAomes reveal sex differences in response to the environment. PLOS Genetics, 15(2), e1007905. 10.1371/journal.pgen.1007905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borne, F. , Kasimatis, K. R. , & Phillips, P. C. (2017). Quantifying male and female pheromone‐based mate choice in Caenorhabditis nematodes using a novel microfluidic technique. PLoS One, 12(12), 1–14. 10.1371/journal.pone.0189679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans . Genetics, 77(1), 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burand, J. P. , Tan, W. , Kim, W. , Nojima, S. , & Roelofs, W. (2005). Infection with the insect virus Hz‐2v alters mating behavior and pheromone production in female Helicoverpa zea moths. Journal of Insect Science, 5(6), 1–6. 10.1673/031.005.0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butlin, R. (2002). The costs and benefits of sex: New insights from old asexual lineages. Nature Reviews Genetics, 3(4), 311–317. 10.1038/nrg749 [DOI] [PubMed] [Google Scholar]
- Chambers, J. , & Hastie, T. (1992). Statistical models in S. Wadsworth & Brooks/Cole. [Google Scholar]
- Chasnov, J. R. , & Chow, K. L. (2002). Why are there males in the hermaphroditic species. Genetics, 160, 983–994. 10.4271/2016-01-0931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasnov, J. R. , So, W. K. , Chan, C. M. , & Chow, K. L. (2007). The species, sex, and stage specificity of a Caenorhabditis sex pheromone. Proceedings of the National Academy of Sciences of the United States of America, 104(16), 6730–6735. 10.1073/pnas.0608050104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. , Franz, C. J. , Jiang, H. , Jiang, Y. , & Wang, D. (2017). An evolutionarily conserved transcriptional response to viral infection in Caenorhabditis nematodes . BMC Genomics, 18(1), 303. 10.1186/s12864-017-3689-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman, S. R. , Lu, J. , Guo, X. , Zhong, J. , Jiang, H. , Broitman‐Maduro, G. , Li, W.‐X. , Lu, R. , Maduro, M. , Ding, S. (2017). Caenorhabditis elegans RIG‐I homolog mediates antiviral RNA interference downstream of dicer‐dependent biogenesis of viral small interfering RNAs. Mbio, 8(2), 1–15. 10.1128/mBio.00264-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, S. J. , Jarrell, T. A. , Brittin, C. A. , Wang, Y. , Bloniarz, A. E. , Yakovlev, M. A. , Nguyen, K. C. Q. , Tang, L.‐H. , Bayer, E. A. , Duerr, J. S. , Bülow, H. E. , Hobert, O. , Hall, D. H. , Emmons, S. W. (2019). Whole‐animal connectomes of both Caenorhabditis elegans sexes. Nature, 571(7763), 63–71. 10.1038/s41586-019-1352-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter, A. D. , Morran, L. T. , & Phillips, P. C. (2019). Males, outcrossing, and sexual selection in Caenorhabditis Nematodes. Genetics, 213(1), 27–57. 10.1534/genetics.119.300244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson, T. (2012). Causes and consequences of skewed sex ratios. Annual Review of Sociology, 38(1), 443–461. 10.1146/annurev-soc-071811-145429 [DOI] [Google Scholar]
- Engelstädter, J. , & Hurst, G. D. D. (2009). The ecology and evolution of microbes that manipulate host reproduction. Annual Review of Ecology, Evolution, and Systematics, 40(1), 127–149. 10.1146/annurev.ecolsys.110308.120206 [DOI] [Google Scholar]
- Félix, M. A. , Ashe, A. , Piffaretti, J. , Wu, G. , Nuez, I. , Bélicard, T. , Jiang, Y. , Zhao, G. , Franz, C. J. , Goldstein, L. D. , Sanroman, M. , Miska, E. A. , Wang, D. (2011). Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biology, 9(1), e1000586. 10.1371/journal.pbio.1000586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix, M.‐A. , & Duveau, F. (2012). Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae . BMC Biology, 10(1), 59. 10.1186/1741-7007-10-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, R. A. (1930). The Genetical Theory of Natural Selection. Oxford: Clarendon Press. [Google Scholar]
- Frézal, L. , Jung, H. , Tahan, S. , Wang, D. , & Félix, M.‐A. (2019). Noda‐like RNA viruses infecting caenorhabditis nematodes: Sympatry, diversity, and reassortment. Journal of Virology, 93(21), e01170‐19. 10.1128/jvi.01170-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, L. R. , LeBoeuf, B. , & Koo, P. (2007). Diversity in mating behavior of hermaphroditic and male‐female Caenorhabditis nematodes. Genetics, 175(4), 1761–1771. 10.1534/genetics.106.068304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein, M. B. , Lu, Z. J. , Van Nostrand, E. L. , Cheng, C. , Arshinoff, B. I. , Liu, T. , Yip, K. Y. , Robilotto, R. , Rechtsteiner, A. , Ikegami, K. , Alves, P. , Chateigner, A. , Perry, M. , Morris, M. , Auerbach, R. K. , Feng, X. , Leng, J. , Vielle, A. , Niu, W. , … Waterston, R. H. (2010). Integrative Analysis of the Caenorhabditis elegans Genome by the modENCODE Project. Science, 330(6012), 1775 LP–1787. 10.1126/science.1196914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein, M. B. , Rozowsky, J. , Yan, K.‐K. , Wang, D. , Cheng, C. , Brown, J. B. , Davis, C. A. , Hillier, L. D. , Sisu, C. , Li, J. J. , Pei, B. , Harmanci, A. O. , Duff, M. O. , Djebali, S. , Alexander, R. P. , Alver, B. H. , Auerbach, R. , Bell, K. , Bickel, P. J. , … Waterston, R. (2014). Comparative analysis of the transcriptome across distant species. Nature, 512(7515), 445–448. 10.1038/nature13424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gipson, S. A. Y. , Jimenez, L. , & Hall, M. D. (2019). Host sexual dimorphism affects the outcome of within‐host pathogen competition. Evolution, 73(7), 1443–1455. 10.1111/evo.13760 [DOI] [PubMed] [Google Scholar]
- Hamilton, W. D. (1980). Sex versus non‐sex versus parasite. Oikos, 35(2), 282–290. 10.2307/3544435 [DOI] [Google Scholar]
- Jaenike, J. (1978). An hypothesis to account for maintainance of sex within populations. Evolutionary Theory, 3, 191–194. [Google Scholar]
- Kaletsky, R. , Moore, R. S. , Vrla, G. D. , Parsons, L. R. , Gitai, Z. , & Murphy, C. T. (2020). C. elegans interprets bacterial non‐coding RNAs to learn pathogenic avoidance. Nature, 586(7829), 445–451. 10.1038/s41586-020-2699-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavaliers, M. , & Choleris, E. (2018). The role of social cognition in parasite and pathogen avoidance. Philosophical Transactions of the Royal Society B: Biological Sciences, 373(1751), 20170206. 10.1098/rstb.2017.0206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesecker, J. M. , Skelly, D. K. , Beard, K. H. , & Preisser, E. (1999). Behavioral reduction of infection risk. Proceedings of the National Academy of Sciences of the United States of America, 96(16), 9165–9168. 10.1073/pnas.96.16.9165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleemann, G. A. , & Basolo, A. L. (2007). Facultative decrease in mating resistance in hermaphroditic Caenorhabditis elegans with self‐sperm depletion. Animal Behaviour, 74(5), 1339–1347. 10.1016/j.anbehav.2007.02.031 [DOI] [Google Scholar]
- Klein, S. L. , & Flanagan, K. L. (2016). Sex differences in immune responses. Nature Reviews Immunology, 16(10), 626–638. 10.1038/nri.2016.90 [DOI] [PubMed] [Google Scholar]
- Kulesa, A. , Krzywinski, M. , Blainey, P. , & Altman, N. (2015). Points of Significance: Sampling distributions and the bootstrap. Nature Methods, 12(6), 477–478. 10.1038/nmeth.3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. , & Salzberg, S. (2013). Fast gapped‐read alignment with Bowtie 2. Nature Methods, 9(4), 357–359. 10.1038/nmeth.1923.Fast [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pen, J. , Jiang, H. , Di Domenico, T. , Kneuss, E. , Kosałka, J. , Leung, C. , Morgan, M. , Much, C. , Rudolph, K. L. M. , Enright, A. J. , O’Carroll, D. , Wang, D. , Miska, E. A. (2018). Terminal uridylyltransferases target RNA viruses as part of the innate immune system. Nature Structural and Molecular Biology, 25(9), 778–786. 10.1038/s41594-018-0106-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBoeuf, B. , Correa, P. , Jee, C. , & García, L. R. (2014). Caenorhabditis elegans male sensory‐motor neurons and dopaminergic support cells couple ejaculation and post‐ejaculatory behaviors. Elife, 3, 1–32. 10.7554/elife.02938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen, J. , Jennions, M. D. , & Kokko, H. (2012). The many costs of sex. Trends in Ecology and Evolution, 27(3), 172–178. 10.1016/j.tree.2011.09.016 [DOI] [PubMed] [Google Scholar]
- Leyva‐Díaz, E. , Stefanakis, N. , Carrera, I. , Glenwinkel, L. , Wang, G. , Driscoll, M. , & Hobert, O. (2017). Silencing of repetitive DNA is controlled by a member of an unusual Caenorhabditis elegans gene family. Genetics, 207(2), 529–545. 10.1534/genetics.117.300134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, P. C. , Sucena, É. , Santos, M. E. , & Magalhães, S. (2008). Rapid experimental evolution of pesticide resistance in C. elegans entails no costs and affects the mating system. PLoS One, 3(11), e3741. 10.1371/journal.pone.0003741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, Z. R. , Penley, M. J. , & Morran, L. T. (2018). Turnover in local parasite populations temporarily favors host outcrossing over self‐fertilization during experimental evolution. Ecology and Evolution, 8(13), 6652–6662. 10.1002/ece3.4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri, L. , Schulte, R. D. , Timmermeyer, N. , Thanisch, S. , Crummenerl, L. L. , Jansen, G. , Michiels, N. K. , Schulenburg, H. (2013). Sex differences in host defence interfere with parasite‐mediated selection for outcrossing during host‐parasite coevolution. Ecology Letters, 16(4), 461–468. 10.1111/ele.12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran, L. T. , Cappy, B. J. , Anderson, J. L. , & Phillips, P. C. (2009). Sexual partners for the stressed: Facultative outcrossing in the self‐fertilizing nematode C. elegans . Evolution, 63(6), 1473–1482. 10.1111/j.1558-5646.2009.00652.x.Sexual [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran, L. T. , Schmidt, O. G. , Gelarden, I. A. , Parrish, R. C. , & Lively, C. M. (2011). Running with the Red Queen: Host‐parasite coevolution selects for biparental sex. Science, 333(6039), 216–218. 10.1126/science.1206360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman, G. A. , Fasseas, M. K. , Koneru, S. L. , Essmann, C. L. , Kyrou, K. , Srinivasan, M. A. , Zhang, G. , Sarkies, P. , Félix, M.‐A. , Barkoulas, M. (2018). Natural infection of C. elegans by an oomycete reveals a new pathogen‐specific immune response. Current Biology, 28(4), 640–648.e5. 10.1016/j.cub.2018.01.029 [DOI] [PubMed] [Google Scholar]
- Paciência, F. M. D. , Rushmore, J. , Chuma, I. S. , Lipende, I. F. , Caillaud, D. , Knauf, S. , & Zinner, D. (2019). Mating avoidance in female olive baboons (Papio anubis) infected by Treponema pallidum . Science Advances, 5(12), 1–8. 10.1126/sciadv.aaw9724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palopoli, M. F. , Rockman, M. V. , TinMaung, A. , Ramsay, C. , Curwen, S. , Aduna, A. , Laurita, J. , Kruglyak, L. (2008). Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature, 454(7207), 1019–1022. 10.1038/nature07171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panek, J. , Gang, S. S. , Reddy, K. C. , Luallen, R. J. , Fulzele, A. , Bennett, E. J. , & Troemel, E. R. (2020). A cullin‐RING ubiquitin ligase promotes thermotolerance as part of the intracellular pathogen response in Caenorhabditis elegans. Proceedings of the National Academy of Sciences, 117(14), 7950–7960. 10.1073/pnas.1918417117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro, J. , Bates, D. , DebRoy, S. , Sarkar, D. , R Core Team . (2021). nlme: Linear and Nonlinear Mixed Effects Models. Retrieved from https://cran.r‐project.org/package=nlme [Google Scholar]
- Pungaliya, C. , Srinivasan, J. , Fox, B. W. , Malik, R. U. , Ludewig, A. H. , Sternberg, P. W. , & Schroeder, F. C. (2009). A shortcut to identifying small molecule signals that regulate behavior and development in Caenorhabditis elegans . Proceedings of the National Academy of Sciences of the United States of America, 106(19), 7708–7713. 10.1073/pnas.0811918106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, K. C. , Dror, T. , Sowa, J. N. , Panek, J. , Chen, K. , Lim, E. S. , Wang, D. , Troemel, E. R. (2017). An intracellular pathogen response pathway promotes proteostasis in C. elegans . Current Biology, 27(22), 3544–3553.e5. 10.1016/j.cub.2017.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, K. C. , Dror, T. , Underwood, R. S. , Osman, G. A. , Desjardins, C. A. , Cuomo, C. A. , Barkoulas, M. , Troemel, E. R. (2019). Antagonistic paralogs control a switch between growth and pathogen resistance in C. elegans . PLoS Path, 15(1), e1007528. 10.1371/journal.ppat.1007528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richaud, A. , Zhang, G. , Lee, D. , Lee, J. , & Félix, M.‐A. (2018). The local coexistence pattern of selfing genotypes in Caenorhabditis elegans natural metapopulations. Genetics, 208(2), 807–821. 10.1534/genetics.117.300564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, M. E. , Phipson, B. , Wu, D. , Hu, Y. , Law, C. W. , Shi, W. , & Smyth, G. K. (2015). Limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Research, 43(7), e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies, P. , Ashe, A. , Le Pen, J. , McKie, M. A. , & Miska, E. A. (2013). Competition between virus‐derived and endogenous small RNAs regulates gene expression in Caenorhabditis elegans . Genome Research, 23(8), 1258–1270. 10.1101/gr.153296.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , Preibisch, S. , Rueden, C. , Saalfeld, S. , Schmid, B. , Tinevez, J.‐Y. , White, D. J. , Hartenstein, V. , Eliceiri, K. , Tomancak, P. , Cardona, A. (2012). Fiji: An open‐source platform for biological‐image analysis. Nature Methods, 9(7), 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C. A. , Rasband, W. S. , & Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Methods, 9(7), 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully, E. P. , Haverfield, J. , Ursin, R. L. , Tannenbaum, C. , & Klein, S. L. (2020). Considering how biological sex impacts immune responses and COVID‐19 outcomes. Nature Reviews Immunology, 20(7), 442–447. 10.1038/s41577-020-0348-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasundar, A. , & Hey, J. (2005). Sampling from natural populations with RNAi reveals high outcrossing and population structure in Caenorhabditis elegans . Current Biology, 15(17), 1598–1602. 10.1016/j.cub.2005.08.034 [DOI] [PubMed] [Google Scholar]
- Smyth, G. K. , & Speed, T. (2003). Normalization of cDNA microarray data. Methods, 31(4), 265–273. 10.1016/S1046-2023(03)00155-5 [DOI] [PubMed] [Google Scholar]
- Sowa, J. N. , Jiang, H. , Somasundaram, L. , Tecle, E. , Xu, G. , Wang, D. , & Troemel, E. R. (2019). The Caenorhabditis elegans RIG‐I Homolog DRH‐1 mediates the intracellular pathogen response upon viral infection. Journal of Virology, 94(2), e01173–e1219. 10.1128/JVI.01173-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, J. , Kaplan, F. , Ajredini, R. , Zachariah, C. , Alborn, H. T. , Teal, P. E. A. , Malik, R. U. , Edison, A. S. , Sternberg, P. W. , Schroeder, F. C. (2008). A blend of small molecules regulates both mating and development in Caenorhabditis elegans . Nature, 454(7208), 1115–1118. 10.1038/nature07168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken, M. G. , Snoek, L. B. , Bosman, K. J. , Daamen, J. , Riksen, J. A. G. , Bakker, J. , Pijlman, G. P. , Kammenga, J. E. (2014). A heritable antiviral RNAi response limits orsay virus infection in Caenorhabditis elegans N2. PLoS One, 9(2), e89760. 10.1371/journal.pone.0089760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken, M. G. , van Sluijs, L. , Wang, Y. A. , Ritmahan, W. , Gultom, M. L. , Riksen, J. A. G. , Volkers, R. J. M. , Snoek, L. B. , Pijlman, G. P. , Kammenga, J. E. (2021). Punctuated loci on chromosome IV determine natural variation in orsay virus susceptibility of Caenorhabditis elegans strains bristol N2 and Hawaiian CB4856. Journal of Virology, 95(12), e02430‐20. 10.1128/JVI.02430-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanguy, M. , Véron, L. , Stempor, P. , Ahringer, J. , Sarkies, P. , & Miska, E. A. (2017). An alternative STAT signaling pathway acts in viral immunity in Caenorhabditis elegans . Mbio, 8(5), 1–16. 10.1128/mBio.00924-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotonio, H. , Carvalho, S. , Manoel, D. , Roque, M. , & Chelo, I. M. (2012). Evolution of outcrossing in experimental populations of Caenorhabditis elegans . PLoS One, 7(4), e35811. 10.1371/journal.pone.0035811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotónio, H. , Manoel, D. , & Phillips, P. C. (2006). Genetic variation for outcrossing among Caenorhabditis elegans isolates. Evolution, 60(6), 1300–1305. 10.1525/aa.1953.55.4.02a00270 [DOI] [PubMed] [Google Scholar]
- Thompson, O. A. , Snoek, L. B. , Nijveen, H. , Sterken, M. G. , Volkers, R. J. M. M. , Brenchley, R. , Van't Hof, A. , Bevers, R. P. J. , Cossins, A. R. , Yanai, I. , Hajnal, A. , Schmid, T. , Perkins, J. D. , Spencer, D. , Kruglyak, L. , Andersen, E. C. , Moerman, D. G. , Hillier, L. W. , Kammenga, J. E. , Waterston, R. H. (2015). Remarkably divergent regions punctuate the genome assembly of the Caenorhabditis elegans Hawaiian strain CB4856. Genetics, 200(3), 975–989. 10.1534/genetics.115.175950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Berg, M. C. W. , Woerlee, J. Z. , Ma, H. , & May, R. C. (2006). Sex‐dependent resistance to the pathogenic fungus Cryptococcus neoformans . Genetics, 173(2), 677–683. 10.1534/genetics.106.056093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sluijs, L. , Bosman, K. , Pankok, F. , Blokhina, T. , Riksen, J. A. G. , Snoek, B. L. , Pijlman, G. P. , Kammenga, J. E. , Sterken, M. G. (2019). Balancing selection shapes the Intracellular Pathogen Response in natural. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Reuss, S. H. , Bose, N. , Srinivasan, J. , Yim, J. J. , Judkins, J. C. , Sternberg, P. W. , & Schroeder, F. C. (2012). Comparative metabolomics reveals biogenesis of ascarosides, a modular library of small‐molecule signals in C. elegans . Journal of the American Chemical Society, 134(3), 1817–1824. 10.1021/ja210202y.Comparative [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegewitz, V. , Schulenburg, H. , & Streit, A. (2008). Experimental insight into the proximate causes of male persistence variation among two strains of the androdioecious Caenorhabditis elegans (Nematoda). BMC Ecology, 8, 1–12. 10.1186/1472-6785-8-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J. Q. , Nicholas, T. J. , Gritton, J. , Truong, L. , Davidson, E. R. , & Jorgensen, E. M. (2007). The sensory circuitry for sexual attraction in C. elegans males. Current Biology, 17(21), 1847–1857. 10.1016/j.cub.2007.09.011 [DOI] [PubMed] [Google Scholar]
- Wickham, H. , Averick, M. , Bryan, J. , Chang, W. , McGowan, L. D. , François, R. , Grolemund, G. , Hayes, A. , Henry, L. , Hester, J. , Kuhn, M. , Pedersen, T. , Miller, E. , Bache, S. , Müller, K. , Ooms, J. , Robinson, D. , Seidel, D. , Spinu, V. , … Yutani, H. (2019). Welcome to the {tidyverse}. Journal of Open Source Software, 4(43), 1686. 10.21105/joss.01686 [DOI] [Google Scholar]
- Woodruff, G. C. , Knauss, C. M. , Maugel, T. K. , & Haag, E. S. (2014). Mating damages the cuticle of C. elegans hermaphrodites. PLoS One, 9(8), 1–5. 10.1371/journal.pone.0104456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahurak, M. , Parmigiani, G. , Yu, W. , Scharpf, R. B. , Berman, D. , Schaeffer, E. , Shabbeer, S. , Cope, L. (2007). Pre‐processing agilent microarray data. BMC Bioinformatics, 8, 1–13. 10.1186/1471-2105-8-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G. , Sachse, M. , Prevost, M.‐C. , Luallen, R. J. , Troemel, E. R. , & Felix, M.‐A. (2016). A large collection of novel nematode‐ infecting microsporidia and their diverse interactions with Caenorhabditis elegans and other related nematodes. PLoS Path, 12(12), e1006093. 10.1371/journal.ppat.1006093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Lu, H. , & Bargmann, C. I. (2005). Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans . Nature, 438(7065), 179–184. 10.1038/nature04216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Figure S7
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Video S1
Data Availability Statement
All custom written scripts and required data for the analyses in R can be accessed via https://git.wur.nl/published_papers/sluijs_etal_2021 Microarray and sRNA sequencing data sets are deposited at ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under accession nos. E‐MTAB‐9561 and E‐MTAB‐10642 respectively. Processed data are included in the supporting tables to support straightforward access.


