Summary
Early life microbiome perturbations can have important effects on host development, physiology and behaviour. In this longitudinal study, we evaluated the impact of early feeding on gut microbiome colonization in neonatal piglets. Early‐fed (EF) piglets had access to a customized fibrous diet from 2 days after birth until weaning in addition to mother's milk, whereas control piglets suckled mother's milk only. Rectal swabs were collected at multiple time points until 6 weeks of age to investigate microbiota development using 16S rRNA gene profiling. The dynamic pre‐weaning microbiota colonization was followed by a relatively stable post‐weaning microbiota, represented by Prevotella, Roseburia, Faecalibacterium, Ruminococcus, Megasphaera, Catenibacterium and Subdoligranulum. EF piglets showed an accelerated microbiota maturation, characterized by increased microbial diversity, pre‐weaning emergence of post‐weaning‐associated microbes and a more rapid decline of typical pre‐weaning microbes. Furthermore, the individual eating behaviour scores of piglets quantitatively correlated with their accelerated microbiome. Importantly, EF piglets displayed a smoother relative weight gain and tended to reach a higher relative weight gain, in addition to reduced diarrhoea scores in the first week post‐weaning. Overall, these findings demonstrate the beneficial impact of early feeding on microbiota development as well as pig health and performance during the weaning transition.
Introduction
The mammalian gastrointestinal tract (GIT) is inhabited by complex and diverse microbial communities that influence host health and disease. The complex ecosystem development initiates by the appearance of individual microbial taxa shortly after birth (derived from the mother and environment) and gradually stabilizes over time (Schokker et al., 2015; Timmerman et al., 2017). In early life, the influence of colonizing gut microbiota on intestinal development is crucial as in this period; the microbiota is essential for appropriate development and programming of the mucosal immune response (Schokker et al., 2015; Zhuang et al., 2019). Based on the plasticity of the mucosal system during this early life development phase, it has been proposed as a ‘window of opportunity’ where perturbations may have long‐lasting impacts on health and welfare (Putignani et al., 2014; Nowland et al., 2019).
The transition from mother's milk to solid food, known as weaning, is normally a gradual process in young mammals. In nature, the weaning process of piglets approaches completion between 17 and 20 weeks of age approximately (Newberry and Wood‐Gush, 1985; Jensen and Stangel, 1992). However, in modern swine industry practice, piglets are weaned abruptly around 3–4 weeks of age, a time period that coincides with the developmental changes of the gastrointestinal system (Moeser et al., 2017). Evidently, early weaning is a highly dynamic and stressful event as the piglets deal with sudden changes in diet and environment, including the separation from their mother and littermates. Weaning stress is typically associated with low feed intake, sub‐optimal weight gain, diarrhoea episodes and maladaptive behaviour, leading to compromised animal health and welfare, increased piglet mortality and economic losses (Le Dividich and Sève, 2000; Bruininx et al., 2002; Heo et al., 2013; Everaert et al., 2017; Gresse et al., 2017; Pluske et al., 2018). Weaning‐associated changes are evident in intestinal physiology and could be broadly divided into two phases: an acute phase occurring immediately after weaning (day 0–5 post‐weaning; acute deterioration of gut structure and function) and an adaptive or maturation phase (day 5–15 post‐weaning; where the intestine adapts to the weaning diet) (Montagne et al., 2007). The first days after weaning thus can be considered as a period where piglets experience maximum weaning stress. Gut microbiota dysbiosis (characterized by microbial imbalance, intestinal inflammation, reduced gut barrier function and increased abundance of potential pathogens) is one of the factors that could contribute to the weaning transition problems, and is considered as one of the leading causes of post‐weaning diarrhoea and associated GIT infections in piglets (Gresse et al., 2017; Pluske et al., 2018).
It is well established that diet is a major factor that can shape the intestinal microbiota in mammals. Dietary fibres, abundant in common plant‐based feedstuffs, pass through the small intestine in an undigested form and are fermented in the distal ileum and colon, stimulating the growth of microbes. Microbial fermentation of undigested fibres usually takes place in the distal part of the GIT, resulting in the formation of short‐chain fatty acids, that are known to influence physiological functioning of the intestines such as formation and protection of the intestinal barrier as well as host defence and inflammatory responses (Den Besten et al., 2013; Furusawa et al., 2013; van der Beek et al., 2017; Xiong et al., 2019).
In the present study, we provided a customized fibrous diet to a group of suckling piglets from 2 days of age, intending to familiarize them with the consumption of solid feed and to investigate the impact on their gut microbiota compared to control piglets that did not receive pre‐weaning solid feed. Both groups of piglets revealed substantial dynamics of the gut microbiome during the pre‐weaning period, characterized by the appearance and disappearance of specific microbial genera over time. Additionally, our findings establish that pre‐weaning fibrous feed can accelerate gut microbiome colonization towards a more mature microbiome, which resembles that of post‐weaning microbial composition. Finally, early life feeding reduced typical weaning stress‐related problems (disrupted relative weight gain and diarrhoea) during the acute phase post‐weaning.
Results
Dynamics of gut microbiome development in early life of piglets
In this study, we first investigated the dynamics of intestinal microbiota development by analysing rectal swab samples collected from 2 days after birth (day 2) to 2 weeks post‐weaning (day +14). All rectal swab samples were used to assess the microbiota development over time, irrespective of their allocation to the intervention group receiving pre‐weaning fibrous feed (EF) or not receiving any additional feed (CON). 16S rRNA Illumina sequencing produced 5 013 336 reads after quality filtering, having a mean depth of 27 009 ± 5019 reads per sample.
Irrespective of the treatment, the gut microbiota showed typical porcine microbiota (Holman et al., 2017; Wang et al., 2019) colonization patterns with Firmicutes being the predominant phyla (60.01%), followed by Bacteroidetes (23.5%), Proteobacteria (8.04%), Actinobacteria (2.77%) and Fusobacteria (1.56%), together capturing 95.9% of total microbiome. Firmicutes and Bacteroidetes together accounted for 64%–97% of total sequences across all ages. From day 2 to day +14 post‐weaning, the relative abundance of Firmicutes increased (44%–67%; P < 0.0001) and Bacteroidetes fluctuated pre‐weaning eventually increasing post‐weaning (19.9%–29.2%; P < 0.0001) (Supplementary Fig. 2). The relative abundance of Fusobacteria significantly dropped from around 10% at day 2 to less than 1% at day 15 (P < 0.05) almost disappearing in the post‐weaning time points (P < 0.0001). During the same period, the microbial (alpha) diversity strongly increased, especially during the pre‐weaning period (Fig. 1A, Supplementary Fig. 3A) and reached an apparent plateau post‐weaning. This is reflected by both the Chao 1 index (richness) that increased from 2 days after birth (day 2) up to day 5 post‐weaning (day +5) as well as the Shannon index (richness and evenness) that increased gradually during the weaning period (day 28) and remained stable afterwards.
Fig. 1.
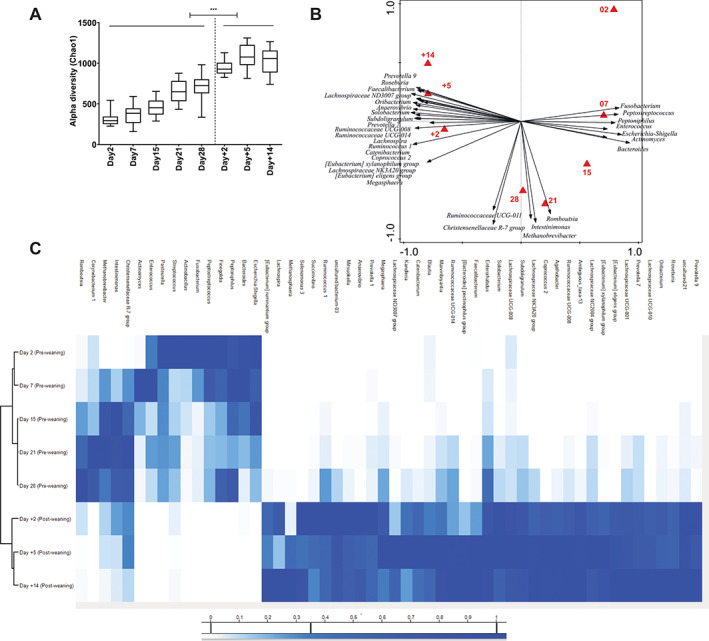
Age‐associated intestinal microbiota dynamics pre‐ and post‐weaning.
A. Alpha diversity (Chao1 bias corrected) displays shifting diversity over time.
B. Redundancy analysis of age at genus level (explained variation = 28.22%, P = 0.002) with associated microbial groups at different ages. Triangles are centroids of all samples of a specific time point. The age‐related microbial groups visualized (black arrows) have minimum 35% fit on horizontal axis with a response score ≥0.7 in the biplot (obtained by projecting the taxa points perpendicular to the axes).
C. Heat map showing normalized relative abundance (see M&M) of the discriminative bacterial genera identified in redundancy analysis of age. Significant differences between time points were assessed by Student t‐test or Mann–Whitney U test (***P < 0.001).
The impact of variables in the experimental setup such as age, litter, gender and weaning on the microbiota composition were investigated by redundancy (RDA) and partial redundancy (pRDA) analyses. The effect of litter on microbiota was assessed by pRDA corrected for other variables (age, treatment, gender) and revealed the expected (Frese et al., 2015) small but significant impact on the intestinal microbiota (explained variation = 4.32%, P = 0.002). The effect of gender was evaluated by pRDA analyses for each time point separately and did not reveal any gender‐related influence on microbiota composition in the present experiment, which is analogous to what has been previously reported (Mach et al., 2015; Ke et al., 2019). A strong age‐related effect (corrected for variables litter, gender and treatment) on microbiome composition at genus level was observed (Fig. 1B; explained variation = 28.22%, P = 0.002). The microbiota composition development as a function of age from day 2 to day +14 displayed a horse‐shoe shaped progression curve over time (Fig. 1B; biplot with red triangles indicating different ages) along with identification of dominant microbial genera (black arrows) associated with different time points. For example, during the pre‐weaning period, microbial groups such as Actinomyces, Fusobacterium, Enterococcus, Peptostreptococcus, Bacteroides and Escherichia‐Shigella were abundant during the first few weeks of age as shown in heatmap representation (Fig. 1C). On the other hand, genera like Romboutsia, Intestinimonas, Methanobrevibacter, Christensellaceae group R‐7 emerged during the third or fourth weeks of age but strikingly disappeared again post‐weaning (Fig. 1C). As expected, a sharp distinction was observed between the pre‐ and post‐weaning microbiota, with several bacterial groups like Prevotella, Faecalibacterium, Lachnospira, Roseburia, Subdoligranulum, Catenibacterium, Succinivibrio, Megasphaera and Coprococcus particularly associated with post‐weaning microbiota composition (Fig. 1B and C, Supplementary Fig. 3B). Hierarchical clustering of age‐related microbial genera (detected by RDA) clearly separated the pre‐ and post‐weaning associated microbiota, which is most probably governed by the sudden dietary shift, from pre‐weaning mother's milk to post‐weaning solid (plant‐based) feed. During the post‐weaning period, the microbiota became more homogeneous among individual animals, which was characterized by comparable alpha diversity scores (Fig. 1A), closely clustered post‐weaning time points (Fig. 1B) and lower Bray–Curtis (PCoA) distance between post‐weaning samples (Supplementary Fig. 3C).
Impact of early life (fibrous) feed intervention on microbiome development
Video observation shows eating behaviour increasing with time
EF litters were provided with fibrous feed from 2 days of age in addition to suckling mother's milk. We observed their eating behaviour around the feed trough using video recordings (0700–1900 h daily; eating time measured in seconds) for 4 weeks pre‐weaning, thereby obtaining a reliable proxy for the amount of feed consumption per piglet (Choudhury et al., 2021) (Experimental procedures). The quantified eating behaviour scores for the EF piglets gradually increased as the piglets became older, reaching the highest in the last week before weaning (Fig. 2A). Eating scores during the first week of life were negligible, with piglets spending no or only a very limited time exploring and eating the feed. However, during the second week of life, all the piglets started exploring and consuming the provided feed, and this behaviour increased rapidly over the third and fourth weeks of age (just before weaning). The individual eating behaviour was also supported by the previously reported increase in feed intake at the litter level during the 4 weeks pre‐weaning (Middelkoop et al., 2020). However, despite this universal and prominent increase in eating behaviour over time, the quantified estimates of eating of individual piglets displayed considerable variation (Middelkoop et al., 2020).
Fig. 2.
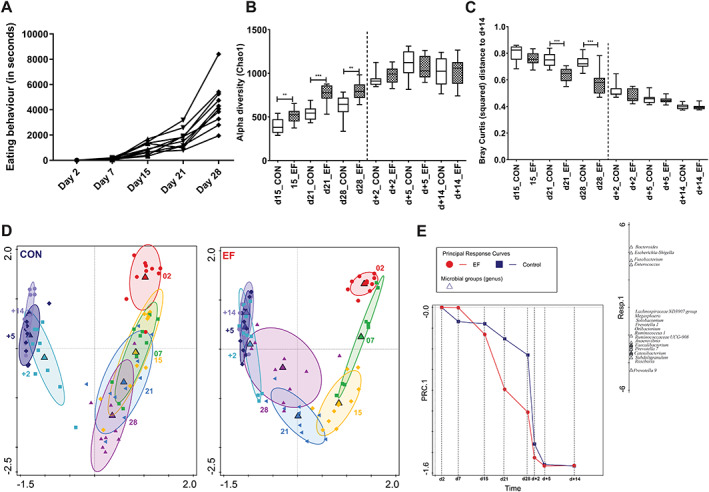
Microbiota colonization development in early‐fed (EF) and control (CON) group.
A. Eating behaviour (total eating seconds per week; n = 10 piglets) in EF litters over time pre‐weaning.
B. Alpha diversity (Chao1 bias corrected) displays diversity differences between the two groups from 15 days onwards.
C. Comparison of squared Bray–Curtis index (distance between different samples with their day +14 time point in individual piglets) between two groups.
D. Principal component analysis of microbiota in all ages (PC1 = 42.4%, PC2 = 12.3%), shown for the CON and EF group separately. The ellipses drawn around each time point represent 66% quantile of the approximated 2D‐Normal density distribution function. Large triangles are centroids of all samples of a specific time point.
E. Principal response curve (PRC) analysis presenting changes in microbiota composition (genus level) across time for EF (red) and CON (green) group. The horizontal axis represents time and the vertical axis denotes PRC score values. Microbial response scores are shown on the right in a one‐dimensional plot. The combination of PRC score values and microbial response scores offers a quantitative interpretation and direction of change at different time points. The effect of treatment (including its interaction with time) was significant according to the Monte Carlo permutation test (P = 0.002). Significant differences between groups were assessed by Student t‐test or Mann–Whitney U test (**P < 0.01; ***P < 0.001).
Early life feeding accelerates microbiome colonization maturation
One of the objectives of our study was to evaluate the impact of early life feeding of fibrous feed on the microbiota composition, compared to the control group of piglets that did not receive any pre‐weaning feed. Therefore, we investigated this effect using the microbiota composition information from 15 days of age onwards, because at the time points prior to day 15, the piglets spent very little time eating solid feed (see above). Remarkably, the period with substantial observed eating behaviour (day 15 and onward; Fig. 2A) appeared to coincide with higher richness levels of the microbiota in EF compared to the CON group (Fig. 2B). Moreover, in EF piglets, squared Bray–Curtis distance (Fig. 2C) of individual samples (day 21 and day 28) was significantly lower from their corresponding day +14, indicating proximity to a more ‘matured’ state. The closeness of the EF group to the post‐weaning microbiome was also reflected in the increased ‘microbiome age’ at pre‐weaning time points (Supplementary Fig. 4A). However, this difference was no longer observed post‐weaning, indicating that the microbiota diversity in the CON group rapidly caught up with that of the EF piglets. These differences between EF and CON groups could be recapitulated in group‐specific (EF or CON) principal component analysis (PCA) analysis (Fig. 2D), where the pre‐weaning time points day 28 and day 21 were considerably closer to the post‐weaning time points in EF piglets, relative to the CON piglets (Fig. 2D). The EF treatment response over time was further investigated by principal response curve (PRC) analysis revealing a significant interaction of early life feeding with time on the first constrained axis (P = 0.002; Fig. 2E). In comparison to the CON group, the microbiota composition of EF group was enriched for several microbial genera (Fig. 2E), including Prevotella 9, Roseburia, Subdoligranulum, Catenibacterium and Faecalibacterium, which are among the genera that are also typically among the microbial groups associated with the post‐weaning microbiota in comparison to the pre‐weaning microbiota (Fig. 1C, Supplementary Fig. 2B). Obviously, day 28 and day 21 emerged as interesting time points to be explored further to evaluate accelerated microbiome colonization in the EF group, which is in agreement with the increased eating scores observed during these later pre‐weaning time points. The strongest discrimination between the EF and CON group was detected by RDA at day 28 (P = 0.002, Fig. 3A), followed by day 21 (P = 0.002; Supplementary Fig. 4B) and day 15 (P = 0.04; Supplementary Fig. 4C). The microbial groups identified as main discriminants between the EF and CON group on day 28 included those identified by PRC analysis, e.g. Prevotella 9, Roseburia, Faecalibacterium, Megasphaera, Succinivibrio, Subdoligranulum, Catenibacterium and Coprococcus, corroborating the enrichment of the post‐weaning associated microbial genera in the EF group pre‐weaning relative to the CON group. The maturation is also illustrated in hierarchical clustering of age‐related microbial genera (detected by RDA), where the EF group at day 28 is clearly separated from the rest of the pre‐weaning clusters (Supplementary Fig. 5). These microbial groups start colonizing EF piglets from 21 and/or 28 days of age (Fig. 3C, Supplementary Fig. 6). For example, Prevotella 9, the dominant genus of the post‐weaning phase, started appearing from day 21 in the EF group and continued to have significantly higher abundance at day 28 (Fig. 3B), compared to the CON group. RDA at earlier time points like day 15 and day 21 (Supplementary Fig. 4B, C) demonstrate the emergence of microbes like Succinivibrio, Ruminococcus 2 (day 15 onwards) and Prevotella 9, Megasphaera, Lachnospira, Subdoligranulum, Coprococcus (day 21 onwards) in EF piglets, which persisted post‐weaning.
Fig. 3.
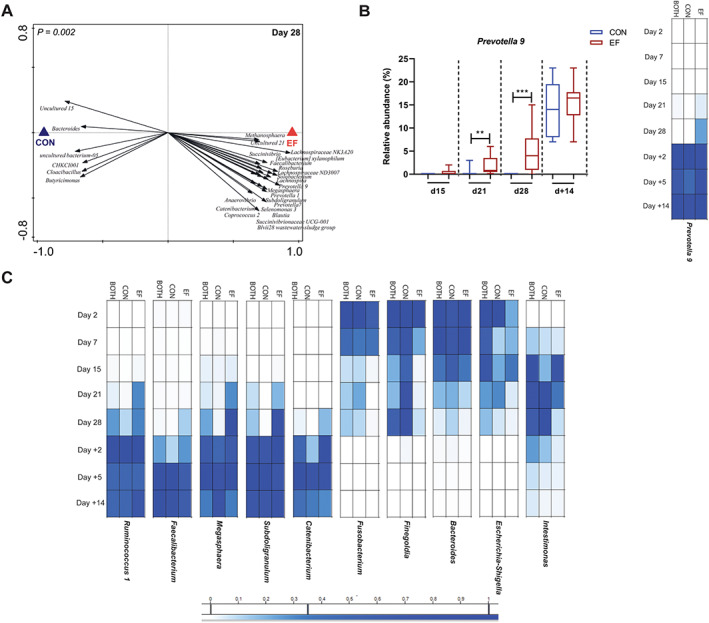
Accelerated microbiota maturation in early‐fed (EF) group illustrated by emergence of post‐weaning associated microbial groups in pre‐weaning period.
A. Redundancy analysis of treatment at day 28 (PC1 = 24.1%, PC2 = 19.7%; P = 0.002) with associated microbial groups in EF and CON groups (response score ≥0.65 on horizontal axis).
B. Changes in relative abundance of Prevotella 9 in EF and CON groups at pre‐ and post‐weaning time points.
C. Heat map of normalized relative abundance for representative individual genera at different ages showing appearance of post‐weaning associated microbial groups and rapid loss of pre‐weaning associated microbes, simultaneously. Significant differences between groups were assessed by Student t‐test or Mann–Whitney U test (**P < 0.01; ***P < 0.001).
Furthermore, the EF group also displayed an accelerated loss of pre‐weaning (day 2–day 7–day 15–day 21–day 28)‐associated microbial genera, like Fusobacterium, Finegoldia, Intestinimonas and Bacteroides (Fig. 3C). Taken together, these results show that early life feeding of fibrous diet accelerates the porcine microbiota colonization, characterized by the earlier appearance and subsequent expansion of post‐weaning associated microbial groups, in parallel with the more rapid disappearance of pre‐weaning associated microbes. The pre‐weaning microbiota differences induced in the EF group appeared to persist somewhat until 5 days post‐weaning (Supplementary Fig. 7A, B) but disappeared 2 weeks post‐weaning (day +14).
Quantitative relation between eating behaviour and microbiota
Since the accelerated microbiota colonization patterns in the EF group coincided with the increased eating behaviour of individual piglets after 2 weeks of age (>15 days), we investigated the quantitative relation between observed eating behaviour and the microbiota composition within the EF group.
Spearman correlation analysis was performed between eating behaviour (summed score per time point) and squared Bray–Curtis distance of individual piglets between a certain time point and their day +14 ‘matured’ time point. It revealed a strong and significant negative correlation between these parameters, for both short term (last day; Supplementary Fig. 8A, last 2 days; r = − 0.77, P < 0.0001; Fig. 4) and longer term (last week; Supplementary Fig. 8B) eating scores prior to the microbiota analysis time point. Similarly, there was a strong positive association between microbiome age and eating behaviour (last 2 days; Supplementary Fig. 8C), fortifying the quantitative relation of eating behaviour and maturing microbiota.
Fig. 4.
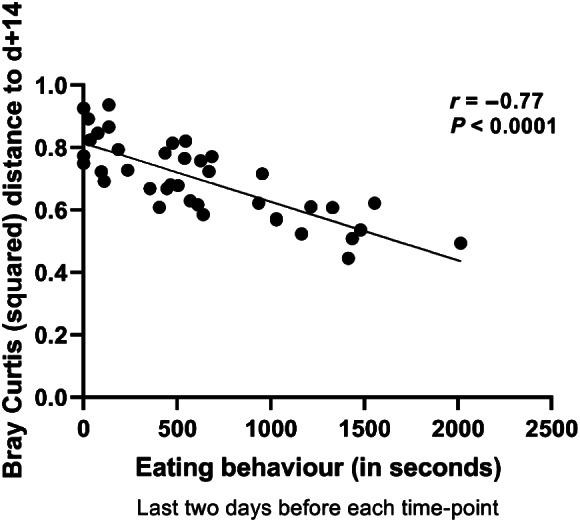
Spearman correlation between eating scores of individual animals (n = 10 EF piglets) and squared Bray–Curtis distance to their day +14 ‘matured’ time point. The last 2 days eating behaviour before each corresponding time point (three time‐points: day 15, day 21 and day 28) was employed (r = −0.77, P < 0.0001).
Impact of early life feeding on relative weight gain and diarrhoea post‐weaning
To assess the impact of early life feeding on post‐weaning performance, bodyweight of piglets was measured at three time points day +2, day +5 and day +14 after weaning. The EF piglets tend to have a higher relative weight gain compared to CON piglets at day +5 post‐weaning (Fig. 5). Notably, we also observed a ‘smoother’ transition for EF piglets in terms of relative weight gain within the first few days post‐weaning, which was further supported by lower coefficients of variation and a consistent lack of weight loss in piglets belonging to the EF group. In addition, we assessed diarrhoea incidence post‐weaning, revealing lower faecal consistency scores (FCS) as well as a shorter duration of diarrhoea during the first week post‐weaning in the EF compared to the CON group (Table 1). However, despite the performance improvements during the acute phase post‐weaning, these differences disappeared at the later post‐weaning phase (day +14; adaptive phase) (Montagne et al., 2007), illustrating the adaptive capacity of young animals.
Fig. 5.
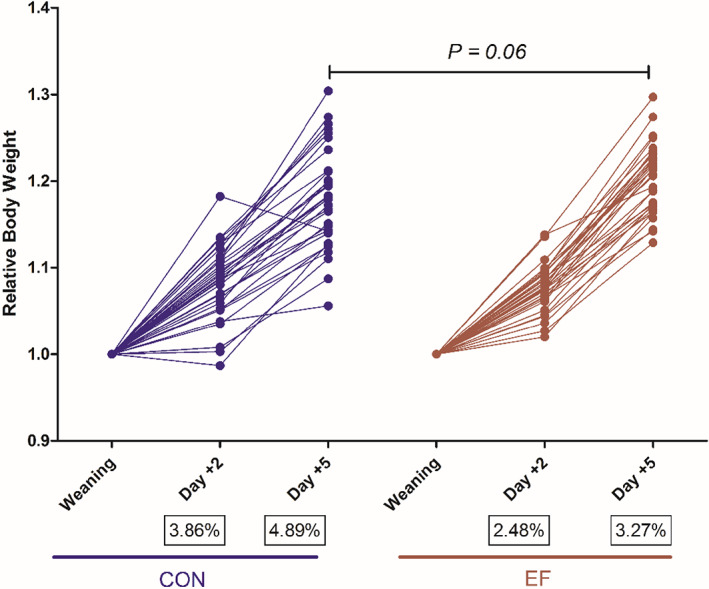
Comparison of relative weight gain between the groups (n = 32 per group) during the weaning transition (till 5 days post‐weaning). Statistical comparisons between the groups were assessed by Student t‐test or Mann–Whitney U test in GraphPad Software 8.1.1. The coefficient of variation (CV%) is indicated in black boxes below the labels.
Table 1.
Post‐weaning diarrhoea measurements at pen level in early‐fed (EF) and control (CON) piglets.
| EF | CON | P‐value | |
|---|---|---|---|
| Faecal consistency score (FCS) | |||
| Day 1–7 | 1.09 ± 0.04 | 1.25 ± 0.06 | 0.04 |
| Day 8–14 | 1.55 ± 0.08 | 1.46 ± 0.08 | 0.45 |
| Diarrhoea, number of days/pen | |||
| Day 1–7 | 0.50 ± 0.19 | 1.38 ± 0.26 | 0.03 |
| Day 8–14 | 3.38 ± 0.65 | 3.25 ± 0.56 | 0.89 |
Discussion
Targeting the early‐life ‘window of opportunity’ to modulate intestinal microbiota can be crucial to improve weaning transition in pigs, which is especially stressful during the ‘acute phase’ immediately post‐weaning (Montagne et al., 2007). In this study, we have assessed the age‐associated microbiota development in young piglets and evaluated the impact of early life feeding (pre‐weaning access to a fibrous feed) on the gut colonization and microbiota maturation over time (until 2 weeks post‐weaning). Our results show that exposure to a fibrous diet in early life accelerates the intestinal microbiota maturation pre‐weaning, which is characterized by an increased microbial diversity, the earlier emergence and expansion of post‐weaning‐associated microbial groups and an enhanced loss of pre‐weaning associated microbes.
We observed a dynamic, age‐related microbiota development with varied microbial groups associated with different time points, and discrete pre‐ and post‐weaning microbiota. This is in line with multiple studies (Alain et al., 2014; Frese et al., 2015; Mach et al., 2015; Niu et al., 2015; Slifierz et al., 2015; Zhao et al., 2015; Guevarra et al., 2018; Li et al., 2018; Ke et al., 2019; Wang et al., 2019) that assessed bacterial communities in early life of piglets, showing age as well as weaning as the driving factors influencing microbiota development. The sudden shift in microbial communities after weaning is likely due to the abrupt dietary change from a highly palatable milk diet to a not‐as‐easily digestible plant‐based solid feed. Firmicutes followed by Bacteroidetes were found to be the dominant phyla across all experimental time points and were particularly abundant post‐weaning, where they accounted for more than 90% of all microbes detected. Although this is consistent with the majority of earlier studies (Kim et al., 2011; Alain et al., 2014; Mach et al., 2015; Niu et al., 2015; Chen et al., 2017; Holman et al., 2017; Ke et al., 2019; Wang et al., 2019), there are a few reports with Bacteroidetes (Lu et al., 2014; Guevarra et al., 2018) or Proteobacteria (Slifierz et al., 2015; Zhao et al., 2015) as the dominant phyla. Analogously, the decreasing abundance of Fusobacteria observed during the first weeks of life has also been reported by several other studies (Alain et al., 2014; Niu et al., 2015; Slifierz et al., 2015; Ke et al., 2019), although there are also studies where this bacterial group was not detected at all (Frese et al., 2015; Guevarra et al., 2018). Such differences in taxonomic identification could, to some extent, be due to the different primers and variable regions of the 16S rRNA gene that were targeted in these metataxonomic investigations, which has been shown to capture different community structures (Albertsen et al., 2015; Holman et al., 2017), disallowing an accurate comparison between different studies. Despite these variations among studies, and the various confounding factors among study designs and (feeding) conditions, including genetics, environmental conditions (farm/rearing hygiene practices), age at weaning, sampling procedures, sample processing (DNA extraction) and sequence analysis methods, there is a striking congruency in the microbiota compositional differences between pre‐ and post‐weaning in most studies (Alain et al., 2014; Frese et al., 2015; Mach et al., 2015; Slifierz et al., 2015). This is largely in agreement with our present study, which pinpoints Bacteroides, Escherichia‐Shigella as early intestinal colonizers and Prevotella as a prominent post‐weaning microbe, along with other typical post‐weaning microbial groups such as Roseburia, Faecalibacterium, Blautia, Subdoligranulum. The drastic increase in relative abundance of Prevotella post‐weaning (from less than 3% to more than 18% of the total community) is likely due to the established capacity of the members of the genus to produce enzymes that can degrade the plant polysaccharides (Flint and Bayer, 2008; Ivarsson et al., 2014), which are prominent constituents of the plant‐based weaner diet. Microbiota metrics like alpha diversity, Bray–Curtis distance, as well as hierarchical clustering clearly separated pre‐ and post‐weaning samples, with the shorter‐term post‐weaning samples (2 and 5 days post‐weaning) being remarkably close to the last post‐weaning time point (14 days post‐weaning). This suggests that the porcine microbiota rapidly converges after weaning, towards an apparently homogeneous and stable microbiome structure within 2 weeks post‐weaning, reflecting the crucial role of diet in dictating the microbiome adaptation. Taken together, the present study confirms and refines various earlier observations of the microbiome development during early life stages in piglets and exemplifies the major impact of the weaning transition that leads to the establishment of a homogenous, rich and stable microbiota composition post‐weaning.
The central aim of the present study was to investigate whether provision of pre‐weaning fibrous diet would impact the microbiome development in suckling piglets. Quantitative eating scores of individual EF piglets rapidly increased over time, reaching the highest score in the last week prior to weaning. This is in agreement with creep feeding studies (Fraser et al., 1994; Bruininx et al., 2002) which provided solid feed pre‐weaning and found that piglets consumed about 60%–80% of total feed in the last week before weaning (4 weeks of age). Since the current study lacks an experimental group fed with traditional creep feed, future research would be necessary to confirm the role of fibres in modulating intestinal microbiota and host physiology. Comparison of different types of creep feed (including liquid milk replacers) could additionally address the question of whether learning to eat solid feed early in life can contribute to the prevention of post‐weaning (psychological/behavioural) stress. Unlike traditional creep feeding diets, in this study, the pre‐weaning diet was designed to contain a mixture of soluble and insoluble fibres that can potentially stimulate the large intestinal microbiota. We demonstrated in the current study that piglets which consumed this pre‐weaning fibrous diet displayed an accelerated microbiota maturation compared to the control (CON) piglets suckling mother's milk only. Acceleration of the microbiota development in the EF piglets was reflected by a higher alpha diversity compared to CON piglets from 15 days of age until weaning, coinciding with the observation of increased eating scores from 15 days of age, thereby suggesting the relation between feed consumption and these microbiota changes. Moreover, the EF piglets' microbiota started to resemble their post‐weaning composition especially during the last 2 weeks of pre‐weaning period, an effect that was not observed in the CON piglets. This effect in the EF piglets was most prominently reflected by the emergence of increasing relative abundances of typical post‐weaning associated genera like Prevotella, Roseburia, Faecalibacterium, Ruminococcus, Megasphaera, Catenibacterium and Subdoligranulum. These typical post‐weaning microbes accumulating in EF piglets were mostly fibre degraders, for example, Prevotella that represents a group of strictly anaerobic bacteria is reported as the dominant genus in the large intestine of pigs (Leser et al., 2002; Liu et al., 2012; Wang et al., 2019), especially after the introduction of fibrous solid feed post‐weaning. Additionally, these EF effects coincided with the accelerated disappearance of typical pre‐weaning associated microbes such as Fusobacterium, Finegoldia, Bacteroides, Escherichia‐Shigella, which did occur at a much slower rate in the CON piglets. Importantly, the degree of resemblance observed was quantitatively correlated to the amount of feed consumed pre‐weaning, strengthening the relation between feed‐consumption and the acceleration in microbiota development (i.e. more eating = more microbiome change). The microbiota differences between the treatment groups disappeared after 2 weeks post‐weaning (day +14), indicating the prominent role of (post‐weaning) diet in convergence of microbiome.
In this study, we also observed the influence of early life feeding on animal performance, especially during the acute phase post‐weaning (i.e. days 0–5 post‐weaning). The EF piglets showed a tendency of higher relative weight gain at 5 days post‐weaning, compared to CON piglets, which was accompanied by a lower variation (CV) and lack of negative trendlines in body weight development. Furthermore, we observed reduced diarrhoea in the first week post‐weaning in EF piglets. These observations support that early life feeding can suppress typical performance and health problems during the acute phase of the weaning transition. In future studies, it would be interesting to investigate whether the short‐lived but favourable effect during early life can influence the health and performance of animals at a later stage of life.
Conclusions
This study illustrates the impact of early life feeding of fibrous diet on gut microbiota, showing an increased microbiome maturation or higher ‘microbiome age’ in EF piglets at pre‐weaning stages. Importantly, the quantitative association between amount of (pre‐weaning) eating and microbiota maturation indicates that the effect may be enhanced by developing pre‐weaning feeding strategies (leading to a higher feed intake). This pre‐weaning intervention has an impact during the acute phase post‐weaning, which is illustrated by a dampening of weaning stress‐related parameters (relative weight gain and diarrhoea). Overall, our results emphasize the potential of early life feed intervention to smoothen the weaning transition, justifying future studies that address the long‐term effects on animal health and performance.
Experimental procedures
Animals and experimental design
The Animal Care and Use Committee of Wageningen University & Research (Wageningen, The Netherlands) approved the protocol of the experiment (AVD104002016515). The protocol is in accordance with the Dutch law on animal experimentation, which complies with the European Directive 2010/63/EU on the protection of animals used for scientific purposes. Ten multiparous Topigs‐20 sows (parity 3–5) housed and inseminated at research facility Carus (Wageningen University & Research, The Netherlands) were divided into two groups (n = 6 litters for early‐fed or EF group; and n = 4 litters for control or CON group) based on sow's parity, body weight and genetic background. Within 2 days after birth, the litter size was set to maximum of 14 piglets per litter (Tempo × Topigs‐20) with no cross‐fostering. The new‐born piglets were cohoused with sow and littermates till weaning (28 days of age) and received ear tags for individual identification and an iron injection, standard to pig husbandry practice. From 2 days of age, the neonatal piglets belonging to the EF group were given access to customized fibrous feed (Supplementary Table 1) ad libitum in addition to suckling sow's milk, whereas the CON group suckled sow's milk only. Briefly, the feed contained 26% dietary non‐starch polysaccharides, mainly originating from sugar beet pulp (4%), oat hulls (4%), inulin (4%), galactooligosaccharides (5%) and high amylose maize starch (4%) as fibrous ingredients. A subset of piglets (n = 64; 32 for each group, EF and CON) were weaned at 4 weeks of age and followed for 2 weeks post‐weaning. At weaning, piglets were mixed within the same treatment group and housed in separate pens with four unfamiliar piglets per pen (i.e. four piglets per pen originating from different litters within treatment). After weaning, all piglets had ad libitum access to a commercial weaner diet (Inno Speen Pro, Coppens Diervoeding, Helmond, The Netherlands). During both pre‐ and post‐weaning periods, precautions were taken to avoid microbiota transfer between pens (via personnel for example) as much as possible. Additional information about the housing and management has been described in detail in a previous study (Middelkoop et al., 2020).
Eating behaviour by video observation
The eating behaviour of piglets was assessed by video recordings from 2 days of age till weaning. For recognition during behavioural observations, piglets were individually numbered on their back using dark permanent hair dye. Eating frequency of individual EF piglets or ‘eating time’ was determined daily from 07:00 to 19:00 h via video observations (continuous sampling) as an estimate for pre‐weaning solid feed intake. From the video observations, the amount of time spent eating or ‘eating time’ was evaluated. When an EF piglet placed its snout into the trough for a minimum of 5 s (s), the behaviour was scored as eating (Pajor et al., 1991; Adeleye et al., 2014). The eating time was categorized into short (5–9 s), medium (10–29 s) and long (≥30 s) eating bouts. The eating bout ended when the snout of the piglet was out of the trough for a minimum of 5 s. When the snout of the piglet went back into the trough within the interval of ≤4 s, the eating bout continued, but these interval seconds were not included to determine eating time. Exploratory behaviour towards the feed trough such as chewing the trough was not scored as eating. Daily/weekly eating time per piglet was (semi‐) quantified by summing the (minimum) number of seconds spent eating (where short, medium and long bouts counted as 5, 10 and 30 s respectively) from 2 days of age to weaning (at 28 days of age). All the observers were trained and instructed with the evaluating criteria to obtain homogeneous and accurate quantification of eating behaviour. Since the eating behaviour observational measurements may have some degree of subjectivity, they were considered as an ‘estimate’ for the amount of eating per piglet. However, analogous to what we have previously reported (Choudhury et al., 2021), eating behavioural scores and the feed‐intake at litter level are strongly correlated (Supplementary Fig. 1), supporting the legitimacy of the eating behaviour as an indicative quantification of eating.
Intestinal microbiota sampling and microbiota metataxonomic analysis
To investigate intestinal microbiota colonization patterns, rectal swab samples were obtained from piglets by inserting a sterile cotton swab (Puritan Medical, Guilford, ME, USA; Cat Number‐25‐3306‐U) 20–30 mm into the rectum and rotating the swab against the bowel wall before placing it into a 5 ml Eppendorf tube. The samples were kept on ice during transport to the laboratory and stored at −20°C until further processing. The selection of piglets for intestinal microbiota sampling (n = 10 per treatment group) was made by the following criteria: (i) no antibiotic treatment, (ii) no pre‐weaning diarrhoea, (iii) close to average weight of the treatment group and (iv) equal male to female ratio. A total of 160 swab samples were collected, repeatedly from the same piglets at five time points pre‐weaning (2, 7, 15, 21, 28 days of age) and three time points post‐weaning (+2, +5 and +14 days of age). At 2 and 7 days of age, samples were collected fixed to their birth date, thereafter all samples were taken on the same day for all piglets fixed to the day of weaning. One sample (from EF group at 7 days of age) was unsuccessful at the sequencing step and therefore could not be included in the analysis. The EF piglets selected for microbiota analysis belonged to four litters before weaning (2–3 piglets selected per litter), and two litters had to be omitted from microbiota analysis due to uterine infection of the sow and subsequent antibiotic treatment during the suckling phase.
DNA extraction from rectal swabs was performed by the repeated bead beating method (Yu and Morrison, 2004) using QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. 500 μl of lysis buffer was added to the 5 ml Eppendorf tube (holding swab) to obtain swab solution, which was used as a starting material for DNA extraction. The quality and quantity of extracted DNA samples were checked by gel electrophoresis (only representative samples) and Nanodrop DeNovix DS‐11 Spectrophotometer (DeNovix, Wilmington, DE, USA) respectively. The DNA template was used for amplifying the V3–V4 region of the bacterial 16S rRNA gene using V3F primer (5′‐CCTACGGGNGGCWGCAG‐3′) and V4R primer (5′‐GACTACHVGGGTATCTAATCC‐3′), 5′‐extended with extension‐PCR‐adapters 5′‐TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG‐3′ and 5′‐GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG‐3′ respectively. PCR Amplification (Bio‐Rad C1000 thermal cycler, Bio‐Rad Laboratories, Veenendaal, The Netherlands) of the 16S rRNA V3–V4 region was completed in a 50 μl reaction volume consisting of 5 μl 10× KOD buffer (Toyobo, Japan), 3 μl MgSO4 (25 mM) 5 μl dNTPs (2 mM each), 1.5 μl V3F primer [10 μM (Eurogentec, Luik, Belgium)], 1.5 μl V4R primer [10 μM, (Eurogentec)], 1.0 μl (0.02 U μl−1) KOD hot start DNA polymerase (Toyobo, Japan) and 10 ng (minimum) of template DNA. The amplification conditions included a single initiation cycle of 95°C for 2 min, followed by 25 amplification cycles encompassing denaturation at 95°C for 20 s, annealing at 55°C for 10 s, and elongation at 70°C for 15 s, and was completed by a single elongation step at 72°C for 5 min. Amplicons were purified using MSB Spin PCRapace (STRATEC Molecular, Germany) and were sequenced at BaseClear BV (Leiden, The Netherlands) using (paired‐end) Illumina MiSeq system. Purified amplicons were subjected to extension‐PCR using barcoded Illumina universal index sequencing adapters prior to sequencing. The Illumina MiSeq system generated FASTAQ sequence files and these sequences were subjected to quality control based on BaseClear in‐house quality/filtering protocol, removing reads containing adapters (up to minimum read length of 50 bp) and/or PhiX control signal, to generate the FASTAQ data file used for microbiota analysis.
Illumina reads were imported into the CLC Genomics Workbench version 11.01 and were processed using the CLC Microbial Genomics Module version 2.5.1 (CLC bio, Arhus, Denmark). The paired‐end reads were merged into one high‐quality representative by CLC Workbench (Mismatch cost = 1, Minimum score = 40, Gap Cost = 4, Maximum unaligned end mismatches = 5). The CLC pipeline was used for primer and quality trimming (Trim using quality scores = 0.05; Trim ambiguous nucleotides: maximum number of ambiguities = 2; Discard reads below length = 5). The remaining high‐quality sequences were clustered into operational taxonomic units (OTUs) at 97% identity threshold using SILVA database v132 (released on Dec. 13, 2017) (Quast et al., 2013). OTUs with less than 2 reads (Minimum combined count = 2) were excluded from the analyses. To achieve even sequencing depth between samples, the OTU table was rarefied to 14 000 reads for calculation of alpha and beta diversity indices. PCA (unsupervised), RDA (supervised) and pRDA (supervised) were performed using CANOCO 5 (Microcomputer Power, Ithaca, NY, USA) according to manufacturer's instructions (ter Braak and Smilauer, 2012). To evaluate the impact of environmental variables like age, litter/pen, gender and treatment separately, RDA and pRDA were performed, the latter analyses (pRDA) allowing us to correct for other covariates in the data as described in Canoco 5 manual (ter Braak and Smilauer, 2012). The litter/pen‐related impact was assessed separately per time point and in pre‐ and post‐weaning phases to avoid confounding age and weaning‐related differences. The microbial groups detected by RDA were selected by their response scores in the biplot (obtained by perpendicular projection of the taxa arrows on the X‐axis) and were further filtered by the condition of having ≥0.1% average relative abundance in one of the time points at least (for biological relevance). The median relative abundance of selected microbial groups was calculated for each of the time points in 20 piglets (assessing age) or 10 piglets (assessing treatment groups). Relative abundance of age‐related microbial groups was visualized by heat maps by normalizing by the highest median of all time points (scaling from 0 to 1) in Perseus (Tyanova et al., 2016). Euclidean distance was utilized to measure the distance and clustering was conducted using the average linkage method. Time‐dependent treatment effects were assessed by using a linear ordination method known as PRCs in Canoco 5, which is based on RDA, adjusted for overall changes in microbial community response over time (Van Den Brink and Ter Braak, 1999). The principal component is plotted against time, yielding a PRC of the community for each treatment. The PRC method also shows microbial response scores by a one‐dimensional plot, thereby enabling quantitative interpretation of treatment effect towards the microbial species level. Statistical significance was evaluated by the Monte Carlo permutation procedure with 499 permutations. Response‐variable‐based case scores (CaseR) was extracted from redundancy analysis of age, which was defined as their ‘microbiome age’ as they correspond to the individual sample position in the progressing‐age‐ordination plot. To evaluate the quantitative relationship between observed eating behaviour and magnitude of the microbiota change, a non‐parametric Spearman correlation was performed between eating scores (in seconds) and squared Bray–Curtis distance of piglets or ‘microbiome age’ between individual time points and their corresponding day +14 time point in GraphPad Software 8.1.1 (CA, USA, www.graphpad.com). Differential abundance analysis between the CON and EF group was implemented in CLC Genomics Workbench (edgeR test) from day 15 onwards, as shown in Supplementary Table 2. Comparison of the specific taxa relative abundance and diversity indices between treatments were performed by Mann–Whitney U test, whereas comparison among time points was assessed by one‐way ANOVA (Kruskal–Wallis statistic) using a Dunnett's test for multiple comparison using GraphPad Software 8.1.1. The level of statistical significance was set at P < 0.05 with a trend defined as 0.1 > P ≥ 0.05.
Post‐weaning performance data analysis
Piglets were weighed individually in all microbiota‐sample‐collection time points pre‐ and post‐weaning. Relative weight gain of piglets (normalized to their weaning weights at day 28) was calculated in the early post‐weaning period to evaluate their body weight development during the weaning transition. To assess differences between the groups, the Mann–Whitney U test and coefficient of variation (CV%) were calculated in GraphPad Software 8.1.1.
Faecal consistency in the weaner pens (n = 8 per group) were scored daily for 14 days post‐weaning based on a faecal classification scale of Pedersen and Toft (2011). Score 1 represented normal (firm/soft shaped) faeces, whereas scores 2 (loose faeces) and 3 (watery faeces) represented diarrhoea. FCSs (per pen) were calculated by averaging the daily score (at pen level) for the first and second week post‐weaning and were subsequently analysed in SAS 9.4 (SAS Institute, Cary, NC, USA) using a general linear mixed model. Furthermore, the number of days with post‐weaning diarrhoea was analysed using a generalized linear mixed model with a Poisson distribution, log link function and an additional multiplicative over‐dispersion parameter. The main effect of pre‐weaning access to a fibrous diet (EF vs. CON) was included in both models.
Supporting information
Appendix S1: Supporting Information.
Supplementary Table 2. Differential abundance analysis of intestinal microbiota in control (CON) and early fed (EF) piglets from day 15 onwards (using EdgeR test in CLC genomics workbench). The microbial genera having ≥0.1% average relative abundance in at least one time‐point are listed here and are sorted by their combined relative abundance across all time‐points.
Acknowledgements
The authors want to thank the technicians and students of the Host‐Microbe Interactomics group and Adaptation Physiology group (Wageningen University & Research) who helped during the animal experiment, sample processing and video observations. We would like to acknowledge the personnel and trainees of the animal research facilities in Wageningen (Carus) for taking care of the animals and for their technical assistance. The authors are grateful to Tamme Zandstra for producing the experimental feed and to FrieslandCampina DOMO (Amersfoort, The Netherlands) for provision of Vivinal GOS.
Data Availability Statement
Raw sequences can be found on SRA‐NCBI (Sequence Read Archive‐National Center for Biotechnology Information) database under the SRA accession number PRJNA684557 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA684557).
References
- Adeleye, O.O. , Guy, J.H. , and Edwards, S.A. (2014) Exploratory behaviour and performance of piglets fed novel flavoured creep in two housing systems. Anim Feed Sci Technol 191: 91–97. [Google Scholar]
- Alain, B.P.E. , Chae, J.P. , Balolong, M.P. , Bum Kim, H. , and Kang, D.K. (2014) Assessment of fecal bacterial diversity among healthy piglets during the weaning transition. J Gen Appl Microbiol 60: 140–146. [DOI] [PubMed] [Google Scholar]
- Albertsen, M. , Karst, S.M. , Ziegler, A.S. , Kirkegaard, R.H. , and Nielsen, P.H. (2015) Back to basics – the influence of DNA extraction and primer choice on phylogenetic analysis of activated sludge communities. PLoS One 10: e0132783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruininx, E.M.A.M. , Binnendijk, G.P. , Schrama, J.W. , Den Hartog, L.A. , Everts, H. , and Beynen, A.C. (2002) Effect of creep feed consumption on individual feed intake characteristics and performance of group‐housed weanling pigs. J Anim Sci 80: 1413–1418. [DOI] [PubMed] [Google Scholar]
- Chen, L. , Xu, Y. , Chen, X. , Fang, C. , Zhao, L. , and Chen, F. (2017) The maturing development of gut microbiota in commercial piglets during the weaning transition. Front Microbiol 8: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury, R. , Middelkoop, A. , de Souza, J.G. , van Veen, L. , Gerrits, W.J. , Kemp, B. , et al. (2021) Impact of early‐life feeding on local intestinal microbiota and digestive system development in piglets. Sci Rep 11: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Besten, G. , Van Eunen, K. , Groen, A.K. , Venema, K. , Reijngoud, D.J. , and Bakker, B.M. (2013) The role of short‐chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54: 2325–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everaert, N. , Van Cruchten, S. , Weström, B. , Bailey, M. , Van Ginneken, C. , Thymann, T. , and Pieper, R. (2017) A review on early gut maturation and colonization in pigs, including biological and dietary factors affecting gut homeostasis. Anim Feed Sci Technol 233: 89–103. [Google Scholar]
- Flint, H.J. , and Bayer, E.A. (2008) Plant cell wall breakdown by anaerobic microorganisms from the mammalian digestive tract. Ann N Y Acad Sci 1125: 280–288. [DOI] [PubMed] [Google Scholar]
- Fraser, D. , Pajor, E.A. , and Feddes, J.J.R. (1994) The relationship between creep feeding behavior of piglets and adaptation to weaning: effect of diet quality. Can J Anim Sci 74: 1–6. [Google Scholar]
- Frese, S.A. , Parker, K. , Calvert, C.C. , and Mills, D.A. (2015) Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome 3: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa, Y. , Obata, Y. , Fukuda, S. , Endo, T.A. , Nakato, G. , Takahashi, D. , et al. (2013) Commensal microbe‐derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504: 446–450. [DOI] [PubMed] [Google Scholar]
- Gresse, R. , Chaucheyras‐Durand, F. , Fleury, M.A. , Van de Wiele, T. , Forano, E. , and Blanquet‐Diot, S. (2017) Gut microbiota dysbiosis in postweaning piglets: understanding the keys to health. Trends Microbiol 25: 851–873. [DOI] [PubMed] [Google Scholar]
- Guevarra, R.B. , Hong, S.H. , Cho, J.H. , Kim, B.R. , Shin, J. , Lee, J.H. , et al. (2018) The dynamics of the piglet gut microbiome during the weaning transition in association with health and nutrition. J Anim Sci Biotechnol 9: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo, J.M. , Opapeju, F.O. , Pluske, J.R. , Kim, J.C. , Hampson, D.J. , and Nyachoti, C.M. (2013) Gastrointestinal health and function in weaned pigs: a review of feeding strategies to control post‐weaning diarrhoea without using in‐feed antimicrobial compounds. J Anim Physiol Anim Nutr (Berl) 97: 207–237. [DOI] [PubMed] [Google Scholar]
- Holman, D.B. , Brunelle, B.W. , Trachsel, J. , and Allen, H.K. (2017) Meta‐analysis to define a core microbiota in the swine gut. mSystems 2: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivarsson, E. , Roos, S. , Liu, H.Y. , and Lindberg, J.E. (2014) Fermentable non‐starch polysaccharides increases the abundance of Bacteroides‐Prevotella‐Porphyromonas in ileal microbial community of growing pigs. Animal 8: 1777–1787. [DOI] [PubMed] [Google Scholar]
- Jensen, P. , and Stangel, G. (1992) Behaviour of piglets during weaning in a seminatural enclosure. Appl Anim Behav Sci 33: 227–238. [Google Scholar]
- Ke, S. , Fang, S. , He, M. , Huang, X. , Yang, H. , Yang, B. , et al. (2019) Age‐based dynamic changes of phylogenetic composition and interaction networks of health pig gut microbiome feeding in a uniformed condition. BMC Vet Res 15: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.B. , Borewicz, K. , White, B.A. , Singer, R.S. , Sreevatsan, S. , Tu, Z.J. , and Isaacson, R.E. (2011) Longitudinal investigation of the age‐related bacterial diversity in the feces of commercial pigs. Vet Microbiol 153: 124–133. [DOI] [PubMed] [Google Scholar]
- Le Dividich, J. , and Sève, B. (2000) Effects of underfeeding during the weaning period on growth, metabolism, and hormonal adjustments in the piglet. Domest Anim Endocrinol 19: 63–74. [DOI] [PubMed] [Google Scholar]
- Leser, T.D. , Amenuvor, J.Z. , Jensen, T.K. , Lindecrona, R.H. , Boye, M. , and Moøller, K. (2002) Culture‐independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl Environ Microbiol 68: 673–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Guo, Y. , Wen, Z. , Jiang, X. , Ma, X. , and Han, X. (2018) Weaning stress perturbs gut microbiome and its metabolic profile in piglets. Sci Rep 8: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Ivarsson, E. , Dicksved, J. , Lundh, T. , and Lindberg, J.E. (2012) Inclusion of chicory (Cichorium intybus L.) in pigs' diets affects the intestinal microenvironment and the gut microbiota. Appl Environ Microbiol 78: 4102–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, X.M. , Lu, P.Z. , and Zhang, H. (2014) Bacterial communities in manures of piglets and adult pigs bred with different feeds revealed by 16S rDNA 454 pyrosequencing. Appl Microbiol Biotechnol 98: 2657–2665. [DOI] [PubMed] [Google Scholar]
- Mach, N. , Berri, M. , Estelle, J. , Levenez, F. , Lemonnier, G. , Denis, C. , et al. (2015) Early‐life establishment of the swine gut microbiome and impact on host phenotypes. Environ Microbiol Rep 7: 554–569. [DOI] [PubMed] [Google Scholar]
- Middelkoop, A. , Choudhury, R. , Gerrits, W.J.J.J. , Kemp, B. , Kleerebezem, M. , and Bolhuis, J.E. (2020) Effects of creep feed provision on behavior and performance of piglets around weaning. Front Vet Sci 7: 879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeser, A.J. , Pohl, C.S. , and Rajput, M. (2017) Weaning stress and gastrointestinal barrier development: implications for lifelong gut health in pigs. Anim Nutr 3: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne, L. , Boundry, G. , Favier, C. , Le Huerou‐Luron, I. , Lallès, J.P. , and Sève, B. (2007) Main intestinal markers associated with the changes in gut architecture and function in piglets after weaning. Br J Nutr 97: 45–57. [DOI] [PubMed] [Google Scholar]
- Newberry, R.C. , and Wood‐Gush, D.G.M. (1985) The suckling behaviour of domestic pigs in a semi‐natural environment. Behaviour 95: 11–25. [Google Scholar]
- Niu, Q. , Li, P. , Hao, S. , Zhang, Y. , Kim, S.W. , Li, H. , et al. (2015) Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci Rep 5: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowland, T.L. , Plush, K.J. , Barton, M. , and Kirkwood, R.N. (2019) Development and function of the intestinal microbiome and potential implications for pig production. Animals 9: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajor, E.A. , Fraser, D. , Pajor, E.A. , Fraser, D. , and Kramer, D.L. (1991) Consumption of solid food by suckling pigs: individual variation and relation to weight gain consumption of solid food by suckling pigs. Appl Anim Behav Sci 32: 139–155. [Google Scholar]
- Pedersen, K.S. , and Toft, N. (2011) Intra‐ and inter‐observer agreement when using a descriptive classification scale for clinical assessment of faecal consistency in growing pigs. Prev Vet Med 98: 288–291. [DOI] [PubMed] [Google Scholar]
- Pluske, J.R. , Turpin, D.L. , and Kim, J.C. (2018) Gastrointestinal tract (gut) health in the young pig. Anim Nutr 4: 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putignani, L. , Del Chierico, F. , Petrucca, A. , Vernocchi, P. , and Dallapiccola, B. (2014) The human gut microbiota: a dynamic interplay with the host from birth to senescence settled during childhood. Pediatr Res 76: 2–10. [DOI] [PubMed] [Google Scholar]
- Quast, C. , Pruesse, E. , Yilmaz, P. , Gerken, J. , Schweer, T. , Yarza, P. , et al. (2013) The SILVA ribosomal RNA gene database project: improved data processing and web‐based tools. Nucleic Acids Res 41: 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schokker, D. , Zhang, J. , Vastenhouw, S.A. , Heilig, H. , Smidt, H. , Rebel, J.M.J. , and Smits, M.A. (2015) Long‐lasting effects of early‐life antibiotic treatment and routine animal handling on gut microbiota composition and immune system in pigs. PLoS One 10: e0116523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slifierz, M.J. , Friendship, R.M. , and Weese, J.S. (2015) Longitudinal study of the early‐life fecal and nasal microbiotasof the domestic pig. BMC Microbiol 15: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Braak, C.J.F. and Smilauer, P. (2012) Canoco reference manual and user's guide: software for ordination, version 5.0, Microcomputer Power.
- Timmerman, H.M. , Rutten, N.B.M.M. , Boekhorst, J. , Saulnier, D.M. , Kortman, G.A.M. , Contractor, N. , et al. (2017) Intestinal colonisation patterns in breastfed and formula‐fed infants during the first 12 weeks of life reveal sequential microbiota signatures. Sci Rep 7: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova, S. , Temu, T. , Sinitcyn, P. , Carlson, A. , Hein, M.Y. , Geiger, T. , et al. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13: 731–740. [DOI] [PubMed] [Google Scholar]
- Van Den Brink, P.J. , and Ter Braak, C.J.F. (1999) Principal response curves: analysis of time‐dependent multivariate responses of biological community to stress. Environ Toxicol Chem 18: 138–148. [Google Scholar]
- van der Beek, C.M. , Dejong, C.H.C. , Troost, F.J. , Masclee, A.A.M. , and Lenaerts, K. (2017) Role of short‐chain fatty acids in colonic inflammation, carcinogenesis, and mucosal protection and healing. Nutr Rev 75: 286–305. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Tsai, T. , Deng, F. , Wei, X. , Chai, J. , Knapp, J. , et al. (2019) Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome 7: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, X. , Tan, B. , Song, M. , Ji, P. , Kim, K. , Yin, Y. , and Liu, Y. (2019) Nutritional intervention for the intestinal development and health of weaned pigs. Front Vet Sci 6: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Z. , and Morrison, M. (2004) Improved extraction of PCR‐quality community DNA from digesta and fecal samples. Biotechniques 36: 808–812. [DOI] [PubMed] [Google Scholar]
- Zhao, W. , Wang, Y. , Liu, S. , Huang, J. , Zhai, Z. , He, C. , et al. (2015) The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS One 10: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, L. , Chen, H. , Zhang, S. , Zhuang, J. , Li, Q. , and Feng, Z. (2019) Intestinal microbiota in early life and its implications on childhood health. Genomics Proteomics Bioinformatics 17: 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information.
Supplementary Table 2. Differential abundance analysis of intestinal microbiota in control (CON) and early fed (EF) piglets from day 15 onwards (using EdgeR test in CLC genomics workbench). The microbial genera having ≥0.1% average relative abundance in at least one time‐point are listed here and are sorted by their combined relative abundance across all time‐points.
Data Availability Statement
Raw sequences can be found on SRA‐NCBI (Sequence Read Archive‐National Center for Biotechnology Information) database under the SRA accession number PRJNA684557 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA684557).


