Abstract
Four axenic bacterial species capable of biodegrading nitroglycerin (glycerol trinitrate [GTN]) were isolated from soil samples taken from a washwater soakaway at a disused GTN manufacturing plant. The isolates were identified by 16S rRNA gene sequence homology as Pseudomonas putida, an Arthrobacter species, a Klebsiella species, and a Rhodococcus species. Each of the isolates utilized GTN as its sole nitrogen source and removed nitro groups sequentially from GTN to produce glycerol dinitrates and mononitrates (GMN), with the exception of the Arthrobacter strain, which achieved removal of only the first nitro group within the time course of the experiment. The Klebsiella strain exhibited a distinct preference for removal of the central nitro group from GTN, while the other five strains exhibited no such regioselectivity. All strains which removed a second nitro group from glycerol 1,2-dinitrate showed regiospecific removal of the end nitro group, thereby producing glycerol 2-mononitrate. Most significant was the finding that the Rhodococcus species was capable of removing the final nitro group from GMN and thus achieved complete biodegradation of GTN. Such complete denitration of GTN has previously been shown only in mixed bacterial populations and in cultures of Penicillium corylophilum Dierckx supplemented with an additional carbon and nitrogen source. Hence, to the best of our knowledge, this is the first report of a microorganism that can achieve complete denitration of GTN.
For most of the 20th century, nitroglycerin (glycerol trinitrate [GTN]) has been a major explosive for both military and civilian applications, and production facilities have been established throughout the world. Synthesis of GTN involves direct nitration of glycerol with nitric acid (with sulfuric acid as a catalyst). The low solubility of GTN in water is exploited in the washing of GTN oil with aqueous media to free the product from residual mineral acids and ions. The washing wastewaters, saturated with GTN (ca. 8 mM) and bearing suspended droplets of GTN, have commonly been transferred to lagoons or soakaways, resulting in actual or potential contamination of soils. However, at the soakaways used for over 80 years at what was once the second largest commercial explosive manufacturing plant in the world, recent extensive site investigation and analysis of soils (10) have shown a remarkable absence of detectable GTN (R. Morris, personal communication).
In recent years, the first discovery and isolation of GTN-degrading bacteria in activated sewage sludge, river water, and soils (18) led to similar work by other workers in Europe and the United States. Thus, bacteria able to utilize GTN as a sole source of nitrogen are now well known (3, 4, 9, 16, 17, 19). Although several of these bacterial strains were capable of removing either one or two nitro groups from GTN to form glycerol dinitrates (GDN) and glycerol mononitrates (GMN), none was able to biodegrade GMN and thus achieve complete mineralization. However, complete biodegradation has been observed in mixed bacterial cultures (1, 15) and fungi (20). Moreover, an analysis of theoretical energy yields, based on entirely reasonable assumptions about the catabolic pathway of GTN, has shown that GTN should be capable of supporting aerobic and anaerobic growths (12).
With these points in mind, we undertook a search for the presence in soakaway soils of bacteria capable of effecting a complete degradation of GTN. The study not only yielded the first known examples of a single strain which could achieve complete denitration of GTN but also expanded the list of known species capable of utilizing GTN as a sole nitrogen source.
MATERIALS AND METHODS
Materials.
A stock solution of GTN (5% [vol/vol] in ethanol) was kindly provided by EXCHEM, Derbyshire, United Kingdom. Unless stated otherwise, all chemicals were obtained from Fisher Scientific, Loughborough, Leicestershire, United Kingdom.
High-purity water (18 MΩ, filtered through 0.22-μm-pore-size filters) from a Milli-Q50 system (Millipore, Watford, United Kingdom) was used for the preparation of aqueous high-pressure liquid chromatography (HPLC) eluents and for rinsing eluent-containing glassware. HPLC grade methanol of far UV grade quality was obtained from Fisher Scientific. Helium and compressed air, for operating the HPLC system, were from BOC, London, United Kingdom.
Taq polymerase and deoxynucleoside triphosphates were obtained from Promega, Southampton, United Kingdom, and New England Biolabs Ltd., Hitchin, United Kingdom, respectively. PCR primers were synthesized by GIBCO BRL, Life Technologies Ltd., Paisley, United Kingdom.
Culture media.
Basal salts medium comprised the following (per liter): K2HPO4, 3.5 g; KH2PO4, 1.5 g; NaCl, 0.5 g; MgSO4, 0.12 g; and 1 ml of trace elements solution. The trace elements solution contained the following (grams per liter): sodium borate, 0.57; FeCl3 · 6H2O, 0.24; CoCl2 · 6H2O, 0.04; CuSO4 · 5H2O, 0.06; MnCl2 · 4H2O, 0.03; ZnSO4 · 7H2O, 0.31; and Na2MoO4 · 2H2O, 0.03. After being autoclaved and cooled, the basal salts medium was amended with 1% (vol/vol) glycerol and GTN (from the ethanolic working stock solution).
Bacteria were also grown and maintained on nutrient broth and nutrient agar, made up according to the manufacturer's instructions (Difco Laboratories, Detroit, Mich.).
Isolation, maintenance, and growth of bacteria.
Bacteria capable of growth at the expense of GTN as the sole nitrogen source were isolated from four soil samples (SP1a, SP1b, SP2, and SP3) taken from a washwater soakaway at a disused GTN manufacturing plant in Somerset West, South Africa. For each enrichment culture, a sample (25 g) of soil was added to 250 ml of basal salts medium containing 1% (vol/vol) glycerol as a carbon source and 0.2 mM GTN as the sole nitrogen source. After incubation at 30°C and shaking at 100 rpm for 2 weeks, a 5-ml portion was diluted into 250 ml of fresh growth medium. A further three serial subcultures were made at approximately weekly intervals, using a 1% (vol/vol) inoculum. Bacterial growth in liquid cultures was monitored by measuring the optical density of the medium at 540 nm. Samples from the final enrichment flask were serially diluted and spread onto nutrient agar plates to allow visible colonies to develop from single bacteria. Single colony types were separated and subcultured onto fresh nutrient agar plates to purity.
Strains were maintained on nutrient agar slopes and plates with regular transfers onto fresh medium. For long-term storage, strains were stored at −70°C in Protect Bacterial Preservers (Technical Service Consultants, Heywood, Lancashire, United Kingdom).
Identification of bacteria.
Bacteria were identified using 16S rRNA gene sequence homology. Genomic DNA was obtained from bacteria grown overnight at 30°C in 5 ml of nutrient broth. Cells were harvested from 1 ml of culture by centrifugation and resuspended in 100 μl of sterile water. The cell suspension was heated at 94°C in a Primus Thermocycler (MWG-Biotech, Ebersberg, Germany) to lyse the cells and release the genomic DNA. Cell debris were removed by centrifugation at maximum speed in a microcentrifuge for 5 min, and the supernatant was utilized as the template in the PCR.
The PCR mixture consisted of 0.2 to 1.0 μl of the genomic DNA template solution, 1× PCR buffer, 1.5 mM MgCl2, 100 ng each of primers 63f (5′ CAG GCC TAA CAC ATG CAA GTC 3′) and 1387r (5′ GGG CGG WGT GTA CAA GGC 3′) (8), 0.2 mM each of dATP, dCTP, dGTP, and dTTP, and 0.25 U of Taq polymerase in a final volume of 25 μl. Thermal cycling was undertaken by initially denaturing the DNA at 96°C for 5 min, followed by 30 cycles of 96°C for 30 s, 55°C for 30 s, and 72°C for 30 s and finally 72°C for 5 min. A portion of the reaction mixture was used to visualize PCR products on 1% (wt/vol) agarose gels. PCR products in the remainder of the reaction mixture were purified using a QIAquick PCR purification kit (QIAGEN, Ltd., Crawley, West Sussex, United Kingdom) and sequenced using an ABI PRISM BigDye terminator cycle ready reaction kit according to the manufacturer's instructions (PE Applied Biosystems, Warrington, United Kingdom). Sequence data obtained were compared with known 16S rDNA sequences using the BLAST algorithm at http://www.ncbi.nlm.nih.gov/BLAST/ (2).
Growth studies.
Each of the six axenic strains was grown at 30°C in basal salts medium containing 1% (vol/vol) glycerol and 0.2 mM GTN to create a starter culture which could be used as an inoculum. Aliquots (1 ml) of each of these starter cultures were transferred to 100 ml of fresh medium, and growth was monitored by measuring the optical density at 540 nm. When necessary, samples (1 ml) were removed at intervals during growth and centrifuged to remove bacterial cells and the supernatant was stored at −70°C for subsequent analysis of nitrate ester content. Concentrations of GTN and its potential biodegradation products, glycerol 1,3-dinitrate (1,3-GDN), glycerol 1,2-dinitrate (1,2-GDN), glycerol 1-mononitrate (1-GMN), and glycerol 2-mononitrate (2-GMN) were measured by HPLC.
HPLC analysis of nitrate esters.
Samples (1 ml) were filtered through 0.2-μm-pore-size Anotop IC filters (Whatman, Maidstone, United Kingdom). HPLC analysis of the filtrate was performed using a DX300 series ion chromatograph unit (Dionex, Camberley, United Kingdom) consisting of an advanced gradient pump (AGP-1) and basic chromatography module with a 50-μl injection loop, linked to a variable-wavelength UV detector via a Lichrosorb ODS column (250 by 4.6 mm, 10-μm bead) fitted with a 10-mm guard column (Phase Separations, Deeside, United Kingdom). Nitrate esters were separated using a programmed gradient of 5% (vol/vol) methanol in water for 5 min, followed by a linear gradient of 5 to 50% (vol/vol) methanol over 30 min, and finally 50% (vol/vol) methanol for 5 min at a flow rate of 1 ml/min. Column effluents were monitored by UV at A217. The system was calibrated using authentic GTN, 1,3-GDN, 1,2-GDN, 1-GMN, and 2-GMN standards (Radian International, Austin, Tex.).
RESULTS
Isolation and identification of bacteria capable of GTN biodegradation.
Standard enrichment techniques initially yielded 7, 8, 5, and 6 different colony types from soils SP1a, SP1b, SP2, and SP3, respectively. Each of the 26 isolates obtained was reinoculated into basal salts medium containing both GTN and glycerol to test the ability of the axenic strains for growth on GTN as the sole source of nitrogen. Growth within 1 to 2 days was observed for six of the axenic strains isolated, namely strains SP1a-2 (i.e., isolate number 2 from sample SP1a), SP1b-5, SP1b-6, SP2-4, SP2-5, and SP3-4. These six axenic strains capable of effecting GTN biodegradation were identified by 16S rRNA gene sequence homology (Table 1). Other strains showed no growth over a period of 5 days.
TABLE 1.
Identification by 16S rRNA gene sequence homology of axenic strains which grow at the expense of GTN as a sole source of nitrogen
| Isolate | Strain | % Identity |
|---|---|---|
| SP1a-2 | P. putida | 98 |
| SP1b-5 | A. ureafaciens | 99 |
| SP1b-6 | P. putida | 98 |
| SP2-4 | Rhodococcus sp. | 97 |
| SP2-5 | K. oxytoca | 98 |
| SP3-4 | Rhodococcus sp. | 98 |
Biodegradation of GTN by axenic strains.
Starter cultures of each of the six axenic strains were prepared by growth for 5 days at 30°C in basal salts medium containing 1% (vol/vol) glycerol and 0.2 mM GTN as the sole source of nitrogen. Aliquots (1 ml) of each starter culture were transferred axenically into 100 ml of fresh, like medium, and the growth and biodegradation of GTN and its metabolites were monitored.
Pseudomonas putida strains SP1a-2 and SP1b-6.
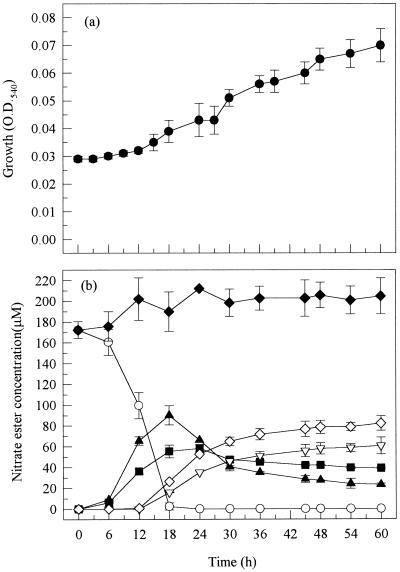
Although the growth of strain SP1a-2 was relatively slow over the 60-h time course studied (Fig. 1), complete disappearance of GTN occurred within the first 18 h of the experiment. GTN was converted into a mixture of 1,3-GDN and 1,2-GDN in a ratio of approximately 1 to 2 (the same ratio as the relative concentrations of central and terminal groups), indicating random removal of either the central or one of the two equivalent terminal groups. Conversion of the dinitrates to mononitrates occurred more slowly, with the 1,2-GDN being converted more rapidly to the mononitrates than was the 1,3-GDN. As a result, the final 1,3-GDN/1,2-GDN/1-GMN/2-GMN ratio in the growth medium was approximately 2 to 1 to 3 to 4 at the end of the experiment (Table 2). The yield of 2-GMN accounted for most of the 1,2-GDN formed, indicating little conversion of the latter to 1-GMN. The total concentration of nitrate esters in the growth medium did not diminish over the time course of the experiment (Fig. 1), showing an inability of this strain to degrade either of the GMN isomers.
FIG. 1.
Biodegradation of GTN by P. putida strain SP1a-2 showing bacterial growth (a) and concentration of nitrate esters (b). Symbols: ○, GTN; ■, 1,3-GDN; ▴, 1,2-GDN; ▿, 1-GMN; ◊, 2-GMN; ⧫, total nitrate esters. O.D.540, optical density at 540 nm.
TABLE 2.
Comparison of strains for ability to biotransform 200 μM GTN
| Strain | Conversion of:
|
GMN degradation time (h)b | Composition of medium at 60 h as % of initial GTN concn
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GTN to GDN
|
GDN to GMN
|
GTN | 1,3-GDN | 1,2-GDN | 1-GMN | 2-GMN | Total recovered | |||||
| Time (h)a | Maximum 1,3-GDN (μM) | Maximum 1,2-GDN (μM) | Maximum 1-GMN (μM) | Maximum 2-GMN (μM) | ||||||||
| P. putida SP1a-2 | 18 | 58 | 66 | 61 | 82 | NDc | 0 | 19.5 | 11.5 | 30.5 | 41 | 102.5 |
| P. putida SP1b-6 | 18 | 57 | 93 | 81 | 98 | ND | 0 | 12 | 0 | 41 | 49 | 102 |
| Rhodococcus strain SP2-4 | 30 | 51 | 95 | 21 | 86 | 45 | 0 | 0 | 0 | 0 | 0 | 0 |
| Rhodococcus strain SP3-4 | 30 | 47 | 94 | 53 | 105 | 45 | 0 | 0 | 0 | 0 | 0 | 0 |
| K. oxytoca SP2-5 | 12 | 140 | 57 | 150 | 58 | ND | 0 | 21.5 | 0 | 75 | 29 | 125.5 |
| A. ureafaciens SP1b-5 | >60 | 35 | 71 | 0 | 0 | ND | 40.5 | 17.5 | 35.5 | 0 | 0 | 93.5 |
Time to >95% completion.
Time to completion.
ND, no degradation observed.
Although identified as the same species, strain SP1b-6 grew slightly faster than SP1a-2 during the middle phase (18 to 36 h) of growth, and this coincided with a more rapid conversion of 1,2-GDN to the mononitrates (data not shown). Apart from this difference, the biodegradation profiles of the two strains were similar, i.e., complete disappearance of GTN within the first 18 h of the experiment, production of 1,3-GDN and 1,2-GDN occurring in a ratio of 1 to 2, the same regioselective removal of the end nitro group from 1,2-GDN, much slower conversion of 1,3-GDN to the mononitrates than that of 1,2-GDN, and the total molar concentration of nitrate esters remaining constant throughout the experiment, indicating inability to degrade the mononitrates.
Rhodococcus sp. strains SP2-4 and SP3-4.
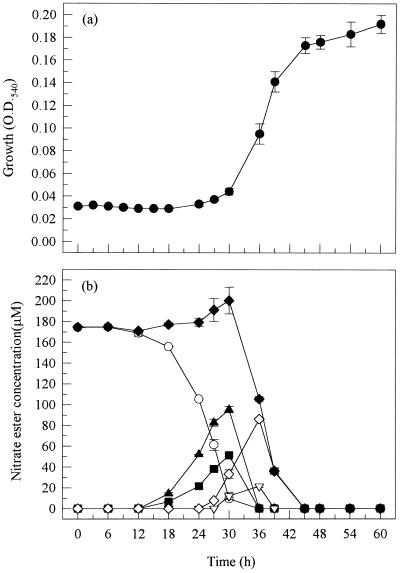
Strain SP2-4 exhibited a long lag period (12 h) before any growth on or biodegradation of GTN (Fig. 2). However, once biodegradation was initiated, GTN was rapidly converted to, first, the dinitrates and then the mononitrates. Removal of the first nitrate group from GTN was again nonspecific, with the ratio of 1,3-GDN to 1,2-GDN being approximately 1 to 2 during their accumulation. The maximum concentration of 2-GMN was almost the same as that of its sole precursor 1,2-GDN, showing that the latter was converted mainly to the 2-GMN isomer. Significantly, this strain was capable of rapidly biodegrading mononitrates, and hence, no nitrate esters were detected in the growth medium at the end of the experiment.
FIG. 2.
Biodegradation of GTN by Rhodococcus sp. strain SP2-4. See the legend to Fig. 1 for key to symbols. O.D.540, optical density at 540 nm.
The growth and biodegradation profile of strain SP3-4 (data not shown) was very similar to that observed for SP2-4 as expected, since both strains were found to belong to the genus Rhodococcus. There was a long lag period before growth on and biodegradation of GTN, but once biodegradation was initiated, this strain completely degraded GTN via di-and mononitrate forms until no nitrate esters could be detected in the growth medium (Table 2). Strains SP2-4 and SP3-4 were probably identical.
Klebsiella oxytoca strain SP2-5.
Of all the strains tested, SP2-5 degraded GTN the most rapidly, with no GTN detectable in the growth medium after 12 h. In contrast to the other strains isolated, this strain produced greater quantities of 1,3-GDN than 1,2-GDN (ratio of 7 to 3). 1,3-GDN was incompletely converted to 1-GMN, such that their concentrations at the end of the experiment were approximately 40 μM and 150 μM, respectively (Table 2). The highest concentration of 1,2-GDN (at 12 h) was comparable to the concentration of 2-GMN at the end of the experiment, indicating that the conversion of 1,2-GDN to the mononitrate form was regiospecific and exhibited a preference for the removal of the end nitrate group. Throughout the experiment, the total concentration of nitrate esters remained constant.
Arthrobacter ureafaciens SP1b-5.
Growth of strain SP1b-5 on GTN as the sole source of nitrogen was extremely slow. GTN was converted to 1,2-GDN and 1,3-GDN, like SP1a-2, in a ratio of approximately 2 to 1 (Table 2). No mononitrates were detected during the course of the experiment, and the total concentration of nitrate esters remained constant.
DISCUSSION
Of the six isolates capable of utilizing GTN as their sole source of nitrogen, two (SP1a-2 and SP1b-6) were identified as P. putida, two (SP2-4 and SP3-4) as Rhodococcus species, one (SP1b-5) as an Arthrobacter species, and one (SP2-5) as a Klebsiella species. When these four species are taken into consideration alongside those strains already known to utilize GTN as a sole nitrogen source, i.e., Agrobacterium radiobacter (19), Enterobacter cloacae (3), P. putida and Pseudomonas fluorescens (4), it seems that the ability to biodegrade GTN is widely distributed. The conversion of GTN to GDN has been shown to involve α/β barrel oxidoreductase flavoproteins (4, 7, 13), which are related to old yellow enzyme (14). Such oxidoreductase flavoproteins are a common family of enzymes, and hence it is not surprising that the ability to biodegrade GTN may also be widely distributed.
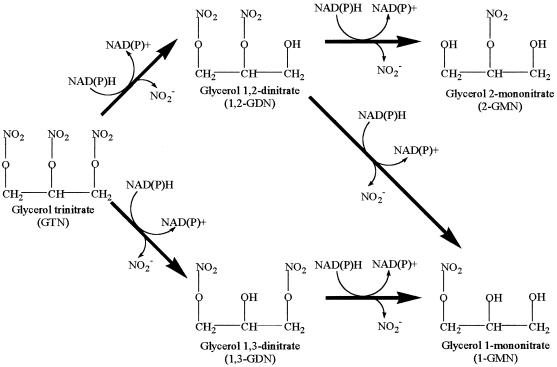
The pathway for the biodegradation of GTN, already established for A. radiobacter (Fig. 3), presents the possibility of regioselectivity in the removal of nitro groups from either the central or one of the terminal carbons of GTN. Of the four species isolated in this study, three (namely P. putida and the Rhodococcus and Arthrobacter species) produced 1,3-GDN and 1,2-GDN in the ratio of 1 to 2 which is the ratio predicted for nonregiospecific removal of the first nitro group from GTN. The P. putida strain isolated by Blehert et al. (4) also exhibited a similar lack of regioselectivity. In contrast, the Klebsiella sp. strain SP2-5 isolated during the present study produced 1,3-GDN and 1,2-GDN in a ratio of 7 to 3. Taking account of the twofold excess of terminal over central groups, this corresponds to a 14 to 3 preference for central attack. In this respect, the Klebsiella strain aligned itself with A. radiobacter (19), P. fluorescens (4), and the fungus Geotrichum candidum (5), all of which exhibited a distinct preference for the removal of the central nitro group of GTN. On the other hand, one fungus, Phanerochaete chrysosporium (11) has been shown to possess a preference for the production of 1,2-GDN from GTN.
FIG. 3.
Generalized pathway for the biodegradation of GTN (adapted from reference 19 with permission from the publisher) based on metabolite profiles for Agrobacterium (19) and Pseudomonas sp. (4, 17) and NAD(P)H dependence of GTN-degrading enzymes liberating NO2− from GTN (4, 7, 13).
In a similar manner, regioselectivity may also arise in the conversion of 1,2-GDN (but not the symmetrical 1,3-isomer) to either 1-GMN or 2-GMN. For P. putida strain SP1a-2, GTN disappearance (and thus production of GDNs) was complete after 18 h in culture; thus analysis of the fate of 1,2-GDN in the post-18-h period was not confounded by its continued formation. In the 18- to 60-h period, 1,2-GDN decreased by 67 μM (Fig. 1), and in the same period 2-GMN increased by 53 μM, a conversion factor for 1,2-GDN to 2-GMN of 80%. Similar calculations for P. putida strain SP1b-6 and K. oxytoca strain SP2-5 gave conversion factors of 73 and 86%, respectively. Thus, these strains all showed a regioselectivity of about 4 to 1 in favor of removal of the terminal nitro group in 1,2-GDN. For the Rhodococcus sp. strains SP2-4 and SP3-4, similar calculations were not possible because both of the GMNs formed were further degraded. Nevertheless, in both strains, the transient maximum concentration of 2-GMN was similar to that of its sole precursor 1,2-GDN, indicating again a strong preference for removal of the terminal rather than the central nitro group. Thus, all of the strains isolated in this study, with the exception of SP1b-5 which produced neither GMN within the time scale of the experiments, exhibited a preference for the removal of the end nitro group from 1,2-GDN to produce 2-GMN. Such a preference has also been shown in A. radiobacter (19) and P. chrysosporium (6, 11).
The predominance of terminal attack on 1,2-GDN contrasts with the regioselectivity for attack on GTN (either central for K. oxytoca strain SP2-5 or nonselective for other strains). This suggests that different enzyme systems are involved in the degradation of GTN and 1,2-GDN, and this is supported, in turn, by the long lag periods (6 to 12 h) between the onset of appearance of the GDNs and that of the GMNs.
The most important and novel discovery from our experiments was that complete biodegradation of GTN occurred in axenic cultures of the Rhodococcus sp. strains SP2-4 and SP3-4. After a long lag period, both isolates achieved rapid biodegradation of GTN, including the removal of nitro groups not only from GTN and GDN but also from GMN, thus attaining full denitration. Complete denitration of GTN has previously been shown to occur only in mixed bacterial populations (1, 15) or in cultures of Penicillium corylophilum Dierckx when grown in the presence of glucose and ammonium nitrate over long periods of time (>300 h) (20). Hence, to the best of our knowledge, this is the first report of a single bacterial species that can achieve complete denitration of GTN and also utilize its products as a sole source of nitrogen.
ACKNOWLEDGMENTS
We thank SRK Consulting and AECI Limited for providing soil samples from wastewater soakaways; EXCHEM, Derbyshire, United Kingdom, for supplies of GTN; the Royal Commission for the Exhibition of 1851 (London) for a Research Fellowship (S.J.M.); and the Royal Society (London) for an equipment grant.
REFERENCES
- 1.Accashian J V, Vinopal R T, Kim B J, Smets B F. Aerobic growth on nitroglycerin as the sole carbon, nitrogen, and energy source by a mixed bacterial culture. Appl Environ Microbiol. 1998;64:3300–3304. doi: 10.1128/aem.64.9.3300-3304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Binks P R, French C E, Nicklin S, Bruce N C. Degradation of pentaerythritol tetranitrate by Enterobacter cloacae PB2. Appl Environ Microbiol. 1996;62:1214–1219. doi: 10.1128/aem.62.4.1214-1219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blehert D S, Knoke K L, Fox B G, Chambliss G H. Regioselectivity of nitroglycerin denitration by flavoprotein nitroester reductases purified from two Pseudomonas species. J Bacteriol. 1997;179:6912–6920. doi: 10.1128/jb.179.22.6912-6920.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ducrocq C, Servy C, Lenfant M. Bioconversion of glyceryl trinitrate into mononitrates by Geotrichum candidum. FEMS Microbiol Lett. 1989;65:219–222. doi: 10.1016/0378-1097(89)90394-7. [DOI] [PubMed] [Google Scholar]
- 6.Ducrocq C, Servy C, Lenfant M. Formation of glyceryl 2-mononitrate by regioselective bioconversion of glyceryl trinitrate—efficiency of the filamentous fungus Phanerochaete chrysosporium. Biotechnol Appl Biochem. 1990;12:325–330. [PubMed] [Google Scholar]
- 7.French C E, Nicklin S, Bruce N C. Sequence and properties of pentaerythritol tetranitrate reductase from Enterobacter cloacae PB2. J Bacteriol. 1996;178:6623–6627. doi: 10.1128/jb.178.22.6623-6627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchesi J R, Sato T, Weightman A J, Martin T A, Fry J C, Hiom S J, Wade W G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng M, Sun W-Q, Geelhaar L A, Kumar G, Patel A R, Payne G F, Speedie M K, Stacy J R. Denitration of glycerol trinitrate by resting cells and cell extracts of Bacillus thuringiensis/cereus and Enterobacter agglomerans. Appl Environ Microbiol. 1995;61:2548–2553. doi: 10.1128/aem.61.7.2548-2553.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morris R. SRK investigates potential nitroglycerine contamination. Borehole Water J. 1998;41:24–25. [Google Scholar]
- 11.Servent D, Ducrocq C, Henry Y, Guissani A, Lenfant M. Nitroglycerin metabolism by Phanerochaete chrysosporium—evidence for nitric oxide and nitrite formation. Biochim Biophys Acta. 1991;1074:320–325. doi: 10.1016/0304-4165(91)90170-l. [DOI] [PubMed] [Google Scholar]
- 12.Smets B F, Vinopal R T, Grasso D, Strevett K A, Kim B-J. Nitroglycerin biodegradation: theoretical thermodynamic considerations. J Energetic Mater. 1995;13:385–398. [Google Scholar]
- 13.Snape J R, Walkley N A, Morby A P, Nicklin S, White G F. Purification, properties, and sequence of glycerol trinitrate reductase from Agrobacterium radiobacter. J Bacteriol. 1997;179:7796–7802. doi: 10.1128/jb.179.24.7796-7802.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stott K, Saito K, Thiele D J, Massey V. Old yellow enzyme—the discovery of multiple isoenzymes and a family of related proteins. J Biol Chem. 1993;268:6097–6106. [PubMed] [Google Scholar]
- 15.Wendt T M, Cornell J H, Kaplan A M. Microbial degradation of glycerol nitrates. Appl Environ Microbiol. 1978;36:693–699. doi: 10.1128/aem.36.5.693-699.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White G F, Snape J R. Bacterial biodegradation of nitrate esters. In: Kaffka A V, editor. Sea-dumped chemical weapons: aspects, problems and solutions. Dordrecht, The Netherlands: Kluwer Academic Press; 1996. pp. 145–156. [Google Scholar]
- 17.White G F, Snape J R, Nicklin S. Bacterial biodegradation of glycerol trinitrate. Int Biodeterior Biodegradation. 1996;38:77–82. doi: 10.1128/aem.62.2.637-642.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White G F, Snape J R, Nicklin S. Proceedings of the 26th International Annual Conference of ICT, Pyrotechnics: basic principles, technology, application. Karlsruhe, Federal Republic of Germany: DWS Werbeagentur und Verlag GmbH; 1995. Bacterial degradation of nitrate ester explosives. [Google Scholar]
- 19.White G F, Snape J S, Nicklin S. Biodegradation of glycerol trinitrate and pentaerythritol tetranitrate by Agrobacterium radiobacter. Appl Environ Microbiol. 1996;62:637–642. doi: 10.1128/aem.62.2.637-642.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y Z, Sundaram S T, Sharma A, Brodman B W. Biodegradation of glyceryl trinitrate by Penicillium corylophilum Dierckx. Appl Environ Microbiol. 1997;63:1712–1714. doi: 10.1128/aem.63.5.1712-1714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]