ABSTRACT
Aberrant expression of circular RNA (circRNA) is involved in the occurrence of various diseases and tumor development, in which plays a vital role, including hepatocellular carcinoma (HCC). Nevertheless, the regulation mechanism and biological function of circITCH in hepatocellular carcinoma (HCC) remain unclear. The expression level of circular RNA itchy E3 ubiquitin protein ligase (circ-ITCH) was identified and validated by real-time polymerase-chain reaction (RT-qPCR) in HCC cell lines. The stability of circITCH was confirmed by Ribonuclease R (RNase R) assay. Subsequently, through silencing and overexpression of circITCH to investigate the functional roles of circITCH in HCC proliferation, invasion, and apoptosis. We also carried out bioinformatics analysis, luciferase reporter assays to define the relationship between microRNA (miR)-184 and circITCH. Moreover, xenograft mouse models and immunohistochemistry were employed to assess the function of circITCH in HCC. CircITCH (hsa_circ_0001141) was a stable circRNA and downregulated in HCC cells. Overexpression of circITCH inhibited cell proliferation, migration, invasion, and promoted apoptosis in vitro and in vivo, whereas knockdown of circITCH had the opposite effects. Mechanistically, miR-184 could be sponged by circITCH, and its overexpression could mitigate the suppressive effects of circITCH overexpression on HCC progression. Through biological website to predict the target genes of miR-184 may be combined. Gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed to investigate mRNAs with significant functional enrichment and pathways, also which its relationship with HCC-related pathway and immune cells. Our findings reveal that circITCH served as a repressor to restrain HCC malignancy via miR-184. Therefore, circITCH may serve as a potential prognostic marker and therapeutic target for HCC.
Abbreviations: HCC: hepatocellular carcinoma; CircRNA: Circular RNA; miRNA: MicroRNA; Circ-ITCH: circular RNA itchy E3 ubiquitin protein ligase; RT-qPCR: real-time polymerase-chain reaction; RNase R: Ribonuclease R; CeRNA: competing endogenous RNAs; SiRNA: small interfering RNA
KEYWORDS: Hepatocellular carcinoma, circular RNA, circRNA, circITCH, miR-184
Introdution
Hepatocellular carcinoma (HCC), as one most common type of primary liver cancer, ranks the third cause of leading cancer-related deaths worldwide [1]. With multiple causes, including HBV infection and liver cirrhosis are the major risk factors for HCC [2]. The development of HCC is a complex multistep process involving persistent inflammatory damage, including hepatocyte necrosis and regeneration associated with fibrotic deposition [3]. Despite the development of numerous new alternative therapies, surgical resection still remains the mainstay of HCC treatment, but the metastasis and recurrence rates of HCC after surgery are still high [4]. Therefore, the most urgent needs are to elucidate the etiology and molecular mechanisms underlying the development and progression of HCC, and to find sensitive markers for the diagnosis and prognosis of HCC [5].
Currently, with the development of high-throughput sequencing techniques, transcriptomics has gradually become a focus of attention. Circular RNAs (circRNAs) are a class of endogenous non-coding RNA which has been found to be commonly expressed in eukaryotic genes [6], a family of covalently closed-loop structure RNAs without 5′-cap and 3′-poly A tail, have been identified to play vital roles in human cancers [7]. The unique circular structure of circRNAs confers inherent resistance to degradation by exonuclease and makes them more stable than their linear parental genes [8–10]. Accumulating evidences about circRNAs have demonstrated that they participate in diverse biological processes including proliferation, invasion, metastasis, apoptosis, and autophagy of malignant tumors [11,12], which implying a great promise of circRNAs as predictive biomarkers and therapeutic targets in cancer [13,14]. Recently, the function and mechanism of circRNAs in HCC have also been preliminary studied. CircFBLIM1 has been shown to act as a ceRNA that promotes the development of hepatocellular cancer by acting as a sponge of miR-346 [15]. Circ-0051443 is transmitted from normal cells to HCC cells via exosomes and suppresses the malignant biological behaviors by promoting cell apoptosis and arresting the cell cycle [16]. Circ-DB promotes HCC growth and reduces DNA damage via the suppression of miR-34a and the activation of deubiquitinating-related USP7 [17].
Circular RNA itchy E3 ubiquitin protein ligase (circ-ITCH) is located on human chromosome 20, 20q11.22, which derived from exons 7–14 of the itchy E3 ubiquitin protein ligase (ITCH) gene. It was demonstrated to function as a tumor suppressor in multiple human cancers, including ovarian cancer [18], bladder cancer [19], breast cancer [20], papillary thyroid cancer [21] and colorectal cancer [22] and so on. Study has found that expression of circ-ITCH in HCC tissues was significantly reduced, and a high level of circITCH was associated with favorable survival of HCC [23]. However, the function and regulatory mechanism of circITCH (circ-0001141) in HCC malignant behaviors remain largely unclear. The discovery of more circRNAs that regulates the development of HCC will provide a more effective choice for targeted therapy.
MicroRNAs (miRNAs) are small, mostly non-coding RNAs containing approximately 22 nucleotides, which are highly conserved across species and function in post-transcriptional regulation of gene expression [24]. The molecular functions of circRNA have been elucidated, such as the regulation of transcription, translation, protein binding. Remarkably, it has been widely reported that circRNA serves as “miRNA sponges” to sequester miRNAs from binding to their target mRNAs [25,26]. Herein, we focus on the underlying mechanisms of circITCH in HCC and identify whether circITCH could affect cell malignant phenotypes through competing endogenous RNAs (ceRNA) regulating cascades “circRNA-miRNA-mRNA”, which enhances the significance of the findings of the present study.
In this study, we confirmed the down-regulation of circITCH in HCC tissues and cells. Circ-0001141, as a ceRNA, inhibited cell proliferation, migration, invasion, and promoted apoptosis by sponging miR-184. The functional complementation analysis was conducted to verify the functions of them and the relationships among the circRNA/miRNA/mRNA networks. This study might provide a novel insights and biomarker for HCC treatment.
Methods
Cell culture
Human normal liver cell lines (LO2) and HCC cell lines (Huh7, HCCLM3, SMMC-7721, MHCC97H, and HepG2) were provided by the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were incubated in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Life Technologies, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS) (Gibco) and antibiotics (1% penicillin/streptomycin, Gibco, USA) in a humidified incubator with 5% CO2 at 37°C.
RNase R treatment
To verify the stability of circITCH molecular structure, RNase R digestion experiment was carried out. 2 μg of total RNA extracted from HCC cells were incubated with 3 U/μg RNase-R (Geneseed Biotech, Guangzhou, China) at 37°C for 15 minutes and 70°C for 10 minutes. Then, the expression levels of linear β-actin and circITCH were determined by qRT-PCR.
RNA extraction and RT-qPCR
Total RNA was extracted from cell lines using TRIzol Reagent. A NanoDrop 2000 microspectrophotometer was used to determine the quality and concentration of total RNA. After spectrophotometric quantification, 1 μg of total RNA in a final volume of 20 μl was used for reverse transcription (RT) with a PrimeScript RT Reagent Kit (Tsingke, China). Then, Fluorescence quantitative PCR was conducted using TB Green Fast qPCR Mix (Tsingke, China). All primers were designed and synthesized by Tsingke (Guangzhou, China). The expression of small nuclear RNA U6 or β-actin genes was used as a control to calibrate the original concentration of miRNA, circRNA respectively. Target gene expression was calculated using the 2-ΔΔCT method. Related primer sequences are shown in Table 1.
Table 1.
Primers for real-time PCR and probe for FISH.
| Genes | Forward (5’-3’) | Reverse (5’-3’) |
|---|---|---|
| CircITCH | AAGGAGCAATGCAGCAGTTT | AGTCACAACTACTTCTTCAACCCAT |
| β-actin | CCTTCCTGGGCATGGAGTC | TGATCTTCATTGTGCTGGGTG |
| MiR-184 | CGCGTGGACGGAGAACTGAT | AGTGCAGGGTCCGAGGTATT |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| circITCH | ACAACTACTTCTTCAACCCATCCAGGTGGCAA |
Cell transfection
Small interfering RNAs (siRNAs) targeting the junction sequence of circITCH (si-circ-1, si-circ-2, and si-circ-3), miRNA-184 mimics, inhibitors and their related negative control oligonucleotides were designed and synthesized by HanBio (Shanghai, China). All transfection operations were performed using EntransterTM -R4000 (Engreen Biosystem, Beijing, China) according to the manufacturer’s instructions. All the sequences used in the present study are listed in Table 2.
Table 2.
Overexpression and silent related sequences for transfection.
| oligo | sense | antisense |
|---|---|---|
| CircITCH-si-1 | GCCACCUGGAUGGGUUGAAGA | UCUUCAACCCAUCCAGGUGGC |
| CircITCH-si-2 | ACCUGGAUGGGUUGAAGAAGU | ACUUCUUCAACCCAUCCAGGU |
| CircITCH-si-3 | AUGGGUUGAAGAAGUAGUUGU | ACAACUACUUCUUCAACCCAU |
| CircITCH-si-NC | UUCUCCGAACGUGUCACGU | ACGUGACACGUUCGGAGAA |
| MiR-184 | UGGACGGAGAACUGAUAAGGGU | ACCCUUAUCAGUUCUCCGUCCA |
| MiR-184-NC | UCACAACCUCCUAGAAAGAGUAGA | UCUACUCUUUCUAGGAGGUUGUGA |
| MiR-184-i | ACCCUUAUCAGUUCUCCGUCCA | |
| MiR-184-i-NC | UCUACUCUUUCUAGGAGGUUGUGA |
Construction of stable cell lines
To establish stable overexpression of circITCH in human HCCLM3 cell lines, we used control lentivirus HBLV-ZsGreen-PURO and purpose lentivirus HBLV-hsa_circ_0001141-Null-ZsGreen-PURO to infect HCCLM3 cells (HanBio, Shanghai, China). In the infected cells after 48 h, by adding and maintaining 1.0 mu g/ml puromycin kill not be infected cells effectively. Thus under puromycin drugs to maintain stable strains of eventually get circITCH stable expression.
Dual luciferase reporter assay
According to the predictions of Circular RNA Interactome (circinteractome) and Starbase online software, circITCH possessed binding sites for miR-184 in the 3ʹ-UTR region. To experimentally verify the possible miRNA target genes and binding sequences, the wild-type and mutant circITCH 3ʹuntranslated region (3ʹUTR) sequences were synthesized and cloned into dual-luciferase reporter plasmids (HanBio, Shanghai, China) containing the psiCheck2 promoter. HCCLM3 cells were inoculated into a 96-well plate and grown to 50%–70% confluence 24 h at 37°C, 5% CO2. They were transfected in combination with the wild-type or mutant circITCH reporter gene plasmids and miR-184 mimic or miR-NC mimic. After 48 h, the activities of both firefly luciferase (LUC) and Renilla luciferase (RLUC) were measured with a Dual-Luciferase Reporter System Kit (Promega, USA).
RNA fluorescence in situ hybridization (FISH)
FISH kit (RiboBio) was used to detect the location of circITCH in HCC cells following the manufacturer’s protocol. Briefly, cells were subjected to pre-hybridization solution for 30 min. Then, cy3-labeled circITCH probe were hybridized at 37°C overnight after pre-hybridization. Next, slides were washed and nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). A confocal microscopy was applied to analyze results.
Cell counting kit-8 (CCK8) assay
For the CCK8 assay, transfected HCC cells in logarithmic growth phase were seeded into a 96-well plate at a density of 1 × 104 cells/well for 6 replicates. The proliferation of cells were monitored every 24 hours for 0, 24, 48,72 h. 10ul of CCK 8 solution (KeyGene, Biotech, China) was added to each well and they were maintained at 37°C for 2 h away from light. The absorbance of each well at 450 nm was measured with a microplate reader.
5-Ethynyl-2′-deoxyuridine (EdU) assay
Cell-Light™ EdU Apollo®567 In Vitro Imaging Kit (RiboBio, Guangzhou, China) was used to detect cell proliferation ability according to the manufacturer’s instructions. The transfected HCC cells (1 × 104 cells for each group) were seeded in 96-well plates and cultured for 24 h, after, a 50 μM EdU solution was added onto plates and incubated for 2 h at 37°C. Then, the cells were immobilized with 4% paraformaldehyde and stained with Apollo Dye Solution and Hoechst 33,342. Finally, the images were acquired using a fluorescence microscope and the ratio of EdU-positive cells was calculated.
Transwell assay
To perform the invasion assay, after 24 h of transfection treatment, HCC cells (1 × 105/well) were prepared into the upper chamber (8-μm pore size; Millipore) with 200 μL serum-free medium, and 800 μL of medium containing 20% fetal bovine serum was added into the bottom transwell chamber. After incubating at 37°C with 5% CO2 for 24 h, we gently scraped the cells on the upper chamber with cotton swabs, whereas the invasive cells on the bottom chamber were fixed with 100% methanol for 20 min and stained with 0.1% crystal violet for 15 min at normal temperature. Five fields were randomly selected under a microscope (200× magnification) to calculate the average number of invasion cells.
Wound healing assay
Wound healing assays were performed to evaluate the migration ability of HCC cells. Transfected and controlled cells (1 × 106 cells/well) were seeded onto 6-well plates and cultured at 37°C, 5% CO2. After the cells were fully grown, cell monolayers were scratched with a 200-μl pipette tip to draw a vertical line in the middle of the cell and then cultured in DMEM. After routine culture for 24 h and 48 h, photographs were taken and the cell migration rate was calculated.
Apoptosis assay
To perform cell apoptosis assay, a FITC Annexin V Apoptosis Assay Kit (BD Biosciences, CA, USA) was used as follows. Cells in logarithmic growth phase were collected, resuspended and evenly spread in a 6-well plate. After 24 h of transfection, a total of 1 × 106 cells were collected and washed twice with pre-cold PBS. Thereafter, 1× binding buffer (100 μL) was used to disperse these cells, 5 μL Annexin V-fluorescein isothiocyanate (Annexin V-FITC) and 5 μL propidine iodide (PI) were added to the cells for 15 min in a dark at room temperature. Subsequently, the apoptosis rate was analyzed by flow cytometry.
In vivo Tumorigenicity Experiments
NOD scid gamma (NSG) mice (4–6 weeks, five mice per group) were purchased from the Nanjing Biomedical Research Institute of Nanjing University, Nanjing, China, and maintained in a specific pathogen-free environment. All mice were treated according to the care guidelines of the Laboratory Animal Center of the First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, China. LM3 cells stably overexpressing circITCH or Vector in PBS were subcutaneously inoculated into the right subaxillary of mice (5 × 106 per injection) for establishing xenograft model. We measured tumor size every five days in vivo until the mice were sacrificed. After four weeks, mice were sacrificed and the tumor was completely dissected and photographed. Then, tumor volume and weight were measured. Tumor tissues were subjected to HE and Ki-67 staining. The enrichments of circITCH, miR-184 in the tumors were detected by qRT-PCR.
Immunohistochemistry
The tumor tissues were fixed with 4% paraformaldehyde, embedded in paraffin, and cut into 5-μm-thick sections. Then the paraffin sections were incubated with primary antibodies against Ki-67(1:100, Geneseed Biotech, Guangzhou, China) overnight at 4°C, followed with HRP labeled secondary antibody (1:300, Boster, Wuhan, China) for 30 minutes at room temperature. After being stained with diaminobenzidine (DAB) and hematoxylin, sections were sealed with neutral resin. Finally, the Immunostaining images were observed with a microscope (Olympus, Tokyo, Japan).
Statistical analysis
All assays were conducted at least three times, quantitative data are presented as the means ± standard deviation (SD). Differences between two groups or among multiple groups were compared via student’s t test (two-tailed) or ANOVA with GraphPad Prism 8.0 (GraphPad Software, La Jolla, CA, USA) was used to analyze the experimental data. Differences were considered statistically significant with *p < 0.05, **p < 0.01, ***p < 0.001.
Results
CircITCH (hsa_circ_0001141) is downregulated in HCC cells
According to the circbase database (http://www.circbase.org/), has-circ-0001141, which is located at chr20:33,095,678–33,095,698[+], is derived from its parental gene ITCH, and its spliced length is 873bp (Figure 1a). To verify the qRT-PCR results of hsa_circ_0001141 in HCC cells, we performed Sanger sequencing on the PCR products of hsa_circ_0001141. The result confirmed the head-to-tail splicing in the PCR products with the expected size and predicted splicing site (Figure 1b). In addition, the stability of circITCH was demonstrated by RNase R treatment. qRT-PCR results showed that the levels of linear β-actin decreased sharply under RNase R treatment, while circITCH showed resistance to RNase R digestion (Figure 1c-d), which confirmed that circITCH has a circular structure. To determine the intracellular distribution of circITCH, RNA FISH assay showed that circITCH mainly located in the cytoplasm (Figure 1e), which might participate in HCC progression. To investigate whether circ-ITCH expression was changed in HCC cell lines, qRT-PCR was utilized to assess circITCH expression. As the data showed in Figure 1f, circ-ITCH expression was remarkably declined in HCC cells (Huh7, HCCLM3, SMMC-7721, and MHCC97H) compared with normal liver cell line (LO2), While circITCH expression of HepG2 in HCC cells was higher than normal liver cells. Thus, we chose HCCLM3 cells with the lowest expression and HepG2 cells with the highest expression for subsequent cell experiment.
Figure 1.
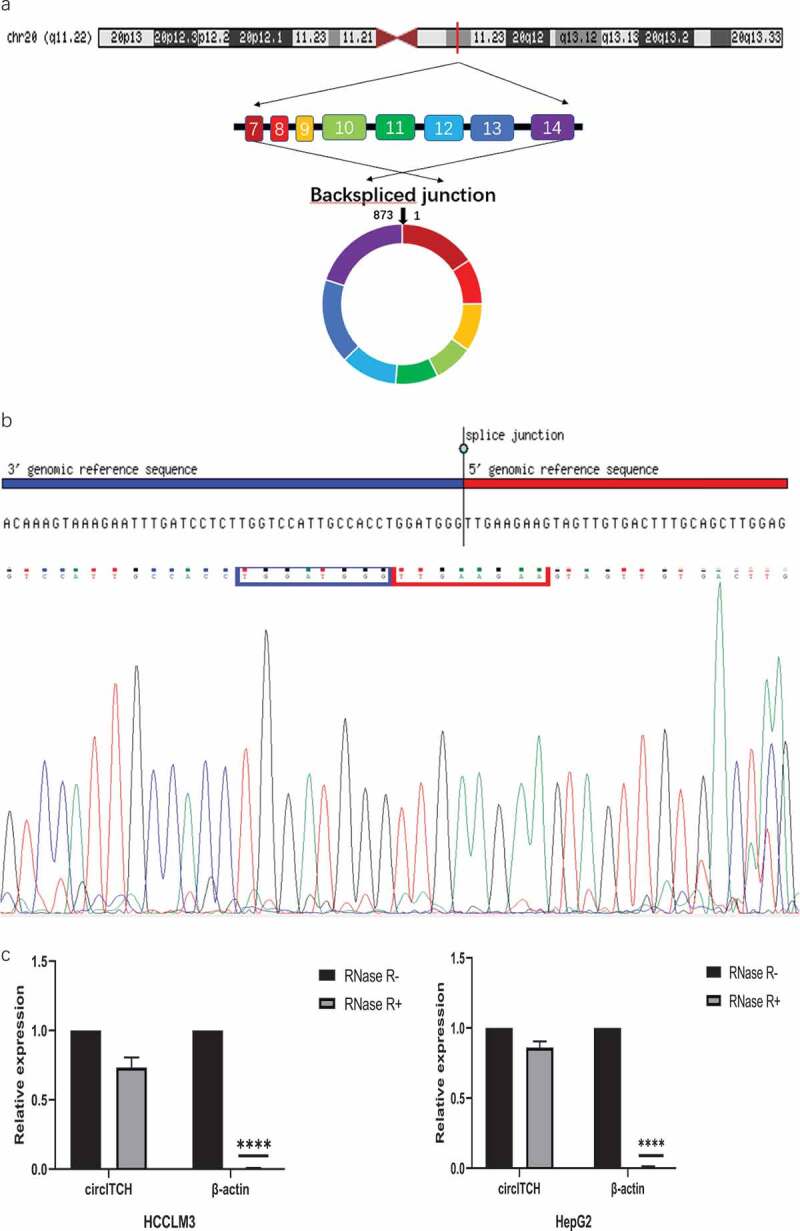
Characterization of circITCH as a circRNA in HCC.
a. Schematic illustration of chromosome localization and the exon linkage structure of circITCH is illustrated as indicated. b. The head-to-tail specific splicing site of circITCH was confirmed by Sanger sequencing. c. qRT-PCR analysis of circITCH and linear β-actin mRNA expression in HCCLM3 and HepG2 cells with or without RNase R treatment. d. Agarose gel electrophoresis analysis were performed to detect the presence of circITCH and linear β-actin from HCC cells using qRT-PCR product.e. RNA FISH analysis of circITCH in HCCLM3 and HepG2 cells. Nuclei were stained with DAPI. Scale bars, 100 µm. The samples were imaged at 1000× magnification. Scale bar = 10 μm.f. The relative expression of circITCH in a normal liver cell line (LO2) and HCC cells (HepG2, MHCC97H, HCCLM3, Huh-7, SMMCC-7721) was measured by RT-qPCR. The data are presented as the means ± SD of at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
Figure 1.
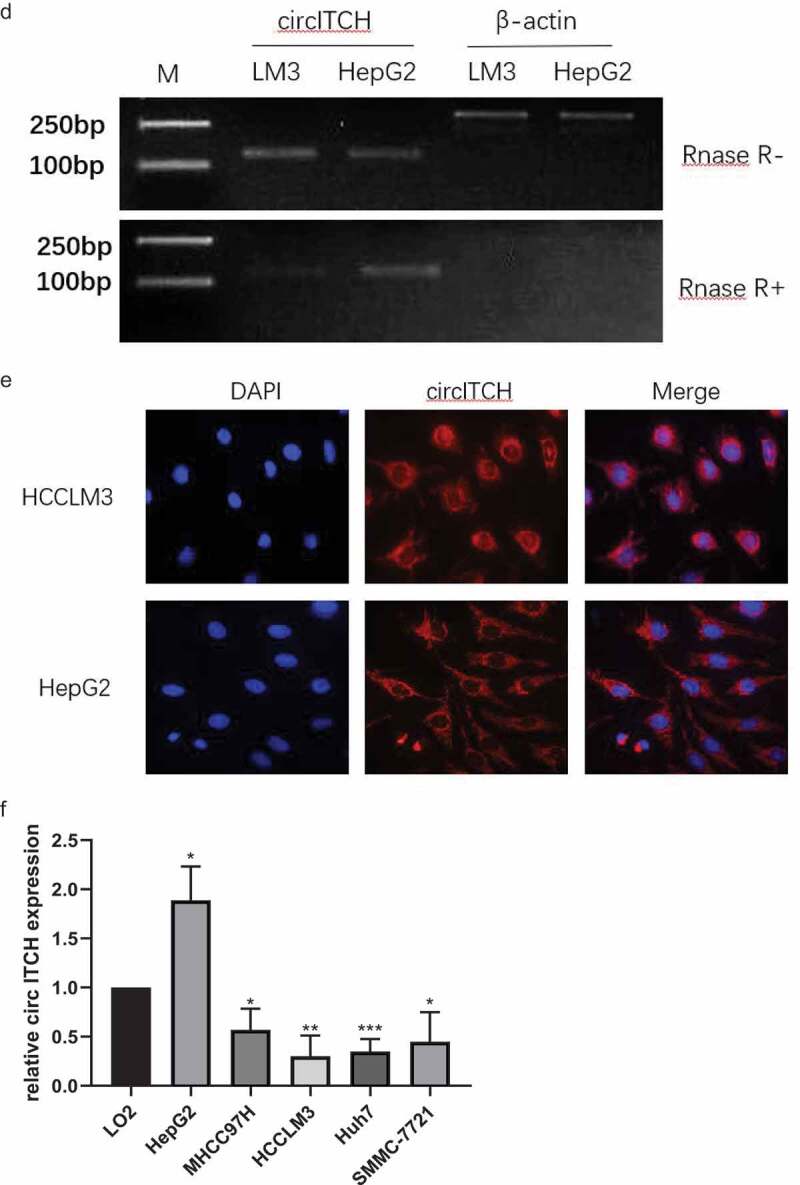
Continued
Function of circITCH in HCC
To study the biological function of circITCH in HCC, the expression of circITCH was downregulated in the HepG2 cell line, three short interfering RNAs targeting the backsplice site of circITCH were constructed and the one that provided the most significant downregulation was chosen to access cell functions. In addition, HCCLM3 cell lines were selected to stabilize overexpression of circITCH. The efficiency and specificity of circITCH knockdown and overexpression in HepG2 and HCCLM3 cells were verified by RT-qPCR (Figure 2a). CCK-8 and EdU assays were carried out to evaluate cell proliferation. As shown in Figure 2b-d, the results showed that circITCH silencing notably promote the vitality and proliferative ability of HepG2 cells, while circITCH upregulation inhibited cell growth. Subsequent wound healing assay and transwell invasion assay also confirmed that mobility of HCC cells was increased in the si-circITCH group and inhibited in the overexpression group (Figure 2e-g). Moreover, the results of flow cytometry showed that silencing circITCH could significantly inhibit cell apoptosis (Figure 2h). Taken together, these data suggest that circITCH exerts a tumor suppressor role in HCC cells.
Figure 2.
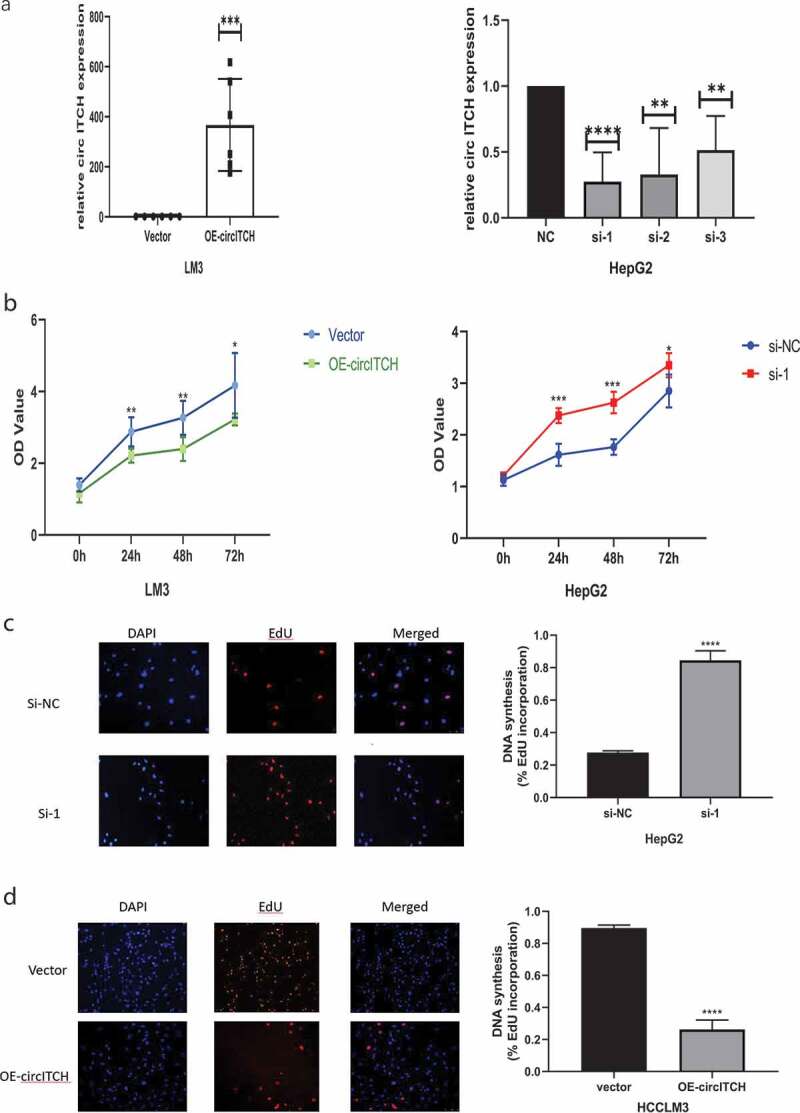
CircITCH suppresses HCC cell proliferation, invasion, and promoted apoptosis of HCC.
a. qRT-PCR was conducted to determine the transfection efficiency of cicrITCH-overexpressing in HCCLM3 cells and siRNAs targeting circITCH (si-circ #1, si-circ #2 and si-circ #3) in HepG2 cells. b-d. CCK-8 and EdU assays was performed to evaluate the influence of circITCH overexpression or knockdown cell proliferation. The samples were imaged at 200× magnification. Scale bar = 50 μme-g. Wound healing assay and transwell experiment was carried out to assess cell invation capacity. The samples were imaged at 100×, 200× magnification. Scale bar = 100um, 50 μm, respectively.h. The apoptosis rate was analyzed via flow cytometry. The data are presented as the means ± S.D. of at least three independent experiments. * P < 0.05, **P < 0.01, ***P < 0.001
Figure 2.
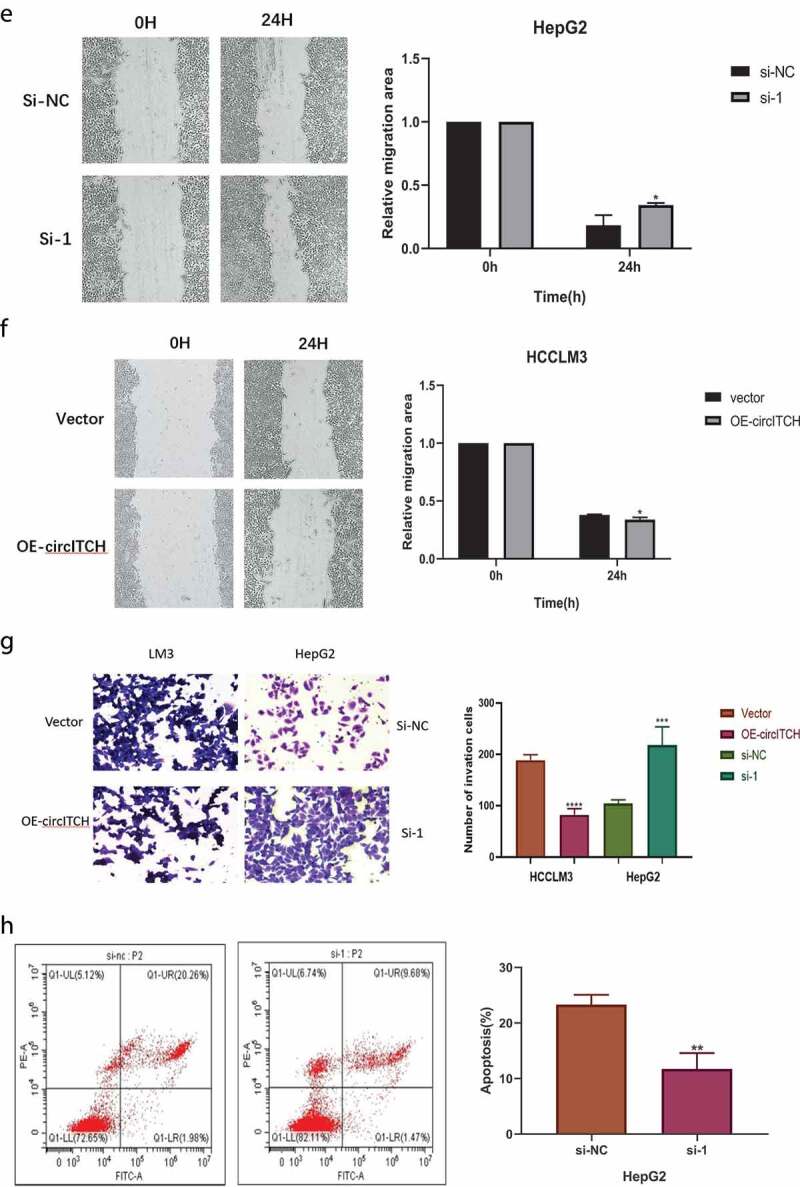
Continued
CircITCH functions as a sponge of miR-184
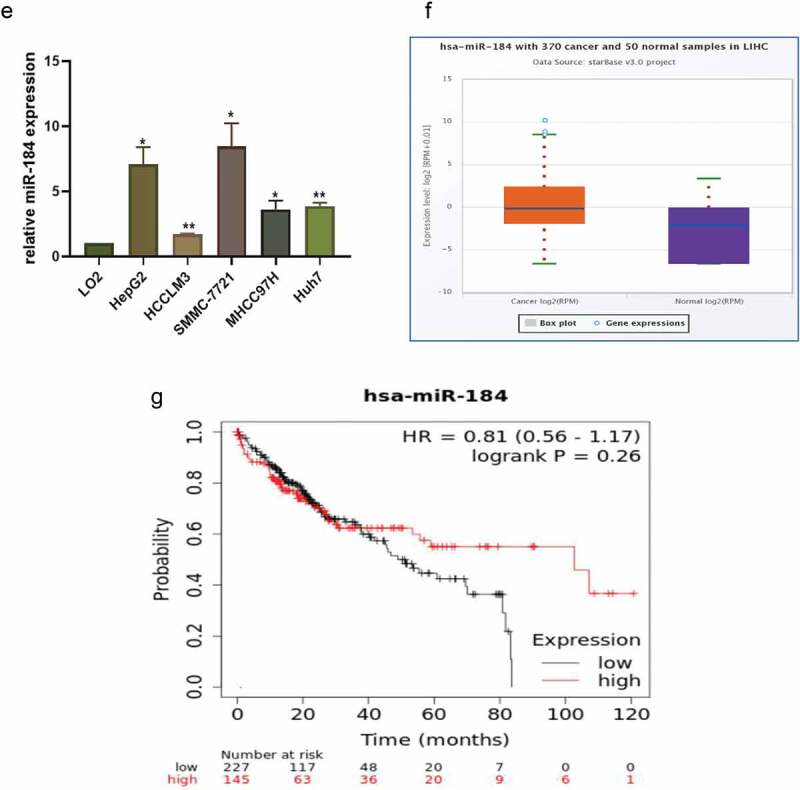
The competing endogenous RNA (ceRNA) mechanism, which mainly regulates gene expression at the post-transcriptional level, is primarily the most widely reported circRNA regulatory mechanism. It has been reported that circRNAs usually act as miRNA “molecular sponges” to regulate the expression of downstream genes [25]. Therefore, we performed a cross-analysis using two databases: CircInteractome (https://circinteractome.irp.nia.nih.gov/) and starBase (http://starbase.sysu.edu.cn/) to predict the potential miRNAs that may bind to circITCH. It was found that hsa-miR-184, hsa-miR-665, hsa-miR-944 from the overlap of this databases (Figure 3a). Among the three candidate miRNAs, only hsa-miR-184 (miR-184) was found to be significantly differentially expressed in HCC, and was reported as a carcinogen. Also, the binding sites between circITCH and miR-184 were predicted by starBase. qRT-PCR results showed that circITCH in HCCLM3 and HepG2 cells negatively regulated the expression of miR-184 after upregulated or downregulated (Figure 3b), suggesting that miR-184 is a miRNA potentially sponged to circITCH. To further determine whether miR-184 could directly target circITCH in HCC cells, we performed a dual luciferase reporter assay. Based on the complementary base pairing prediction with the “seed” region of miR-184, plasmids containing the wild-type sequence (circITCH-WT) and the mutant-binding sequence (circITCH-MUT) were constructed (Figure 3c). HepG2 cells were cotransfected with miR-184 mimic or miR-NC. As shown in Figure 3d, the luciferase activity in miR-184 mimics with circITCH-WT groups was significantly lower than other groups. However, miR-184 mimics or miR-NC showed no significant difference in luciferase activity when cotransfected with mutant reporter. These results confirmed that circITCH can directly and specifically interact with miR-184. Additionally, we investigated the expression of miR-184 in HCC cell lines, in contrast to circITCH, miR-184 was notably increased in HCC cell lines (Figure 3e). According to the Starbase database (http://starbase.sysu.edu.cn/), the expression of miR-184 in HCC tissues was significantly increased (Figure 3f). The Kaplan–Meier Plotter database (http://kmplot.com/analysis/index.php) revealed that the low expression of miR-184 had a significant advantage in the short-term survival rate of patients with HCC, while the high expression of miR-184 was associated with the long-term survival rate of patients, but with no significant statistical significance (p = 0.26) (Figure 3g). Allover,the above results revealed that circITCH functions as a “molecular sponge” for miR-184 might participate in HCC progression.
Figure 3.
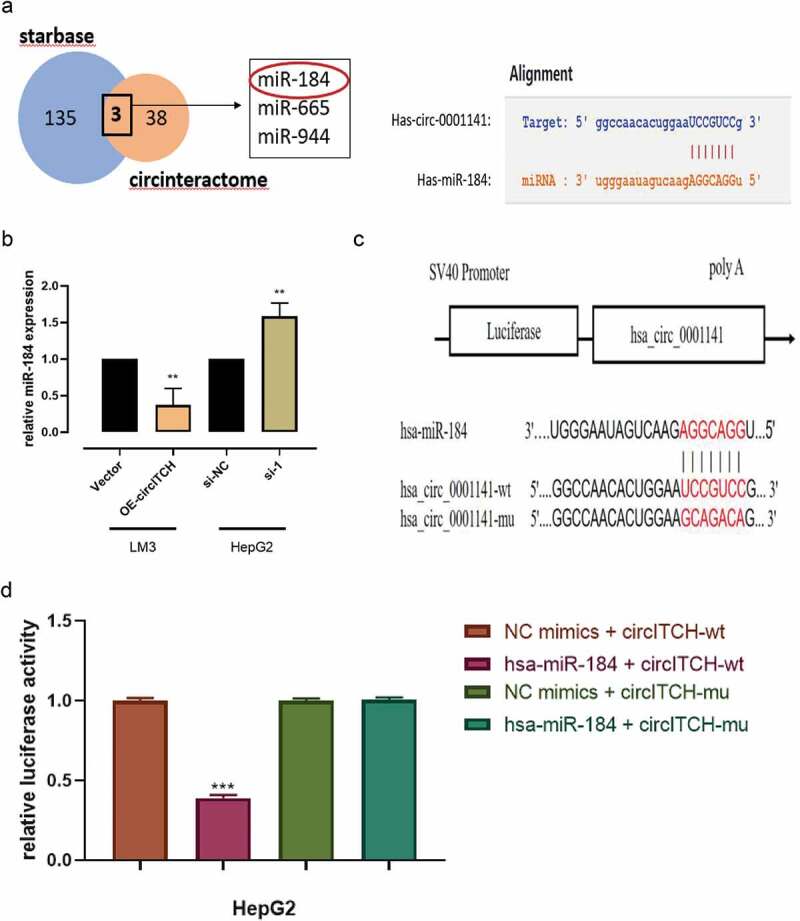
CircITCH acts as a sponge of miR-184 in HCC.
a. Venn diagram showing the overlap of the target miRNAs of circITCH predicted by circinteractome and starbase online databases. A schematic model shows potential-binding sites for miR-184 and 3ʹUTR of circITCH. b. The relative expression of miR-184 in HCCLM3 and HepG2 cells transfected with circWAC or vector and si-circITCH or control was detected by RT-qPCR. c. A schematic of miR-184 and wild-type/mutant circITCH binding site sequences with luciferase reporter vectors. d. Luciferase activity of the circITCH luciferase reporter vector (wt or mut) in HepG2 cells cotransfected with the miR-184 mimic or control. e. RT-qPCR analysis of the relative expression levels of miR-184 in LO2 cells and HCC cell lines. f. The relative expression of miR-184 in HCC tissues and adjacent nontumor tissues was analyzed by starBase Pan-Cancer Analysis Platform. g. Kaplan–Meier survival analysis of TCGA cohort with HCC stratified by the correlation between the overall survival and miR-184 expression. The data are presented as the means ± SD of at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
Figure 3.
Continued
MiR-184 reverses the antitumor effects of circITCH on HCC progression
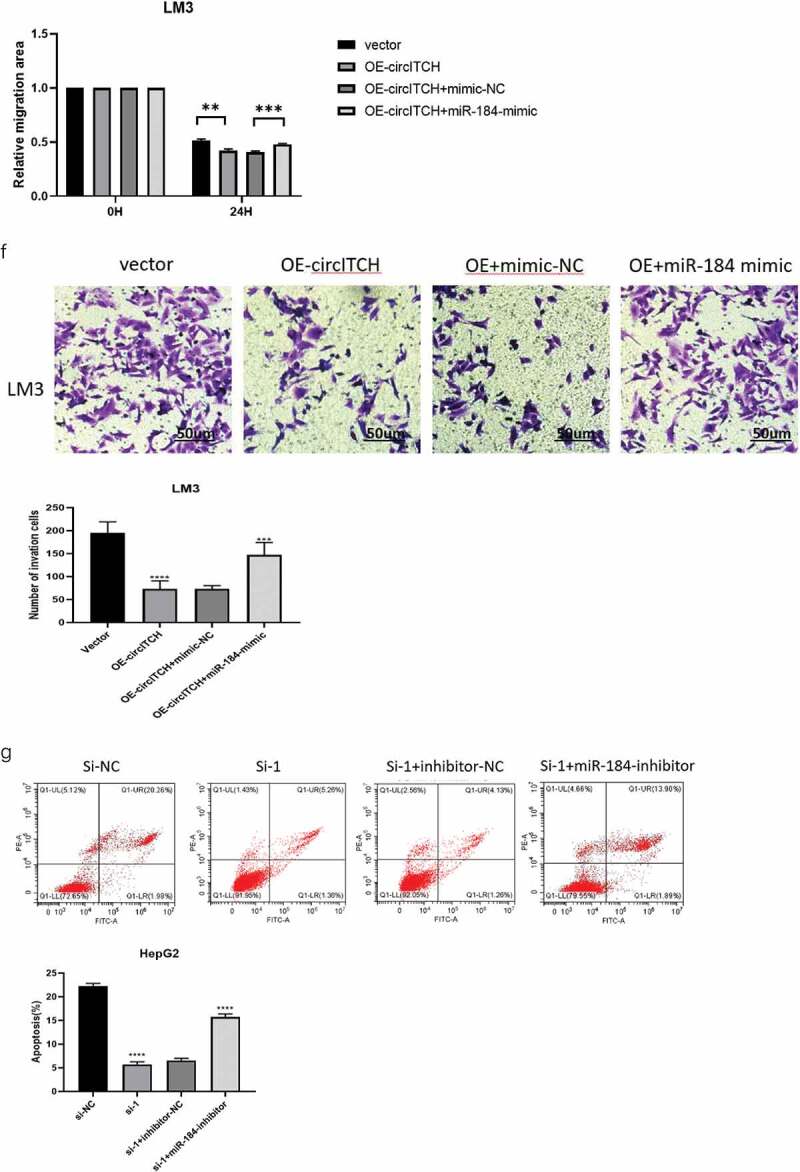
To elucidate whether circITCH functions by sponging miR-184 in HCC progression, rescue experiments were conducted with cotransfection of miR-184 mimics or inhibitor with OE-circITCH or si-circITCH. The efficiency of miR-184 mimics and inhibitor on its expression level in HepG2 cells was verified by RT-qPCR (Figure 4a). Then used to verify the expression of circITCH and miR-184 after transfection. The results demonstrated that after adding miR-184 mimics on the basis of circITCH overexpression, the expression level of miR-184 was significantly increased while the expression level of circITCH was not affected (Figure 4b). As shown in Figure 4c-f, miR-184 mimics partly reversed the suppressive effects on the proliferation, migration, and invasion induced by circITCH upregulation in CCK-8, EdU, wound healing, and transwell assays. In addition, miR-184 inhibitors reversed the inhibition of apoptosis induced by down-regulation of circITCH (Figure 4g). Collectively, these data suggested that circITCH inhibited HCC proliferation, invasion and promoted cell apoptosis, at least partly depend on miR-184.
Figure 4.
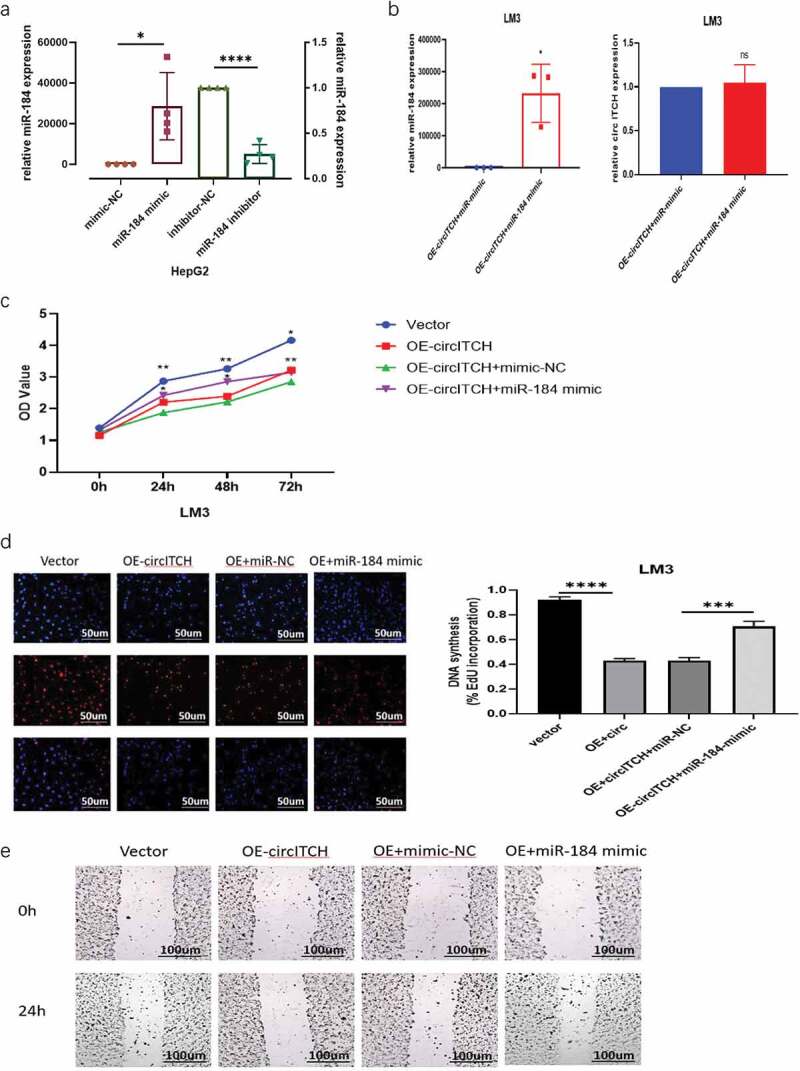
MiR-184 reverses the suppresses effect of circITCH in HCC.
a. The expression of miR-184 in HepG2 cells transfected with miR-184 mimics, inhibitor, and corresponding NC was detected by RT-qPCR. b. The expression of circITCH and miR-184 in HCCLM3 cells cotransfected with circITCH + miR-NC, or circITCH +miR-184 mimic was verified by qRT-PCR. c–f. HCCLM3 cells were divided into four groups (vector, OE-circITCH, OE-circITCH+mimic-NC, OE-circITCH+miR-184 mimic). The proliferation and invasion ability was analyzed through CCK-8, EdU, wound healing assays and transwell. The EdU and transwell samples were imaged at 200× magnification. Scale bar = 50 μm. The wound healing images taken at 100× magnification. Scale bar = 100 μm.g. Flow cytometry were conducted to examine apoptosis rates in HepG2 cells transfected with si-circITCH and miR-184 inhibitor. The data are presented as the means± S.D. of at least three independent experiments. * P < 0.05, **P < 0.01, ***P < 0.001
Figure 4.
Continued
Overexpression of circITCH depressed tumorigenesis of HCC In Vivo
To further investigate the biological roles of circITCH in HCC in vivo, we injected normal LM3 cells and LM3 cells stably overexpressing circITCH into the NSG mice. Compared with those of control xenografts (Vector group), the tumor growth curves and tumor weight demonstrated circITCH obviously suppressed tumor growth in mice (Figures 5A–c). Moreover, we used RT-qPCR to measure the expression of circITCH and miR-184 in the tumors and found that miR-184 were significantly reduced while circITCH was upregulated in the OE-circITCH group (Figure 5d-e). Congruously, tumor tissues were harvested for HE and IHC staining in Figures 5F-g. Ki-67 assay evidenced that overexpression of circITCH repressed Ki-67 positive cells in vivo. Altogether, these suggested that overexpression of circITCH suppressed HCC tumorigenesis in vivo. Collectively, the in vivo mice experiments suggested that circITCH inhibited tumor formation by regulating the miR-184.
Figure 5.
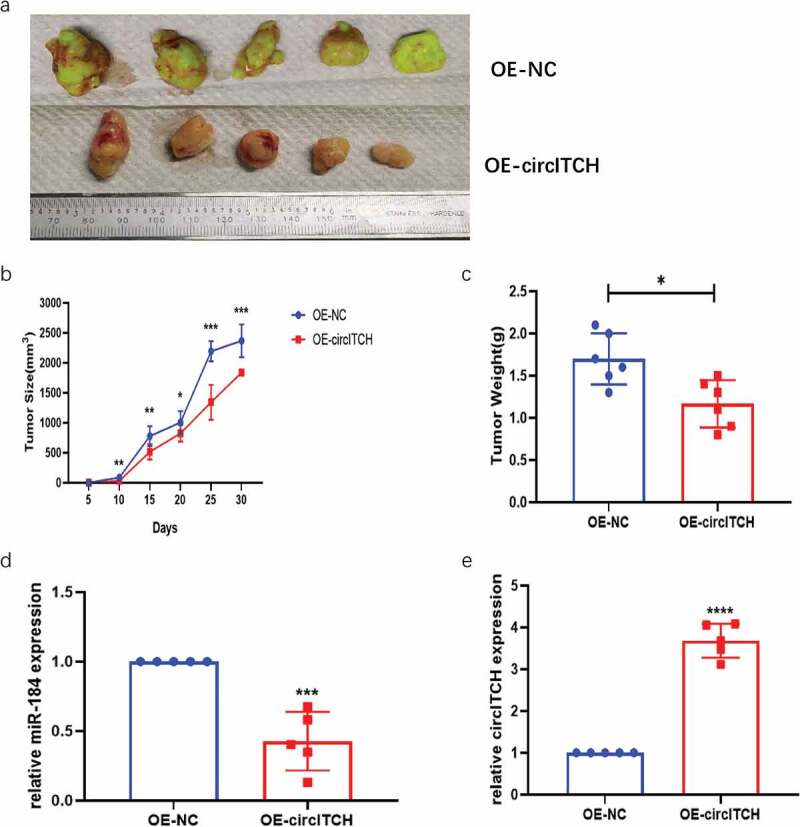
Overexpression of circITCH Depressed Tumorigenesis of HCC In Vivo. 4–6 week NSG mice were injected with LM3 cells infected with OE-circITCH or OE-NC. Five mice were used in each group.
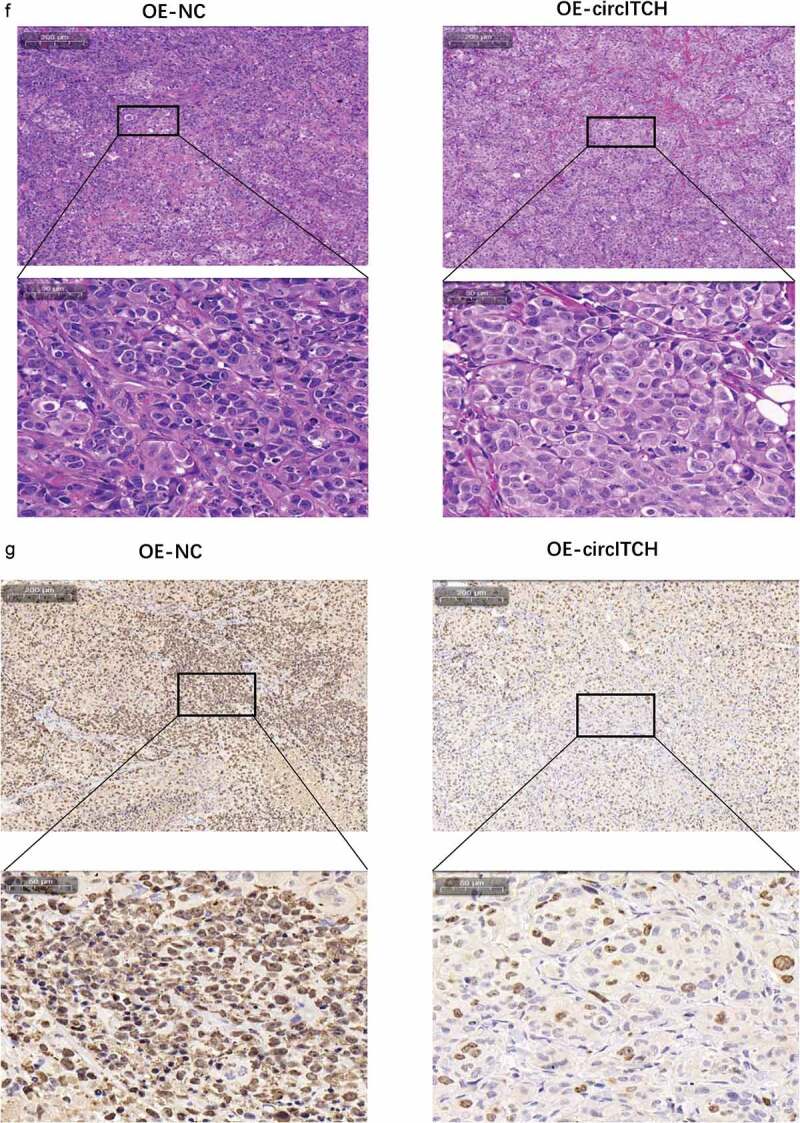
a. Representative images of the HCC tumor bearing NSG mice. b. Comparison of the tumor volume between OE-circITCH and OE-NC. c. Comparison of the tumor weight between OE-circITCH and OE-NC. d and e. The expression of circITCH and miR-184 in tumors measured by RT-qPCR. f. H & E staining.g. IHC staining of Ki-67 in tumor tissues. *p < 0.05, **p < 0.01, ***P < 0.001
Figure 5.
Continued
Prediction and discovery of circITCH/miR-184/mRNA related regulatory networks in HCC
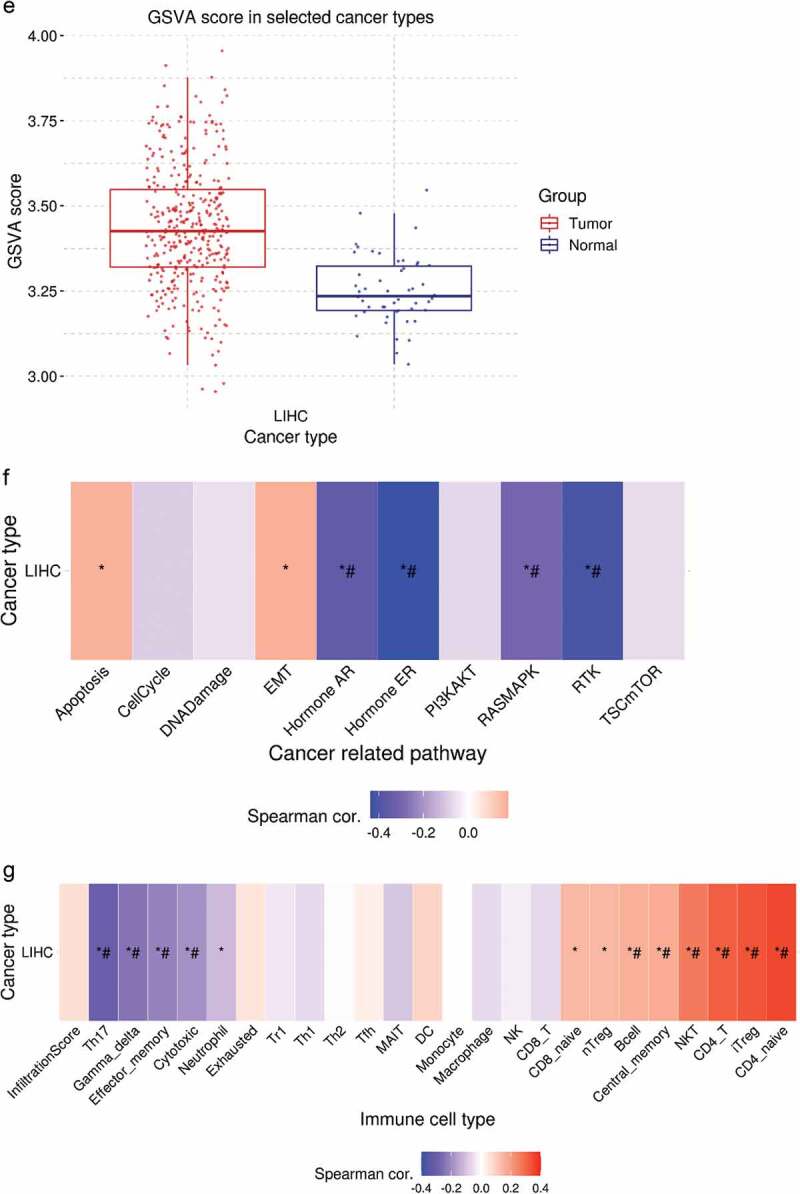
CircRNA/miRNA/mRNA regulatory networks have been extensively reported in the literature and play an important role in various diseases and cancers. Therefore, we searched for and predicted the downstream targets of miR-184 from various biological information websites to determine the main functions and roles of relevant molecules in HCC. RNAInter database [27] shows the top 100 mRNA molecules that miR-184 may bind to (Figure 6a). The WebGestalt (WEB-based Gene SeT AnaLysis Toolkit) (https://webgestalt.org/) predicted 785 target genes of miR-184, and the related functions and effects of these genes enrichment are shown in Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. The results showed that many enriched GO terms were associated with biological regulation and metabolic process in biological process, membrane and nucleus in celluar component, protein binging and ion binging in molecular function (Figure 6b). According to KEGG analysis, the highly enriched pathway was regulation of membrane permeability, which related to the mechanism of materials in and out of the cell, whereas the most differentially expressed pathway was cell–cell signaling by Wnt, which may impact cell proliferation, cell fate decisions, cell polarity, and stem cell maintenance (Figure 6c-d). We then validated the association between these molecules and HCC against the Gene Set Cancer Analysis (GSCA) database [28]. For GSVA (Gene Set Enrichment Analysis) analysis, GSCA estimates variation of gene set activity (represents as GSVA score) over a specific cancer’s sample population in an unsupervised manner. This module provides differential gene set activity analysis between tumor and normal samples. The GSVA score represents the integrated level of the expression of gene set, which is positively correlate with the expression of gene set. Therefore, As shown in Figure 6e-g, the GSVA score in tumor group is higher than that in adjacent group, it can indicate that the overall expression of the gene set in the tumor group is higher. The correlation between GSVA score and activity of cancer-related pathways indicated that GSVA score were highly expressed in Apoptosis and EMT pathways, while the expression levels were decreased in Hormone AR, Hormone ER, RASMAPK, and RTK pathways. As for immune cells, GSVA score was negatively correlated with the expression of Th17, effector memory cells, cytotoxic cells, and neutrophils, while positively correlated with the expression of naive CD8 cells, naive CD4 cells, n/iTreg cells, NKT, CD4-T, B cells, and central memory cells.
Figure 6.
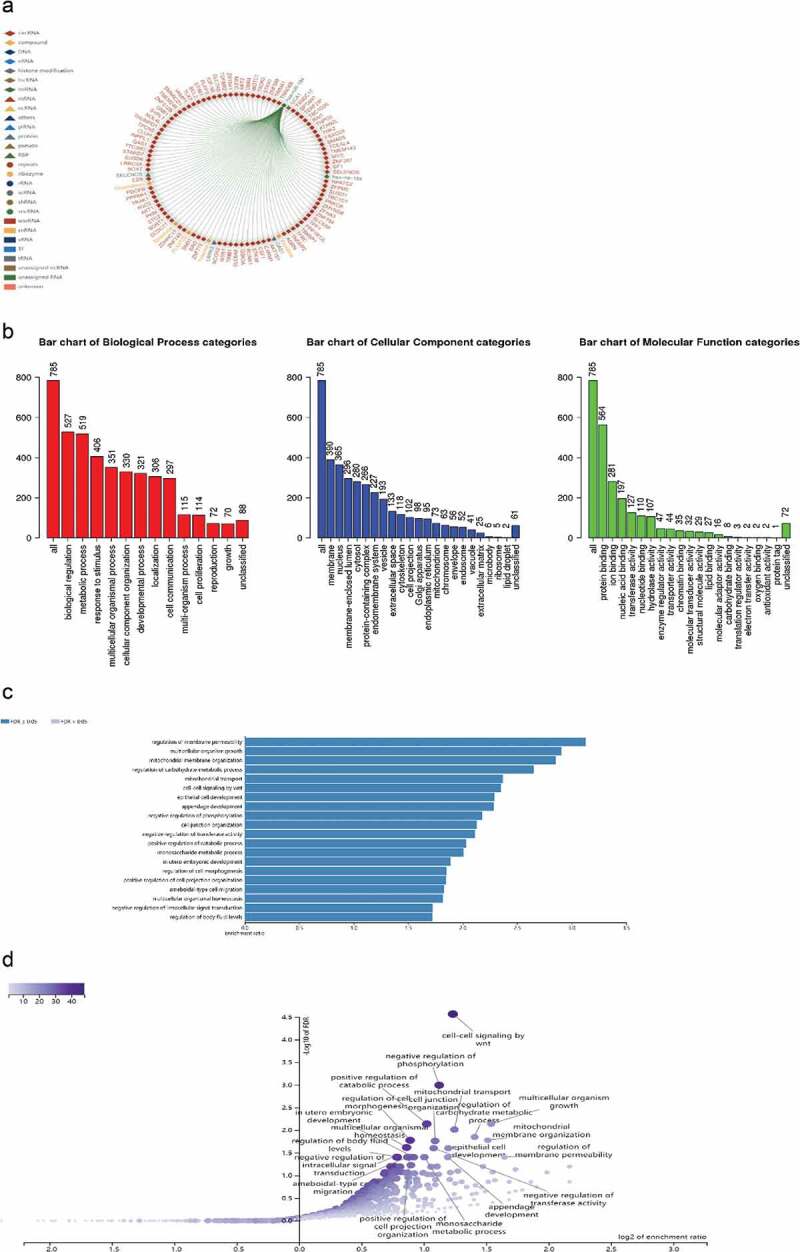
CeRNA regulatory networks and relative functions in HCC.
a. The top 100 target genes bound to miR-184 were shown in the RNAInter database. b–d. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis show the related enrichment genes function in biological process and are involved in major functional pathways including bar charts and volcanic charts. e. GSVA (Gene Set Enrichment Analysis) analysis provides differential gene set activity analysis between HCC tumor and normal samples. f. The correlation between GSVA score and activity of cancer-related pathways. g. The relationship between GSVA score and immune cells.*: P value < 0.05; #: FDR < 0.05
Figure 6.
Continue
Discussion
As novel noncoding molecules, increasing quality of circRNAs have been discovered and identified through next-generation sequencing technology (NGS) [29,30]. It has been shown that circRNAs play important roles in the occurrence and development of multiple cancers and involved in pleiotropic regulation of cell functions [8].
Previous reports have identified a number of abnormally expressed circRNAs in HCC that are upregulated or downregulated in HCC tissues and cells function as oncogenes or suppressor genes might be prognostic and diagnostic biomarkers. For example, the circARPP21/miR-543/LIFR axis suppresses the proliferation, invasion, and migration of hepatocellular carcinoma cells [31]. circMEMO1 can promote the demethylation and expression of TCF21 and can be considered a crucial epigenetic modifier in HCC progression [32]. Hsa_circ_0110102 acted as a sponge for miR-580-5p and inhibited CCL2 secretion into tumor microenvironment by decrease the expression of PPARα in HCC cells, then inhibited the pro-inflammatory cytokine release from macrophages by regulating the COX-2/PGE2 pathway [33].
In this study, we demonstrated circITCH as a significantly downregulated circRNA in HCC cell lines. For loss-and gain-of-function experiments, we revealed that overexpression of circITCH inhibits the proliferation, migration and invasion, promotes apoptosis of HCC cells in vitro, while the experimental group with silenced circITCH showed the opposite phenotype. Overall, these results elucicate the potential of circITCH as a new biomarker and tumor-suppressor molecule in hepatocellular carcinoma. The classic regulatory mechanisms of circRNA include ceRNA, bind to specific proteins to influence their functions and functional proteins coding [34]. Accumulating evidence has revealed that circRNAs primarily function as miRNA sponges. Considering that circITICH was primarily localized in the cytoplasm, we assume that circITCH may perform by sponging miRNA. To verify miRNAs targeted by circITCH, candidate miRNAs with sequences complementary to circITCH were screened through bioinformatic databases (starbase and circinteractome). We observed that miR-184 was the most highly enriched in the binding sites with circITCH, and its expression was also negatively regulated by circITCH in HCC cells. Furthermore, dual-luciferase reporter assays indicated that circITCH could directly bind to the seed region of miR-184. In a rescue experiment, miR-184 blocked the suppression of tumor proliferation and invation induced by overexpression circITCH. In addition, current research indicates that miR-184 acts as an oncogene in many types of cancers [35–37]. In short, our work suggests that circITCH might alleviate HCC progression via the circITCH/miR-184 axis (Figure 7).
Figure 7.
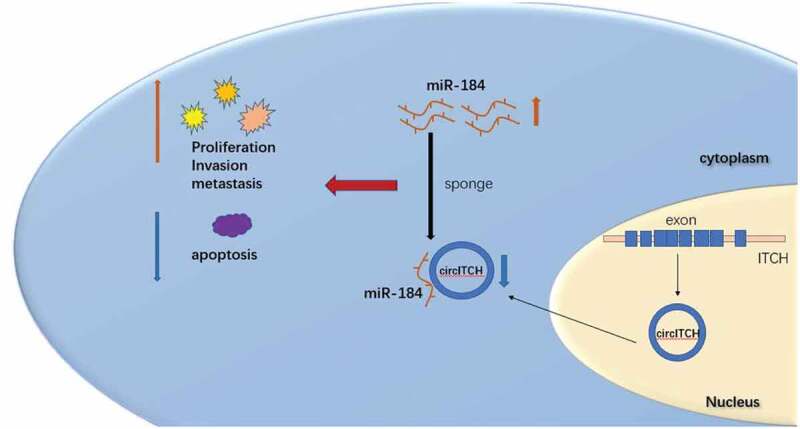
The schematic diagram illustrate the mechanism of circITCH in HCC.
Finally, to further elucidate the underlying mechanism of miR-184, bioinformatic analyses were performed to identify target genes regulated by miR-184. The results identifying target gene set that miR-184 might bind to was analyzed by biological function analysis. GO functional annotation and KEGG enrichment analysis showed that the enriched function and pathway were mainly related to biological process of cancer. GSVA score showed that gene sets were mainly involved in tumor apoptosis and EMT process. In addition, it may influence the immune process by regulating the expression of relevant immune cells. However, the specific relationship between miR-184 and its target gene needs to be further illustrated to explore a brand-new idea for circRNA biogenesis regulation. We have also described for the first time the relationship between miR-184 and circITCH. These findings offer a promising new therapeutic approach for treatment of HCC.
Trustily, there are several weaknesses in this research due to experimental condition limitations and schematic design insufficiency. First, we need clinical samples to further verify our study. Second, this study lacks the in vivo verification link of animal experiments. Finally, the regulatory network of ceRNA is not perfect, and the downstream further experiments are needed to provide valuable theoretical support for this finding. Hence, further research is essential to acquire a deep understanding of operation of molecular mechanisms in HCC biology.
Conclusion
Here, our findings reveal that circITCH served as a repressor to restrain HCC malignancy via miR-184. This novel finding not only improves our knowledge of the molecular mechanisms underlying HCC tumorigenesis, but also points to a promising therapeutic strategy against HCC.
Acknowledgments
We sincerely appreciate all participants in the study.
Funding Statement
This study was supported by The General Program of National Natural Science Foundation of China (No. 81774261); Science and Technology Program of Guangzhou, China (No. 202102080232); Project of Administration of Traditional Chinese Medicine of Guangdong Province China (No. 20191007).
Authors’ contributions
XG were responsible for performing the experiments and drafted the manuscript. XG and CjW were responsible for designing the experiments and supervising the study. All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author details
1. School of Medicine, South China University of Technology, Guangzhou, 510,006, China
2. Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Guangzhou, 510,080, China
3. South China University of Technology, Guangzhou, 510,515, China
4. Guangzhou University of Chinese Medicine, Guangzhou, 510,405, China
5. First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, 510,080, China
References
- [1].Bray F,F, Soerjomataram J, Siegel I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- [2].Yang JD,H, P GGJ, Amadou A, et al. A Global View of Hepatocellular Carcinoma: trends, Risk, Prevention and Management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Forner A, Reig M, Bruix J.. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. [DOI] [PubMed] [Google Scholar]
- [4].Colquhoun SD. Hepatocellular carcinoma: the current role of surgical intervention. Crit Rev Oncog. 2016;21(1–2):93–103. [DOI] [PubMed] [Google Scholar]
- [5].Zhou F, Shang W, Yu X, et al. Glypican-3: a Promising Biomarker for Hepatocellular Carcinoma Diagnosis and Treatment. Med Res Rev. 2018;38(2):741–767. [DOI] [PubMed] [Google Scholar]
- [6].Petkovic S, Müller S.. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43(4):2454–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang Y, Mo Y, Gong Z, et al. Circular RNAs in human cancer. Mol Cancer. 2017;16(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li J, Sun D, Pu W, et al. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6(4):319–336. [DOI] [PubMed] [Google Scholar]
- [9].Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. [DOI] [PubMed] [Google Scholar]
- [11].Li X, Diao H. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR1 and CDR1. J Cell Physiol. 2019;234(8):13807–13819. [DOI] [PubMed] [Google Scholar]
- [12].Liu L, Yang X, Li NF, et al. Circ_0015756 promotes proliferation, invasion and migration by microRNA-7-dependent inhibition of FAK in hepatocellular carcinoma. Cell Cycle. 2019;18(21):2939–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Arnaiz E, Sole C, Manterola L, et al. CircRNAs and cancer: biomarkers and master regulators. Semin Cancer Biol. 2019;58:90–99. [DOI] [PubMed] [Google Scholar]
- [14].Su M, Xiao Y, Ma J, et al. Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. 2019;18(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bai N, Peng E, Qiu X, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Cancer Res. 2018;37:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wei C, Yingyao Q, Shaoyi F, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–128. [DOI] [PubMed] [Google Scholar]
- [17].Zhang H, Deng T, Ge S, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38(15):2844–2859. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [18].Luo L, Gao Y, Sun X. Circ-ITCH correlates with small tumor size, decreased FIGO stage and prolonged overall survival, and it inhibits cells proliferation while promotes cells apoptosis in epithelial ovarian cancer. Cancer Biomark. 2018;23:505–513. [DOI] [PubMed] [Google Scholar]
- [19].Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang ST, Liu LB, Li XM, et al. Circ-ITCH regulates triple-negative breast cancer progression through the Wnt/beta-catenin pathway. Neoplasma. 2019;66:232–239. [DOI] [PubMed] [Google Scholar]
- [21].Wang M, Chen B, Ru Z, et al. CircRNA circ-ITCH suppresses papillary thyroid cancer progression through miR-22-3p/CBL/betacatenin pathway. Biochem Biophys Res Commun. 2018;504:283–288. [DOI] [PubMed] [Google Scholar]
- [22].Huang G, Hua Z, Shi Y, et al. cir-ITCH Plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PLoS ONE. 2015;10(6):e0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wenzhi G, Jiakai Z, Dongyu Z, et al. Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma.[J]. Oncotarget. 2017;8:48169–48177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, et al. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234(5):5451–5465. [DOI] [PubMed] [Google Scholar]
- [25].Panda AC. Circular RNAs Act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79. [DOI] [PubMed] [Google Scholar]
- [26].Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14(8):1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lin Y, Liu T, Cui T. RNAInter in 2020: RNA interactome repository with increased coverage and annotation. Nucleic Acids Res. 2020;48(D1):D189–D197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liu CJ, Hu FF, Xia M, et al. GSCALite: a Web Server for Gene Set Cancer Analysis. Bioinformatics. 2018;34(21):3771–3772. [DOI] [PubMed] [Google Scholar]
- [29].Lopez-Jimenez E, Rojas AM, Andres-Leon E. RNA sequencing and Prediction Tools for Circular RNAs Analysis. Adv Exp Med Biol. 2018;1087:17–33. [DOI] [PubMed] [Google Scholar]
- [30].Hsiao KY, Sun HS, Tsai SJ. Circular RNA - New member of noncoding RNA with novel functions. Exp Biol Med (Maywood, NJ). 2017;242:1136–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yichao G, Fan W, Hao W, et al. Circular RNA circARPP21 Acts as a Sponge of miR-543 to Suppress Hepatocellular Carcinoma by Regulating LIFR.[J]. Onco Targets Ther. 2021;14:879–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhao-Ru D, Ai-Wu K, Tao L, et al. CircMEMO1 modulates the promoter methylation and expression of TCF21 to regulate hepatocellular carcinoma progression and sorafenib treatment sensitivity.[J]. Mol Cancer. 2021;20(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Xinxing W, Wei S, Tao X, et al. CircRNA hsa_circ_0110102 inhibited macrophage activation and hepatocellular carcinoma progression via miR-580-5p/PPARα/CCL2 pathway.[J]. Aging (Albany NY). 2021;13(8):11969–11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–490. [DOI] [PubMed] [Google Scholar]
- [35].Jingdong L, Youmei Z, Wanling W, et al. Long Noncoding RNA MEG3 Inhibits Cell Proliferation and Metastasis in Chronic Myeloid Leukemia via Targeting miR-184. Oncol Res. 2018;26:297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yin Y, Hong L, Chunhua W, et al. Circ_0021087 acts as a miR-184 sponge and represses gastric cancer progression by adsorbing miR-184 and elevating FOSB expression.[J]. Eur J Clin Invest. 2021undefined: e13605; 5111: 10.1111/eci.13605 [DOI] [PubMed] [Google Scholar]
- [37].Wenjie H, Yueming Z. Circ_0025033 promotes the progression of ovarian cancer by activating the expression of LSM4 via targeting miR-184.[J]. Pathol Res Pract. 2021;217:153275. [DOI] [PubMed] [Google Scholar]


