ABSTRACT
Diabetic retinopathy (DR) as a frequent diabetic microvascular complication shows signs in one-third of diabetic patients. Long non-coding RNAs (lncRNAs) have drawn increasing attention because of their regulatory roles in DR. LncRNA plasmacytoma variant translocation 1 (PVT1) is documented to be upregulated in diabetes-related diseases, while its effects in DR remains unexplored. ARPE-19 cells under the treatment of high-glucose (HG) were used as DR cell models. The gene expression in ARPE-19 cells was examined using RT-qPCR. The viability and apoptosis of ARPE-19 cells were determined by MTT and TUNEL assays. The levels of inflammation-associated proteins or mRNA were measured using western blot. Luciferase reporter assay and RNA pull down assay were conducted for the exploration of the underlying mechanism of PVT1. PVT1 was revealed to be upregulated in DR cell models. Silencing of PVT1 promoted the viability and inhibited apoptosis of HG-stimulated ARPE-19 cells. The results revealed that PVT1 can bind with miR-1301-3p. PVT1 negatively modulated miR-1301-3p expression. Additionally, KLF7 was targeted by miR-1301-3p. PVT1 upregulated KLF7 expression by binding with miR-1301-3p. The silenced PVT1-mediated influence on cell viability and cell apoptosis was rescued by overexpression of KLF7. PVT1 suppresses proliferation and promotes apoptosis of ARPE-19 cells treated with HG in vitro by binding with miR-1301-3p to upregulate KLF7.
KEYWORDS: PVT1, miR-1301-3p, KLF7
Introduction
A series of complications can be induced in the development of diabetes [1,2]. More than 30% of patients with diabetes have a pathological phenotype of diabetic retinopathy (DR) [3,4]. Inhibition of angiogenic factors, regenerative therapy, and topical therapy are promising for DR treatment [3]. Long-term exposure to high glucose (HG) environment, albuminuria, poor glycemic control and hypertension are risk factors for DR [5,6]. HG-induced retinal damage is the main pathological phenotype of DR, and ARPE-19 cells under the HG treatment were previously used as an in vitro model of DR [7–9].
Long noncoding RNAs (lncRNAs) are noncoding transcripts longer over 200 nucleotides and possess no ability to code proteins [10,11]. Studies have revealed that lncRNAs act as critical regulators in various diseases, including DR. In DR, lncRNA Hotair promotes the dysfunction of retinal endothelial cells [12]. LncRNA AK077216 downregulates miR-383 to inhibit the apoptosis of retinal pigment epithelial cells [13]. LncRNA H19 suppresses the endothelial–mesenchymal transition process of human retinal endothelial cells [14].
LncRNA plasmacytoma variant translocation 1 (PVT1) is a critical regulator in many diabetes-related diseases. For example, PVT1 is suggested to be a key factor of population susceptibility to diabetes-induced end-stage renal disease [15]. PVT1 knockdown ameliorates podocyte impairment and apoptosis in diabetic nephropathy [16]. PVT1 regulates the PI3K/AKT pathway to exert a protective effect on diabetic peripheral neuropathy [17]. SP1-induced PVT1 mediates the miR-214-3p/MMP2 axis to regulate lens epithelial cell proliferation and apoptosis in diabetic cataract [18]. Importantly, PVT1 has been widely reported to serve as a competitive endogenous RNA (ceRNA) in diabetes-related diseases [19–21]. The ceRNA pattern refers to that lncRNAs competitively bind with microRNAs (miRNAs) to prevent the degradation of target mRNAs caused by miRNAs [22]. In recent years, more and more attention has been paid to the investigation of ceRNA mechanism in diabetes-related diseases. For instance, SPAG5-AS1 serves as a ceRNA against miR-769-5p to elevate the level of SPAG5, inhibiting autophagy and promoting apoptosis of podocytes, predictors of diabetic nephropathy [23]. LncRNA SNHG16 regulates miR-141-3p and CCND1 to facilitate proliferation and fibrogenesis in diabetic nephropathy [24]. MEG3 suppresses HG-stimulated retinal pigment epithelium cell apoptosis by modulating the miR-93/Nrf2 axis [25].
The study aimed to explore the function and underlying mechanism of PVT1 in DR. We assumed that PVT1 acted as a ceRNA in DR and investigated its downstream regulatory mechanism in DR cell models, which might provide new light on DR treatment.
Materials and methods
Cell line and cell culture
The human retinal pigment epithelial cell line (ARPE-19, catalog number: CRL-2302) was provided by the American Type Culture Collection (ATCC, Manassas, VA, USA). Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Rockville, MD, USA) containing 10% fetal bovine serum (FBS, HyClone, South Logan, UT), 1/100 penicillin/streptomycin was used for incubating ARPE-19 cells at 37°C with 5% CO2. Different concentrations (0, 0.1, 0.5, 1, 5, 10, 20, 50 mM) of glucose (D-glucose, Thermo Fisher Scientific) were added to culture ARPE-19 cells for 24 h.
Transfection
The short hairpin RNA targeting PVT1 (sh-PVT1) with corresponding negative control (sh-NC), miR-1301-3p mimics with control NC mimics were provided by GenePharma (Shanghai, China). The full length of KLF7 was inserted into pcDNA3.1 to construct KLF7 overexpression vector (pcDNA3.1/KLF7) with empty pcDNA3.1 as control. Lipofectamine 2000 (Invitrogen) was used to transfect these plasmids into ARPE-19 cells.
RT-qPCR analysis
TRIzol reagent (Sigma-Aldrich, Shanghai, China) was used to isolate RNA in ARPE-19 cells. Then cDNAs were synthesized using a Script High Fidelity One Step RT-PCR Kit (Vazyme) for PVT1 and KLF7 and an RNA to cDNA EcoDry Premix Kit (Vazyme) for miR-1301-3p. The quantitative PCR was performed with VeriQuest SYBR Green qPCR Master Mix (Thermo Fisher Scientific, Waltham, MA) under an Applied Biosystems 7500 system. The levels of RNAs were calculated with the 2−ΔΔCt method normalized to GAPDH and U6. This assay was repeated three times. The primer sequences were presented:
PVT1:
F: 5’-AGAATTAAGAGTGTGGGCAC-3’,
R: 5’-GAATGCCCTCTTCTTAGGG-3’;
miR-1301-3p:
F: 5’-TTGCAGCTGCCTGGGA-3’,
R: 5’-CTCTACAGCTATATTGCCAGCCAC-3’;
KLF7:
F: 5’-AGCTACAACTTGTCCACGA-3’,
R: 5’-ATTCAAGGCATGTCTGCTG-3’;
GAPDH:
F: 5’-CATTTCCTGGTATGACAACGA-3’,
R: 5’-GGGTCTTACTCCTTGGAGG-3’;
U6:
F: 5’-TGCTATCACTTCAGCAGCA-3’,
R: 5’-GAGGTCATGCTAATCTTCTCTG-3’.
MTT assay
The cell viability in different groups was assessed by an MTT assay. Transfected ARPE-19 cells were seeded into 96-well plates. After 48 h of culturing, 20 μL of MTT solution was added into each well and underwent another 4 h incubation. After removing the supernatant, dimethyl sulfoxide (DMSO, 100 μL, Sigma) was used to dissolve the formazan. The absorbance was measured at 490 nm with a microplate reader (Molecular Devices, USA). This experiment was performed three times.
TUNEL assay
The paraformaldehyde (4%) was used to fix ARPE-19 cells (1 × 104) after indicated transfection on culture slides. A TUNEL Detection Kit (Roche, Basel, Switzerland) was used to examine the cell apoptosis. After incubating at 37°C for 60 min, nuclei of ARPE-19 cells were stained using DAPI. Then, a microscope (Olympus, Tokyo, Japan) was used to count the TUNEL positive cells.
Western blot analysis
A RIPA lysis buffer (Sigma-Aldrich) with a protease inhibitor was used to lyse ARPE-19 cells and extract proteins inside. Proteins were subjected to SDS-PAGE and then were transferred to PVDF membranes. Next, 5%-skim milk was used to block the membranes, which were cultured with primary antibodies against GAPDH (ab9485, Abcam, Shanghai, China), KLF7 (ab197690), ICAM (ab53013), VEGF (ab150375) and TNF-α (ab183218) overnight at 4°C. The secondary antibodies were cultured with membranes for 1 h in the dark. GAPDH served as the internal control. Thereafter, an enhanced chemiluminescent reagent (Millipore, Plano, TX, USA) was applied to capture the protein bands. This experiment was conducted independently three times.
Subcellular fractionation assay
Total RNA was extracted using a mirVana™ PARIS™ RNA and Native Protein Purification Kit according to the manufacturer’s instructions. ARPE-19 cells placed into cell fractionation buffer were subjected to centrifugation. Next, we collected the supernatant, and rinsed the remaining lysates with cell fractionation buffer following lysing cell nuclei by cell disruption buffer. Thereafter, the lysate was incubated with the supernatant, ethanol and 2 × lysis/binding solution. Finally, expressions of cytoplasmic and nuclear RNAs were detected by RT-qPCR.
RNA immunoprecipitation assay
An EZ Magna RNA immunoprecipitation Kit (Millipore, USA) was used based on the manufacturer’s instructions. Briefly, ARPE-19 cells were lysed in RIPA lysis buffer. Magnetic beads were pre-incubated with antibodies including anti-IgG and anti-Ago2 for 30 min at 27°C. Subsequently, cell lysates were immunoprecipitated with beads for 6 h at 4°C. Then, RNA was purified and detected by RT-qPCR.
RNA pull down assay
MiR-1301-3p probe-no biotin and miR-1301-3p probe-biotin were purchased from Sangon (Shanghai, China). After culture for 48 h, cells were lysed. Cell lysate, bio-probes, and Dynabeads M-280 streptavidin beads (Invitrogen) were mixed. The relative enrichment of PVT1 or mRNAs in ARPE-19 cells was subject to RT-qPCR analysis. This assay was performed three times.
Luciferase reporter assay
The fragments of KLF7 with wild type or mutant-binding sequences for miR-1301-3p were subcloned into the pmirGLO vectors (Promega, Madison WI, USA), which were subsequently cotransfected with miR-1301-3p mimics or NC mimics by Lipofectamine 2000 (Invitrogen) into ARPE-19 cells for 2 days. The luciferase activities of the indicated reporters were accessed using the luciferase reporter assay system (Promega). This assay was conducted independently three times.
Statistical analysis
Data are exhibited as the mean values ± standard deviation (SD). The analysis of the results was performed with SPSS 19.0 (SPSS, Chicago, IL). Each experiment was conducted three times. The two-group comparisons were performed with student’s t-tests and comparisons in multiple groups were subject to one-way analysis of variance (ANOVA) followed by Tukey’s post hoc analysis. p < 0.05 was regarded as statistically significant.
Results
PVT1 knockdown enhanced viability and suppressed cell apoptosis of HG-stimulated ARPE-19 cells
First, human retinal pigment epithelial cell line (ARPE-19) received treatment of different concentrations (0, 0.1, 0.5, 1, 5, 10, 20, 50 mM) of D-glucose (glucose), and RT-qPCR analysis revealed the increased PVT1 expression along with elevated glucose concentrations. Glucose concentration at 50 mM was used for the following assays (Figure 1(a)). The level of PVT1 was assessed in HG treated ARPE-19 cells using RT-qPCR analysis. PVT1 expression was higher in HG group, and the knockdown efficiency of PVT1 was verified (Figure 1(b)). Silenced PVT1 reversed the decreased ARPE-19 cell viability induced by HG (Figure 1(c)). Results from the TUNEL assay indicated that suppression of PVT1 rescued the HG-caused elevation in apoptosis of ARPE-19 cells (Figure 1(d)). Furthermore, the expression of proinflammatory proteins (ICAM, VEGF and TNF-α) was examined. The levels of ICAM, VEGF and TNF-α were increased in ARPE-19 cells under HG treatment, while silenced PVT1 offset the HG-mediated proinflammatory effects on ARPE-19 cells (Figure 1(e)).
Figure 1.
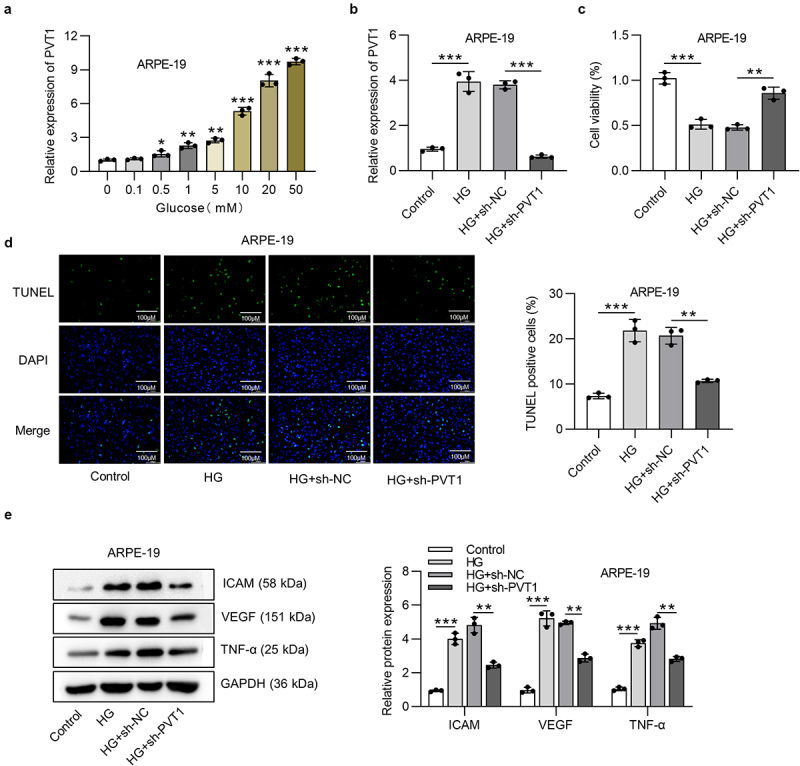
PVT1 inhibition promoted viability and suppressed cell apoptosis in HG-stimulated ARPE-19 cells. (a) The PVT1 expression in ARPE-19 cells stimulated by D-glucose (glucose) at concentrations of 0, 0.1, 0.5, 1, 5, 10, 20 and 50 mM for 24 h. (b) The expression of PVT1 under the HG treatment and knockdown efficiency of PVT1 was assessed using RT-qPCR analysis. (c) The viability of ARPE-19 cells under HG stimulation and posttransfection of sh-PVT1 was measured using MTT assay. (d) TUNEL assay was performed to access the ARPE-19 cell apoptosis. (e) The western blot analysis showed the protein expression of ICAM, VEGF and TNF-α in HG-stimulated ARPE-19 cells after the transfection of sh-PVT1. *p < 0.05, **p < 0.01, ***p < 0.001.
MiR-1301-3p was negatively regulated by PVT1
Abundant data have revealed that lncRNAs act as ceRNAs absorbing miRNAs to upregulate mRNAs in various diseases [26,27]. We supposed that PVT1 is involved in the development of DR by the ceRNA network. On the starBase website under the condition of strict stringency (≥5), three miRNAs that can bind with PVT1 were found. The following assays were conducted in HG-stimulated ARPE-19 cells. RT-qPCR analysis indicated the highest expression of miR-1301-3p in ARPE-19 cells transfected with sh-PVT1 compared with other miRNAs (Figure 2(a)). The miR-1301-3p was expressed at a low level in HG-stimulated ARPE-19 cells, and the overexpression efficiency of miR-1301-3p was validated (Figure 2(b)). The binding sequences between PVT1 and miR-1301-3p were presented (Figure 2(c)). Moreover, miR-1301-3p expression was increased after PVT1 silencing. RNA pull down assay revealed that PVT1 was abundantly enriched in the complex pulled by miR-1301-3p probe-biotin (Figure 2(d)).
Figure 2.
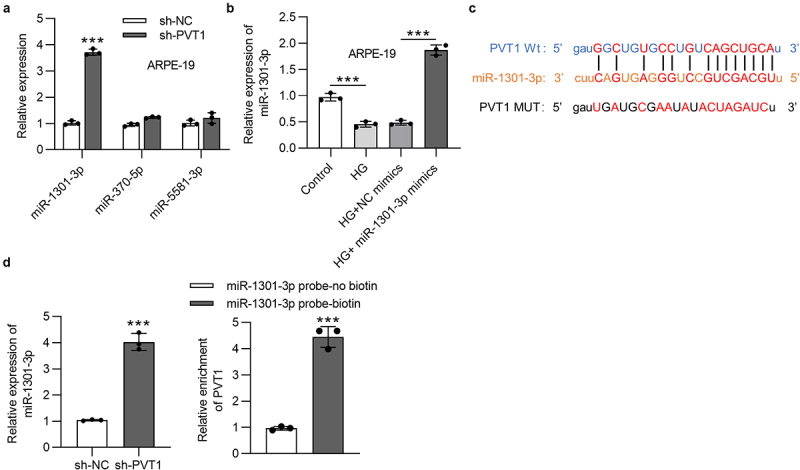
PVT1 interacted with miR-1301-3p in ARPE-19 cells. (a) The expression of three miRNAs (miR-1301-3p, miR-370-5p and miR-5581-3p) in ARPE-19 cells posttransfection of sh-PVT1. (b) The miR-1301-3p expression in ARPE-19 cells posttransfection of miR-1301-3p mimics and stimulated by HG. (c) The binding sequences between PVT1 and miR-1301-3p. (d) The miR-1301-3p level in ARPE-19 cells with silenced PVT1. The interaction between miR-1301-3p and PVT1 was investigated using RNA pull down assay. ***p < 0.001.
KLF7 was the downstream target gene of miR-1301-3p
The Venn diagram showed that there are 11 mRNAs, which can bind with miR-1301-3p after searching on the microT, miRmap and PicTar websites under screening condition of pan-Cancer≥8 (Figure 3(a)). Then the following assays were conducted in ARPE-19 cells treated with HG. The expression of KLF7 was abundantly enriched in miR-1301-3p probe biotin group compared with other mRNAs (Figure 3(b)). The KLF7 was expressed at a high level at the mRNA and protein levels in HG-treated ARPE-19 cells (Figure 3(c)). Luciferase reporter assay demonstrated that the overexpression of miR-1301-3p reduced the luciferase activity of KLF7-Wt reporters but did not affect that of KLF7-Mut reporters (Figure 3(d)). The expression of KLF7 was decreased at the mRNA and protein levels by miR-1301-3p overexpression or PVT1 knockdown in ARPE-19 cells (Figure 3(e,f)). PVT1, miR-1301-3p, KLF7 were majorly located in the cytoplasm of ARPE-19 cells (Figure 3(g)). An Ago2 RIP assay revealed that PVT1, miR-1301-3p, KLF7 were enriched in Ago2 precipitated products, indicating the co-existence of PVT1, miR-1301-3p, KLF7 in RNA-induced silence complexes (Figure 3(h)).
Figure 3.
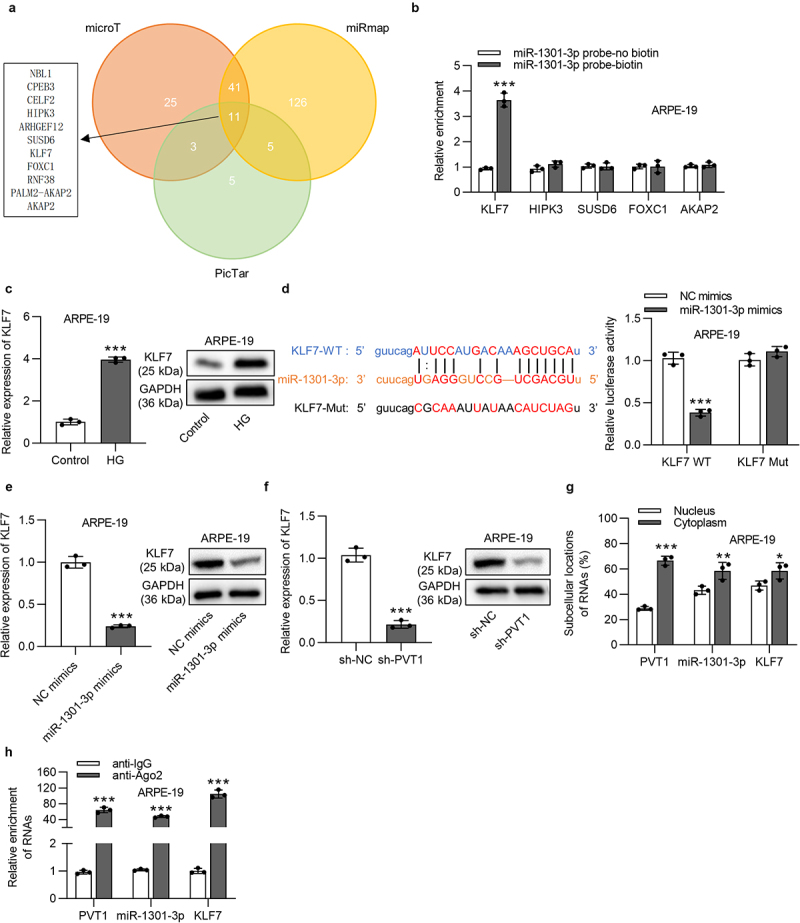
KLF7 was targeted by miR-1301-3p. (a) The Venn diagram of potential mRNAs possessing the binding site with miR-1301-3p on the microT, PicTar, miRmap websites under the condition of pan-cancer ≥8. (b) The enrichment of KLF7, HIPK3, SUSD6, FOXC1 and AKAP2 for miR-1301-3p was examined by RNA pull down assay. (c) The KLF7 level in HG-stimulated ARPE-19 cells was detected by RT-qPCR and western blotting assays. (d) The binding site and relationship between KLF7 and miR-1301-3p were examined using a luciferase reporter assay. (e-f) KLF7 level in ARPE-19 cells with overexpressed miR-1301-3p or silenced PVT1 was detected by RT-qPCR and western blotting assays. (g) After subcellular fraction, PVT1, miR-1301-3p, KLF7 expression was detected by RT-qPCR. (h) Enrichment of PVT1, miR-1301-3p, KLF7 in Ago2 precipitated products was detected by RIP assay followed by RT-qPCR and was normalized to IgG. *p < 0.05, **p < 0.01, ***p < 0.001.
KLF7 reversed the effect induced by silenced PVT1 on viability and apoptosis in DR cell model
To explore whether PVT1 regulates DR progression by modulating KLF7 expression, rescue assays were done. First, the overexpression efficiency of KLF7 was verified in HG-stimulated ARPE-19 cells (Figure 4(a)). PVT1 deficiency-mediated increase in cell viability was counteracted by KLF7 overexpression (Figure 4(b)). The apoptosis of ARPE-19 cells treated with HG was reduced after silencing PVT1, while the overexpression of KLF7 reversed this effect (Figure 4(c,d)). The protein levels of ICAM, VEGF and TNF-α were reduced after PVT1 knockdown, while the upregulation of KLF7 neutralized the suppressive effect caused by PVT1 knockdown on proinflammatory proteins (Figure 4(e)).
Figure 4.
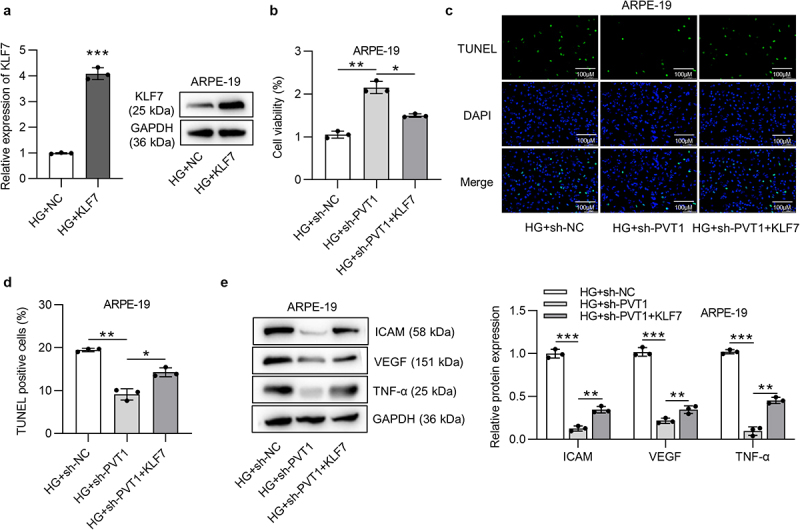
PVT1 regulated ARPE-19 cell viability and apoptosis by upregulating KLF7. (a) The overexpression efficiency of KLF7 was verified by RT-qPCR. (B) The viability of ARPE-19 cells in HG+sh-NC, HG+sh-PVT1 and HG+sh-PVT1+ KLF7 groups was measured using MTT assay. (c-d) The ARPE-19 cell apoptosis in HG+sh-NC, HG+sh-PVT1 and HG+sh-PVT1+ KLF7 groups was detected by TUNEL assay. (e) The protein expression of ICAM, VEGF and TNF-α in HG+sh-NC, HG+sh-PVT1 and HG+sh-PVT1+ KLF7 groups was detected by western blotting assay. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
LncRNAs have been revealed to be important regulators in DR [12–14], among which PVT1 has been identified as a key regulator in diabetes-related diseases [16–18]. In our study, we used HG-treated ARPE-19 cells as the in vitro model of PD. Our findings revealed that HG treatment inhibited viability and promoted apoptosis of ARPE-19 cells. Chronic inflammation is a significant pathogenic factor of DR and diabetes-induced retinal inflammation may result in endothelium impairment, pericyte loss, increased capillary degeneration, and vascular leakage in DR [28,29]. We found that HG induced the upregulation of proinflammatory proteins and cytokines including ICAM, VEGF, TNF-α in ARPE-19 cells. PVT1 was upregulated in ARPE-19 cells under HG treatment. PVT1 knockdown elevated cell viability and suppressed the apoptosis and inflammatory response of HG-stimulated ARPE-19 cells.
We hypothesized that PVT1 participates in the regulation of DR by the ceRNA pattern, and the downstream miRNA for PVT1 was searched. Based on the starBase website, PVT1 possesses a binding site with miR-1301-3p. The current work revealed that PVT1 interacted with miR-1301-3p in ARPE-19 cells and negatively modulated the level of miR-1301-3p. Subsequently, KLF7 was identified the downstream target gene of miR-1301-3p.
KLF7 is a member of the KLF family of transcription factors. Previous studies found that KLF7 is involved in diabetes-associated diseases. The minor A-allele of KLF7 rs7568369 is closely associated with the protection against obesity in the Danish population [30]. KLF7 overexpression leads to a significant suppression of adipogenesis in 3T3-L1 adipocytes, indicating KLF7 as candidate for conferring genetic susceptibility to type 2 diabetes [31]. KLF7 is involved in TGF-β1 regulated DNA methylation, which causes damage to kidney glomerular mesangial cells and contributes to the progression of diabetic nephropathy [32]. KLF7 promotes type II diabetes progression by damaging the insulin biosynthesis and secretion in pancreatic beta-cells and attenuating the insulin sensitivity of peripheral tissues [33]. In our work, it was confirmed that miR-1301-3p inhibited KLF7 expression at the mRNA and protein levels by targeting the 3ʹUTR of KLF7. PVT1 upregulated KLF7 expression in ARPE-19 cells by binding with miR-1301-3p. Rescue assays indicated that PVT1 knockdown-induced influence on cell viability, inflammatory response and cell apoptosis were rescued by KLF7 overexpression.
In conclusion, our study demonstrated that silencing of PVT1 promotes viability and suppresses apoptosis and inflammatory response of HG-stimulated ARPE-19 cells. Mechanistically, PVT1 binds with miR-1301-3p to upregulate KLF7 and further to exert its functions in HG-stimulated ARPE-19 cells. The findings in our study indicate that targeting PVT1 is promising for DR treatment.
Funding Statement
The work was supported by Program for the Scientific Activities of Selected Returned Overseas Professionals in Shanxi Province (Fund Program for the Scientific Activities of Selected Returned Overseas Professionals in Shanxi Province 20200041) and Research Project Supported by Shanxi Scholarship Council of China (2020-188).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
Jianjin Guo conceived and designed research; Jianjin Guo, Feng Xiao, Wei Ren, and Yikun Zhu performed the research; Jianjin Guo, Qiujing Du, Qian Li, and Xing Li analyzed the data; Jianjin Guo wrote the paper; Jianjin Guo and Feng Xiao edited the manuscript. The final version of the manuscript was approved by all authors.
Data availability statement
Data generated in our study are available from the corresponding author on reasonable request.
References
- [1].Yau JW, Rogers SL, Kawasaki R, et al. Global prevalence and major risk factors of diabetic retinopathy. Diabetes Care. 2012. Mar;35(3):556–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Karaa A, Goldstein A.. The spectrum of clinical presentation, diagnosis, and management of mitochondrial forms of diabetes. Pediatr Diabetes. 2015. Feb;16(1):1–9. [DOI] [PubMed] [Google Scholar]
- [3].Cheung N, Mitchell P, Wong TY.. Diabetic retinopathy. Lancet. 2010. Jul 10;376(9735):124–136. [DOI] [PubMed] [Google Scholar]
- [4].Moreno A, Lozano M, Salinas P. Diabetic retinopathy. Nutr Hosp. 2013. Mar;28 Suppl 2:53–56. [DOI] [PubMed] [Google Scholar]
- [5].Ghamdi AHA. Clinical predictors of diabetic retinopathy progression; A systematic review. Curr Diabetes Rev. 2020;16(3):242–247. [DOI] [PubMed] [Google Scholar]
- [6].Win Tin ST, Kenilorea G, Gadabu E, et al. The prevalence of diabetes complications and associated risk factors in Pacific Islands countries. Diabetes Res Clin Pract. 2014. Jan;103(1):114–118. [DOI] [PubMed] [Google Scholar]
- [7].Tenconi PE, Bermúdez V, Oresti GM, et al. High glucose-induced phospholipase D activity in retinal pigment epithelium cells: new insights into the molecular mechanisms of diabetic retinopathy. Exp Eye Res. 2019. Jul;184:243–257. [DOI] [PubMed] [Google Scholar]
- [8].Chen P, Miao Y, Yan P, et al. MiR-455-5p ameliorates HG-induced apoptosis, oxidative stress and inflammatory via targeting SOCS3 in retinal pigment epithelial cells. J Cell Physiol. 2019. Dec;234(12):21915–21924. [DOI] [PubMed] [Google Scholar]
- [9].Yang Z, Hu H, Zou Y, et al. miR-7 reduces high glucose induced-damage via HoxB3 and PI3K/AKT/mTOR signaling pathways in retinal pigment epithelial cells. Curr Mol Med. 2020;20(5):372–378. [DOI] [PubMed] [Google Scholar]
- [10].Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46. [DOI] [PubMed] [Google Scholar]
- [11].Zhu J, Fu H, Wu Y, et al. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci China Life Sci. 2013. Oct;56(10):876–885. [DOI] [PubMed] [Google Scholar]
- [12].Zhao D, Zhao Y, Wu L, et al. Long noncoding RNA Hotair facilitates retinal endothelial cell dysfunction in diabetic retinopathy. Clin Sci (Lond). 2020. Aug 19;134:2419–2434. [DOI] [PubMed] [Google Scholar]
- [13].Zhu L, Zhang Q, Li S, et al. Interference of the long noncoding RNA CDKN2B-AS1 upregulates miR-181a-5p/TGFβI axis to restrain the metastasis and promote apoptosis and senescence of cervical cancer cells. Cancer Med. 2019. Apr;8(4):1721–1730. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [14].Thomas AA, Biswas S, Feng B, et al. lncRNA H19 prevents endothelial-mesenchymal transition in diabetic retinopathy. Diabetologia. 2019. Mar;62(3):517–530. [DOI] [PubMed] [Google Scholar]
- [15].Millis MP, Bowen D, Kingsley C, et al. Variants in the plasmacytoma variant translocation gene (PVT1) are associated with end-stage renal disease attributed to type 1 diabetes. Diabetes. 2007. Dec;56(12):3027–3032. [DOI] [PubMed] [Google Scholar]
- [16].Liu DW, Zhang JH, Liu FX, et al. Silencing of long noncoding RNA PVT1 inhibits podocyte damage and apoptosis in diabetic nephropathy by upregulating FOXA1. Exp Mol Med. 2019. Aug 1;51(8):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chen L, Gong HY, Xu L. PVT1 protects diabetic peripheral neuropathy via PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2018. Oct;22(20):6905–6911. [DOI] [PubMed] [Google Scholar]
- [18].Yang J, Zhao S, Tian F. SP1-mediated lncRNA PVT1 modulates the proliferation and apoptosis of lens epithelial cells in diabetic cataract via miR-214-3p/MMP2 axis. J Cell Mol Med. 2020. Jan;24(1):554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhong W, Zeng J, Xue J, et al. Knockdown of lncRNA PVT1 alleviates high glucose-induced proliferation and fibrosis in human mesangial cells by miR-23b-3p/WT1 axis. Diabetol Metab Syndr. 2020;12:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yu FR, Xia YW, Wang SB, et al. Long noncoding RNA PVT1 facilitates high glucose-induced cardiomyocyte death through the miR-23a-3p/CASP10 axis. Cell Biol Int. 2021. Jan;45(1):154–163. [DOI] [PubMed] [Google Scholar]
- [21].Yu D, Yang X, Zhu Y, et al. Knockdown of plasmacytoma variant translocation 1 (PVT1) inhibits high glucose-induced proliferation and renal fibrosis in HRMCs by regulating miR-23b-3p/early growth response factor 1 (EGR1). Endocr J. 2021. Jan 7;68:519–529. [DOI] [PubMed] [Google Scholar]
- [22].Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011. Aug 5;146(3):353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Xu J, Deng Y, Wang Y, et al. SPAG5-AS1 inhibited autophagy and aggravated apoptosis of podocytes via SPAG5/AKT/mTOR pathway. Cell Prolif. 2020. Feb;53(2):e12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jiang X, Ru Q, Li P, et al. LncRNA SNHG16 induces proliferation and fibrogenesis via modulating miR-141-3p and CCND1 in diabetic nephropathy. Gene Ther. 2020. Jun 5;27(12):557–566. [DOI] [PubMed] [Google Scholar]
- [25].Luo R, Jin H, Li L, et al. Long noncoding RNA meg3 inhibits apoptosis of retinal pigment epithelium cells induced by high glucose via the miR-93/Nrf2 axis. Am J Pathol. 2020. Sep;190(9):1813–1822. [DOI] [PubMed] [Google Scholar]
- [26].Qi X, Zhang DH, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015. Oct;52(10):710–718. [DOI] [PubMed] [Google Scholar]
- [27].Smillie CL, Sirey T, Ponting CP. Complexities of post-transcriptional regulation and the modeling of ceRNA crosstalk. Crit Rev Biochem Mol Biol. 2018. Jun;53(3):231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tang J, Kern TS. Inflammation in diabetic retinopathy. Prog Retin Eye Res. 2011. Sep;30(5):343–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tomić M, Ljubić S, Kastelan S. The role of inflammation and endothelial dysfunction in the pathogenesis of diabetic retinopathy. Coll Antropol. 2013. Apr;37(Suppl 1):51–57. [PubMed] [Google Scholar]
- [30].Zobel DP, Andreasen CH, Burgdorf KS, et al. Variation in the gene encoding Krüppel-like factor 7 influences body fat: studies of 14 818 Danes. Eur J Endocrinol. 2009. Apr;160(4):603–609. [DOI] [PubMed] [Google Scholar]
- [31].Kanazawa A, Kawamura Y, Sekine A, et al. Single nucleotide polymorphisms in the gene encoding Krüppel-like factor 7 are associated with type 2 diabetes. Diabetologia. 2005. Jul;48(7):1315–1322. [DOI] [PubMed] [Google Scholar]
- [32].Wang B, Ji G, Naeem H, et al. The use of targeted next generation sequencing to explore candidate regulators of TGF-β1ʹs impact on kidney cells. Front Physiol. 2018;9:1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kawamura Y, Tanaka Y, Kawamori R, et al. Overexpression of Kruppel-like factor 7 regulates adipocytokine gene expressions in human adipocytes and inhibits glucose-induced insulin secretion in pancreatic beta-cell line. Mol Endocrinol. 2006. Apr;20(4):844–856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data generated in our study are available from the corresponding author on reasonable request.


