Abstract
Background and purpose
We investigated plasma neurofilament light chain concentration (pNfL) as a biomarker for neuroaxonal damage and disease activity using data from Phase 3 trials of ozanimod in relapsing multiple sclerosis (RMS).
Methods
pNfL was measured before and after ozanimod 0.46 mg or 0.92 mg daily or interferon β‐1a 30 µg weekly in the randomized, double‐blind SUNBEAM and RADIANCE trials. In these post hoc analyses, we investigated relationships between pNfL (at baseline and median percentage change from baseline to Month 12 [SUNBEAM] or 24 [RADIANCE]) and clinical and magnetic resonance imaging outcomes.
Results
Median (Q1, Q3) baseline pNfL, available in 1244 of 1346 SUNBEAM participants, was 14.70 (10.16, 23.26) pg/ml and in 1109 of 1313 RADIANCE participants was 13.35 (9.42, 20.41) pg/ml. Baseline gadolinium‐enhancing (GdE) and T2 lesion counts increased and brain volume decreased with increasing baseline pNfL. Baseline pNfL was higher in those with versus without on‐treatment relapse. Median percentage reduction in pNfL at 12 months in SUNBEAM (n = 1238) and 24 months in RADIANCE (n = 1088) was greater for ozanimod (20%–27%) than interferon β‐1a (13%–16%; p < 0.01). Greater pNfL reduction was associated with fewer GdE lesions, fewer new/enlarging T2 lesions per scan, less loss of brain volume, lower annualized relapse rate (ARR), and no evidence of disease activity. The following models predicted ARR: 0.5111 + 0.0116 × ΔNfL at 12 months (SUNBEAM) and 0.4079 + 0.0088 × ΔNfL at 24 months (RADIANCE).
Conclusions
pNfL was associated with clinical and radiologic measures of disease and treatment effects in RMS, supporting its use as a biomarker.
Keywords: blood biomarkers, multiple sclerosis, neurofilament light, relapse, treatment outcome
Higher plasma neurofilament light chain concentration (pNfL) at baseline is related to higher baseline T2 and gadolinium‐enhancing magnetic resonance imaging (MRI) lesion counts and smaller brain volume, and is predictive of on‐treatment relapse in patients with relapsing multiple sclerosis. Ozanimod causes greater dose‐dependent reductions in pNfL from baseline compared with interferon β‐1a, and these reductions relate to annualized relapse rate (ARR), number of MRI brain lesions, and rate of brain volume loss. We developed a model by which change in pNfL can be used to predict ARR.

INTRODUCTION
Neurofilament light chain is a structural component of the neuron and axon cytoskeleton. Neurofilament light chain is released into the cerebrospinal fluid (CSF) and bloodstream after neuronal injury and degeneration in various neurodegenerative disorders, including multiple sclerosis (MS), amyotrophic lateral sclerosis, Alzheimer disease, Guillain–Barré syndrome, and Huntington disease [1, 2, 3, 4, 5, 6, 7].
Blood (serum and plasma) and CSF neurofilament light chain concentrations (NfLs) correlate with each other [1, 8, 9] and with MS disease activity, including relapse rate, disability worsening, magnetic resonance imaging (MRI) activity, and brain volume loss [1, 2, 10, 11, 12, 13, 14, 15, 16]. Several studies demonstrated that disease‐modifying therapies (DMTs) reduce NfL in CSF and blood [9, 11, 13, 17, 18, 19, 20], and some reported correlations between reductions in NfL during DMT use and fewer new/enlarging T2 lesions and/or gadolinium‐enhancing (GdE) lesions on MRI [11, 12, 19, 20]. Based on these findings, NfL was proposed as a biomarker for neurologic damage and disease activity in relapsing MS (RMS) [1, 2, 8, 11, 14]. Here, NfL was evaluated as a biomarker in RMS using data from two Phase 3 trials of ozanimod, an oral sphingosine 1‐phosphate receptor 1 and 5 modulator that reduces lymphocyte migration into the central nervous system [21] and has proven efficacy in MS [22, 23]. Post hoc exploratory analyses evaluated the effect of ozanimod versus interferon (IFN) β‐1a on plasma NfL (pNfL) in patients with RMS, as well as the relationships between pNfL (baseline and median percentage change during treatment), and baseline and on‐treatment clinical and radiologic outcomes.
METHODS
Phase 3 studies
As previously reported, SUNBEAM (clinicaltrials.gov identifier: NCT02294058) and RADIANCE (clinicaltrials.gov identifier: NCT02047734) were multicenter, randomized, double‐blind, double‐dummy, active‐controlled, Phase 3 trials of ozanimod 0.92 and 0.46 mg (equivalent to ozanimod HCl 1 and 0.5 mg, respectively) compared with intramuscular IFN β‐1a 30 µg weekly in patients with RMS [22, 23]. The primary efficacy endpoint in the Phase 3 trials was annualized relapse rate (ARR).
Brain MRI scans were performed at baseline, Month 6, and Month 12 in SUNBEAM, and at baseline, Month 12, and Month 24 in RADIANCE [22, 23]. An independent MRI analysis center (NeuroRx, Montreal, Quebec, Canada) with no knowledge of treatment assignment or outcomes assessed and scored all MRI scans [22, 23]. Whole brain volume and cortical gray matter volume at baseline were measured using SienaX, and thalamic volume was measured using ThalamicVolume software [22, 23]. Percentage change in whole brain volume was established using SIENA in Phase 3 RADIANCE and Jacobian atrophy software using longitudinal Jacobian integration for whole brain volume in SUNBEAM [22, 23]. Percentage change in cortical gray matter and thalamic volumes was calculated with Jacobian atrophy software using longitudinal Jacobian integration in both trials [22, 23].
Ethical considerations
The institutional review board or ethics committee at each site approved the protocol and informed consent (Table S1). All participants provided written informed consent, and the trials conformed with the World Medical Association Declaration of Helsinki. Funding for the trials was provided by Celgene International II.
Study procedures and outcomes
In this exploratory, post hoc analysis, pNfL was measured in heparinized plasma samples obtained at baseline and Month 12 in both trials, and at Month 24 in RADIANCE. Specimens were sent to Quanterix Corporation (Lexington, MA) for analysis. The Simoa NF‐light Advantage Kit, which is a two‐step, digital, immunoassay, quantified total pNfL using the Simoa HD‐1 Analyzer and Single Molecule Array (Simoa) technology [24]. The precision and sensitivity of the Simoa immunoassay at the subfemtomolar level in serum samples have been established previously [25]. The lower limit of detection was 0.152 pg/ml, the lower limit of quantification was 0.696 pg/ml, and the average coefficient of variation was 3.4% in SUNBEAM and 4.0% in RADIANCE.
Relationships between baseline pNfL and number of T2 and GdE brain lesions at baseline; whole brain, cortical gray matter, and thalamic volumes at baseline; and number of relapses during ozanimod treatment were evaluated. In addition, the probability of having one or more relapses during treatment with ozanimod versus IFN β‐1a was assessed according to baseline pNfL. The median pNfL value and median percentage change in pNfL from baseline to Month 12 (SUNBEAM) and Month 24 (RADIANCE) were determined and analyzed by treatment group. Relationships between ARR and number of GdE lesions over the study period, and between ARR and new/enlarging T2 lesions per scan were evaluated. In addition, how the median percentage change in pNfL from baseline related to ARR, number of GdE lesions, number of new/enlarging T2 lesions, changes in brain volume (whole brain, cortical gray matter, and thalamic volume), and no evidence of disease activity (NEDA‐3) at Month 12 in SUNBEAM and Month 24 in RADIANCE were assessed. NEDA‐3 was defined as no relapses, no Expanded Disability Status Scale progression, no new/enlarging T2 lesions, and no GdE lesions. Finally, models to predict the number of relapses over a 12‐month (SUNBEAM) or 24‐month (RADIANCE) period based on median percentage change in pNfL from baseline were developed. Medians were used, rather than arithmetic or geometric means, due to the presence of outliers and the skewness of the data.
Statistics
This exploratory, post hoc analysis was hypothesis‐generating. All fitted models were descriptive and exploratory, and there was no adjustment for multiplicity of testing. The relationships between baseline T2 and GdE lesion counts and log(baseline pNfL) were explored with Poisson generalized linear models, with log(baseline pNfL) as the predictor. The relationship between baseline brain volume (whole brain, cortical gray matter, and thalamic volume) and log(baseline pNfL) was explored in each study via robust linear regression analysis using MM estimation with bisquare weight functions and 85% Gaussian efficiency [26] with log(baseline pNfL) as the predictor. The relationship between risk of one or more on‐treatment relapses and log(baseline pNfL) was explored via logistic regression model and contained terms for log(baseline pNfL) and treatment group. Robust linear models were used to investigate between‐treatment differences in postbaseline (Month 12, SUNBEAM; Month 24, RADIANCE) pNfL. Baseline and postbaseline pNfLs were log‐transformed, and MM estimation with bisquare weight functions and 85% Gaussian efficiency was used [26].
The number of T2 lesions per scan was estimated by negative binomial model adjusted for treatment group, with an offset for number of scans. Median T2 lesions per scan in this analysis differ from the median values in the primary publications, because this analysis did not adjust for baseline GdE count, as it is related to pNfL.
To examine the relationships at the study level, between median percentage change in pNfL from baseline and clinical and radiologic outcomes, we arranged the data such that relevant measures were in a single row for each patient, used stratified bootstrap sampling to resample rows by treatment group (thus retaining the relationships between columns), computed summary statistics, and then repeated this algorithm 1000 times. The bootstrap procedure allowed us to estimate the treatment effects on pNfL, ARR, MRI outcomes, and NEDA‐3 as if the studies had been run many independent times, and, therefore, allowed us to examine the relationships between the outcomes in each treatment group across hypothetical repetitions of the studies.
A simple least squares regression was used to predict ARR as the response and median percentage change from baseline pNfL (ΔNfL) as the explanatory variable. The model coefficients can be used to predict the number of relapses seen in a future study given a known median reduction in pNfL, and the residual standard error can be used to produce a prediction interval, within which a future observation will fall for such an estimate.
For each relationship, participants who were missing data for one or both of the relevant outcomes were excluded, such that only those with paired data were analyzed. For example, if a participant had data available for GdE lesions and relapse, but not NfL, that participant was included in analyses of the relationship between GdE and relapse, but not in analyses of the relationship between NfL and either GdE lesions or relapse.
R version 4.0.2 (2020‐06‐22; R Core Team 2020) was used for all analyses. The Tidyverse suite of packages [27] was used for various aspects of data restructuring and production of graphs. The mgcv package [28] was used for negative binomial modeling [29].
RESULTS
Disposition and baseline demographics
pNfL was available at baseline for 1244 of 1346 (92.4%) SUNBEAM study [22] participants and for 1109 of 1313 (84.5%) RADIANCE study [23] participants. A total of 1238 (92.0%) and 1088 (82.9%) participants had paired pNfL at baseline and Month 12 (SUNBEAM) or Month 24 (RADIANCE), respectively.
Demographics and baseline disease characteristics, including categorical distribution of T2 and GdE lesion counts, were similar for participants with pNfL at baseline in both studies (Table 1). Most participants were white women with a mean age of approximately 36 years.
TABLE 1.
Baseline demographics and disease characteristics among participants in SUNBEAM and RADIANCE with baseline plasma neurofilament light chain assessments
| Characteristic | SUNBEAM, N = 1244 | RADIANCE, N = 1109 |
|---|---|---|
| Age, years, mean (SD) | 35.7 (9.3) | 35.6 (8.9) |
| Sex, % | ||
| Male | 34 | 33 |
| Female | 66 | 67 |
| Race, % | ||
| White | 99.6 | 99.3 |
| Other | 0.4 | 0.7 |
| Number of GdE lesions, % a | ||
| 0 | 54 | 58 |
| 1 | 18 | 15 |
| 2 | 9 | 7 |
| 3–5 | 10 | 12 |
| >5 | 9 | 9 |
| Number of T2 lesions, % a | ||
| 0–10 | 7 | 6 |
| 11–20 | 11 | 13 |
| 21–50 | 38 | 44 |
| 51–75 | 19 | 19 |
| >75 | 24 | 18 |
Abbreviations: GdE, gadolinium‐enhancing; SD, standard deviation.
Percentages may not add up to 100% due to rounding.
pNfL at baseline and after treatment
Baseline pNfL was similar in both studies: median (Q1, Q3) baseline pNfL was 14.70 (10.16, 23.26) pg/ml in SUNBEAM and 13.35 (9.42, 20.41) pg/ml in RADIANCE. At Month 12 in SUNBEAM, the median percentage change in pNfL was −13.4% with IFN β‐1a, −22.8% with ozanimod 0.46 mg (p = 0.0003 vs IFN β‐1a, derived based on approximate asymptotic z‐tests), and −26.9% with ozanimod 0.92 mg (p < 0.0001 vs IFN β‐1a; Table S2). Similar results were obtained at Month 24 in the RADIANCE trial, with median pNfL reductions of −15.5%, −19.7% (p = 0.0024 vs IFN β‐1a), and −23.5% (p = 0.0001 vs IFN β‐1a), respectively (Table S2).
Relationships between pNfL and MRI metrics
Baseline counts of GdE lesions were higher with higher baseline pNfL (p < 0.0001; Figure 1A, Table 2). Treatment groups with fewer GdE lesions over the study period had a greater median percentage reduction in pNfL (Figure 1B).
FIGURE 1.
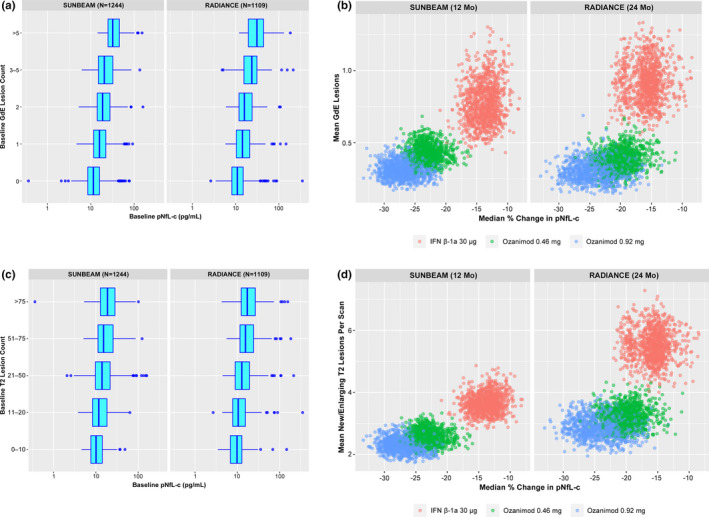
Relationships between brain lesions and plasma neurofilament light chain concentration (pNfL). (a) Baseline pNfL and gadolinium‐enhancing (GdE) lesion counts at baseline. (b) On‐treatment pNfL reduction and adjusted mean numbers of GdE lesions over the study period (SUNBEAM, 12 months; RADIANCE, 24 months). (c) Baseline pNfL and T2 lesion counts at baseline. (d) On‐treatment pNfL reduction and adjusted mean numbers of new/enlarging T2 lesions per scan. (a, c) The relationship between baseline GdE and T2 brain lesion counts and baseline pNfL was explored via Poisson generalized linear models, with log(baseline pNfL) as the predictor. (b, d) Adjusted mean numbers of GdE lesions and new/enlarging T2 lesions per scan were estimated from a negative binomial regression model adjusted for treatment group, with an offset for number of scans. Relationship between median percentage change from baseline in pNfL and lesion counts was based on bootstrap sampling. Individual dots on each plot represent the individual simulations from the bootstrap procedure. IFN, interferon
TABLE 2.
Relationship between imaging parameters and baseline plasma neurofilament light chain concentrations a
| SUNBEAM | RADIANCE | |||||
|---|---|---|---|---|---|---|
| Intercept (SE) | Slope (SE) | p | Intercept (SE) | Slope (SE) | p | |
| Brain lesion counts at baseline | ||||||
| GdE lesions b | −3.37 (0.10) | 1.28 (0.03) | <0.0001 | −2.62 (0.09) | 1.06 (0.03) | <0.0001 |
| T2 lesions c | 3.22 (0.02) | 0.27 (0.01) | <0.0001 | 3.17 (0.02) | 0.26 (0.01) | <0.0001 |
| Brain volume at baseline | ||||||
| Whole brain volume | 1491.01 (10.58) | −14.63 (3.74) | <0.0001 | 1475.77 (10.89) | −9.70 (3.95) | 0.0141 |
| Cortical gray matter volume | 550.12 (5.89) | −8.81 (2.09) | <0.0001 | 547.00 (5.72) | −5.94 (2.08) | 0.0042 |
| Thalamic volume | 17.23 (0.27) | −0.68 (0.09) | <0.0001 | 17.04 (0.26) | −0.50 (0.09) | <0.0001 |
Abbreviations: GdE, gadolinium‐enhancing; SE, standard error.
Based on log(baseline plasma neurofilament light chain concentration).
Indicative of ongoing disease activity.
Indicative of cumulative disease (magnetic resonance imaging lesion) burden.
Baseline counts of T2 lesions were higher with higher baseline pNfL in both studies (p < 0.0001; Figure 1C, Table 2). Groups with fewer new/enlarging T2 lesions per scan also had a greater median percentage reduction in pNfL (Figure 1D).
Whole brain volume, cortical gray matter volume, and thalamic volume at baseline were smaller in participants who had higher baseline pNfL (p < 0.05 for all measures; Table 2; Figure S1A–C). During treatment, participants with a greater median percentage reduction from baseline in pNfL exhibited less loss of whole brain volume, cortical gray matter volume, and thalamic volume (Figure S1D–F). Greater differences between IFN β‐1a and ozanimod were observed in cortical gray matter volume and thalamic volume compared with whole brain volume.
Relationship between MRI lesions and ARR
Treatment groups with lower ARR had fewer GdE lesions at Months 12 and 24 (Figure S2A). Groups with lower ARR also had fewer new/enlarging T2 lesions per scan over 12 and 24 months (Figure S2B).
Relationship between pNfL and relapse
A trend was observed suggesting that baseline pNfL was higher in those who relapsed during treatment compared with those who did not relapse (Figure 2A). Treatment groups with a greater median percentage reduction from baseline in pNfL during treatment had lower adjusted ARR (Figure 2B).
FIGURE 2.
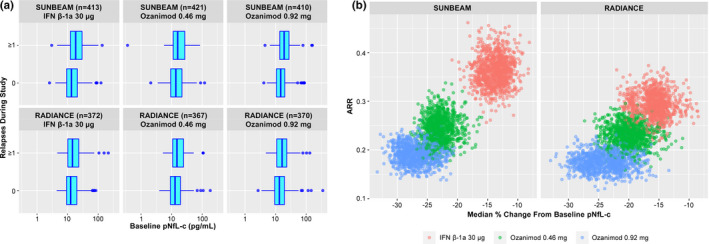
Relationships between relapse and (a) baseline plasma neurofilament light chain concentration (pNfL) and (b) change in pNfL on‐treatment. (a) The relationship between risk of one or more relapses during treatment and log(baseline pNfL) was explored via logistic regression model. (b) Relationship between adjusted annualized relapse rate (ARR) over the treatment period (SUNBEAM, 12 months; RADIANCE, 24 months) and median percentage change from baseline in pNfL based on bootstrap sampling. ARR was based on a Poisson generalized linear regression model as a function of treatment group, with an offset for duration. Individual dots on each plot represent the individual simulations from the bootstrap procedure. IFN, interferon
Relationship between pNfL and NEDA‐3
NEDA‐3 rates at Month 12 in SUNBEAM were 22.5% with IFN β‐1a, 25.9% with ozanimod 0.46 mg, and 26.8% with ozanimod 0.92 mg. Corresponding NEDA‐3 rates at Month 24 in RADIANCE were 12.9%, 17.3%, and 18.2%. In both studies, NEDA‐3 was associated with larger reductions in pNfL during treatment (Figure S3).
Predicting relapse based on change in pNfL
Probability of having one or more relapses in the next 12 months (SUNBEAM) or 24 months (RADIANCE) increased with increasing baseline pNfL and was numerically lower with ozanimod 0.92 mg than with IFN β‐1a (Table 3).
TABLE 3.
Probability of one or more relapses in the next 12 months (SUNBEAM) or 24 months (RADIANCE) based on baseline pNfL
| Baseline pNfL, pg/ml | SUNBEAM Probability (95% CI) of ≥1 relapse in next 12 months | RADIANCE probability (95% CI) of ≥1 relapse in next 24 months | ||||
|---|---|---|---|---|---|---|
| IFN β‐1a | Ozanimod 0.46 mg | Ozanimod 0.92 mg | IFN β‐1a | Ozanimod 0.46 mg | Ozanimod 0.92 mg | |
| 1 | 0.07 (0.04–0.13) | 0.05 (0.03–0.09) | 0.04 (0.02–0.07) | 0.16 (0.10–0.26) | 0.13 (0.07–0.21) | 0.11 (0.06–0.18) |
| 5 | 0.17 (0.13–0.22) | 0.11 (0.08–0.15) | 0.10 (0.07–0.14) | 0.26 (0.20–0.32) | 0.20 (0.15–0.26) | 0.17 (0.13–0.23) |
| 10 | 0.24 (0.19–0.28) | 0.16 (0.13–0.20) | 0.14 (0.11–0.18) | 0.31 (0.26–0.36) | 0.24 (0.20–0.29) | 0.21 (0.17–0.26) |
| 20 | 0.32 (0.27–0.37) | 0.23 (0.19–0.27) | 0.20 (0.16–0.25) | 0.36 (0.31–0.41) | 0.29 (0.24–0.34) | 0.25 (0.21–0.30) |
| 50 | 0.45 (0.37–0.52) | 0.34 (0.27–0.42) | 0.30 (0.24–0.38) | 0.44 (0.36–0.52) | 0.36 (0.29–0.44) | 0.32 (0.25–0.39) |
| 100 | 0.55 (0.45–0.65) | 0.44 (0.33–0.55) | 0.40 (0.30–0.51) | 0.50 (0.39–0.61) | 0.42 (0.32–0.53) | 0.37 (0.27–0.48) |
Abbreviations: CI, confidence interval; IFN, interferon; pNfL, plasma neurofilament light chain concentration.
The following model was developed to predict relapse based on change in pNfL using 95% prediction intervals:
SUNBEAM: ARR = 0.5111 + 0.0116 × ΔNfL at 12 months; residual standard error = 0.035
RADIANCE: ARR = 0.4079 + 0.0088 × ΔNfL at 24 months; residual standard error = 0.037
Predictive modeling showed that the median percentage change from baseline pNfL appears to be approximately linearly related to ARR (Figure 3). The modeling estimated that a 25% reduction in pNfL (similar to that observed with ozanimod 0.92 mg in SUNBEAM and RADIANCE) predicts an ARR (standard error [SE]) of 0.22 (0.04) and 0.19 (0.04) based on SUNBEAM and RADIANCE, respectively. A 13% pNfL reduction (similar to that observed with IFN β‐1a in SUNBEAM and RADIANCE) predicts an ARR (SE) of 0.36 (0.04) and 0.29 (0.04), respectively.
FIGURE 3.
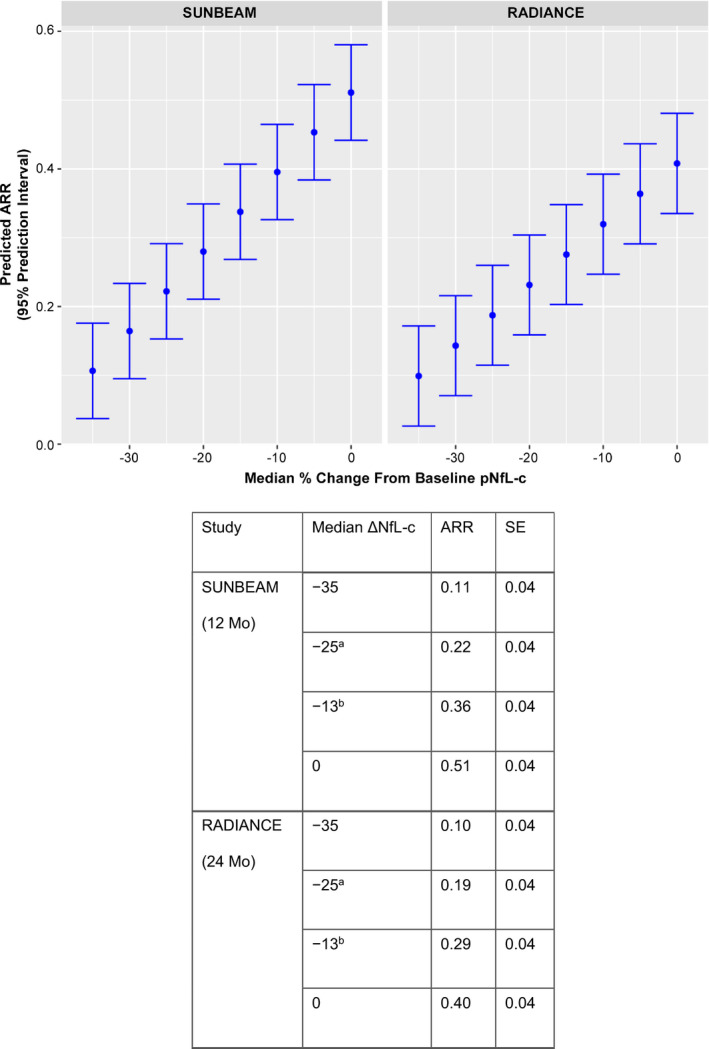
Prediction of relapse based on plasma neurofilament light chain concentration (pNfL). Prediction of annualized relapse rate (ARR) is based on median percentage change from baseline pNfL. The model is based on a least squares regression model with ARR as the response and median percentage change from baseline NfL as the explanatory variable. A 25% reduction in plasma NfL was similar to that observed in participants treated with ozanimod 0.92 mg in SUNBEAM and RADIANCE. A 13% reduction in plasma NfL was similar to that observed in participants treated with interferon β‐1a in SUNBEAM and RADIANCE. aA 25% reduction in plasma NfL was similar to that observed in participants treated with ozanimod 0.92 mg in SUNBEAM and RADIANCE. bA 13% reduction in plasma NfL was similar to that observed in participants treated with IFN β‐1a in SUNBEAM and RADIANCE.
DISCUSSION
This exploratory, post hoc analysis of the Phase 3 SUNBEAM and RADIANCE trials of ozanimod supports further evaluation of pNfL as a biomarker to monitor and predict disease activity and neurologic damage in patients with RMS and is the first publication describing the impact of ozanimod on pNfL. In SUNBEAM and RADIANCE participants, pNfL was reduced by treatment with ozanimod to a greater extent than IFN β‐1a. At the study level, relationships were found between treatment‐related reduction in pNfL and reductions in clinical relapse and radiologic disease activity (GdE and new/enlarging T2 lesion counts). Furthermore, median change in pNfL was related to magnitude of brain volume loss and NEDA‐3 across the study population.
These results add to literature providing support for pNfL as a biomarker for RMS disease activity and treatment response. Reductions of NfL in CSF and blood were observed following treatment with IFN β‐1a [11, 12], natalizumab [17, 18, 30, 31], rituximab [32, 33], cladribine [34], dimethyl fumarate [35], alemtuzumab [36], and fingolimod [9, 13, 19, 20, 37]. These reductions in NfL corresponded to reductions in MS activity. For example, pNfL was measured over a 1‐year or 2‐year period in 589 patients with relapsing–remitting MS enrolled in the Phase 3, double‐blind, randomized, controlled fingolimod studies (TRANSFORMS and FREEDOMS) [13]. High baseline pNfL was associated with presence of GdE lesions at baseline (p < 0.0001) and predicted an increased number of new/enlarging T2 lesions (p = 0.0006), higher ARR (p < 0.0001), and greater rate of brain volume loss (p < 0.0001) over 24 months in FREEDOMS [13]. Patients with new/enlarging T2 lesions during the studies had higher pNfL at the end of treatment. In addition, fingolimod reduced pNfL to a greater extent than IFN β‐1a [13]; thus, both fingolimod and ozanimod produce greater reductions in pNfL than IFN β‐1a.
Other studies also suggest differential effects of DMTs on pNfL. Two large studies found that patients receiving higher efficacy DMTs (e.g., natalizumab, ocrelizumab, alemtuzumab, fingolimod, dimethyl fumarate, mitoxantrone) had a larger relative decrease in NfL compared with no DMT, IFNs, or glatiramer acetate [14, 38]. In addition, a study of 1139 patients with RMS who were initiating newer DMTs and had pNfL over two time points found the largest NfL reduction in the alemtuzumab‐treated group and the least reduction in the teriflunomide treated‐group, consistent with differences in clinical efficacy [39]. Thus, NfL in serum/plasma or CSF is a therapeutic response biomarker in RMS that may be related to anti‐inflammatory activity, and a consequent prevention of neurologic damage, by DMTs.
Currently, due to lack of an accepted biomarker, serial MRI scans are standard of care to assess MS disease activity and treatment response despite their high cost [15, 40]. Compared with MRI, pNfL has some advantages for monitoring for subclinical disease activity, including lower costs, accessibility, and a single assessment that includes both spinal cord and brain pathology [16]. CSF and serum NfL (sNfL) were previously found to have similar long‐term predictive value as MRI measures, suggesting complementary use [12]. In a study of 94 MS patients, sNfL was significantly higher (estimated 35% increase) within 90 days of a GdE‐positive lesion [41]. In addition, significantly elevated sNfL was observed within 3 months of clinical relapse only when associated with a GdE lesion. The present study found similar relationships of pNfL with clinical and radiologic disease activity, although relationships with MRI endpoints were assessed only at the study level.
A recent analysis of Phase 2 and 3 fingolimod studies and their extensions concluded that longitudinal measurements of NfL over 12 or 24 months add prognostic value for 10‐year disability outcomes when used in combination with both clinical measures and conventional MRI [42]. The frequency of sampling sufficient for prediction of relapsing activity has not been established but may need to be more frequent than annually [38].
This analysis has a number of strengths. The two prospective Phase 3 clinical trials included large cohorts of patients treated for 1 and 2 years, an active comparator, and clinical relapse as the primary outcome. In addition, the highly sensitive Simoa assay was used to quantify pNfL [43]. Use of a robust regression and median versus means was intended to reduce the undue influence of outliers in the data. One key limitation is the exploratory, post hoc nature of this analysis. Although a bootstrap procedure was used to estimate the study‐level relationship between the changes from baseline in pNfL and on‐treatment outcomes, two studies are too few to estimate between‐study variances in this relationship. Also, the two studies were of different durations and included only three treatment groups and two drugs. The effect of confounding factors known to affect NfL in plasma or serum (e.g., neurologic comorbidities [4, 5, 6, 7], age [44]) was not determined, and may have limited detection of the full range of associations and predictive values.
CONCLUSIONS
These findings support the potential use of pNfL to characterize RMS at a specific time point and to serve as a biomarker to monitor and predict disease activity and treatment response. Ozanimod caused dose‐dependent reductions in pNfL from baseline compared with IFN β‐1a, and these reductions were related to ARR, number of MRI brain lesions, and rate of brain volume loss at the study level. These findings, coupled with the primary analyses from the ozanimod Phase 3 trials, support the use of pNfL as a biomarker in RMS.
CONFLICT OF INTEREST
S.H. is an employee of Bristol Myers Squibb. G.C. reports compensation for consulting and/or speaking activities from Almirall, Biogen, Celgene, EXCEMED, Forward Pharma, Genzyme, Merck, Novartis, Roche, Sanofi, and Teva. B.A.C.C. reports personal compensation for consulting for Akili, Alexion, Autobahn, EMD Serono, Novartis, Sanofi, TG Therapeutics, and Therini, and has received grant support from Genentech. D.L.A. reports personal fees for consulting and/or grants from Albert Charitable Trust, Biogen, Celgene, F. Hoffmann‐La Roche, Frequency Therapeutics, MedDay, Merck Serono, Novartis, and Sanofi‐Aventis, and an equity interest in NeuroRx Research. L.S. reports consulting for AbbVie, Atreca, Celgene, Novartis, Teva, Tolerion, and EMD Serono, and research support from Atara, Biogen, and Celgene. J.K.S. is an employee of Bristol Myers Squibb. H.S. has received compensation from Aptus Clinical, BresMed Health Solutions, Bristol Myers Squibb, Celgene, CytomX, F2G, GlaxoSmithKline, and Gossamer Bio. L.K.'s institution (University Hospital Basel) has received in the past 3 years the following, which was used exclusively for research support: steering committee, advisory board, and consultancy fees, and support of educational activities from Actelion, Allergan, Almirall, Baxalta, Bayer, Biogen, Celgene, CSL Behring, Desitin, EXCEMED, Eisai, Genzyme, Japan Tobacco, Merck, Minoryx, Novartis, Pfizer, F. Hoffmann‐La Roche, Sanofi Aventis, Santhera, and Teva, and license fees for Neurostatus‐UHB products; and the research of the MS Center in Basel has been supported by grants from Bayer, Biogen, the European Union, Innosuisse, Novartis, Roche Research Foundations, the Swiss MS Society, and the Swiss National Research Foundation. J.A.C. reports personal compensation for consulting for Adamas, Atara, Bristol Myers Squibb, Convelo, MedDay, and Mylan, and for serving as an editor of Multiple Sclerosis Journal.
AUTHOR CONTRIBUTIONS
Sarah Harris: Data curation (equal), writing–original draft (equal), writing–review & editing (equal). Giancarlo Comi: Conceptualization (equal), data curation (equal), supervision (equal), writing–original draft (equal), writing–review & editing (equal). Bruce A. C. Cree: Conceptualization (equal), data curation (equal), supervision (equal), writing–original draft (equal), writing–review & editing (equal). Douglas L. Arnold: Conceptualization (equal), data curation (equal), supervision (equal), writing–original draft (equal), writing–review & editing (equal). Lawrence Steinman: Conceptualization (equal), data curation (equal), supervision (equal), writing–original draft (equal), writing–review & editing (equal). James K. Sheffield: Data curation (equal), writing–original draft (equal), writing–review & editing (equal). Harry Southworth: Data curation (equal), formal analysis (equal), validation (equal), writing–original draft (equal), writing–review & editing (equal). Ludwig Kappos: Conceptualization (equal), data curation (equal), supervision (equal), writing–original draft (equal), writing–review & editing (equal). Jeffrey A. Cohen: Conceptualization (equal), data curation (equal), supervision (equal), writing–original draft (equal), writing–review & editing (equal).
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
Support for third‐party writing assistance for this article was provided by Peloton Advantage, an OPEN Health company, and was funded by Bristol Myers Squibb.
Harris S, Comi G, Cree BAC, et al; on behalf of the Ozanimod Study Investigators . Plasma neurofilament light chain concentrations as a biomarker of clinical and radiologic outcomes in relapsing multiple sclerosis: Post hoc analysis of Phase 3 ozanimod trials. Eur J Neurol. 2021;28:3722–3730. 10.1111/ene.15009
Funding information
The Phase 3 SUNBEAM and RADIANCE studies were sponsored by Celgene International II.
DATA AVAILABILITY STATEMENT
Bristol Myers Squibb policy on data sharing may be found at https://www.bms.com/researchers‐and‐partners/independent‐research/data‐sharing‐request‐process.html
REFERENCES
- 1. Disanto G, Barro C, Benkert P, et al. Serum neurofilament light: a biomarker of neuronal damage in multiple sclerosis. Ann Neurol. 2017;81(6):857‐870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Novakova L, Axelsson M, Khademi M, et al. Cerebrospinal fluid biomarkers as a measure of disease activity and treatment efficacy in relapsing‐remitting multiple sclerosis. J Neurochem. 2017;141(2):296‐304. [DOI] [PubMed] [Google Scholar]
- 3. Varhaug KN, Torkildsen O, Myhr KM, Vedeler CA. Neurofilament light chain as a biomarker in multiple sclerosis. Front Neurol. 2019;10:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gaiottino J, Norgren N, Dobson R, et al. Increased neurofilament light chain blood levels in neurodegenerative neurological diseases. PLoS One. 2013;8(9):e75091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu CH, Macdonald‐Wallis C, Gray E, et al. Neurofilament light chain: a prognostic biomarker in amyotrophic lateral sclerosis. Neurology. 2015;84(22):2247‐2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mattsson N, Andreasson U, Zetterberg H, Blennow K. Association of plasma neurofilament light with neurodegeneration in patients with Alzheimer disease. JAMA Neurol. 2017;74(5):557‐566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Byrne LM, Rodrigues FB, Blennow K, et al. Neurofilament light protein in blood as a potential biomarker of neurodegeneration in Huntington's disease: a retrospective cohort analysis. Lancet Neurol. 2017;16(8):601‐609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cai L, Huang J. Neurofilament light chain as a biological marker for multiple sclerosis: a meta‐analysis study. Neuropsychiatr Dis Treat. 2018;14:2241‐2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Piehl F, Kockum I, Khademi M, et al. Plasma neurofilament light chain levels in patients with MS switching from injectable therapies to fingolimod. Mult Scler. 2018;24(8):1046‐1054. [DOI] [PubMed] [Google Scholar]
- 10. Lycke JN, Karlsson JE, Andersen O, Rosengren LE. Neurofilament protein in cerebrospinal fluid: a potential marker of activity in multiple sclerosis. J Neurol Neurosurg Psychiatry. 1998;64(3):402‐404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Varhaug KN, Barro C, Bjørnevik K, et al. Neurofilament light chain predicts disease activity in relapsing‐remitting MS. Neurol Neuroimmunol Neuroinflamm. 2018;5(1):e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuhle J, Plavina T, Barro C, et al. Neurofilament light levels are associated with long‐term outcomes in multiple sclerosis. Mult Scler. 2019;26(13):1691‐1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuhle J, Kropshofer H, Haering DA, et al. Blood neurofilament light chain as a biomarker of MS disease activity and treatment response. Neurology. 2019;92(10):e1007‐e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bittner S, Steffen F, Uphaus T, et al. Clinical implications of serum neurofilament in newly diagnosed MS patients: A longitudinal multicentre cohort study. EBioMedicine. 2020;56:102807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thebault S, Abdoli M, Fereshtehnejad SM, Tessier D, Tabard‐Cossa V, Freedman MS. Serum neurofilament light chain predicts long term clinical outcomes in multiple sclerosis. Sci Rep. 2020;10(1):10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manouchehrinia A, Stridh P, Khademi M, et al. Plasma neurofilament light levels are associated with risk of disability in multiple sclerosis. Neurology. 2020;94(23):e2457‐e2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Amor S, van der Star BJ, Bosca I, et al. Neurofilament light antibodies in serum reflect response to natalizumab treatment in multiple sclerosis. Mult Scler. 2014;20(10):1355‐1362. [DOI] [PubMed] [Google Scholar]
- 18. Gunnarsson M, Malmeström C, Axelsson M, et al. Axonal damage in relapsing multiple sclerosis is markedly reduced by natalizumab. Ann Neurol. 2011;69(1):83‐89. [DOI] [PubMed] [Google Scholar]
- 19. Kuhle J, Disanto G, Lorscheider J, et al. Fingolimod and CSF neurofilament light chain levels in relapsing‐remitting multiple sclerosis. Neurology. 2015;84(16):1639‐1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Novakova L, Axelsson M, Khademi M, et al. Cerebrospinal fluid biomarkers of inflammation and degeneration as measures of fingolimod efficacy in multiple sclerosis. Mult Scler. 2017;23(1):62‐71. [DOI] [PubMed] [Google Scholar]
- 21. Scott FL, Clemons B, Brooks J, et al. Ozanimod (RPC1063) is a potent sphingosine‐1‐phosphate receptor‐1 (S1P1) and receptor‐5 (S1P5) agonist with autoimmune disease‐modifying activity. Br J Pharmacol. 2016;173(11):1778‐1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Comi G, Kappos L, Selmaj KW, et al. Safety and efficacy of ozanimod versus interferon beta‐1a in relapsing multiple sclerosis (SUNBEAM): a multicentre, randomised, minimum 12‐month, phase 3 trial. Lancet Neurol. 2019;18(11):1009‐1020. [DOI] [PubMed] [Google Scholar]
- 23. Cohen JA, Comi G, Selmaj KW, et al. Safety and efficacy of ozanimod versus interferon beta‐1a in relapsing multiple sclerosis (RADIANCE): a multicentre, randomised, 24‐month, phase 3 trial. Lancet Neurol. 2019;18(11):1021‐1033. [DOI] [PubMed] [Google Scholar]
- 24.Simoa NF‐light® Advantage Kit. 2018. www.quanterix.com. Accessed February 8, 2021.
- 25. Rissin DM, Kan CW, Campbell TG, et al. Single‐molecule enzyme‐linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28(6):595‐599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maronna RA, Martin RD, Yohai VJ. Linear Regression 2. Robust Statistics: Theory and Methods. John Wiley & Sons; 2006:115‐173. [Google Scholar]
- 27.Tidyverse suite of packages. 2020. https://www.tidyverse.org/packages/. Accessed September 23, 2020.
- 28. Wood SN. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J R Statist Soc B. 2011;73(1):3‐36. [Google Scholar]
- 29. Epskamp S, Cramer AOJ, Waldorp LJ, Schmittmann VD, Borsboom D. qgraph: network visualizations of relationships in psychometric data. J Stat Softw. 2012;48(4):1‐18. [Google Scholar]
- 30. Kuhle J, Malmeström C, Axelsson M, et al. Neurofilament light and heavy subunits compared as therapeutic biomarkers in multiple sclerosis. Acta Neurol Scand. 2013;128(6):e33‐36. [DOI] [PubMed] [Google Scholar]
- 31. Mellergård J, Tisell A, Blystad I, et al. Cerebrospinal fluid levels of neurofilament and tau correlate with brain atrophy in natalizumab‐treated multiple sclerosis. Eur J Neurol. 2017;24(1):112‐121. [DOI] [PubMed] [Google Scholar]
- 32. Alvarez E, Piccio L, Mikesell RJ, et al. Predicting optimal response to B‐cell depletion with rituximab in multiple sclerosis using CXCL13 index, magnetic resonance imaging and clinical measures. Mult Scler J Exp Transl Clin. 2015. doi: 10.1001/10.1177/2055217315623800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Flon P, Gunnarsson M, Laurell K, et al. Reduced inflammation in relapsing‐remitting multiple sclerosis after therapy switch to rituximab. Neurology. 2016;87(2):141‐147. [DOI] [PubMed] [Google Scholar]
- 34. Yildiz O, Mao Z, Adams A, et al. Disease activity in progressive multiple sclerosis can be effectively reduced by cladribine. Mult Scler Relat Disord. 2018;24:20‐27. [DOI] [PubMed] [Google Scholar]
- 35. Sejbaek T, Nielsen HH, Penner N, et al. Dimethyl fumarate decreases neurofilament light chain in CSF and blood of treatment naive relapsing MS patients. J Neurol Neurosurg Psychiatry. 2019;90(12):1324‐1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hyun JW, Kim Y, Kim G, Kim SH, Kim HJ. Longitudinal analysis of serum neurofilament light chain: a potential therapeutic monitoring biomarker for multiple sclerosis. Mult Scler. 2020;26(6):559–667. [DOI] [PubMed] [Google Scholar]
- 37. Sehr T, Akgün K, Proschmann U, Bucki R, Zendzian‐Piotrowska M, Ziemssen T. Early central vs. peripheral immunological and neurobiological effects of fingolimod‐a longitudinal study. J Mol Med (Berl). 2019;97(9):1263‐1271. [DOI] [PubMed] [Google Scholar]
- 38. Cantó E, Barro C, Zhao C, et al. Association between serum neurofilament light chain levels and long‐term disease course among patients with multiple sclerosis followed up for 12 years. JAMA Neurol. 2019;76(11):1359‐1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Delcoigne B, Manouchehrinia A, Barro C, et al. Blood neurofilament light levels segregate treatment effects in multiple sclerosis. Neurology. 2020;94(11):e1201‐e1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Uher T, Schaedelin S, Srpova B, et al. Monitoring of radiologic disease activity by serum neurofilaments in MS. Neurol Neuroimmunol Neuroinflamm. 2020;7(4):e714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rosso M, Gonzalez CT, Healy BC, et al. Temporal association of sNfL and gad‐enhancing lesions in multiple sclerosis. Ann Clin Transl Neurol. 2020;7(6):945‐955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Häring DA, Kropshofer H, Kappos L, et al. Long‐term prognostic value of longitudinal measurements of blood neurofilament levels. Neurol Neuroimmunol Neuroinflamm. 2020;7(5):e856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuhle J, Barro C, Andreasson U, et al. Comparison of three analytical platforms for quantification of the neurofilament light chain in blood samples: ELISA, electrochemiluminescence immunoassay and Simoa. Clin Chem Lab Med. 2016;54(10):1655‐1661. [DOI] [PubMed] [Google Scholar]
- 44. Khalil M, Pirpamer L, Hofer E, et al. Serum neurofilament light levels in normal aging and their association with morphologic brain changes. Nat Commun. 2020;11(1):812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
Bristol Myers Squibb policy on data sharing may be found at https://www.bms.com/researchers‐and‐partners/independent‐research/data‐sharing‐request‐process.html


