Abstract
Porcine circovirus 3 (PCV‐3) was discovered in 2015 using next‐generation sequencing (NGS) methods. Since then, the virus has been detected worldwide in pigs displaying several clinical–pathological outcomes as well as in healthy animals. The objective of this review is to critically discuss the evidence existing so far regarding PCV‐3 as a swine pathogen. In fact, a significant number of publications claim PCV‐3 as a disease causal infectious agent, but very few of them have shown strong evidence of such potential causality. The most convincing proofs of disease association are those that demonstrate a clinical picture linked to multisystemic lymphoplasmacytic to lymphohistiocytic perivascular inflammation and presence of viral nucleic acid within these lesions. Based on these evidence, individual case definitions for PCV‐3‐reproductive disease and PCV‐3‐systemic disease are proposed to standardize diagnostic criteria for PCV‐3‐associated diseases. However, the real frequency of these clinical–pathological conditions linked to the novel virus is unknown, and the most frequent outcome of PCV‐3 infection is likely subclinical based on its worlwide distribution.
Keywords: case definition, disease causality, porcine circovirus 3 (PCV‐3), reproductive disease, systemic disease
1. INTRODUCTION
Porcine circovirus 3 (PCV‐3) was discovered in the United States (2015) by next‐generation sequencing (NGS) methods in swine affected by respiratory and neurological signs, cardiac and multisystemic inflammation, reproductive failure and a porcine dermatitis and nephropathy syndrome (PDNS)‐like condition (Palinski et al., 2017; Phan et al., 2016). Since then, the virus has been detected in Suidae species (domestic swine and wild boar) from many countries all over the world as well as occasionally in some non‐Suidae species (Czyżewska‐Dors et al., 2020; Franzo et al., 2019; Jiang et al., 2019; Sun et al., 2019; Wang, Sun, et al., 2019).
Following the first descriptions, PCV‐3 has been detected in pigs displaying several clinical–pathological outcomes, such as respiratory disease (Kedkovid, Woonwong, Arunorat, Sirisereewan, Sangpratum, Lumyai et al., 2018; Qi et al., 2019; Shen et al., 2018; Zhai et al., 2017), digestive disorders (Qi et al., 2019; Zhai et al., 2017), congenital tremors (Chen et al., 2017), rectal prolapse (Phan et al., 2016), reproductive problems (Arruda et al., 2019; Deim et al., 2019; Faccini et al., 2017) and multisystemic inflammation (Arruda et al., 2019; Phan et al., 2016). Additionally, this novel virus has been detected in healthy animals of different ages and countries (Franzo, Legnardi, et al., 2018; Klaumann et al., 2019; Klaumann, Correa‐Fiz, et al., 2018; Klaumann, Franzo, et al., 2018; Saporiti, Huerta, et al., 2020; Stadejek et al., 2017; Ye et al., 2018; Zhai et al., 2017).
After those initial and further studies on PCV‐3 detection in pigs with different pathological outcomes (Klaumann et al., 2018), it was rapidly accepted as a potential swine pathogen worldwide by the veterinary community. This is in sharp contrast with the history of another porcine circovirus, PCV‐2, whose causal association with a deadly condition (the so‐called post‐weaning multisystemic wasting syndrome, PMWS) was debated for a long time (Segalés et al., 2013). In fact, only the advent of PCV‐2 vaccines almost a decade after PMWS description served to close the debate; vaccination was extremely efficient in counteracting PMWS, afterwards called PCV‐2‐systemic disease (Segalés, 2015). It is tempting to speculate that the previous experience with PCV‐2 as a pathogen favoured the acceptance of PCV‐3 as a such.
There are several parallelisms between PCV‐2 and PCV‐3, since both are very similar from a molecular organization point of view (Franzo et al., 2018) and they have been detected retrospectively many years before their first identification/report in potential association with disease (Jacobsen et al., 2009; Rodrigues et al., 2020). In turn, significant differences also apply, since PCV‐2 evolves much more rapidly than PCV‐3 (Franzo, Segalés, et al., 2018), a remarkably higher genomic variability has been detected in PCV‐2 compared to PCV‐3 (Franzo et al., 2020) and PCV‐2 was discovered in the context of a new disease with epidemic proportions worldwide, while that has not been the case with PCV‐3 (Opriessnig et al., 2020). Morover, with regards the amino‐acid (aa) similarity, PCV‐3 is far distant from PCV‐2, with an identity of Cap and Rep proteins around 26%–37% and 48%, respectively (Palinski et al., 2017; Phan et al., 2016).
Therefore, the objective of this review is to critically discuss the evidence existing so far regarding PCV‐3 as a swine pathogen and to propose a disease case definition for these conditions that show a rather strong causal relationship.
2. DISEASE ASSOCIATION
2.1. Naturally occurring PCV‐3 infection
Presence of PCV‐3 genome in a sick animal does not entail that this virus is the cause of the clinical signs or lesions. Moreover, most of the studies on PCV‐3 detection lack of proper negative controls (age‐matched, healthy pigs) to compare with, further complicating the interpretation of the causal association between viral infection and disease.
Literature linking PCV‐3 with different disorders is extensive already, mainly regarding respiratory, digestive, reproductive and neurological signs. However, most of these studies do not provide information on viral genome detection in healthy pigs from the same farms; a number of them only offer PCV‐3 DNA detection by means of molecular methods, without showing evidence of viral presence in the lesions of affected animals. Only few reports combine the detection of PCV‐3 within the lesions, using mainly in situ hybridization (ISH) (Arruda et al., 2019; Kim, Park, et al., 2018; Phan et al., 2016; Saporiti et al., 2021; Vargas‐Bermúdez et al., 2021; Williamson et al., 2021). In these studies, myocarditis, periarteritis and/or encephalitis were the most significant lesions associated with the presence of PCV‐3 nucleic acid in fetuses, stillborn and weak‐born piglets, while myocarditis, systemic periarteritis and/or dermatopathy associated to necrotizing vasculitis were observed in weaned pigs, respectively. Also, one study pointed out that PCV‐3 genome was found in a case resembling proliferative necrotizing pneumonia, but pictures offered in the publication are not indicative of this pathological condition (Kedkovid, Woonwong, Arunorat, Sirisereewan, Sangpratum, Lumyai et al., 2018).
Tables 1, 2, 3, 4, 5 summarize current studies, performed in different countries, that have detected PCV‐3 DNA in tissues from different clinical–pathological conditions in domestic swine, indicating the production phase of affected pigs, tested samples, proportion of PCV‐3 PCR detection in sick animals and in healthy control animals (when available). Besides, Table 6 includes the studies that detected PCV‐3 genome in healthy animals.
TABLE 1.
PCV‐3 PCR detection in pigs suffering from respiratory disorders
| % (and proportion) of PCR positivity | ||||||
|---|---|---|---|---|---|---|
| Clinical signs/lesions | Production phase | Tested samples | Diseased animals | Healthy animals | Country | Reference |
| Respiratory disease with dyspnea / diffuse moderate lymphohistiocytic interstitial pneumonia and acute bronchitis | Suckling/Nursery | Tissues* | 100.0% (3/3)** | NI | USA | Phan et al., 2016 |
| Respiratory disease | NA | Lung homogenate/oral fluid/nasal swab | 12.5% (34/271) | NI | USA | Palinski et al., 2017 |
| Suckling | Lung tissues | 26.6% (25/94) | 0.0% (0/42)*** | China | Qi et al., 2019 | |
| Severe respiratory disease | Nursery | Sera | 63.7% (51/80) | 1.85% (4/216) | China | Zhai et al., 2017 |
| Mild respiratory disease | Nursery | Sera | 13.1% (23/175) | 1.85% (4/216) | China | |
| Abdominal breathing/lung swelling and congestion | Nursery | Tissues/sera | NA**** | NI | China | Shen et al., 2018 |
| Respiratory disease/interstitial pneumonia, suppurative bron‐ chopneumonia, pleuritis and fibrinous‐necrotizing pneumonia | Nursery/growing | Sera | 6.2% (8/129) | 6.6% (4/60) | Spain | Saporiti, Cruz, et al., 2020 |
| Porcine respiratory disease complex related signs | Growing | Sera | 60.0% (15/25) | 28.0% (7/25) | Thailand | Kedkovid, Woonwong, Arunorat, Sirisereewan, Sangpratum, Lumyai, et al., 2018 |
| Porcine respiratory disease complex/bronchointerstitial pneumonia | Growing | Lung and lymph node tissues | 62.5% (5/8) | NI | Thailand | |
| Respiratory distress/bronchointerstitial pneumonia and infiltrating lymphocytes | Growing | Tissues | 100% (2/2) | NI | South Korea | Kim, Park, et al., 2018 |
NI: not included in the published manuscript; NA: non‐available information in the published manuscript.
*Not specified.
**NGS results.
***The type of samples analyzed in control animals (feces) was different from the ones used in diseased pigs (lung tissues).
****Number of tested samples not included in the published manuscript.
TABLE 2.
PCV‐3 PCR detection in pigs suffering from enteric disorders
| % (and proportion) of PCR positivity | ||||||
|---|---|---|---|---|---|---|
| Clinical signs/lesions | Production phase | Tested samples | Diseased animals | Healthy animals | Country | Reference |
| Diarrhea | Nursery | Fecal samples | 17.14% (6/35) | 2.86% (1/35) | China | Zhai et al., 2017 |
| Diarrhea/vomiting | Suckling | Intestinal tissues/fecal samples | 10.4% (50/480) | 0.0% (0/42)* | China | Qi et al., 2019 |
| Digestive disorders/catarrhal en‐ teritis with or without villi atrophy and fusion, and catarrhal colitis | Nursery/growing | Sera | 5.5% (7/126) | 6.6% (4/60) | Spain | Saporiti, Cruz, et al., 2020 |
*The type of samples analyzed in control animals (feces) was different from the ones used in diseased pigs (intestinal tissues) .
TABLE 3.
PCV‐3 PCR detection in pigs suffering from reproductive disorders
| % (and proportion) of PCR positivity | ||||||
|---|---|---|---|---|---|---|
| Clinical signs/lesions | Production phase | Tested samples | Diseased animals | Healthy animals | Country | Reference |
| Reproductive failure | Gestation | Sera from sows | 45.9% (39/85) | 21.9% (23/105) | China | Zou et al., 2018 |
| Pool of tissues from aborted fetuses/Pool of tissues from stillborn piglets | 100.0% (2/2) | 100.0% (2/2) | Italy | Faccini et al., 2017 | ||
| Tissues from mummified fetuses | 97.0% (270/276) | NI | Brazil | Dal Santo et al., 2020 | ||
| Sow mortality and reproductive failure (aborted mummified fetuses) | Sow tissues/fetal tissues | NA | NI | USA | Palinski et al., 2017 | |
| Sows delivering stillbirth piglets | Pool of sera from sows | 100.0% (2/2) | 0.0% (0/2) | Brazil | Tochetto et al., 2018 | |
| Sera sows | 67.4% (31/46) | 60.5% (26/43) | Brazil | Tochetto et al., 2020 | ||
| Acute losses in neonatal piglets/increased rate of stillborn/sow mortality | Stillborn/tissues/ semen/sera | 34.7% (77/222) | NI | China | Ku et al., 2017 | |
| Reproductive losses/abortion and stillborn piglets | Pool of tissues from aborted fetuses or stillborn piglets | 33.9% (18/53) | NI | Spain | Saporiti et al., 2021 | |
| Reproductive losses/abortion and stillborn piglets | Sera from sows and thoracic/abdominal fluid from fetuses and spleen |
10% (sow sera) 100% (fluid samples) 70% (spleen) |
NI | Russia | Yuzhakov et al., 2018 | |
| Abortion/death of suckling piglets | Gestation/suckling | Tissues from aborted fetuses/weak suckling piglets | 36.4% (8/22) | NI | South Korea | Kim, Nazki, et al., 2018 |
| Acute loss of neonatal piglets | Tissues from aborted fetuses/stillborn/weak‐born piglets | 89.0% (49/55) | NI | Hungary | Deim et al., 2019 | |
| Reproductive failure/weak‐born neonatal piglets/myocarditis/encephalitis | Tissues from fetuses/suckling/weaning | 100.0% (25/25) | NI | USA | Arruda et al., 2019 | |
NA: non‐available information in the published manuscript; NI: not included in the published manuscript.
TABLE 4.
PCV‐3 PCR detection in pigs suffering from neurological disorders
| % (and proportion) of PCR positivity | ||||||
|---|---|---|---|---|---|---|
| Clinical signs/lesions | Production phase | Tested samples | Diseased animals | Healthy animals | Country | Reference |
| Neurological signs | Suckling | Tissue pool | 100.0% (1/1)* | NI | USA | Phan et al., 2016 |
| Congenital tremors | Suckling | Brain | 100.0% (7/7) | NI | China | Chen et al., 2017 |
| Congenital tremors, neurological signs in piglets after birth and multisystemic inflammation/non‐suppurative encephalomyelitis | Suckling | Brain, other tissues | 100.0% (3/3) | NI | UK | Williamson et al., 2021 |
| Tremors, weak‐born neonatal piglets/myocarditis, encephalitis, gliosis and lymphocytic perivascular cuffing | Suckling | Brain, other tissues | 100.0% (2/2) | NI | USA | Arruda et al., 2019 |
*NGS results.
NA: non‐available information in the published manuscript; NI: not included in the published manuscript.
TABLE 5.
PCV‐3 PCR detection in pigs suffering from other conditions not listed in previous tables
| % (and proportion) of PCR positivity | ||||||
|---|---|---|---|---|---|---|
| Clinical signs/lesions | Production phase | Tested samples | Diseased animals | Healthy animals | Country | Reference |
| Myocarditis/periarteritis | Suckling/nursery/fattening | Several tissues | 100.0% (3/3)* | NI | USA | Phan et al., 2016 |
| PDNS | NA | Several tissues | 93.8% (45/48) | NI | USA | Palinski et al., 2017 |
| PDNS | Sows | Pooled tissues | NA* | NI | USA | Palinski et al., 2017 |
| PDNS/acute deaths/myocarditis/arteritis/periarteritis | Nursery | Several tissues | 100.0% (11/11) | NI | USA | Arruda et al., 2019 |
| PDNS/systemic inflammation | Nursery and fattening | Kidney and spleen | 40–50% (depending on tested tissue) | NI | Russia | Yuzhakov et al., 2018 |
| Myocarditis/arteritis/periarteritis | Nursery | Several tissues | 100% (4/4) | 100% (2/2)** | Portugal | Alomar et al., 2021 |
| Arthrogryposis | Stillborn piglets | Several tissues | 100.0% (4/4) | NI | UK | Williamson et al., 2021 |
*NGS results. ** Viral load was higher in sick animals; by in situ hybridization, only diseased animals were positive.
NA: non‐available information in the published manuscript; NI: not included in the published manuscript.
TABLE 6.
PCV‐3 PCR detection in healthy pigs
| Clinical signs/lesions | Production phase | Tested samples | % (and proportion) of PCR positivity | Country | Reference |
|---|---|---|---|---|---|
| Asymptomatic | Weaning/growing/finishing | Oral fluids | 43.4% (142/327) | South Korea | Kwon et al., 2017 |
| Asymptomatic | Sows/fetuses | Tissues | 59.5% (132/222) | China | Zheng et al., 2017 |
| Asymptomatic | Sows (in lactation) | Sera | 47.3% (18/38) | Thailand | Kedkovid, Woonwong, Arunorat, Sirisereewan, Sangpratum, Kesdangsakonwut, et al., 2018 |
| Asymptomatic | Sows | Sera | 15.7% (19/121) | Spain | Saporiti, Martorell, et al., 2020 |
| Asymptomatic | Sows and fetuses | Tissues (brain and lung) | 33.7% (86/255) | ||
| Asymptomatic | Different production phases | Tissues and sera | 56.4% (44/78) | Denmark | Franzo, Legnardi, et al., 2018 |
| Asymptomatic | Different production phases | Tissues, sera and nasal swabs | 37.4% (37/99) | Italy | |
| Asymptomatic | Different production phases | Pool of sera | 15.0% (14/94) | Spain | |
| Asymptomatic | Non‐available | Lymph node tissues | NA | Sweden | Ye et al., 2018 |
| Asymptomatic | Growing | Tissues, serum and nasal swabs | 5.9% (5/90) | Poland | Stadejek et al., 2017 |
| Asymptomatic | Nursery/finishing | Sera | 10% (7/73) | Spain | Klaumann, Franzo, et al., 2018 |
| Asymptomatic | Nursery/finishing | Sera | 6.4% (7/110) | Spain | Saporiti, Huerta, et al., 2020 |
| Asymptomatic | Nursery/finishing | Sera | 13.0% (13/100) | Belgium | |
| Asymptomatic | Nursery/finishing | Sera | 10.4% (7/67) | France | |
| Asymptomatic | Nursery/finishing | Sera | 6.3% (5/80) | Germany | |
| Asymptomatic | Nursery/finishing | Sera | 4.5% (3/67) | Italy | |
| Asymptomatic | Nursery/finishing | Sera | 6.3% (5/80) | Denmark | |
| Asymptomatic | Nursery/finishing | Sera | 14.0% (7/50) | The Netherlands | |
| Asymptomatic | Nursery/finishing | Sera | 4.0% (2/50) | Ireland | |
| Asymptomatic | Nursery/finishing | Sera | 15.0% (3/20) | Sweden |
2.2. PCV‐3 in co‐infection with other pathogens
Being a worldwide spread virus, PCV‐3 has been found in many studies in co‐infection with other pathogens such the ones listed in Table 7. The existence of such mixed infections in diseased animals emphasizes the need to study the differential pathogenicity of PCV‐3 alone or in co‐infection . PCV‐2 co‐infection with other pathogens has been proven to lead to more severe disease presentation under field (Opriessnig & Halbur, 2012) and experimental (Tomás et al., 2008) conditions. The impact of PCV‐3 co‐infection with other agents is currently unknown and a potential and vast field of further research.
TABLE 7.
List of pathogens found concomitantly with the presence of PCV‐3 in domestic swine
| Pathogen | Country | % (and proportion) of PCR positivity for PCV‐3 | Reference |
|---|---|---|---|
| PCV‐2 | China | 15.8% (35/222) | Ku et al., 2017 |
| 39.4% (52/132) | Zheng et al., 2017 | ||
| 30.0% (3/10) | Sun et al., 2018 | ||
| 70.0% (28/40) | Zhao et al., 2018 | ||
| 1.9% (3/159) | Chen et al., 2019 | ||
| 6.78% (32/472) | Xia et al., 2019 | ||
| South Korea | 28.3% (13/46) | Kim et al., 2017 | |
| 19.3% (11/57) | Kim, Nazki et al., 2018 | ||
| Thailand | 20.0% (1/5) | Kedkovid, Woonwong, Arunorat, Sirisereewan, Sangpratum, Lumyai, et al., 2018 | |
| Poland | 4.8% (8/166) | Woźniak et al., 2019 | |
| USA | 5.4% (115/2125) | Wang, Noll et al., 2019 | |
| European countries | 2.6% (16/624) | Saporiti, Huerta et al., 2020 | |
| Brazil | 78.3% (216/276) | Dal Santo et al., 2020 | |
| Colombia | 24.0% (12/50) | Vargas‐Bermúdez et al., 2021 | |
| Spain | 1.9% (1/53) | Saporiti et al., 2021 | |
| Porcine reproductive and respiratory syndrome virus (PRRSV) | Thailand | 20.0% (1/5) | Kedkovid, Woonwong, Arunorat, Sirisereewan, Sangpratum, Lumyai, et al., 2018 |
| South Korea | 100.0% (2/2) | Kim, Park et al., 2018 | |
| 43.8% (25/57) | Kim, Nazki, et al., 2018 | ||
| China | 0.6% (1/159) | Chen et al., 2019 | |
| Spain | 3.8% (2/53) | Saporiti et al., 2021 | |
| Porcine parvovirus (PPV) | China | 20.0% (8/40) | Zhao et al., 2018 |
| Brazil | 58.7% (162/276) | Dal Santo et al., 2020 | |
| Classical swine fever virus (CSFV) | China | 90.0% (9/10) | Sun et al., 2018 |
| 2.5% (1/40) | Zhao et al., 2018 | ||
| Porcine epidemic diarrhea virus (PEDV) | China | NA | Chen et al., 2017 |
| Atypical porcine pestivirus (APPV) | China | NA | Chen et al., 2017 |
| UK | 42.8.% (3/7) | Williamson et al., 2021 | |
| Porcine kobuvirus (PKV) | China | NA | Chen et al., 2017 |
| Porcine pseudorabies virus (PRV) | China | NA | Chen et al., 2017 |
| 5.0% (2/40) | Zhao et al., 2018 | ||
| Porcine sapelovirus (PSV) | China | NA | Chen et al., 2017 |
| Porcine bocavirus (PBoV) | China | NA | Chen et al., 2017 |
| Torque teno sus virus (TTSuV1 and 2) | China | 50% (66/132) | Zheng et al., 2018 |
| Streptococcus spp | USA | NA | Phan et al., 2016 |
| Thailand | 20.0% (1/5) | Kedkovid, Woonwong, Arunorat, Sirisereewan, Sangpratum, Lumyai, et al., 2018 | |
| South Korea | 100.0% (2/2) | Kim, Park, et al., 2018 | |
| Glaeserella parasuis | USA | NA | Phan et al., 2016 |
| Mycoplasma hyorhinis | USA | NA | Phan et al., 2016 |
| Mycoplasma hyopneumoniae | South Korea | 100.0% (2/2) | Kim, Park, et al., 2018 |
| Pasteurella multocida | Thailand | NA | Kedkovid, Woonwong, Arunorat, Sirisereewan, Sangpratum, Lumyai, et al., 2018 |
| Leptospira spp | Brazil | 9.4% (26/276) | Dal Santo et al., 2020 |
NA: non‐available information in the published manuscript.
2.3. Experimental infections with PCV‐3
The pathogenesis of PCV‐3 infection is still a mystery. Several limitations account for such paucity of knowledge, including the very scarce availability of virus isolates (Mora‐Díaz et al., 2020; Oh & Chae, 2020), the lack of serologically and virologically free pigs and the widespread nature of the virus, that make very difficult to get suitable tools, animals and conditions to perform experimental infections. Some laboratory reagents have been generated for intra‐laboratory use only (Li et al., 2018; Zhang et al., 2019), but they have not been apparently validated by other research groups.
In consequence, only three experimental infections have been published in the literature to date, all of them using nursery aged pigs (4‐ to 8‐week‐old). Two of them were done by the same research group in the United States by means of a cell culture propagated virus (1 mL intranasal [IN] and 1 mL intramuscular [IM] of 6.6 × 1010 genomic copies/mL) (Mora‐Díaz et al., 2020) or tissue homogenate containing high PCV‐3 load (2 mL IN of 3.38 × 1012 genomic copies/mL and 2 mL IM of 1.04 × 1011 genomic copies/mL and re‐inoculated after 7 days through the same routes and with the same doses), with or without keyhole limpet hemocyanin emulsified in incomplete Freund's adjuvant (KLH/ICFA) (Temeeyasen et al., 2021). The third one was performed by a Chinese research group, using a PCV‐3 infectious clone (2 mL IN of 106.53 TCID50/mL), with or without KLH/ICFA (Jiang et al., 2018). The group inoculated with KLH/ICFA also received a infectious booster after 4 days (Jiang et al., 2018).
The first two studies used caesarean‐derived, colostrum‐deprived piglets and no clinical signs upon inoculation occurred. However, mild‐to‐moderate lesions consisting of multisystemic inflammation and perivasculitis were observed, associated with a low to moderate amount of PCV‐3 genome detected by ISH within the lesions (Mora‐Díaz et al., 2020; Temeeyasen et al., 2021). These data mirror the limited number of studies on PCV‐3 naturally infection cases (A; Arruda et al., 2019; Phan et al., 2016; Saporiti et al., 2021), in which myocarditis and periarteritis were the dominant histological findings. Importantly, the experimental inoculation of the virus caused a detectable antibody response around 7–10 days post‐challenge (DPC) in both studies, but with different profiles; IgM response dominated in one study (Mora‐Díaz et al., 2020) and IgG in the other (Temeeyasen et al., 2021). The virus was detected in serum as soon as 3 days after inoculation until the end of the experiment at 42 DPC (Temeeyasen et al., 2021).
The Chinese study used specific pathogen‐free animals inoculated with a PCV‐3 infectious clone together or not with KLH/ICFA (Jiang et al., 2018). In contrast to previous studies, fever was observed in the challenged pigs, which showed anorexia, coughing, sneezing, diarrhoea and respiratory signs; also, skin lesions consisting of multifocal papules were observed by 15 DPC until the end of the experiment (28 DPC). Although the authors indicated that PDNS was reproduced (Jiang et al., 2018), the reported kidney histopathological lesions were not compatible with systemic necrotizing vasculitis and fibrino‐necrotizing glomerulonephritis, the well‐known microscopic lesions of PDNS (Segalés, 2012). Therefore, based on current evidence, it is not possible to claim that PDNS has been reproduced by means of a PCV‐3 experimental inoculation. Importantly, detection of this virus in tissues was attempted by immunohistochemistry, but published images (Jiang et al., 2018) are difficult to be interpreted.
2.4. Proposal of case definition for PCV‐3 associated diseases
The sole detection of an endemic virus in tissues or other biological samples is not enough to establish a causal association or to establish disease diagnoses (Arruda et al., 2019). Most studies so far published on PCV‐3 detection in diseased animals have been based on molecular methods, with very few of them reporting macro‐ or microscopic lesions and even less using methods detecting the genome of the virus in the observed lesions (Arruda et al., 2019; Kim, Park, et al., 2018; Phan et al., 2016; Saporiti et al., 2021). Therefore, the latter studies associating the presence of the viral genome with the lesions provide the strongest evidence of this virus in association with pathological conditions.
The establishment of disease diagnosis criteria for widespread pathogens is not an easy task. A good example of such a scenario would be a relative of PCV‐3, PCV‐2. Three major criteria were proposed to establish the diagnosis of PCV‐2‐systemic disease (Segalés & Domingo, 1999; Sorden, 2000): (1) presence of compatible clinical signs, mainly wasting, (2) observation of moderate‐to‐severe histological lesions in lymphoid tissues (lymphocytic infiltration and histiocytic infiltration) and (3) detection of moderate to high amount of PCV‐2 within such lesions. Such criteria were crucial to provide an ordered, concise and systematic approach for diagnosing a disease that was considered new by the end of the 1990s and early 2000s. Based on the general reluctance to accept that PCV‐2 was truly pathogenic for swine at that time (Segalés et al., 2013), such demanding case definition guaranteed the necessary strictness and was found acceptable for most members of veterinary and scientific communities. The description of a novel PCV (PCV‐3) almost 20 years after the report of PMWS was taken with more caution, and despite the lack of an associated severe and globally distributed disease, veterinarians and scientists have been more open‐minded to accept its potential disease causality.
In any case, and following the path paved by PCV‐2 diagnostic approach (Segalés, 2012), the existence of PCV‐3 associated disease (PCV‐3‐AD) diagnostic criteria would help in placing the novel virus into the general context of swine disorders. Therefore, based on existing information that provide clinical, pathological and virological assessments of PCV‐3 infection cases (Arruda et al., 2019; Kim, Park, et al., 2018; Phan et al., 2016; Saporiti et al., 2021), the authors would like to propose two major disease outcomes related with PCV‐3 infection: PCV‐3‐reproductive disease (PCV‐3‐RD) in sows and fetuses/neonatal piglets and PCV‐3‐systemic disease (PCV‐3‐SD) in pre‐ and post‐weaning pigs (Table 8, Figure 1). The authors consider that PDNS, which has been linked with PCV‐3 infection by some studies (Jiang et al., 2018; Palinski et al., 2017; Yuzhakov et al., 2018), does not fulfil so far specific criteria demonstrating a putative etiological association with PCV‐3 based on clinical, pathological, virological and epidemiological facts.
TABLE 8.
Proposed diagnostic criteria for the individual case definition of PCV‐3 associated diseases (PCV‐3‐AD)
| PCV‐3‐AD proposed name (acronym) | Main clinical sign | Individual diagnostic criteria |
|---|---|---|
| PCV‐3‐reproductive disease (PCV‐3‐RD) | Late abortion, malformations, mummified fetuses, stillborn fetuses, weak‐born piglets |
|
| PCV‐3‐systemic disease (PCV‐3‐SD) | Wasting, weight loss, ill thrift or poor‐doers, neurological signs |
|
FIGURE 1.
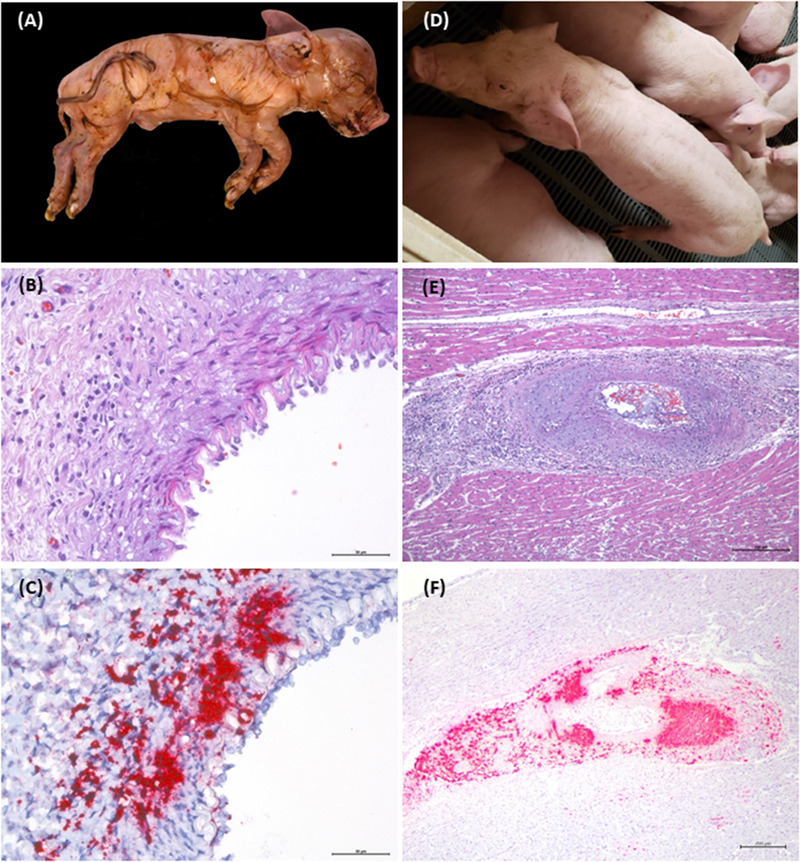
Proposed diagnostic criteria for the individual case definition of PCV‐3‐associated reproductive (A, B, C) and systemic disease (D, E, F). PCV‐3‐reproductive disease: (A) stillborn piglet from a litter with a late reproductive problem characterized by increased percentage of stillborn and weak‐born piglets, (B) mild‐to‐moderate mononuclear inflammatory infiltrates in the arterial wall of the fetal spleen and (C) moderate to high amount of PCV‐3 nucleic acid in the damaged arterial area. PCV‐3‐systemic disease: (D) clinical picture of a pig showing wasting, (E) moderate‐to‐severe non‐suppurative arteritis in the heart and (F) high amount of PCV‐3 genome in the damaged artery
3. DISCUSSION
Traditionally, swine veterinarians have dealt with overt diseases, with the main task of counteracting them and improving the profitability of farms. Several decades ago, the most important diseases affecting pigs were considered mostly ‘unifactorial’, in which the unique presence of the infectious agent was sufficient to cause disease or production losses. However, the current worldwide swine disease scenario is dominated by disorders that are considered of ‘multifactorial’ nature, since the mere presence of the agent is not sufficient to induce the disease (Segalés, 2013). Moreover, most of the new swine pathogens discovered in the last 20 years are infectious agents that (1) had been circulating in pigs for extended periods but remained undetected until recently or (2) infectious agents that had newly emerged in swine because of host species jump and further evolution (Fournié et al., 2015). Detection of these novel pathogens has been driven by advances in diagnostic methods such as broad‐range PCR and NGS methods (Blomström, 2011), as well as increased surveillance efforts and particular research interests (Fournié et al., 2015). PCV‐3 is an excellent example of a virus discovered through NGS that has been circulating for an extended period before its first detection (Rodrigues et al., 2020) and for which the surveillance efforts, mainly linked to research, have remarkably increased in the last 5 years (Opriessnig et al., 2020).
In contrast with PCV‐2, PCV‐3 was not discovered because of the emergence/identification of a new disease with severe impact on swine production, but as an extra‐diagnostic effort on cases with different clinical outcomes and lack of etiologic diagnosis (Palinski et al., 2017; Phan et al., 2016). This starting point prompted the search for PCV‐3 employing molecular methods, which was soon demonstrated to be a widespread virus in the swine population (Klaumann, Correa‐Fiz et al., 2018). Unlike PCV‐2, isolation of PCV‐3 in cell culture was unsuccessful (Faccini et al., 2017; Palinski et al., 2017) until recently (Mora‐Díaz et al., 2020; Oh & Chae, 2020), and the availability of virus isolates or other reagents is extremely restricted still today. Therefore, the progress made on PCV‐3 pathogenesis knowledge, immunity and diagnostic technique development is still very limited.
Nevertheless, evidence of PCV‐3 involvement in certain pathological conditions is expanding. The presence of a particular infectious agent within certain histopathological lesions of animals showing overt disease, when consistently detected, is probably the strongest evidence of potential disease causality. In such regards, a laboratory technique such as ISH has ultimately allowed detecting PCV‐3 nucleic acid within lesions of diseased animals. More specifically, the viral genome has been detected at moderate/high amounts in fetuses and stillborn/weak‐born piglets from cases of reproductive disorders as well as in pre‐ and post‐weaning pigs with wasting, sudden death or neurological signs showing multisystemic inflammatory infiltrates, mainly at perivascular level (Arruda et al., 2019; Kim, Park, et al., 2018; Phan et al., 2016; Saporiti et al., 2021; Williamson et al., 2021). Therefore, the existing combination of clinical, pathological and virological data provides a potential diagnostic framework for PCV‐3‐AD case definition.
In summary, compiled data on PCV‐3 knowledge so far points it out as a virus with pathogenic potential, implying the need to standardize diagnostic criteria for at least reproductive and pre‐/post‐weaning disorders. Such proposal is independent of the frequency, geographic distribution or economic impact of PCV‐3‐AD, which are rather unknown at present. While the PCV‐3‐SD in pre‐ and post‐weaning pigs has been scarcely diagnosed at a global level to date (Arruda et al., 2019; Williamson et al., 2021), PCV‐3‐RD (Arruda et al., 2019; Saporiti et al., 2021; Williamson et al., 2021) seems to occur more often. So far, however, the most frequent presentation of this viral infection is likely subclinical, and its potential health and economic impact on the swine industry worldwide is to be determined.
CONFLICT OF INTEREST
None of the contributing authors has any conflict of interest.
ETHICAL STATEMENT
Non‐applicable; this study did not include sample collection or questionnaires from animals or humans.
ACKNOWLEDGEMENTS
The authors thank the funding by E‐RTA2017‐00007‐00‐00 INIA Project from the Instituto Nacional de Investigación y Tecnologia Agraria y Alimentaria (Spanish Government) and the CERCA Programme/Generalitat de Catalunya.
Saporiti, V. , Franzo, G. , Sibila, M. , & Segalés, J . (2021). Porcine circovirus 3 (PCV‐3) as a causal agent of disease in swine and a proposal of PCV‐3 associated disease case definition. Transbound. Emerg. Dis, 68, 2936–2948. 10.1111/tbed.14204
Authors Marina Sibila and Joaquim Segalés contributed equally.
DATA AVAILABILITY STATEMENT
Data used in this review is available in the different journals referenced.
REFERENCES
- Alomar, J. , Saporiti, V. , Pérez, M. , Gonçalves, D. , Sibila, M. , & Segalés, J. (2021). Porcine circovirus 3 associated wasting in postweaning pigs. Unpublished manuscript.
- Arruda, B. , Piñeyro, P. , Derscheid, R. , Hause, B. , Byers, E. , Dion, K. , Long, D. , Sievers, C. , Tangen, J. , Williams, T. , & Schwartz, K. (2019). PCV3‐associated disease in the United States swine herd. Emerging Microbes and Infections, 8(1), 684–698. 10.1080/22221751.2019.1613176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomström, A. L. (2011). Viral metagenomics as an emerging and powerful tool in veterinary medicine. Veterinary Quarterly, 31(3), 107–114. 10.1080/01652176.2011.604971 [DOI] [PubMed] [Google Scholar]
- Chen, G. H. , Mai, K. J. , Zhou, L. , Wu, R. T. , Tang, X. Y. , Wu, J. L. , He, L. L. , Lan, T. , Xie, Q. M. , Sun, Y. , & Ma, J. Y. (2017). Detection and genome sequencing of porcine circovirus 3 in neonatal pigs with congenital tremors in South China. Transboundary and Emerging Diseases, 64(6), 1650–1654. 10.1111/tbed.12702 [DOI] [PubMed] [Google Scholar]
- Chen, N. , Huang, Y. , Ye, M. , Li, S. , Xiao, Y. , Cui, B. , & Zhu, J. (2019). Co‐infection status of classical swine fever virus (CSFV), porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circoviruses (PCV2 and PCV3) in eight regions of China from 2016 to 2018. Infection, Genetics and Evolution, 68(November 2018), 127–135. 10.1016/j.meegid.2018.12.011 [DOI] [PubMed] [Google Scholar]
- Czyżewska‐Dors, E. , Núñez, J. I. , Saporiti, V. , Huerta, E. , Riutord, C. , Cabezón, O. , Segalés, J. , & Sibila, M. (2020). Detection of porcine circovirus 3 in wildlife species in Spain. Pathogens, 9(5), 7–12. 10.3390/pathogens9050341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Santo, A. C. , Cezario, K. C. , Bennemann, P. E. , Machado, S. A. , & Martins, M. (2020). Full‐genome sequences of porcine circovirus 3 (PCV3) and high prevalence in mummified fetuses from commercial farms in Brazil. Microbial Pathogenesis, 141, 104027. 10.1016/j.micpath.2020.104027 [DOI] [PubMed] [Google Scholar]
- Deim, Z. , Dencso, L. , Erdélyi, I. , Valappil, S. K. , Varga, C. , Pósa, A. , Makrai, L. , & Rákhely, G. (2019). Porcine circovirus type 3 detection in a Hungarian pig farm experiencing reproductive failures. Veterinary Record, 185(3), 84. 10.1136/vr.104784 [DOI] [PubMed] [Google Scholar]
- Faccini, S. , Barbieri, I. , Gilioli, A. , Sala, G. , Gibelli, L. R. , Moreno, A. , Sacchi, C. , Rosignoli, C. , Franzini, G. , & Nigrelli, A. (2017). Detection and genetic characterization of porcine circovirus type 3 in Italy. Transboundary and Emerging Diseases, 64(6), 1661–1664. 10.1111/tbed.12714 [DOI] [PubMed] [Google Scholar]
- Fournié, G. , Kearsley‐Fleet, L. , Otte, J. , & Pfeiffer, D. U. (2015). Spatiotemporal trends in the discovery of new swine infectious agents. Veterinary Research, 46(1), 1–9. 10.1186/s13567-015-0226-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzo, G. , Legnardi, M. , Hjulsager, C. K. , Klaumann, F. , Larsen, L. E. , Segales, J. , & Drigo, M. (2018). Full‐genome sequencing of porcine circovirus 3 field strains from Denmark, Italy and Spain demonstrates a high within‐Europe genetic heterogeneity. Transboundary and Emerging Diseases, 65(3), 602–606. 10.1111/tbed.12836 [DOI] [PubMed] [Google Scholar]
- Franzo, G. , Delwart, E. , Fux, R. , Hause, B. , Su, S. , Zhou, J. Y. , & Segalés, J. (2020). Genotyping porcine circovirus 3 (PCV‐3) nowadays: Does it make sense? Viruses, 12(3), 1–15. 10.3390/v12030265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzo, G. , Grassi, L. , Tucciarone, C. M. , Drigo, M. , Martini, M. , Pasotto, D. , Mondin, A. , & Menandro, M. L. (2019). A wild Circ‐ulation: High presence of Porcine circovirus 3 in different mammalian wild hosts and ticks. Transboundary and Emerging Diseases, 66(4), 1548–1557. 10.1111/tbed.13180 [DOI] [PubMed] [Google Scholar]
- Franzo, G. , Segales, J. , Tucciarone, C. M. , Cecchinato, M. , & Drigo, M. (2018). The analysis of genome composition and codon bias reveals distinctive patterns between avian and mammalian circoviruses which suggest a potential recombinant origin for porcine circovirus 3. PLoS ONE, 13(6), e0199950. 10.1371/journal.pone.0199950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, B. , Krueger, L. , Seeliger, F. , Bruegmann, M. , Segalés, J. , & Baumgaertner, W. (2009). Retrospective study on the occurrence of porcine circovirus 2 infection and associated entities in Northern Germany. Veterinary Microbiology, 138(1–2), 27–33. 10.1016/j.vetmic.2009.02.005 [DOI] [PubMed] [Google Scholar]
- Jiang, H. , Wang, D. , Wang, J. , Zhu, S. , She, R. , Ren, X. , Tian, J. , Quan, R. , Hou, L. , Li, Z. , Chu, J. , Guo, Y. , Xi, Y. , Song, H. , Yuan, F. , Wei, L. , & Liu, J. (2018). Induction of porcine dermatitis and nephropathy syndrome in piglets by infection with porcine circovirus type 3. Journal of Virology, 93(4), e02045‐18. 10.1128/JVI.02045-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S. , Zhou, N. , Li, Y. , An, J. , & Chang, T. (2019). Detection and sequencing of porcine circovirus 3 in commercially sourced laboratory mice. Veterinary Medicine and Science, 5(2), 176–181. 10.1002/vms3.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedkovid, R. , Woonwong, Y. , Arunorat, J. , Sirisereewan, C. , Sangpratum, N. , Kesdangsakonwut, S. , Tummaruk, P. , Teankum, K. , Assavacheep, P. , Jittimanee, S. , & Thanawongnuwech, R. (2018). Porcine circovirus type 3 (PCV3) shedding in sow colostrum. Veterinary Microbiology, 220(April), 12–17. 10.1016/j.vetmic.2018.04.032 [DOI] [PubMed] [Google Scholar]
- Kedkovid, R. , Woonwong, Y. , Arunorat, J. , Sirisereewan, C. , Sangpratum, N. , Lumyai, M. , Kesdangsakonwut, S. , Teankum, K. , Jittimanee, S. , & Thanawongnuwech, R. (2018). Porcine circovirus type 3 (PCV3) infection in grower pigs from a Thai farm suffering from porcine respiratory disease complex (PRDC). Veterinary Microbiology, 215, 71–76. 10.1016/j.vetmic.2018.01.004 [DOI] [PubMed] [Google Scholar]
- Kim, H. R. , Park, Y. R. , Lim, D. R. , Park, M. J. , Park, J. Y. , Kim, S. H. , Lee, K. K. , Lyoo, Y. S. , & Park, C. K. (2017). Multiplex real‐time polymerase chain reaction for the differential detection of porcine circovirus 2 and 3. Journal of Virological Methods, 250, 11–16. 10.1016/j.jviromet.2017.09.021 [DOI] [PubMed] [Google Scholar]
- Kim, S. C. , Nazki, S. , Kwon, S. , Juhng, J. H. , Mun, K. H. , Jeon, D. Y. , Jeong, C. G. , Khatun, A. , Kang, S. J. , & Kim, W. Il . (2018). The prevalence and genetic characteristics of porcine circovirus type 2 and 3 in Korea. BMC Veterinary Research, 14(1), 294. 10.1186/s12917-018-1614-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. H. , Park, J. Y. , Jung, J. Y. , Kim, H. Y. , Park, Y. R. , Lee, K. K. , Lyoo, Y. S. , Yeo, S. G. , & Park, C. K. (2018). Detection and genetic characterization of porcine circovirus 3 from aborted fetuses and pigs with respiratory disease in Korea. Journal of Veterinary Science, 19(5), 721–724. 10.4142/jvs.2018.19.5.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaumann, F. , Franzo, G. , Sohrmann, M. , Correa‐Fiz, F. , Drigo, M. , Núñez, J. I. , Sibila, M. , & Segalés, J. (2018). Retrospective detection of porcine circovirus 3 (PCV‐3) in pig serum samples from Spain. Transboundary and Emerging Diseases, 65(5), 1290–1296. 10.1111/tbed.12876 [DOI] [PubMed] [Google Scholar]
- Klaumann, F. , Correa‐Fiz, F. , Franzo, G. , Sibila, M. , Núñez, J. I. , & Segalés, J. (2018). Current knowledge on porcine circovirus 3 (PCV‐3): A novel virus with a yet unknown impact on the swine industry. Frontiers in Veterinary Science, 5, 315. 10.3389/fvets.2018.00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaumann, F. , Correa‐Fiz, F. , Sibila, M. , Núñez, J. I. , & Segalés, J. (2019). Infection dynamics of porcine circovirus type 3 in longitudinally sampled pigs from four Spanish farms. Veterinary Record, 184(20), 619. 10.1136/vr.105219 [DOI] [PubMed] [Google Scholar]
- Ku, X. , Chen, F. , Li, P. , Wang, Y. , Yu, X. , Fan, S. , Qian, P. , Wu, M. , & He, Q. (2017). Identification and genetic characterization of porcine circovirus type 3 in China. Transboundary and Emerging Diseases, 64(3), 703–708. 10.1111/tbed.12638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, T. , Yoo, S. J. , Park, C. K. , & Lyoo, Y. S. (2017). Prevalence of novel porcine circovirus 3 in Korean pig populations. Veterinary Microbiology, 207, 178–180. 10.1016/j.vetmic.2017.06.013 [DOI] [PubMed] [Google Scholar]
- Li, X. , Bai, Y. , Zhang, H. , Zheng, D. , Wang, T. , Wang, Y. , Deng, J. , Sun, Z. , & Tian, K. (2018). Production of a monoclonal antibody against porcine circovirus type 3 cap protein. Journal of Virological Methods, 261, 10–13. 10.1016/j.jviromet.2018.07.014 [DOI] [PubMed] [Google Scholar]
- Mora‐Díaz, J. , Piñeyro, P. , Shen, H. , Schwartz, K. , Vannucci, F. , Li, G. , Arruda, B. , & Giménez‐Lirola, L. (2020). Isolation of PCV3 from perinatal and reproductive cases of PCV3‐associated disease and in vivo characterization of PCV3 replication in CD/CD growing pigs. Viruses, 12(2), 219. 10.3390/v12020219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, T. , & Chae, C. (2020). First isolation and genetic characterization of porcine circovirus type 3 using primary porcine kidney cells. Veterinary Microbiology, 241, 108576. 10.1016/j.vetmic.2020.108576 [DOI] [PubMed] [Google Scholar]
- Opriessnig, T. , & Halbur, P. G. (2012). Concurrent infections are important for expression of porcine circovirus associated disease. Virus Research, 164(1–2), 20–32. 10.1016/j.virusres.2011.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opriessnig, T. , Karuppannan, A. K. , Castro, A. M. M. G. , & Xiao, C. T. (2020). Porcine circoviruses: Current status, knowledge gaps and challenges. Virus Research, 286, 198044. 10.1016/j.virusres.2020.198044 [DOI] [PubMed] [Google Scholar]
- Palinski, R. , Piñeyro, P. , Shang, P. , Yuan, F. , Guo, R. , Fang, Y. , Byers, E. , & Hause, B. M. (2017). A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. Journal of Virology, 91(1), e01879‐16. 10.1128/JVI.01879-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, T. G. , Giannitti, F. , Rossow, S. , Marthaler, D. , Knutson, T. , Li, L. , Deng, X. , Resende, T. , Vannucci, F. , & Delwart, E. (2016). Detection of a novel circovirus PCV3 in pigs with cardiac and multi‐systemic inflammation. Virology Journal, 13(1), 184. 10.1186/s12985-016-0642-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, S. , Su, M. , Guo, D. , Li, C. , Wei, S. , Feng, L. , & Sun, D. (2019). Molecular detection and phylogenetic analysis of porcine circovirus type 3 in 21 provinces of China during 2015–2017. Transboundary and Emerging Diseases, 66(2), 1004–1015. 10.1111/tbed.13125 [DOI] [PubMed] [Google Scholar]
- Rodrigues, I. L. F. , Cruz, A. C. M. , Souza, A. E. , Knackfuss, F. B. , Costa, C. H. C. , Silveira, R. L. , & Castro, T. X. (2020). Retrospective study of porcine circovirus 3 (PCV3) in swine tissue from Brazil (1967–2018). Brazilian Journal of Microbiology, 51(3), 1391–1397. 10.1007/s42770-020-00281-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saporiti, V. , Cruz, T. F. , Correa‐Fiz, F. , Núñez, J. I. , Sibila, M. , & Segalés, J. (2020). Similar frequency of Porcine circovirus 3 (PCV‐3) detection in serum samples of pigs affected by digestive or respiratory disorders and age‐matched clinically healthy pigs. Transboundary and Emerging Diseases, 67(1), 199–205. 10.1111/tbed.13341 [DOI] [PubMed] [Google Scholar]
- Saporiti, V. , Huerta, E. , Correa‐Fiz, F. , Grosse Liesner, B. , Duran, O. , Segalés, J. , & Sibila, M. (2020). Detection and genotyping of Porcine circovirus 2 (PCV‐2) and detection of Porcine circovirus 3 (PCV‐3) in sera from fattening pigs of different European countries. Transboundary and Emerging Diseases, 67(6), 2521–2531. 10.1111/tbed.13596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saporiti, V. Martorell, S. , Cruz, T. F. , Klaumann, F. , Correa‐Fiz, F. , Balasch, M. , Sibila, M. , & Segalés, J. (2020). Frequency of detection and phylogenetic analysis of porcine circovirus 3 (PCV‐3) in healthy primiparous and multiparous sows and their mummified fetuses and stillborn. Pathogens, 9(7), 533. 10.3390/pathogens9070533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saporiti, V. , Valls, L. , Maldonado, J. , Perez, M. , Correa‐Fiz, F. , Segalés, J. , & Sibila, M. (2021). Porcine circovirus 3 detection in aborted fetuses and stillborn piglets from swine reproductive failure cases. Viruses, 13(2), 264. 10.3390/v13020264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segalés, J. & Domingo, M. (1999). Clinical and pathological findings of PMWS cases in Europe. In Proceedings of the 26th Annual Allen D. Leman Swine Conference , Brooklyn Park, Minnesota, USA. [Google Scholar]
- Segalés, J. (2013). The threat of emerging and re‐emerging diseases in pigs. Pig Journal, 69, 7–16 [Google Scholar]
- Segalés, J. (2012). Porcine circovirus type 2 (PCV2) infections: Clinical signs, pathology and laboratory diagnosis. Virus Research, 164(1–2), 10–19 10.1016/j.virusres.2011.10.007 [DOI] [PubMed] [Google Scholar]
- Segalés, J. (2015). Best practice and future challenges for vaccination against porcine circovirus type 2. Expert Review of Vaccines, 14(3), 473–487. 10.1586/14760584.2015.983084 [DOI] [PubMed] [Google Scholar]
- Segalés, J. , Kekarainen, T. , & Cortey, M. (2013). The natural history of porcine circovirus type 2: From an inoffensive virus to a devastating swine disease? Veterinary Microbiology, 165(1–2), 13–20. 10.1016/j.vetmic.2012.12.033 [DOI] [PubMed] [Google Scholar]
- Shen, H. , Liu, X. , Zhang, P. , Wang, L. , Liu, Y. , Zhang, L. , Liang, P. , & Song, C. (2018). Genome characterization of a porcine circovirus type 3 in South China. Transboundary and Emerging Diseases, 65(1), 264–266. 10.1111/tbed.12639 [DOI] [PubMed] [Google Scholar]
- Sorden, S. (2000). Update on porcine circovirus and postweaning multisystemic wasting syndrome (PMWS). Swine Health Production, 8(3), 133–136 [Google Scholar]
- Stadejek, T. , Woźniak, A. , Miłek, D. , & Biernacka, K. (2017). First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transboundary and Emerging Diseases, 64(5), 1350–1353. 10.1111/tbed.12672 [DOI] [PubMed] [Google Scholar]
- Sun, J. , Wei, L. , Lu, Z. , Mi, S. , Bao, F. , Guo, H. , Tu, C. , Zhu, Y. , & Gong, W. (2018). Retrospective study of porcine circovirus 3 infection in China. Transboundary and Emerging Diseases, 65(3), 607–613. 10.1111/tbed.12853 [DOI] [PubMed] [Google Scholar]
- Sun, W. , Wang, W. , Xin, J. , Cao, L. , Zhuang, X. , Zhang, C. , Zhu, Y. , Zhang, H. , Qin, Y. , Du, Q. , Han, Z. , Lu, H. , Zheng, M. , & Jin, N. (2019). An epidemiological investigation of porcine circovirus 3 infection in dogs in the Guangxi Province from 2015 to 2017, China. Virus Research, 270, 197663. 10.1016/j.virusres.2019.197663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temeeyasen, G. , Lierman, S. , Arruda, B. L. , Main, R. , Vannucci, F. , & Gimenez‐, L. G . (2021). Pathogenicity and immune response against porcine circovirus type 3 infection in caesarean‐deprived pigs. Jounal of General Virology, 102(1). 10.1099/jgv.0.001502 [DOI] [PubMed] [Google Scholar]
- Tochetto, C. , Lima, D. A. , Varela, A. P. M. , Loiko, M. R. , Paim, W. P. , Scheffer, C. M. , Herpich, J. I. , Cerva, C. , Schmitd, C. , Cibulski, S. P. , Santos, A. C. , Mayer, F. Q. , & Roehe, P. M. (2018). Full‐genome sequence of porcine circovirus type 3 recovered from serum of sows with stillbirths in Brazil. Transboundary and Emerging Diseases, 65(1), 5–9. 10.1111/tbed.12735 [DOI] [PubMed] [Google Scholar]
- Tochetto, Caroline , de Lima, D. A. , Varela, A. P. M. , Ortiz, L. C. , Loiko, M. R. , Scheffer, C. M. , Paim, W. P. , Cibulski, S. P. , Cerva, C. , Herpich, J. , Schmidt, C. , Franco, A. C. , Mayer, F. Q. , & Roehe, P. M. (2020). Investigation on porcine circovirus type 3 in serum of farrowing sows with stillbirths. Microbial Pathogenesis, 149, 104316. 10.1016/j.micpath.2020.104316 [DOI] [PubMed] [Google Scholar]
- Tomás, A. , Fernandes, L. T. , Valero, O. , & Segalés, J. (2008). A meta‐analysis on experimental infections with porcine circovirus type 2 (PCV2). Veterinary Microbiology, 132(3–4), 260–273. 10.1016/j.vetmic.2008.05.023 [DOI] [PubMed] [Google Scholar]
- Vargas‐Bermúdez, D. S. , Vargas‐pinto, M. A. , Mogollón, J. D. , & Jaime, J. (2021). Field infection of a gilt and its litter demonstrates vertical transmission and effect on reproductive failure caused by porcine circovirus type 3 (PCV3). BMC Veterinary Research, 17(1), 150. 10.1186/s12917-021-02862-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Sun, W. , Cao, L. , Zheng, M. , Zhu, Y. , Li, W. , Liu, C. , Zhuang, X. , Xing, J. , Lu, H. , Luo, T. , & Jin, N. (2019). An epidemiological investigation of porcine circovirus 3 infection in cattle in Shandong province, China. BMC Veterinary Research, 15(1), 60. 10.1186/s12917-019-1793-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Noll, L. , Lu, N. , Porter, E. , Stoy, C. , Zheng, W. , Liu, X. , Peddireddi, L. , Niederwerder, M. , & Bai, J. (2019). Genetic diversity and prevalence of porcine circovirus type 3 (PCV3) and type 2 (PCV2) in the Midwest of the USA during 2016–2018. Transboundary and Emerging Diseases, 67(3), 1284–1294. 10.1111/tbed.13467 [DOI] [PubMed] [Google Scholar]
- Williamson, S. , Wilson, L. , Collins, R. , Floyd, T. , Dastjerdi, A. , Saporiti, V. , Sibila, M. , & Segalés, J. (2021). Stillbirths, arthrogryposis and preweaned nervous disease: Evidence for Porcine circovirus 3 (PCV‐ 3) involvement. In Proceedings of 12th European Symposium of Porcine Health Management. VVD‐PP‐56, 253.
- Woźniak, A. , Miłek, D. , Bąska, P. , & Stadejek, T. (2019). Does porcine circovirus type 3 (PCV3) interfere with porcine circovirus type 2 (PCV2) vaccine efficacy? Transboundary and Emerging Diseases, 66(4), 1454–1461. 10.1111/tbed.13221 [DOI] [PubMed] [Google Scholar]
- Xia, D. , Huang, L. , Xie, Y. , Zhang, X. , Wei, Y. , Liu, D. , Zhu, H. , Bian, H. , Feng, L. , & Liu, C. (2019). The prevalence and genetic diversity of porcine circovirus types 2 and 3 in Northeast China from 2015 to 2018. Archives of Virology, 164(10), 2435–2449. 10.1007/s00705-019-04336-4 [DOI] [PubMed] [Google Scholar]
- Ye, X. , Berg, M. , Fossum, C. , Wallgren, P. , & Blomström, A. L. (2018). Detection and genetic characterisation of porcine circovirus 3 from pigs in Sweden. Virus Genes, 54(3), 466–469. 10.1007/s11262-018-1553-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzhakov, A.G. , Raev, S.A. , Alekseevm K.P., Grebennikova, T.V. , Verkhovsky, O.A. , Zaberezhny, A.D. & Aliper, T.I. (2018) First detection and full genome sequence of porcine circovirus type 3 in Russia. Virus Genes, 54, 608–611. 10.1007/s11262-018-1582-z [DOI] [PubMed] [Google Scholar]
- Zhai, S. L. , Zhou, X. , Zhang, H. , Hause, B. M. , Lin, T. , Liu, R. , Chen, Q. L. , Wei, W. K. , Lv, D. H. , Wen, X. H. , Li, F. , & Wang, D. (2017). Comparative epidemiology of porcine circovirus type 3 in pigs with different clinical presentations. Virology Journal, 14(1), 222. 10.1186/s12985-017-0892-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. , Wang, D. , Jiang, Y. , Li, Z. , Zou, Y. , Li, M. , Yu, H. , Huang, K. , Yang, Y. , & Wang, N. (2019). Development and application of a baculovirus‐expressed capsid protein‐based indirect ELISA for detection of porcine circovirus 3 IgG antibodies. BMC Veterinary Research, 15(1), 79. 10.1186/s12917-019-1810-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D. , Wang, X. , Gao, Q. , Huan, C. , Wang, W. , Gao, S. , & Liu, X. (2018). Retrospective survey and phylogenetic analysis of porcine circovirus type 3 in Jiangsu province, China, 2008 to 2017. Archives of Virology, 163(9), 2531–2538. 10.1007/s00705-018-3870-2 [DOI] [PubMed] [Google Scholar]
- Zheng, S. , Shi, J. , Wu, X. , Peng, Z. , Xin, C. , Zhang, L. , Liu, Y. , Gao, M. , Xu, S. , Han, H. , Yu, J. , Sun, W. , Cong, X. , Li, J. , & Wang, J. (2018). Presence of Torque teno sus virus 1 and 2 in porcine circovirus 3‐positive pigs. Transboundary and Emerging Diseases, 65(2), 327–330. 10.1111/tbed.12792 [DOI] [PubMed] [Google Scholar]
- Zheng, S. , Wu, X. , Zhang, L. , Xin, C. , Liu, Y. , Shi, J. , Peng, Z. , Xu, S. , Fu, F. , Yu, J. , Sun, W. , Xu, S. , Li, J. , & Wang, J. (2017). The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transboundary and Emerging Diseases, 64(5), 1337–1341. 10.1111/tbed.12667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, Y. , Zhang, N. , Zhang, J. , Zhang, S. , Jiang, Y. , Wang, D. , Tan, Q. , Yang, Y. , & Wang, N. (2018). Molecular detection and sequence analysis of porcine circovirus type 3 in sow sera from farms with prolonged histories of reproductive problems in Hunan, China. Archives of Virology, 163(10), 2841–2847. 10.1007/s00705-018-3914-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data used in this review is available in the different journals referenced.


