Abstract
Aberrant lipid metabolism has recently been recognized as a new hallmark of malignancy, but the characteristics of fatty acid metabolism in breast cancer stem cells (BCSC) and potential interventions targeting this pathway remain to be addressed. Here, by using the in vitro BCSC models, mammosphere‐derived MCF‐7 cells and HMLE‐Twist‐ER cells, we found that the cells with stem cell‐like properties exhibited a very distinct profile of fatty acid metabolism compared with that of their parental cancer cells, characterized by increased lipogenesis, especially the activity of stearoyl‐CoA desaturase 1 (SCD1) responsible for the production of monounsaturated fatty acids, and augmented synthesis and utilization of the omega‐6 arachidonic acid (AA). Suppression of SCD1 activity by either enzyme inhibitors or small interfering RNA (siRNA) knockdown strikingly limited self‐renewal and growth of the BCSC, suggesting a key role for SCD1 in BCSC proliferation. Furthermore, elevated levels of SCD1 and other lipogenic enzymes were observed in human breast cancer tissues relative to the noncancer tissues from the same patients and correlated with the pathological grades. Interestingly, treatment of BCSC with omega‐3 fatty acids, eicosapentaenoic acid and docosahexaenoic acid, effectively downregulated the expression of the lipogenic enzymes and markedly suppressed BCSC self‐renewal and growth. Dietary supplementation of nude mice bearing BCSC‐derived tumors with omega‐3 fatty acids also significantly reduced their tumor load. These findings have demonstrated that increased lipogenesis is critical for self‐renewal and growth of BCSC, and that omega‐3 fatty acids are effective in targeting this pathway to exert their anticancer effect.
Keywords: breast cancer, cancer stem cells, growth inhibition, proliferation, self‐renewal
Breast cancer stem cells (BCSC) have a distinct fatty acid profile due to the aberrant expressions of lipogenic enzymes (eg, FAS, SCD1, and FADS1/2), which are highly involved in sustaining their self‐renewal, and proliferative capabilities. Omega‐3 polyunsaturated fatty acids can effectively suppress the self‐renewal and growth of BCSC by downregulation of the lipogenic enzymes, especially SCD1.
Significance statement.
Dysregulated lipid metabolism is proved to be associated with the growth of cancer stem cells (CSC); however, the potential interventions targeting this pathway remain to be elucidated. This study has demonstrated that breast CSC have a distinct fatty acid profile due to the aberrant expressions of lipogenic enzymes, which are highly involved in sustaining their self‐renewal and proliferative capabilities, and that omega‐3 polyunsaturated fatty acids (PUFA) can effectively suppress the self‐renewal and growth of breast CSC by downregulation of the lipogenic enzymes, especially SCD1. These findings provide a potential therapeutic application of omega‐3 PUFA by targeting fatty acid metabolism in breast CSC.
1. INTRODUCTION
Breast cancer is the most common type of cancer in women worldwide, which represents 25% of new cases and 15% of deaths from all cancers. 1 , 2 Surgical resection and a combination of radiotherapy and chemotherapy are now the primary treatments for breast cancer. Even through these available treatments improve survival for breast cancer patients, nearly 30% to 40% of patients diagnosed with early stage or locally invasive breast cancer eventually develops disseminated diseases, 3 indicating the poor prognosis in these advanced‐stage of breast cancers.
Cumulative evidence from breast cancer studies suggests that the underlying presence of a small, unique subpopulation of undifferentiated cells, also termed cancer stem cells (CSC) or tumor‐initiating cells, plays a key role in tumor progression, metastasis, acquired resistance to conventional therapies, and tumor recurrence. 4 , 5 , 6 These CSC are selectively capable of tumor initiation, self‐renewal, and giving rise to bulk populations of nontumorigenic cancer cell progeny through differentiation. 7 , 8 , 9 Thus, targeting molecular pathways against CSC self‐renewal and differentiation become essential in treating cancers. Nowadays, metabolic reprogramming such as dysregulated lipid metabolism is a newly recognized hallmark of malignancy 10 , 11 and elevated lipogenesis is found in a variety of cancers, which is believed to satisfy the demands of rapid tumor growth. 10 , 12 , 13 An emerging role of lipid metabolism in CSC has been reported recently. 14 , 15 It is proposed that these cells are reliant on the activity of enzymes involved in lipogenesis, such as fatty acids synthase (FAS), and several fatty acid desaturases, including SCD1 and fatty acid desaturase 1 and 2 (FADS1 and FADS2). 14 , 16 , 17 Particularly, an increased level of monounsaturated fatty acids (MUFA) as a result of a higher SCD1 activity has been shown to be important for maintaining cancer stemness and promoting cancer cell survival in some cancer types. 18 , 19 , 20 Owning to the metabolic flexibility of CSC, it is difficult to eliminate these cells by targeting a single lipid metabolic pathway. Therefore, understanding the intrinsic cellular fatty acid metabolic properties of breast CSC and targeting lipid metabolism pathways to eliminate CSC population hold the promise of developing more effective treatments for breast cancer patients.
Omega‐3 polyunsaturated fatty acids (n‐3 PUFA), including alpha‐linolenic acid (C18:3), eicosapentaenoic acid (EPA) (C20:5), and docosahexaenoic acid (DHA) (C22:6), are increasingly being recognized as important modulators of multiple biological pathways that affect health and disease. 21 , 22 , 23 , 24 , 25 , 26 A growing number of studies have shown that n‐3 PUFA and their lipid derivatives exhibit remarkable anticancer effects in various types of cancers, including breast cancer. 27 , 28 , 29 , 30 , 31 The anticancer effects of n‐3 PUFA involve a variety of distinct mechanisms, including the suppression of nuclear factor‐κB, activation of AMPK/SIRT1, inhibition of cyclooxygenase‐2 (COX2) activity, and upregulation of novel anti‐inflammatory lipid mediators. 26 , 32 , 33 , 34 , 35 Moreover, it has been demonstrated that n‐3 PUFA can improve the efficacy and tolerability of chemotherapy. 36 , 37 , 38 Even though many lines of evidence have shown that n‐3 PUFA possess anticancer properties, whether n‐3 PUFA have a suppressive effect on cancer stem cells remains to be addressed.
It has been well documented that n‐3 PUFA are an effective inhibitor of SCD1 and can markedly suppress lipogenesis through downregulating the expression of lipogenic enzymes. 39 , 40 , 41 , 42 In this context, we hypothesized that n‐3 PUFA might be highly effective in suppressing the self‐renewal and growth of breast CSC due to their potent inhibitory effect on lipogenesis and lipid metabolism, which are intrinsically enhanced in CSC. To test this hypothesis, we first characterized the fatty acid profile and the activity of lipid metabolism of breast CSC (mammosphere‐derived MCF‐7 cells and HMLE‐Twist‐ER cells) and then examined whether n‐3 PUFA could alter the phenotype and tumorigenicity of breast CSC. Our results demonstrated that breast CSC displayed enhanced lipogenesis while n‐3 PUFA significantly inhibited the expression of several key lipid metabolic enzymes and thereby suppressed the proliferation and tumorigenicity of breast CSC. This study not only uncovers a new mechanism for the anticancer effect of n‐3 PUFA by modulating CSC lipid metabolism, but also provides new insight into the potential utility of n‐3 PUFA as a therapeutic agent for patients with breast cancer.
2. MATERIALS AND METHODS
All chemicals, unless otherwise stated, were purchased from Sigma‐Aldrich.
2.1. Cell culture
Human mammary epithelial cells, HMLE‐Twist‐ER cells, were generously provided by Dr Robert A. Weinberg (Whitehead Institute for Biomedical Research, Massachusetts Institute of Technology, Cambridge, Massachusetts) and cultured in serum‐free mammary epithelial cell growth medium (CC‐3150; Lonza) at 37°C under 5% CO2. For epithelial‐mesenchymal transition (EMT) induction, HLME‐Twist‐ER cells were exposed to 4‐hydroxytamoxifen at 20 nM for 12 days as previously described. 43 MCF‐7 cells were purchased from ATCC (Manassas, Virginia) and cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Gibco, Invitrogen, Grand Island, New York) and penicillin/streptomycin (100 U/mL). To determine the effect of exogenous PUFA on cancer stem cell growth, cells were treated with EPA, DHA, or arachidonic acid (AA) at a final concentration of 20 μM and ethanol was used as a vehicle for control cells. Forty‐eight hours later, the cells were harvested for further analysis.
2.2. Human sample collection
Eighty breast cancer formalin‐fixed paraffin‐embedded (FFPE) tissues and 40 tumor‐adjacent normal FFPE tissues were obtained during surgery for immunohistochemistry analysis. The clinicopathological features of the patients were summarized in Table S4. All tissues were collected from the Clinical Biobank of Collaborative Innovation Center for Medical Molecular Diagnostics of Guangdong Province, The Affiliated Jiangmen Hospital of Sun Yat‐sen University (Guangdong, China) between January 2015 and December 2017. Patients were diagnosed based on clinical and pathological evidence, and the proportions of tumor vs nontumor in H&E‐stained tissue samples were evaluated by two independent professional pathologists. The use of these clinical materials for research was obtained the consent of patients and human tissues used for this experiment was approved by of the Institutional Research Ethics Committee.
2.3. Mammosphere‐formation assay
Breast CSC can be enriched in vitro by expanding multicellular spheroids (mammospheres [MS]) from breast cancer surgical specimens or cell lines. 44 Briefly, MCF‐7 cells (2.5 × 104 cells/mL) were seeded at in ultralow adherence six‐well plates in serum‐free DMEM/F12 media supplemented with 5 μg/mL bovine insulin, 20 ng/mL recombinant epidermal growth factor, 20 ng/mL basic fibroblast growth factor (Gibco), B27 supplement (Invitrogen), 0.5 μg/mL cortisone, and 1% penicillin/streptomycin (Gibco). Mammospheres were collected by gentle centrifugation (800 rpm) after 7 to 10 days, dissociated enzymatically into single cell, and then used for the following indicated treatments.
2.4. Small‐molecule inhibitor treatment and siRNA transfection
FAS inhibitor (C75), SCD1 inhibitor (CAY‐10566), and D6D inhibitor (SC‐26196) were purchased from Cayman. Compounds were dissolved in DMSO and stored at −20°C. For the treatment with small‐molecule inhibitors, cells were exposed to different agents at a final concentration of 20 μM for 72 hours and cells were collected for further analysis. For siRNA transfection, Silencer Pre‐Designed siRNA for SCD1 (AM16708), Silencer Negative Control No. 1 siRNA (AM4611), and Lipofectamine RNAiMAX transfection reagent were purchased form Life Technologies. The SCD1 siRNA sequence was shown as follow, sense: 5′‐GCA AGU AGA UUG UCU CGA GTT‐3′; antisense: 5′‐CUC GAG ACA AUC UAC UUG CTC‐3′. The transfection procedures were performed following the manufacturer instructions. After transfection for 72 hours, cells were collected and used for further analysis. The values were presented as means ± SEM from one representative experiment performed in triplicate and similar outcomes were obtained at least three times.
2.5. CellTiter‐Blue cell viability assay
Celltiter‐Blue assay provides a homogeneous, fluorescent method for monitoring cell viability, which is based on the ability of living cells to convert a redox dye into a fluorescent end‐product. In this study, we used Celltiter‐Blue to assess the cell viability in MCF‐7 mammospheres and HMLE‐Twist‐ER cells with the indicated treatments according to the manufacturer protocol. Briefly, cells were incubated with 4:1 of cell culture medium and CellTiter‐Blue reagent, the plates were shaken for 10 seconds, and then, incubated in dark under standard cell culture conditions for 2 hours. Prior to reading the fluorescence intensity with excitation and emission wavelengths of 560 and 590 nm, the plate was shaken again for 10 seconds.
2.6. Fatty acid profile analysis
Fatty acid profiles of cells were analyzed by gas chromatography (GC) as described previously. 45 Briefly, 5 × 105 cells were subjected to fatty acid methylation by mixing with 1.5 mL of hexane and 1.5 mL of 14% boron trifluoride/methanol at 100°C for 1 hour. Fatty acid methyl esters were analyzed using a fully automated 6890N network GC system equipped with a flame‐ionization detector (Agilent Technologies, Palo Alto, California). The individual fatty acid peak was identified by their relative retention times compared to the reference standard GLC461 (Nu‐Chek Prep, Elysian, Minnesota), and the area percentage of each peak was analyzed using GC Chemstation software.
2.7. Flow cytometry analysis
Cells were detached using trypsin/EDTA and the cell number was counted by trypan blue. A single cell suspension was washed with cold FACS staining buffer (2% FCS + 1% sodium azide) and centrifuged with 1000 rpm for 5 minutes at 4°C. A total of 106 cells were resuspended in 100 μL of FACS staining buffer containing PE‐Cy5 CD44 and primary CD24 antibodies and incubated for 60 minutes in the dark at 4°C. After washing, cells were incubated with the secondary anti‐human FITC‐conjugated antibody for 30 minutes on ice. 105 cells were used for cell sorting by LSR II flow cytometer (BD Biosciences) and the results were analyzed using FlowJo_V10 software.
2.8. Gene expression measurement
Total RNAs were extracted from cells using TRIzol reagent (Invitrogen Life Technologies, Grand Island, New York), cDNAs were synthesized from 1.5 μg of RNA using iScrip Reverse Transcription Supermix (Bio‐Rad). qPCR was performed using cDNA in an Mx3000P qPCR system (Stratagene, Agilent Technologies, Santa Clara, California) with the specific primers and iTaq Universal SYBR Green Supermix (Stratagene). GAPDH was used as internal standard control and the relative abundance of expression was calculated by the 2−ΔΔCt method. All the procedures were followed by the manufacturer's instructions and the primer sequences were listed in Table S6.
2.9. Immunoblot analysis
Protein extracts of cells were prepared using RIPA buffer containing protease inhibitor cocktail, and protein concentrations were measured using a BCA kit (Pierce). One hundred micrograms of protein was resolved by electrophoresis in 8% SDS‐PAGE gel and blotted to PVDF membranes (Invitrogen). Membranes were incubated with indicated primary antibodies overnight at 4°C and then with an anti‐rabbit HRP‐conjugated secondary antibody (1:3000; Santa Cruz Biotechnology) for 2 hours at room temperature. The primary antibodies used in this study were rabbit anti‐FAS (1:1000; Cell signaling), rabbit anti‐SCD1 (1:500; Cell signaling), rabbit anti‐FADS2 (1:1000; Abcam), rabbit anti‐COX2 (1:1000; Abcam), and anti‐β‐actin (1:2000; Santa Cruz Biotechnology) as a loading control. HRP activity was measured using Supersignal West Pico Chemiluminescent Substrate kit (Thermo Fisher Scientific).
2.10. Immunohistochemistry staining
The total of 120 FFPE samples were subjected to immunohistochemical analysis for lipid metabolism‐related proteins, following the standard protocol as described previously. 46 Briefly, the slides were incubated overnight at 4°C in a humidified chamber with anti‐SCD1 antibody (1:400; proteintech), anti‐FAS antibody (1:400; proteintech), and anti‐FADS1 (D5D) antibody (1:400; proteintech), respectively. Scores given by two independent investigators were averaged for further comparative evaluation of protein expression mentioned above. The tumor cell proportion was scored as follows: 0 (no positive tumor cells); 1 (<10% positive tumor cells); 2 (10%‐35% positive tumor cells); 3 (35%‐70% positive tumor cells); and 4 (>70% positive tumor cells). The staining intensity was graded as follows: 0 (no staining); 1 (weak staining, light yellow); 2 (moderate staining, yellow brown); and 3 (strong staining, brown). The staining index (SI) was calculated as the product of the staining intensity score and the proportion of positive tumor cells. Based on this assessment, the expression of each lipid metabolism‐related proteins in breast carcinoma samples was evaluated by the SI, with scores of 0, 1, 2, 3, 4, 6, 8, 9, or 12.
2.11. Animal study
All animal procedures were performed in compliance with the guidelines approved by Massachusetts General Hospital (MGH) (IACUC). Male athymic nude (J:NU) mice purchased from Jackson Laboratory (Bar Harbor, Maine) were housed in SPF area in animal facility, MGH and were fed irradiated chow diet and autoclaved DI water ad libitum. The mice were randomly assigned to two groups, Control (n = 3) and n‐3 PUFA (n = 4), receiving 100 μL of corn oil (control) and omega‐3 oil (containing 90% of EPA + DHA) through oral gavage, respectively. Total duration of oil intervention included 30 days before and 45 days after tumor implantation. Prior to xenograft induction, MCF‐7‐CSC were grown in vitro in the presence of mammosphere‐formation medium described in method of cell culture. A cell suspension of 1.5 × 107 MCF‐7‐CSC in 150 μL of PBS was subcutaneously injected into the lower right flank. Following the xenograft induction, the mice were monitored for tumor growth and tumor size was measured using digital caliper in longest and shortest dimension. Formula = (longest length × shorted length2)/2 was applied to calculate the tumor size.
2.12. Statistical analysis
Except human results plotted as minimum to maximum, all data were presented as means ± SEM. Statistical analysis was carried out using GraphPad Prism 6.0 (GraphPad software Inc, La Jolla, California). Statistical differences between two groups were evaluated using Student's t test. One‐way ANOVA with Tukey test was used to determine statistical differences between multiple groups. The chi‐square test was used to analyze the relationship between lipid metabolism‐related gene and clinicopathological characteristics. P < .05 was considered statistically significant.
3. RESULTS
3.1. Characteristics of lipid metabolism in breast CSC
First, we established breast CSC‐like cells modeled by mammosphere‐derived MCF‐7 cells (referred to as MCF‐7‐CSC), which were generated from MCF‐7 cell line under conditioned medium culture, and HMLE‐Twist‐ER cells (referred to as EMT‐CSC), which were derived from EMT by exposing to an ER ligand, 4‐hydroxytamoxifen. 43 Both MCF‐7‐CSC and EMT‐CSC were then confirmed to have typical phenotypes of breast CSC, as shown by a remarkable increase in the expression of stem cell‐markers (such as CD44, Sox2, Nanog, BMI, and Oct4), the mesenchymal markers (eg, FN‐1 and N‐Cad), and the proportion of cells with CD44high/CD24low by flow cytometry analysis (Figure S1A,B,E‐H). In addition, both MCF‐7‐CSC and EMT‐CSC exhibited higher viability and proliferation rate than their parental cell lines (MCF‐7 and HMLE) (Figure S1C,D), indicating another property of CSC‐like cells.
Next, we analyzed the fatty acid profiles of MCF‐7‐CSC, EMT‐CSC, and their parental cell lines by GC to identify potential changes of lipid metabolism in the CSC‐like cells. We found a remarkable difference in cellular fatty acid composition between the CSC‐like cells and their parent cells (Table S1). Relative to their parental cell lines, both MCF‐7‐CSC and EMT‐CSC exhibited higher levels of the MUFA, palmitoleic acid (C16:1n‐7) and oleic acid (C18:1n‐9), and lower levels of the saturated fatty acids (SFA), palmitic acid (C16:0) and stearic acid (C18:0) (Figure 1C,D), leading to significantly higher ratios of C16:1n‐7/C16:0 (MCF‐7: 0.14 vs MCF‐7‐CSC: 0.43; HLME: 0.80 vs EMT‐CSC: 1.28) and C18:1n‐9/C18:0 (MCF‐7: 1.09 vs MCF‐7‐CSC: 1.66; HLME: 2.73 vs EMT‐CSC: 4.12), which reflect an increased conversion (desaturation) of SFA to MUFA by SCD1 in both MCF‐7‐CSC and EMT‐CSC. Another notable difference between the CSC‐like cells and their parental cells was an increase in the level of AA (C20:4n‐6) with a decrease in the level of linoleic acid (C18:2n‐6), indicating an enhanced synthesis of C20:4n‐6 (catalyzed by the key desaturases, FADS1 and FADS2) in both MCF‐7‐CSC and EMT‐CSC compared with their parental cell lines (Figure 1C,D).
FIGURE 1.
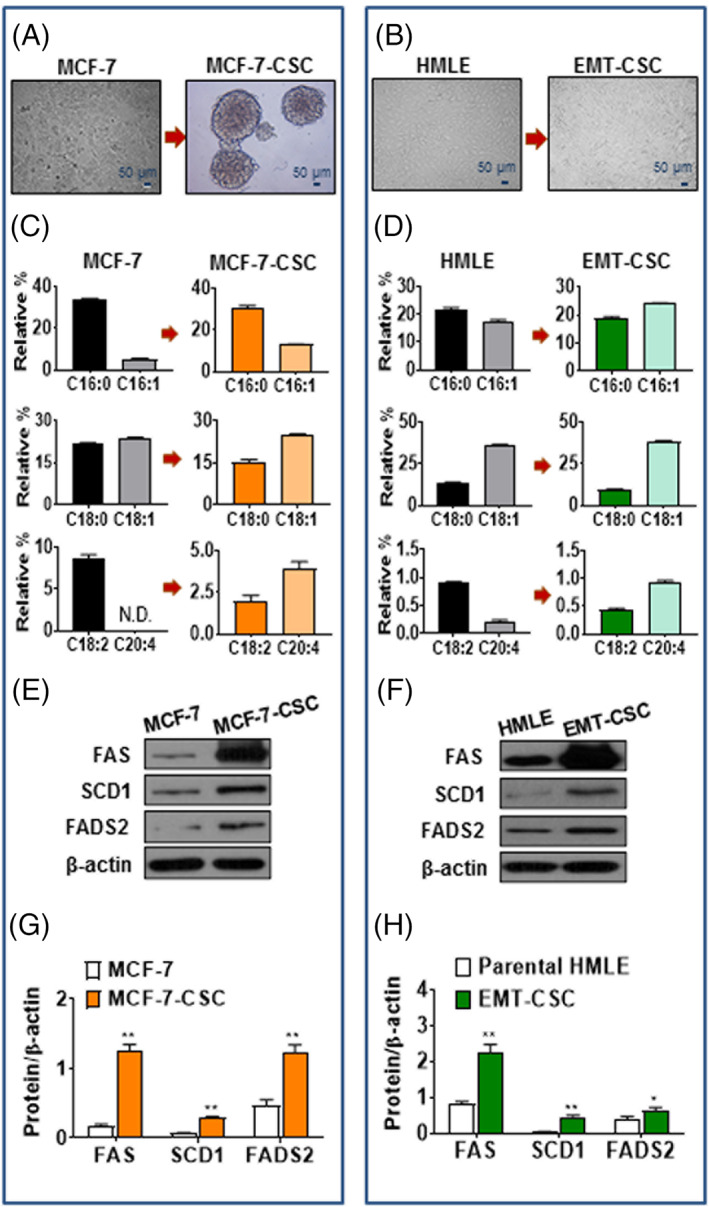
Characteristics of lipid metabolism in breast cancer stem cells. A,B, Representative images of mammosphere cells (MCF‐7‐CSC) derived from MCF‐7 cell line (A) and EMT‐CSC induced from HMLE‐Twist‐ER cells (B). C,D, Comparison of the profiles of SFA and MUFA (top and middle), and of C18:2n‐6 and C20:4n‐6 PUFA (bottom) in MCF‐7‐CSC (C) and EMT‐CSC (D) compared with their parental cells, respectively. E,F, Representative Western blot for protein expressions of FAS, SCD1, and FADS2 in MCF‐7‐CSC (E) and EMT‐CSC (F) compared with their parental cells, respectively. G,H, Relative densiometric bar graphs of FAS, SCD1, and FADS2 in MCF‐7‐CSC (G) and EMT‐CSC (H) compared with their parental cells, respectively. *P < .05; **P < .01 vs parental cells. All the experiments were repeated at least three times. CSC, cancer stem cells; EMT, epithelial‐mesenchymal transition; MUFA, monounsaturated fatty acids; PUFA, polyunsaturated fatty acids; SFA, saturated fatty acids
In the context of the observed fatty acid profiles, we speculated that the CSC‐like cells might upregulate the genes responsible for fatty acid synthesis. To verify the speculation, we examined the expression of the key genes involved in fatty acid metabolism in MCF‐7‐CSC, EMT‐CSC, and their parental cell lines. As shown in Figure 1, the protein levels of three key lipogenic enzymes (FAS, SCD1, and FADS2) were significantly elevated in both MCF‐7‐CSC and EMT‐CSC relative to their parental cell lines (MCF‐7 and HMLE). The gene expression data are well consistent with the fatty acid profiles, suggesting an upregulation of fatty acid synthesis in breast CSC (Figure S1I,J).
3.2. Role of lipogenic enzymes in self‐renewal and growth of breast CSC
To evaluate the role of the lipogenic enzymes (FAS, SCD1, and FADS2) in the proliferation and tumorigenicity of breast CSC, we examined the effect of suppressing the activity of these enzymes, by using either enzyme inhibitors or siRNA knockdown of the gene expression, on the growth of MCF‐7‐CSC and EMT‐CSC. As shown in Figure 2A‐D, treatment of the CSC‐like cells with specific inhibitors of SCD1 (CAY‐10566), FAS (C75), and FADS2 (SC‐26196) dramatically reduced the number and size of MCF‐7‐CSC mammospheres and the viability of EMT‐CSC. Notably, the most significant effect on cell growth and fatty acid profile was observed by the inhibitor of SCD1 (CAY‐10566), suggesting that SCD1 is a more critical factor for breast CSC to maintain the proliferative phenotype. Next, we used SCD1 siRNA to knock down the expression of SCD1 gene in MCF‐7‐CSC and EMT‐CSC, as demonstrated by a marked reduction of the enzyme protein (Figure 2E,F) and functional activity (C16:1/C16:0 and C18:1/C18:0) of SCD1 in the cells (Figure S2C,D; Table S2). The knockdown of SCD1 dramatically inhibited mammosphere formation of MCF‐7‐CSC and reduced the viability of EMT‐CSC (Figure 2G,H). These results indicate that the lipogenic enzymes, especially SCD1, play a key role in governing the self‐renewal and growth of breast CSC.
FIGURE 2.
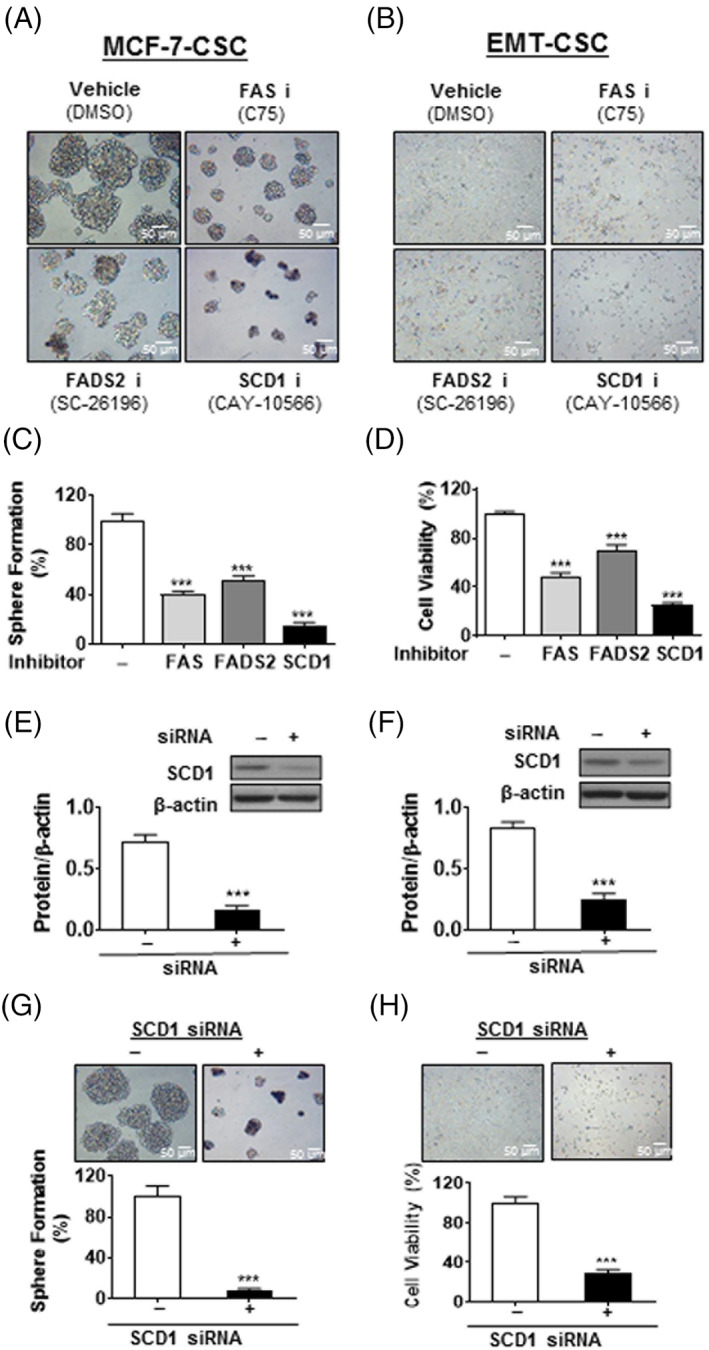
Increased lipogenesis is critical for self‐renewal and growth of breast cancer stem cells. A,B, Representative cell morphology of MCF‐7‐CSC (A) and EMT‐CSC (B) were shown the inhibition of cell growth following the indicated treatments for 48 hours. C, Quantitation of sphere formation in (A). D, Quantitation of cell viability in (B). E,F, Representative image of immunoblotting and relative densiometric bar graphs of SCD1 expression in MCF‐7‐CSC (E) and EMT‐CSC (F). G,H, Decreased cell viability was observed in MCF‐7‐CSC (G) and EMT‐CSC (H) after the treatment of SCD1‐specific siRNA. ***P < .001 vs vehicle‐treated control cells. All the experiments were repeated at least three times. CSC, cancer stem cells; EMT, epithelial‐mesenchymal transition; siRNA, small interfering RNA
3.3. Increased expression of the lipogenic enzymes in human breast cancer tissues
To follow up the in vitro observations, we further investigated whether there was any correlation between the expression of these lipogenic enzymes (FAS, SCD1, and FADS) and cancer pathological status in clinical breast cancer biospecimens. Immunohistological staining of the enzymes in 80 breast cancer tissue samples and 40 tumor‐adjacent normal tissue samples showed that the protein levels of FAS, SCD1, and FADS1 were markedly higher in tumor tissues relative to the corresponding tumor‐adjacent normal tissues (Figure 3A,C,E). Statistical analysis revealed a positive correlation between the protein level of each enzyme and pathological grades. That was, more advanced histological grades of breast cancer exhibited higher protein levels of FAS, SCD1, and FADS1 (Figure 3B,D,F). These results are consistent with the in vitro data and further suggest that these lipogenic enzymes (FAS, SCD1, and FADS1/2) may be critical for the growth of CSC and promote the progression of breast tumors.
FIGURE 3.
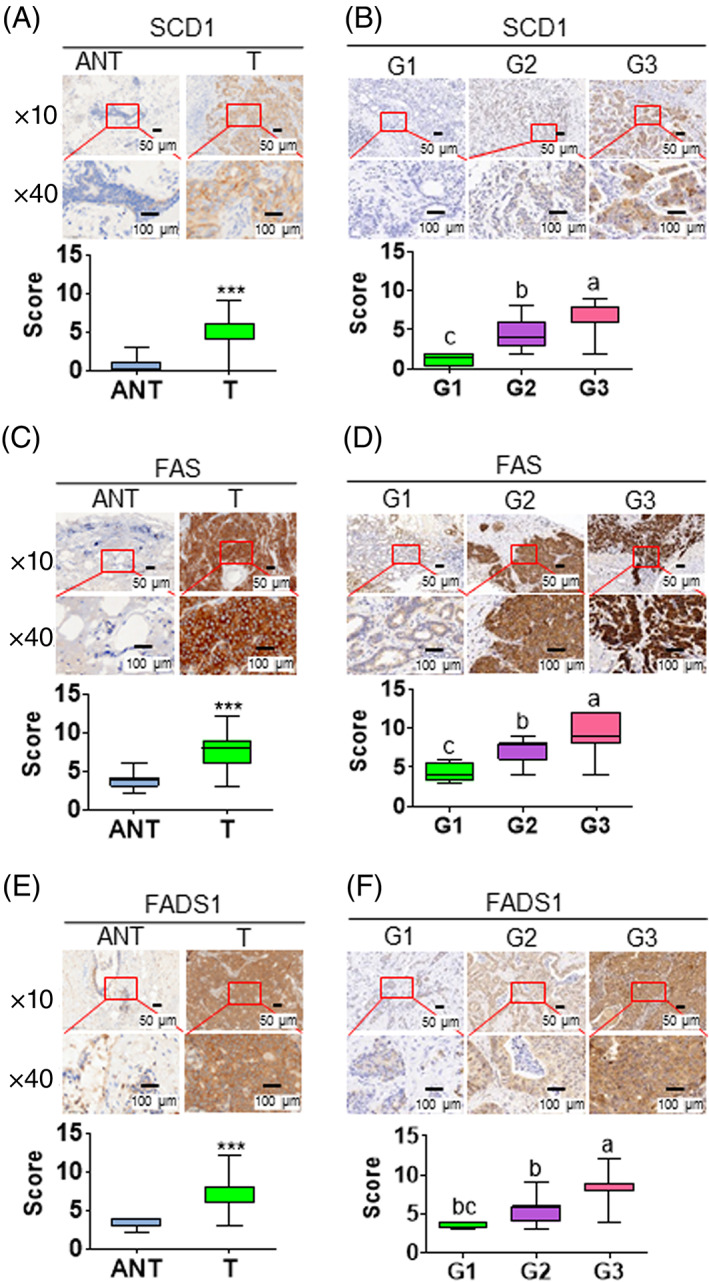
Increased expression of the lipogenic enzymes in human breast cancer tissues. A,C,E, Representative image of IHC analysis for SCD1 (A), FAS (C), and FADS1 (E) expressions in normal and tumor tissues. ANT, tumor‐adjacent normal tissue; T, tumor. B,D,F, Representative image of immunohistochemistry (IHC) analysis for SCD1 (B), FAS (D), and FADS1 (F) expressions between various clinical and tumor grades (G1‐3). ***P < .001 vs vehicle‐treated control cells. Alphabet letters were used to show the significant difference between groups. Each subfigure contains representative image (top) and quantitative results (bottom). The quantitative results were plotted by the values of minimum to maximum
3.4. Effect of omega‐3 fatty acids on the growth of breast CSC in vitro and in vivo
Given the known inhibitory effect of n‐3 PUFA on lipogenesis, we examined if n‐3 PUFA are effective in downregulating the expression of the lipogenic enzymes in breast CSC and suppressing their growth and tumorigenicity. Treatment of MCF‐7‐CSC and EMT‐CSC with n‐3 PUFA (EPA and DHA) and n‐6 PUFA (AA) at a final concentration of 20 μM for 48 hours showed that EPA and DHA, but not AA, dramatically inhibited the mammosphere formation of MCF‐7‐CSC (Figure 4A,C) and reduced the viability of EMT‐CSC (Figure 4B,D). Notably, the n‐3 PUFA, especially EPA, significantly suppressed both protein and mRNA expression of SCD1 and FAS in both MCF‐7‐CSC (Figures 4E,G and S3A) and EMT‐CSC (Figure 4F,H and S3B). The most profound effect was found on SCD1, while the effect on FADS2 were moderate. AA had much less or no effect on the expression of these enzymes (Figure 4E‐H and S3). These results are consistent with the functional activity of SCD1 (the ratios of C16:1/C16:0 and C18:1/C18:0; Table S3) and further demonstrate that n‐3 PUFA can effectively suppress the upregulation of the lipogenic enzymes and growth of breast CSC.
FIGURE 4.
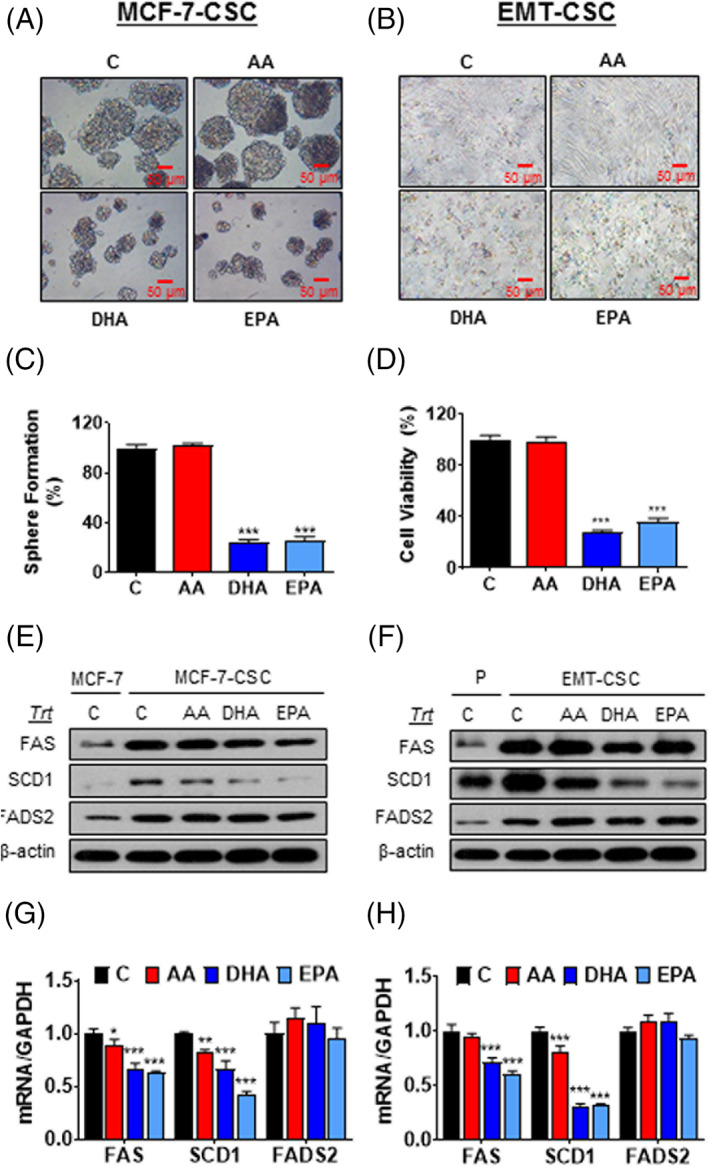
n‐3 PUFA suppress the growth of breast cancer stem cells by blocking lipid metabolism in vitro. A,B, Representative cell morphology of MCF‐7‐CSC (A) and EMT‐CSC (B) treated with 20 μM of indicated fatty acids for 48 hours. C, Quantitation of sphere formation in (A). D, Quantitation of cell viability in (B). E,F, Representative Western blot for protein expressions of the lipogenic enzymes in MCF‐7‐CSC (E) and EMT‐CSC (F) treated with indicated fatty acids. G,H, mRNA expressions of the lipogenic enzymes in MCF‐7‐CSC (G) and EMT‐CSC (H) after the treatment of indicated fatty acids. *P < .05; **P < .01; ***P < .001 vs either vehicle‐treated control cells or their corresponding parental cells. All the experiments were repeated at least three times. CSC, cancer stem cells; EMT, epithelial‐mesenchymal transition; PUFA, polyunsaturated fatty acids
We next extended our investigation to mouse xenograft model using MCF‐7‐CSC heterotopically implanted in nude mice. As shown in Figure 5A‐C, supplementation of MCF‐7‐CSC‐implanted mice with n‐3 PUFA (EPA + DHA) remarkably decreased tumor growth. To evaluate the association between tumor fatty acid profile and indices of tumor growth, we analyzed fatty acid composition of the tumor tissues from the mice with and without n‐3 PUFA supplementation. As shown in Figure 5D, tumor tissues from n‐3 PUFA‐supplemented mice had significantly higher levels of n‐3 PUFA, including EPA, DPA, and DHA, with a nearly sixfold reduction in tumor n‐6/n‐3 PUFA ratio compared to those from control animals. Furthermore, we found that both mRNA and protein levels of SCD1, FADS2, and COX2 were significantly suppressed in the tumor tissues of n‐3 PUFA‐supplemented group compared with those from control group. (Figure 5G,H). Accordingly, the enzymatic activities of SCD1 and FADS2, reflected by the fatty acid profiles (C16:1 + C18:1)/(C16:0 + C18:0) ratio and C20:4n‐6/C18:2n‐6, respectively, were markedly decreased in n‐3 PUFA‐supplemented group relative to control group. (Figure 5E,F; Table S5). Taken together, these results indicate that n‐3 PUFA can exert an inhibitory effect on the growth and tumorigenicity of breast CSC through downregulating lipogenic enzymes, particularly SCD1.
FIGURE 5.
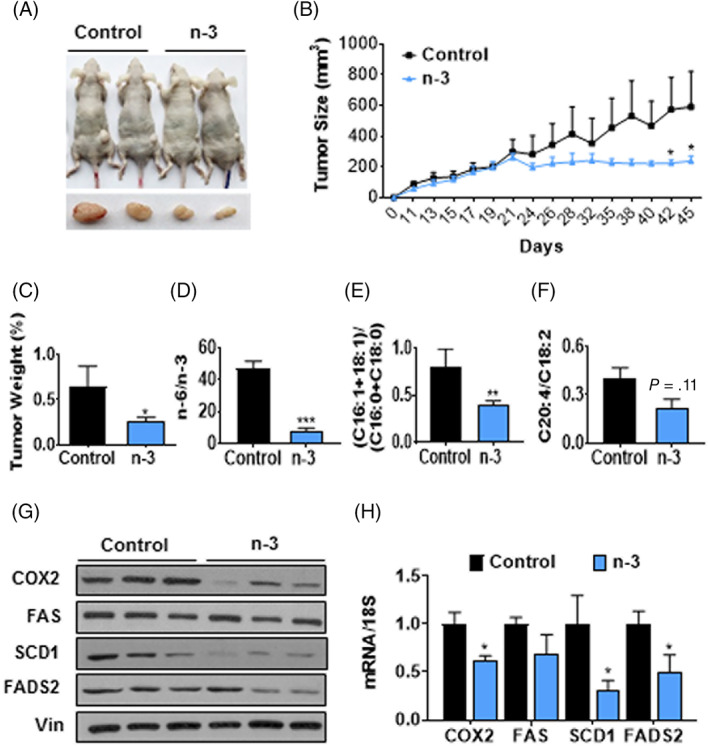
n‐3 PUFA inhibit tumor growth of breast cancer stem cells in xenograft mouse model. A, Representative image of tumor‐bearing mice. Prior to implanting 1.5 × 107 of MCF‐7‐CSC to nude mice, these mice were orally gavaged by either fish oil (n‐3 PUFA) or corn oil (control) for a month and continued giving oils until the end of the experiment. B, Tumor growth of the nude mice in each group. C, Relative tumor weight by body weight. D, The ratio of n‐6/n‐3 PUFA. E, The ratio of MUFA (C16:1 + C18:1)/SFA (C16:0 + C18:0). F, The ratio of C20:4/C18:2. G,H, Protein levels (G) and mRNA levels (H) of SCD1, FAS, FADS2, and COX2 between control and n‐3 PUFA groups. n = 3 to 4. *P < .05; **P < .01; ***P < .001 vs control. MUFA, monounsaturated fatty acids; PUFA, polyunsaturated fatty acids; SFA, saturated fatty acids
4. DISCUSSION
Breast cancer is the most common cancer and ranks the second among all cancer‐related deaths in women. The main cause of death in breast cancer patients is metastasis development. To date, the prevailing model of metastasis suggests that a small subpopulation of cancer cells acquires CSC traits characterized by a mesenchymal cell phenotype with enhanced migratory and metastatic capabilities. 47 , 48 A growing body of studies has shown that the EMT process is essential for distant metastases of epithelial tumor cells including breast cancer cells and also facilitates the generation of CSC. 49 , 50 Thus, therapeutic approaches targeting CSC hold a great promise for more efficient cancer treatments than conventional therapies aiming at killing the actively proliferating bulk tumor cells. Prior to developing new CSC‐targeted therapies, it is crucial to improve our understanding of CSC's properties and the underlying molecular mechanisms.
Metabolic reprogramming has emerged as one of the hallmarks in cancers, which establishes tumor favorable microenvironment such as Warburg effect (enhanced glycolysis/aerobic glycolysis) and lipid dysregulation (aberrant lipogenesis) to support their rapid growth and proliferation. 51 , 52 Recently, accumulating evidence has indicated that the plastic metabolic state of CSC not only governs their tumor‐initiating ability but also contributes to their resistance to conventional therapies and tumor recurrence. 53 , 54 , 55 Therefore, targeting aberrant cancer metabolism may constitute a basis for developing new therapeutic strategies to eliminate CSC. So far, many efforts have been devoted to glucose metabolism‐targeted therapies, particularly the inhibition of glycolysis in cancer cells; however, the overall outcomes remain unsatisfactory. 56 In addition to glucose metabolism, lipid biosynthesis (lipogenesis) has recently been recognized to play an important role in CSC self‐renewal and survival. In this context, targeting endogenous lipogenesis may impact the CSC cellular state in the heterogeneous cancer cell populations. However, little is known about whether there is any difference in lipid metabolism between CSC and non‐CSC.
In the present study, we characterized the fatty acid profile of two well‐established breast CSC‐like cell lines (MCF‐7‐CSC and EMT‐CSC) and compared it with that of their parental non‐CSC cell lines. Our results have identified two major characteristics in lipid profile of the breast CSC: (a) higher levels of C16:1n‐7 and C18:1n‐9 with higher ratios of C16:1n‐7/C16:0 and C18:1n‐9/ C18:0, reflecting an increased synthesis of MUFA from SFA catalyzed by SCD1; and (b) increased level of C20:4n‐6 with a higher ratio of C20:4n‐6/C18:2n‐6, suggesting an enhanced synthesis of C20:4n‐6 from C18:2n‐6 catalyzed by the key desaturases, FADS1 and FADS2 (a rate‐limiting enzyme of the pathway). Our analysis of gene expression verified that SCD1 and FADS2 were indeed upregulated in the breast CSC‐like cells (MCF‐7‐CSC and EMT‐CSC), consistent with the observation that these cells exhibited higher levels of MUFA and C20:4n‐6. Examination of human breast cancer tissues with antibodies against FAS, SCD1, FADS1 also revealed higher levels of these enzyme proteins in cancer tissues relative to noncancer tissues and a positive correlation between the enzyme level and tumor pathological grade. All these findings together support the notion that breast CSC possess aberrant lipid metabolism characterized by increased lipogenesis.
The next question that our study has addressed is about the importance of the aberrant lipid metabolism in the self‐renewal and growth of breast CSC. Our experiments using both enzyme inhibitors and siRNA knockdown approaches have demonstrated that suppression of the activity of the lipogenic enzymes, particularly SCD1, dramatically reduce the mammosphere formation of MCF‐7‐CSC and the viability of EMT‐CSC, suggesting that the aberrant lipid metabolism (increased lipogenesis) is critical for breast CSC to sustains their self‐renewal and growth.
Elucidation of the metabolic characteristics of cancer cells, especially CSC, is extremely important and helpful for identifying cancer biomarkers, discovering new drug targets, and developing effective solutions for cancer prevention and treatment. The findings of the present study provide new insight into the aberrant lipid metabolism in breast CSC and highlight the importance of increased production of certain fatty acids that serve as building blocks and support the tumor microenvironment to meet proliferation and metastasis requirements. Particularly, the enhanced synthesis of MUFA by the upregulation of the lipogenic enzyme, SCD1, seems to be a key mechanism for CSC to produce building materials (ie, components of cell membrane) to meet their need for rapid self‐renewal and growth. 57 Suppression of SCD1 is also shown to inhibit the growth of colon cancer stem cells via Notch/Wnt signaling pathways 58 and enhances the efficacy of chemotherapy for treating lung cancer stem cells through autophagy as well as ER stress. 59 This notion is also supported by other studies showing that SCD1 is overexpressed in various types of malignant cells and that selective inhibition of this enzyme exerts anticancer effects. 60 , 61 Furthermore, the augmented production of the omega‐6 PUFA, AA (C20:4n‐6), through the upregulation of FADS2, a rate‐limiting enzyme of the pathway, may be another critical mechanism for breast CSC to generate bioactive lipid mediators that enable their aggressive tumorigenicity. It is well established that AA (C20:4n‐6) and its eicosanoid metabolites (eg, PGE2, LTB4, TXA2, HETEs, etc) generated through cyclooxygenases (COX), lipooxygenases (LOX), and cytochrome P450 (CYP 450) metabolic pathways have potent pro‐inflammatory, pro‐angiogenic, and pro‐proliferative effects, and thereby promote tumor development. 26 Our previous observations that inhibiting FADS2 activity dramatically reduced tumor growth in mice 17 and that an increase in both FADS2 activity and AA synthesis was found in human breast cancer tissues relative to noncancer tissues 62 support a key role played by the augmented AA synthesis in promoting the tumorigenicity of CSC. Although further research is needed to elucidate the details of the SCD1 and FADS2 pathways in breast CSC, the findings of the present study suggest that increased activities of both SCD1 and FADS2 may be a hallmark of breast CSC.
Another important discovery of the present study is that omega‐3 fatty acids (EPA and DHA) are highly effective in suppressing the expression and/or activity of the lipogenic enzymes (SCD1, FAS, FADS2) in breast CSC and reducing the cell growth in vitro and the tumorigenicity in vivo. Thus, this study has uncovered a new mechanism for the anticancer effect of omega‐3 fatty acids by downregulating lipogenesis in CSC. It has been well documented that omega‐3 fatty acids possess anti‐inflammatory, antiangiogenic, and antiproliferative properties, all of which may contribute to their anticancer effect. 26 One of the underlying mechanisms for the multiple beneficial effects of omega‐3 fatty acids is their ability to suppress the production of AA (C20:4n‐6)‐derived eicosanoids by competing with the AA‐metabolizing enzymes (eg, COX‐2, FADS2, etc). 17 , 30 , 63 Notably, our findings that omega‐3 fatty acids can effectively suppress the upregulation of lipogenesis in breast CSC, together with the known inhibitory effect on AA metabolism, indicate that omega‐3 fatty acids may be a very promising anticancer agent as they can, on one hand, limit the resource of building blocks (ie, inhibition of fatty acid synthesis) and, on the other hand, reduce the availability of the lipid mediators (ie, reduction of eicosanoid production) that stimulate cell growth and support tumor microenvironment. Indeed, numerous previous studies have shown various beneficial effects of omega‐3 fatty acids in patients with cancer. 63 , 64 , 65 , 66 Given the multiple benefits of omega‐3 fatty acids as safe and natural nutrients, they hold great promise to be used as healthy food, supplements, and drugs for the prevention and treatment of cancer.
In summary, our findings have demonstrated that breast CSC have a unique fatty acid composition due to the aberrant expression of lipogenic enzymes, which play a critical role in sustaining their self‐renewal and survival capabilities, and that omega‐3 PUFA can effectively suppress the self‐renewal and growth of breast CSC by, at least in part, downregulation of the lipogenic enzymes, especially SCD1. These findings open new avenues for exploring novel therapeutic approaches by targeting fatty acid metabolism in breast CSC and suggest potential utilities of omega‐3 fatty acids for the prevention and treatment of breast cancer. CSC is a key driver for tumor recurrence and metastasis and controlling fatty acid metabolism could establish unfavorable tumor microenvironment for developing parental cells to CSC.
CONFLICT OF INTEREST
The authors declared no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
H.L.: designed the study, performed the experiments, analyzed data, drafted the manuscript; C.‐Y.C., X.L.: designed the study, performed the experiments, drafted the manuscript; C.‐W.S., T.C., L.H., Y.L., M.W.: performed the experiments, contributed to the analysis of data; X.Z.: collected clinical specimens, conducted protein detection and analysis; J.X.K.: conceived, designed, and supervised the study, prepared the manuscript.
Supporting information
Appendix S1: Supporting Information
ACKNOWLEDGMENTS
The authors would like to thank Kophu Chiang for his technical assistance. This research was supported by Fortune Education Foundation (New York, USA), and Sunsan Life Science (Hong Kong, China).
Luo H, Chen C‐Y, Li X, et al. Increased lipogenesis is critical for self‐renewal and growth of breast cancer stem cells: Impact of omega‐3 fatty acids. Stem Cells. 2021;39(12):1660‐1670. doi: 10.1002/stem.3452
Haiqing Luo, Chih‐Yu Chen, and Xiangyong Li contributed equally to this work.
Funding information Fortune Education Foundation; Sunsan Life Science
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9‐29. [DOI] [PubMed] [Google Scholar]
- 2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87‐108. [DOI] [PubMed] [Google Scholar]
- 3. Early Breast Cancer Trialists' Collaborative Group (EBCTCG) , Peto R, Davies C, et al. Comparisons between different polychemotherapy regimens for early breast cancer: meta‐analyses of long‐term outcome among 100,000 women in 123 randomised trials. Lancet. 2012;379(9814):432‐444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem‐like cells that self‐renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10(2):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guo W, Keckesova Z, Donaher JL, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148(5):1015‐1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu S, Wicha MS. Targeting breast cancer stem cells. J Clin Oncol. 2010;28(25):4006‐4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hermann PC, Huber SL, Herrler T, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313‐323. [DOI] [PubMed] [Google Scholar]
- 8. Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105‐111. [DOI] [PubMed] [Google Scholar]
- 9. Ponti D, Costa A, Zaffaroni N, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65(13):5506‐5511. [DOI] [PubMed] [Google Scholar]
- 10. Long J, Zhang CJ, Zhu N, et al. Lipid metabolism and carcinogenesis, cancer development. Am J Cancer Res. 2018;8(5):778‐791. [PMC free article] [PubMed] [Google Scholar]
- 11. Sevinsky CJ, Khan F, Kokabee L, Darehshouri A, Maddipati KR, Conklin DS. NDRG1 regulates neutral lipid metabolism in breast cancer cells. Breast Cancer Res. 2018;20(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo D, Bell EH, Chakravarti A. Lipid metabolism emerges as a promising target for malignant glioma therapy. CNS Oncol. 2013;2(3):289‐299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rohrig F, Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat Rev Cancer. 2016;16(11):732‐749. [DOI] [PubMed] [Google Scholar]
- 14. Visweswaran M, Arfuso F, Warrier S, Dharmarajan A. Aberrant lipid metabolism as an emerging therapeutic strategy to target cancer stem cells. Stem Cells. 2020;38(1):6‐14. [DOI] [PubMed] [Google Scholar]
- 15. Yi M, Li J, Chen S, et al. Emerging role of lipid metabolism alterations in cancer stem cells. J Exp Clin Cancer Res. 2018;37(1):118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li J, Condello S, Thomes‐Pepin J, et al. Lipid desaturation is a metabolic marker and therapeutic target of ovarian cancer stem cells. Cell Stem Cell. 2017;20(3):303‐314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He C, Qu X, Wan J, et al. Inhibiting delta‐6 desaturase activity suppresses tumor growth in mice. PLoS One. 2012;7(10):e47567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pisanu ME, Maugeri‐Saccà M, Fattore L, et al. Inhibition of Stearoyl‐CoA desaturase 1 reverts BRAF and MEK inhibition‐induced selection of cancer stem cells in BRAF‐mutated melanoma. J Exp Clin Cancer Res. 2018;37(1):318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tracz‐Gaszewska Z, Dobrzyn P. Stearoyl‐CoA desaturase 1 as a therapeutic target for the treatment of cancer. Cancer. 2019;11(7):948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mukherjee A, Kenny HA, Lengyel E. Unsaturated fatty acids maintain cancer cell stemness. Cell Stem Cell. 2017;20(3):291‐292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thomas J, Thomas CJ, Radcliffe J, et al. Omega‐3 fatty acids in early prevention of inflammatory neurodegenerative disease: a focus on Alzheimer's disease. Biomed Res Int. 2015;2015:172801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Walz CP, Barry AR, Koshman SL. Omega‐3 polyunsaturated fatty acid supplementation in the prevention of cardiovascular disease. Can Pharm J. 2016;149(3):166‐173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weylandt KH, Serini S, Chen YQ, et al. Omega‐3 polyunsaturated fatty acids: the way forward in times of mixed evidence. Biomed Res Int. 2015;2015:143109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Calder PC. Marine omega‐3 fatty acids and inflammatory processes: effects, mechanisms and clinical relevance. Biochim Biophys Acta. 2015;1851(4):469‐484. [DOI] [PubMed] [Google Scholar]
- 25. Simopoulos AP. The importance of the ratio of omega‐6/omega‐3 essential fatty acids. Biomed Pharmacother. 2002;56(8):365‐379. [DOI] [PubMed] [Google Scholar]
- 26. Kang JX, Liu A. The role of the tissue omega‐6/omega‐3 fatty acid ratio in regulating tumor angiogenesis. Cancer Metastasis Rev. 2013;32(1‐2):201‐210. [DOI] [PubMed] [Google Scholar]
- 27. Liu J, Abdelmagid SA, Pinelli CJ, et al. Marine fish oil is more potent than plant‐based n‐3 polyunsaturated fatty acids in the prevention of mammary tumors. J Nutr Biochem. 2018;55:41‐52. [DOI] [PubMed] [Google Scholar]
- 28. Zou Z, Bellenger S, Massey KA, et al. Inhibition of the HER2 pathway by n‐3 polyunsaturated fatty acids prevents breast cancer in fat‐1 transgenic mice. J Lipid Res. 2013;54(12):3453‐3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li J, Chen CY, Arita M, et al. An omega‐3 polyunsaturated fatty acid derivative, 18‐HEPE, protects against CXCR4‐associated melanoma metastasis. Carcinogenesis. 2018;39(11):1380‐1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xia S, Lu Y, Wang J, et al. Melanoma growth is reduced in fat‐1 transgenic mice: impact of omega‐6/omega‐3 essential fatty acids. Proc Natl Acad Sci U S A. 2006;103(33):12499‐12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yun EJ, Song KS, Shin S, et al. Docosahexaenoic acid suppresses breast cancer cell metastasis by targeting matrix‐metalloproteinases. Oncotarget. 2016;7(31):49961‐49971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fluckiger A, Dumont A, Derangere V, et al. Inhibition of colon cancer growth by docosahexaenoic acid involves autocrine production of TNFα. Oncogene. 2016;35(35):4611‐4622. [DOI] [PubMed] [Google Scholar]
- 33. Kim N, Jeong S, Jing K, et al. Docosahexaenoic acid induces cell death in human non‐small cell lung cancer cells by repressing mTOR via AMPK activation and PI3K/Akt inhibition. Biomed Res Int. 2015;2015:239764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rovito D, Giordano C, Plastina P, et al. Omega‐3 DHA‐ and EPA‐dopamine conjugates induce PPARγ‐dependent breast cancer cell death through autophagy and apoptosis. Biochim Biophys Acta. 2015;1850(11):2185‐2195. [DOI] [PubMed] [Google Scholar]
- 35. Wan XH, Fu X, Ababaikeli G. Docosahexaenoic acid induces growth suppression on epithelial ovarian cancer cells more effectively than eicosapentaenoic acid. Nutr Cancer. 2016;68(2):320‐327. [DOI] [PubMed] [Google Scholar]
- 36. Chauvin L, Goupille C, Blanc C, et al. Long chain n‐3 polyunsaturated fatty acids increase the efficacy of docetaxel in mammary cancer cells by downregulating Akt and PKCɛ/δ‐induced ERK pathways. Biochim Biophys Acta. 2016;1861(4):380‐390. [DOI] [PubMed] [Google Scholar]
- 37. Sheng H, Chen X, Liu B, Li P, Cao W. Omega‐3 polyunsaturated fatty acids enhance cisplatin efficacy in gastric cancer cells by inducing apoptosis via ADORA1. Anticancer Agents Med Chem. 2016;16(9):1085‐1092. [DOI] [PubMed] [Google Scholar]
- 38. Freitas RDS, Campos MM. Protective effects of omega‐3 fatty acids in cancer‐related complications. Nutrients. 2019;11(5):945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hofacer R, Magrisso IJ, Jandacek R, et al. Omega‐3 fatty acid deficiency increases stearoyl‐CoA desaturase expression and activity indices in rat liver: positive association with non‐fasting plasma triglyceride levels. Prostaglandins Leukot Essent Fatty Acids. 2012;86(1‐2):71‐77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li J, Li FR, Wei D, et al. Endogenous ω‐3 polyunsaturated fatty acid production confers resistance to obesity, dyslipidemia, and diabetes in mice. Mol Endocrinol. 2014;28(8):1316‐1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang LL, Wan JB, Wang B, et al. Suppression of acute ethanol‐induced hepatic steatosis by docosahexaenoic acid is associated with downregulation of stearoyl‐CoA desaturase 1 and inflammatory cytokines. Prostaglandins Leukot Essent Fatty Acids. 2013;88(5):347‐353. [DOI] [PubMed] [Google Scholar]
- 42. Huang W, Wang B, Li X, Kang JX. Endogenously elevated n‐3 polyunsaturated fatty acids alleviate acute ethanol‐induced liver steatosis. Biofactors. 2015;41(6):453‐462. [DOI] [PubMed] [Google Scholar]
- 43. Mani SA, Guo W, Liao MJ, et al. The epithelial‐mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704‐715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sansone P, Storci G, Giovannini C, et al. p66Shc/Notch‐3 interplay controls self‐renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells. 2007;25(3):807‐815. [DOI] [PubMed] [Google Scholar]
- 45. Kang JX, Wang J. A simplified method for analysis of polyunsaturated fatty acids. BMC Biochem. 2005;6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang X, Zhang L, Lin B, et al. Phospholipid phosphatase 4 promotes proliferation and tumorigenesis, and activates Ca2+‐permeable cationic channel in lung carcinoma cells. Mol Cancer. 2017;16(1):147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Adorno‐Cruz V, Kibria G, Liu X, et al. Cancer stem cells: targeting the roots of cancer, seeds of metastasis, and sources of therapy resistance. Cancer Res. 2015;75(6):924‐929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baccelli I, Trumpp A. The evolving concept of cancer and metastasis stem cells. J Cell Biol. 2012;198(3):281‐293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bian Y, Chang X, Liao Y, et al. Promotion of epithelial‐mesenchymal transition by Frizzled2 is involved in the metastasis of endometrial cancer. Oncol Rep. 2016;36(2):803‐810. [DOI] [PubMed] [Google Scholar]
- 50. Foroni C, Broggini M, Generali D, Damia G. Epithelial‐mesenchymal transition and breast cancer: role, molecular mechanisms and clinical impact. Cancer Treat Rev. 2012;38(6):689‐697. [DOI] [PubMed] [Google Scholar]
- 51. Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7(10):763‐777. [DOI] [PubMed] [Google Scholar]
- 52. Currie E, Schulze A, Zechner R, et al. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18(2):153‐161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Corominas‐Faja B, Cuyas E, Gumuzio J, et al. Chemical inhibition of acetyl‐CoA carboxylase suppresses self‐renewal growth of cancer stem cells. Oncotarget. 2014;5(18):8306‐8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Feng W, Gentles A, Nair RV, et al. Targeting unique metabolic properties of breast tumor initiating cells. Stem Cells. 2014;32(7):1734‐1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Peiris‐Pages M, Martinez‐Outschoorn UE, Pestell RG, et al. Cancer stem cell metabolism. Breast Cancer Res. 2016;18(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Galluzzi L, Kepp O, Vander Heiden MG, et al. Metabolic targets for cancer therapy. Nat Rev Drug Discov. 2013;12(11):829‐846. [DOI] [PubMed] [Google Scholar]
- 57. Noto A, Raffa S, De Vitis C, et al. Stearoyl‐CoA desaturase‐1 is a key factor for lung cancer‐initiating cells. Cell Death Dis. 2013;4:e947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yu Y, Kim H, Choi S, et al. Targeting a lipid desaturation enzyme, SCD1, selectively eliminates colon cancer stem cells through the suppression of Wnt and NOTCH signaling. Cells. 2021;10(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pisanu ME, Noto A, De Vitis C, et al. Blockade of Stearoyl‐CoA‐desaturase 1 activity reverts resistance to cisplatin in lung cancer stem cells. Cancer Lett. 2017;406:93‐104. [DOI] [PubMed] [Google Scholar]
- 60. Mason P, Liang B, Li L, et al. SCD1 inhibition causes cancer cell death by depleting mono‐unsaturated fatty acids. PLoS One. 2012;7(3):e33823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Peck B, Schug ZT, Zhang Q, et al. Inhibition of fatty acid desaturation is detrimental to cancer cell survival in metabolically compromised environments. Cancer Metab. 2016;4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pender‐Cudlip MC, Krag KJ, Martini D, et al. Delta‐6‐desaturase activity and arachidonic acid synthesis are increased in human breast cancer tissue. Cancer Sci. 2013;104(6):760‐764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Song M, Ou FS, Zemla TJ, et al. Marine omega‐3 fatty acid intake and survival of stage III colon cancer according to tumor molecular markers in NCCTG Phase III trial N0147 (alliance). Int J Cancer. 2019;145(2):380‐389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fabian CJ, Kimler BF, Hursting SD. Omega‐3 fatty acids for breast cancer prevention and survivorship. Breast Cancer Res. 2015;17(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Song M, Zhang X, Meyerhardt JA, et al. Marine ω‐3 polyunsaturated fatty acid intake and survival after colorectal cancer diagnosis. Gut. 2017;66(10):1790‐1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zheng JS, Hu XJ, Zhao YM, Yang J, Li D. Intake of fish and marine n‐3 polyunsaturated fatty acids and risk of breast cancer: meta‐analysis of data from 21 independent prospective cohort studies. BMJ. 2013;346:f3706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


