Abstract
Directed cell migration is essential for cells to efficiently migrate in physiological and pathological processes. While migrating in their native environment, cells interact with multiple types of cues, such as mechanical and chemical signals. The role of chemical guidance via chemotaxis has been studied in the past, the understanding of mechanical guidance of cell migration via durotaxis remained unclear until very recently. Nonetheless, durotaxis has become a topic of intensive research and several advances have been made in the study of mechanically guided cell migration across multiple fields. Thus, in this article we provide a state of the art about durotaxis by discussing in silico, in vitro and in vivo data. We also present insights on the general mechanisms by which cells sense, transduce and respond to environmental mechanics, to then contextualize these mechanisms in the process of durotaxis and explain how cells bias their migration in anisotropic substrates. Furthermore, we discuss what is known about durotaxis in vivo and we comment on how haptotaxis could arise from integrating durotaxis and chemotaxis in native environments.
Keywords: biomechanics, chemotaxis, directed cell migration, durotaxis, haptotaxis, mechanosensing, mechanotransduction
In this article, we provide a state‐of‐the‐art overview of durotaxis, which can be defined as thedirected motion or growth of cells based on variations in the stiffness oftheir extracellular matrix. Here, we discuss in silico, in vitro and in vivo data about durotaxis and we commenton how cells may need to integrate durotaxis and chemotaxis to navigate innative environments.
Abbreviations
- CCM
collective cell migration
- ECM
extracellular matrix
- FA
focal adhesion
- FAK
focal adhesion kinase
- MSCs
mesenchymal stem cells
- PM
plasma membrane
- RhoA
Ras homolog family member A
- ROCK
Rho kinase
- SAC
stretch‐activated channels
Introduction
When moving in their native environments, migrating cells and clusters requireto move in a directional and persistent manner by performing directed cellmigration (Box 1; Fig. 1). Migratory cells have been shown to reply to a plethora of extracellular cues that bias their migration in a given direction (extensively reviewed in Ref. [1, 2]). Among these cues, elastic properties of the migratory substrate have been shown to bias single and collective cell migration (CCM) to perform a process known as durotaxis [3, 4, 5, 6, 7] (see Box 2: elastic properties definition and an updated operational definition of durotaxis). Here, we discuss different aspects of durotaxis, providing examples of in silico, in vitro and in vivo durotaxis.
Box 1. Cell polarity and directed cell motion.
When moving in their native environments, migrating cells and clusters require to move in a directional and persistent manner. To directionally migrate, cells polarize and orient their intrinsic polarity and motility machinery towards an extrinsic biasing cue [8, 9]. These extrinsic cues vary in nature as biochemical, electrical, mechanical cues or even variations in substrate topology have been proposed to direct cell migration (explained below) and extensively discussed in Ref. [1].
Polarity achievement and protrusion formation are key events that allow cells to directionally migrate, and their establishment is activated by cell‐intrinsic molecular signalling but oriented by external factors [1] (Fig. 1). For instance, actin polymerization at the leading edge is driven by small GTPase activity such as Rac1 or Cdc42 [10, 11, 12, 13, 14, 15]. During lamellipodia formation, Rac1 activates actin nucleator proteins from the WAVE and phosphoinositide families, known to be required for the activity of actin‐nucleating proteins such as N‐WASP‐ARP2/3 complex [13, 14, 16, 17, 18]. Unlike lamellipodia, filopodia of the leading edge are formed by Cdc42‐induced actin polymerization [10, 12, 19]. At the cell rear, RhoA activity induces retraction of the back inhibiting protrusion formation [16, 19, 20, 21] (Fig. 1B). Retraction has been proposed to be mediated by stress fibres formed by actomyosin contractility in a force‐dependent manner [19, 20, 21]. Here, RhoA kinase (ROCK) activates myosin II through phosphorylation, inducing contractility and protrusion disassembly [19, 20, 21, 22, 23]. Data suggest that this mechanism could be mediated by ROCK activation of the phosphoinositide phosphatase PTEN at the back of the cell, which is known to inactivate PI3K, and thus inhibiting adhesion and protrusion formation [23, 24, 25, 26]. Thereby, to achieve polarity at signalling levels, different GTPases have to change their distribution within the cell. How cells achieve this is poorly understood, but some relevant molecules have been discovered. For instance, at the leading edge, p190RhoGAP RAC‐dependent activation inhibits RhoA activity, while in the back ROCK‐induced serine phosphorylation of FilGAP leads to Rac inhibition [27, 28, 29]. Some evidence also shows that RhoA is present at the leading edge, to depolymerize actin in the sites of FA dynamics [30, 31]. These observations unveil the complexity of the initial simplified model of RhoA/Rac polarization and reveal the necessity of further investigation to understand the mechanisms that establish cell polarity.
As stated before, these mechanisms of intrinsic cell polarity are essential for cells to migrate. Nonetheless, in order to navigate across their native environment, cells require ‘reading’ a combination of cues that will bias their intrinsic polarity machinery towards their final positions [1]. The process by which intrinsic polarity is biased by external cues in order to orient cells is named directed cell migration [32]. Thus, cells with low intrinsic polarity capabilities would poorly reply to environmental biasing cues. These cells require further exploration of their surroundings in order to find their way and will exhibit a type of migration named ‘random walk’. On the other hand, cells with a higher degree of intrinsic polarity will efficiently move towards external cues to migrate with high directionality and persistence [9, 33]. How a cell can have high or low intrinsic directionality have been extensively studied. However, despite being extensively investigated, there are still many open questions, and most studied point to the subfamily of Rho GTPases: the Rac proteins. For instance, it has been observed that changes in total Rac activity can be used as a switch between random and directional migration. In this article, fibroblast cultured in 3D or 2D environment exhibit variations in Rac activity responsible for changes in the intrinsic directionality of cell migration [34]. Thus, the control of Rac activity by the migratory environment is one of the mechanisms that determine the degree of intrinsic directionality which cells display when persistently and directionally migrating.
Fig. 1.
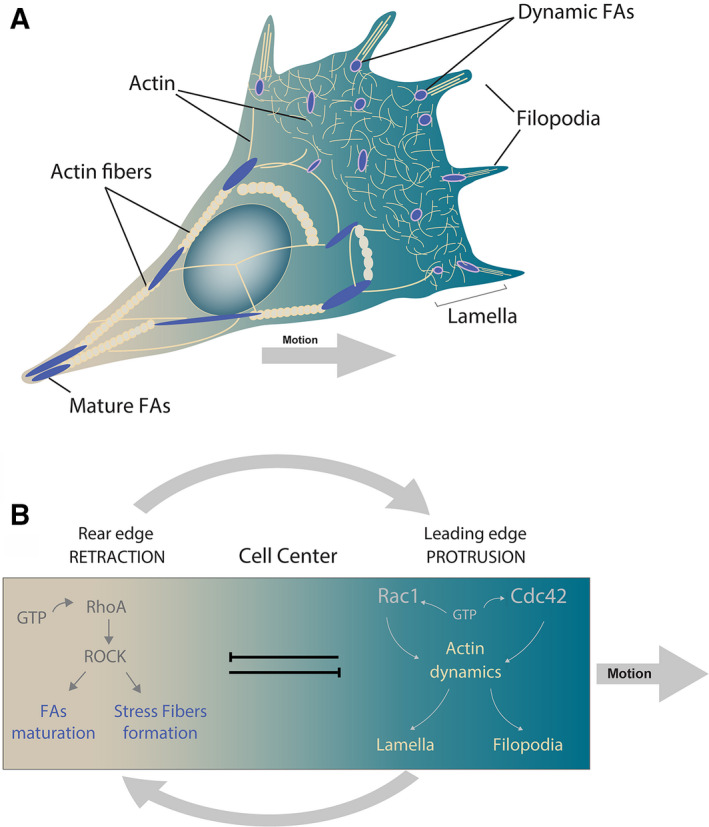
Cell polarity and directed cell motion.
Box 2. Definitions of biological and physical terms.
Durotaxis
In the light of recent discoveries, durotaxis can be defined as the directed motion or growth of cells based on variations in the stiffness of their extracellular matrix (ECM) [132].
Cell leading edge (front)
The closest border of the cell to the direction of the movement, normally where protrusions are formed.
Cell rear (back)
The farthest border of the cell to the direction of movement. A region of high actomyosin contractility.
Directionality
The displacement of a cell divided by the total length of the migrated distance. Thus, randomly migrating cells display lower directionality than cells that persistently migrate in the same direction.
Lamella
Flat and large projections of cell membrane that typically, but not exclusively, form in the leading edge of a cell that migrates with a mesenchymal migratory mode.
Filopodia
Long and thin cytoplasmic protrusions that extend beyond the extended lamella.
Elastic properties
The property of some materials to deform upon a stress and then return to their original shape after the input is removed.
Stress
Physical quantity used to express the internal force that surroundings exert over a cross‐sectional area of a given object. In mechanobiology, this term can be used as the force required to deform a biological material.
Strain
Correspond to the rate of deformation a material exhibit when exposed to mechanical stress.
Tension
According to Newton's laws, every time a force is applied over an object, the object simultaneously exerts a force of equal magnitude and opposite direction over the origin of the force. Thus, tension is referred as the reaction force applied over a source of mechanical stress.
Durotaxis was named after Latin durus (translated as ‘hard’) and the Greek word taxis (used for ‘logical arrangement’). One of the first studies suggesting that substrate mechanics can regulate biological responses was conducted in chick neurons [35, 36, 37]. These authors observed that the application of a mechanical stimulus resulted in axon elongation [35]. A decade later, the transition from nonpolarized cell state to a polarized migratory state was also demonstrated to be influenced by mechanical stimuli [38]. Following on from this result, Lo et al. finally coined the term durotaxis, at the start of the 21st century. In their seminal article, the authors applied an anisotropic strain to hydrogels and showed that substrate mechanics can bias the direction of cell motion (see Box 2: strain definition). They also found that cells exert higher traction and increase their areas when exposed to stiffer surfaces. At the time, these authors speculated that cells may dynamically alter their contractility in order to withstand the mechanical properties of their migratory substrate [3]. To date, a range of cell types have been shown to bias their directionality (see Box 2: directionality definition) when exposed to a stiffness gradient, by using multiple strategies that we discuss further in this article. This framework about durotaxis in different cell types would not be possible without the use of hydrogels carrying stiffness gradients, defined as ‘a chemically or physically cross‐linked polymeric network swollen by water’ [39, 40].
Although initially described in single cells, durotaxis has been also shown to guide the migration of cellular cluster during CCM. A seminal article showed that epithelial cells, when plated on stiffness gradients, exhibited directional migration towards stiffer regions of the gels [7]. In addition, these authors showed that substrate rigidity biased collective migration by modulating cadherin‐based contacts in a myosin II‐dependent mechanism [41]. More recently, another group of authors showed that durotactic behaviour is an emergent property of a collective of cells. In this study, Sunyer et al. compared the response of isolated human mammary epithelial cells (MCF‐10A) and clusters of the same cell type, plated on a stiffness gradient, observing that multicellular cluster exhibited durotaxis even when isolated cells did not. Interestingly, the same behaviour was observed in Madin‐Darby canine kidney (MDCK) epithelial cells. Mechanistically, Sunyer et al. [4] showed that collective durotaxis involves long‐range force transmission across the cluster via cadherin‐based contacts.
In addition to the role of durotaxis in physiological processes, in vitro evidence has suggested that durotaxis is also involved in fibrosis [42] and cancer [43, 44]. It has been well established that cancer cells plated on stiff substrates display more aggressive phenotypes than those plated on soft substrates [43, 44]. However, recent evidence has demonstrated that cultured U87‐MG (‘U87’), T98G, MDA‐MB‐231 and HT‐1080 human cancer cell lines are able to undergo durotaxis [43]. In a more mechanistic approach, another group showed that Cdc42 GTPase‐activating protein, a Rac1 and Cdc42 GTPase that specifically localize in the cell–substrate adhesion points, is required for U2OS osteosarcoma cells to migrate along stiffness gradients [45]. On the other hand, pancreatic stellate cells (PSCs), involved in pancreatic fibrosis, are also able to durotax [46]. Furthermore, in another study, it was observed that hepatic stellate cells (HSCs), known profibrogenic cells in the liver, follow stiffness gradients by using a mechanism involving FA kinase (FAK) activation, which, in turn, promotes YAP1‐mediated downstream signalling [47]. Durotaxis was also observed in ex vivo studies of spheroids of human epidermoid carcinoma cells (A431) demonstrating that durotaxis has the potential to operate in challenging 3D in vivo environments. Together, these findings highlight the importance of dissecting the mechanisms underlying durotaxis in order to further expand our understanding of cancer, fibrosis and other pathological conditions, which are normally studied from a molecular perspective.
Translating mechanical inputs into a cellular response
How do cells change their behaviour when exposed to a mechanically challenging environment? Addressing this question is a milestone in our quest to elucidate how a cell or group of cells durotax in a stiffness gradient. In this section, we describe the macromolecular structures and mechanisms that cells use to sense and translate mechanical inputs into a cellular response. We initially describe work performed in isotropic hydrogels carrying a defined value of stiffness (not in a durotactic gradient) to then further explain how these structures and mechanisms operate in a concerted manner to drive durotaxis. In this last section, we refer just to work performed in anisotropic hydrogels carrying a stiffness gradient. This is an important point as durotaxis should involve displacement or growth along a stiffness gradient and studying in gels of isotropic stiffness values can potentially lead to misconceptions in the field. Nonetheless, we believe that studies performed in isotropic gels have paved the way to achieve our current level of understanding about durotaxis.
In order to adapt to the extracellular environment, migratory cells are first required to detect a mechanical stimulus and subsequently to respond to this cue. The mechanisms by which cells achieve this have been named as mechanosensation and mechanotransduction, respectively [48]. Since these concepts are highly interdependent, the concepts of mechanosensation and mechanotransduction are still under refinement, and literature sometimes tends to be misleading when establishing a proper definition for molecules that may play a role in sensing or transducing mechanical inputs. This is due to the fact that many molecules involved in the response to a mechanical cue can act as sensors and/or transducers [49] (Figs 2A,B and 3). For instance, focal adhesions (FAs) sense a mechanical stimulus and activate internal signalling pathways, which in turn promote their own reinforcement and dynamics. Thus, these structures act as sensors, transducers and the responsible element that executes the response to mechanics (we described this example in more detail below). Likewise, in several cases it is challenging to identify whether a protein or structure plays a role in sensing or transducing the mechanical environment of a cell. Hence, in order to establish a definition that allows us to distinguish between the processes of mechanosensing and mechanotransduction, we believe it is essential for authors to start considering the time scales in which these processes operate, as it is starting to be revealed [50]. Additionally, it may prove to be important to consider the submodules within a cellular structure, as in the case of FAs, where mechanosensing and mechanotransduction modules are starting to be dissected [51]. For example, mechanosensing must occur in a rather short temporal regime (in the order of milliseconds) and it is characterized by changes in the structure of specific molecules, the ‘sensors’. Since other structural changes have been reported to involve modulation of actomyosin contractility [52], protein stretching [53] or mechanically activated ion channel opening [54, 55], it may result interesting to also evaluate the existence of substructural modules and temporal regimes that may be in charge of differentially sense or transduce a mechanical stimuli in these cellular components.
Fig. 2.
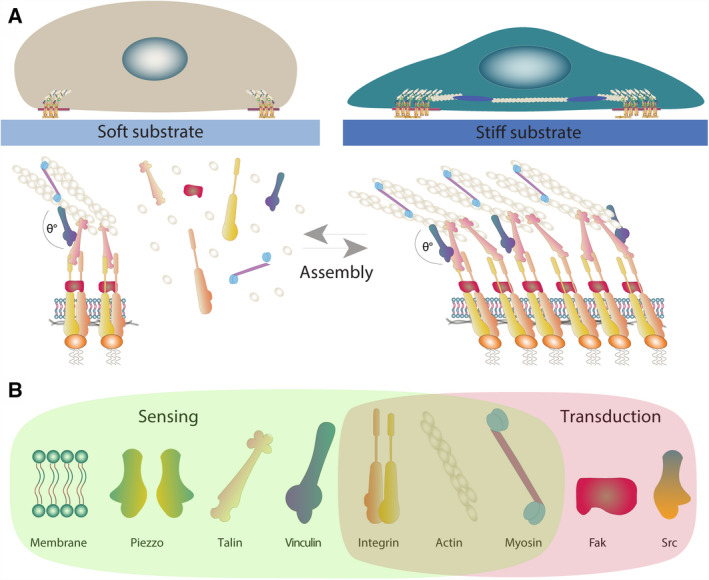
Mechanosensing in soft and stiff substrates. (A) Mechanosensing: Mechanical cues of the environment, such as substrate rigidity, can modulate the composition and dimensions of the FAs. In turn, FAs promote the spatial reorganization of the actomyosin cytoskeleton, thereby mediating i tension (represented by FA angle) over the same FA and the cell cortex. Thus, cells plated on soft substrates have small FA complexes, with low degree of assembly, which experience low levels of tension, while cells plated on stiff substrates present a higher degree of FA assembly and experience greater levels of tension. (B) Some of the most common elements of the mechanical response to substrate rigidity including sensing and transduction modules. Here, it should be noted that some of the elements, such as integrin or some cytoskeleton elements, can exhibit both functions.
Fig. 3.
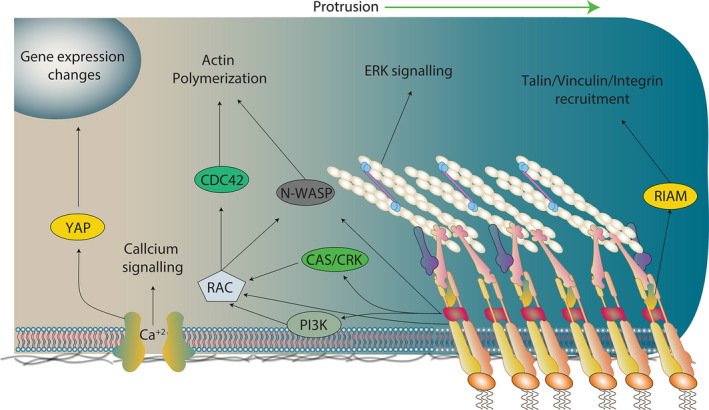
Mechanotransduction. Once assembled, FAs can act as a signalling hub that can activate several pathways, leading to the modulation of cell behaviour in response to the stiffness. However, FAs are not the only sensitive or transduction hub in the cell. For instance, there are mechanosensitive channels such as Piezo1, or even the membrane itself can act as a mechanosensitive/mechanotransduction element (discussed in this review). Thus, the figure presented here corresponds to a simplification of the mechanotransduction phenomena, just for general guidance purposes.
Mechanosensing
Several cellular structures have been proposed to work as mechanical sensors, including FAs and the actomyosin network (Fig. 2A). Additionally, in certain contexts, there are examples in which the plasma membrane (PM) and cell‐to‐cell contact molecules can also act as mechanosensors [56, 57]. In this section, we will analyse some of the mechanosensitive structures, described above, and discuss some of the latest advances in this field.
Focal adhesion sensing module
Focal adhesions are structures that can be part of either the mechanosensing or mechanotransduction process. In this section, we will focus on the role of the FAs as sensors of a mechanical input, while their role as transducers will be described further later.
Focal adhesions are structures that connect cells to the ECM of their migratory substrate [58]. The strength of adhesion and traction that adhering cells exert on their migratory substrates depends almost exclusively on integrin‐based signalling. Integrins are transmembrane proteins that bind to proteins from the ECM and recruit many regulatory and adaptor proteins. The recruitment of proteins to integrins facilitates linkage to the cytoskeleton to promote changes in cell shape and downstream signalling [59]. As the major force‐bearing structure between cells and the ECM, integrins play a central role in determining how cells sense and respond to the mechanical properties of their migratory substrate [58, 60]. Other FA proteins that have been shown to be relevant for mechanosensing are the adaptors vinculin and talin. In the FA, these proteins are the molecular bridges that transmit the actin‐based force to the integrin in the adhesion sites and vice versa [61, 62]. Interestingly, the recruitment of vinculin and talin to the FA is force‐dependent [63, 64]. Furthermore, when migrating in stiffer substrates FAs are stretched and with that vinculin and talin expose new binding domains to promote cytoskeletal dynamics via downstream signalling [65]. For instance, upon stretching, talin exhibits binding sites for several proteins such as integrins, vinculin and actin, thus reinforcing FA formation and dynamics [66].
Actomyosin network
The actomyosin cytoskeleton adapts and polarizes depending on mechanical environmental cues [52, 60, 67]. For instance, migratory B16F1 mouse melanoma cells present differential cytoskeletal arrangements when cultured on substrates of various dimensions and geometries [68]. Furthermore, it has been shown that ordering of the actin cytoskeleton also depends on substrate rigidity [69]. In this work, the authors showed that the dynamic mechanical properties of the actin cytoskeleton change from fluid‐like to solid‐like in response to substrates of different stiffnesses [69].
Mechanosensing has not only been tested at the level of actin fibres. For instance, it has been suggested that myosin II isoforms cooperate to work as mechanosensors. In this context, it has been observed that stiffness can induce accumulation of MIIB at the rear of the cell, primed by MIIA assembly [67]. In a more in vivo approach, it was suggested that geometrical and mechanical constraints are required to orient the cytoskeleton and the axis of tension during ventral furrow formation in the Drosophila embryo [70] (see Box 2: tension definition). In this work, they showed that anisotropic cortical tension at apical cell junctions is sufficient to drive tissue elongation, and this differential distribution of tension is dependent on myosin II accumulation. Taken together, these findings have allowed to formulate a model where mechanosensing through the actomyosin network emerges from the collective dynamics of the cortex [50, 71].
Plasma membrane
The PM must be able to accommodate for the fluctuations in tension arising from interior and exterior of the cell. This serves a dual role, preventing PM rupture and maintaining cell homeostasis in response to environmental changes [72, 73, 74]. Thus, as a structure, which is able to adapt in response to mechanical cues, the PM has been postulated to function as a mechanosensory structure. Notably, physical changes in the PM can induce conformational changes in key signalling membrane components [75]. These changes range from lipid rearrangements [74], ion channel opening [76], conformational changes in G proteins and translocation of signalling proteins [77]. One of the most studied cases of how physical changes in cell membrane can affect cell behaviour is related to changes in mechanically gated channels [54]. Mechanosensitive channels are directly activated by membrane stress, or indirectly by other elements associated with the membrane (see Box 2: stress definition). Some channels have been directly related to mechanosensing during cell migration [78, 79]. One of the most well‐studied mechanosensitive channel is Piezo1, an unspecific cation channel from the family of membrane stretch‐activated channels (SACs) [80, 81]. The PM can be stretched in several ways, when a cell experiences compressive forces. Interestingly, it has been shown that Piezo1 is a channel that is sensitive to compressive forces [82]. In this article, they observe that the changes in the migratory modes that are exhibited by Dictyostelium in response to increases in confinement are not present in Piezo1 mutants. The mechanism proposed here is that bleb‐mediated migration requires calcium influx, facilitated by Piezo1 channels. It is suggested that this could be related to an increment of myosin II at the cortex by an unknown mechanism [82]. Some other mechanosensitive channels have been related to decision making and cell navigation in complex environments [83] and in cancer metastatic spreading [84]. Their role in durotaxis is also starting to be studied [85].
Another way by which PM modulates migratory processes is through lipid signalling [75]. In this line, it has been observed that phosphatidylinositol 3,4‐bisphosphate is present in cancer protrusions, positively regulating cancer aggressiveness through lamellipodia maturation, podosomes and invadopodia formation and also playing a critical role in FA dynamics [86, 87] (see Box 2: lamella and filopodia definitions). On the other hand, lipid droplets from the PM have been shown to disrupt mechanosensing in hepatocellular carcinoma. In this work, the authors found that the presence of small lipid droplets can result in a decrease in stiffness‐induced cell spreading, disrupting FA and stress fibres. These data suggest that lipid droplets can impair the ability of the hepatocyte to sense its underlying matrix stiffness [88].
Mechanotransduction
Once a mechanical input is sensed, this signal is translated into a biochemical signalling cascade that will, in turn, promote a molecular and cellular response to mechanics (Figs 2B and 3). The process by which cells achieve this is referred to here as mechanotransduction. In general, mechanotransduction can be mediated by rapid protein modifications that in turn modify the cytoskeleton and cell behaviour to eventually generate a transcriptional response. While protein modifications occur in seconds to minutes (midterm), a transcriptional response requires minutes to hours to occur (long term). Here, we discuss some of the most studied signalling cascades that are activated by the different aforementioned mechanosensory mechanisms to transduce a mechanical stimulus into a cellular response.
As discussed above, FAs are not only mechanosensitive structures. Indeed, the proteins recruited into FA complexes are also able to mediate downstream signalling via modulation of signalling molecules such as kinases, phosphatases and scaffold proteins, as part of the mechanotransductive machinery (Fig. 2B). Thus, FA components act both as sensors and as transducers of mechanical cues [51]. For example, integrin engagement can activate Rac1 and Cdc42 to promote cell spreading [89]. In this work, the authors showed that integrin‐dependent adhesion leads to rapid activation of p‐21‐activated kinase, a downstream effector of Rac1 and Cdc42, which is implicated in cytoskeleton remodelling and cell motility [89, 90]. In line with this, β1 integrins induce Rac1 activation [91, 92], while α1 integrin reinforces adhesion by Ras homolog family member A (RhoA)‐mDIA activation pathway [92]. Remarkably, Rho kinase (ROCK)‐dependent activation of the c‐Fos/c‐Jun transcription complex has been shown to underpin an increase in α6 integrin in fibroblasts cultured on stiff substrates [93]. Also, it is known that stiff substrates promote ROCK activation in fibroblasts [94]. Furthermore, it has been shown that cells can adapt to new substrates by modulating the type of integrin that they express in their FAs and with that modulate traction and motility [95]. Cancer cells also modulate the amount and type of integrins in stiff substrates, leading to increase in aggressiveness [96]. Together, these last examples suggest that FAs not only sense and transduce but also can rapidly respond to mechanical cues by modulating cell adhesion and traction.
In addition to integrins, many other members of the FA complexes have been shown to promote downstream signalling upon exposure to mechanical stimuli. One of the most studied proteins is FAK. Several signalling molecules have been identified downstream FAK, and most of these pathways are involved in promoting cell migration, that is, FAK can form a complex with Src to phosphorylate p130CAS [97]. Phosphorylated p130CAS in turn associates with Cas/Crk complex, which is known to play a role in migration by mediating Rac1 activation at the leading edge of motile cells [98, 99] (see Box : cell leading edge definition). p130CAS activity has been directly demonstrated to promote downstream signalling upon mechanical stress [53]. In this seminal study, the authors developed a battery of tools to stretch the protein itself. Using these tools, the authors observed that tyrosine residues within the CAS substrate domain (CasSD) became increasingly phosphorylated upon the application of mechanical stretch to the protein. Tyrosine phosphorylation of CasSD triggered the activation of downstream signalling cascades. Notably, activated CasSD localizes to the periphery of cells where normally tractions are exerted [53]. These results suggested for the first time that a molecule could act as a sensor that would transduce stretch into a mechanical response. A few years later, it was shown that another member of the CAS family of proteins, known as Nedd9, is required for avian neural crest cell migration in vivo by controlling actin dynamics [100]. More recently, it was shown that Nedd9 regulates cell polarity and the migration of the neural crest through modulation of RhoA activity, via the association with the RhoGAP DLC1 [101]. In support of our recent findings where we demonstrated that neural crest cells are a mechanosensitive cell population [102], an interesting possibility is that CAS proteins can work as mechanosensors and/or transducers in the neural crest, an embryonic and highly migratory cell population. Lastly, p130CAS does not only play a role in cell migration, but also play a role in survival and proliferation via regulation of ERK and JNK signalling [103].
Vinculin and talin can also transduce force across the FA [19, 104, 105]. For instance, in mouse embryonic fibroblasts, talin has been shown to transmit force via unfolding its rod domain [105]. More precisely, it was shown that DLC1, which is a negative regulator of RhoA, directly binds to the R8 subdomain within the talin rod domain. Interestingly, this force‐dependent interaction is required to localize DLC1 to the FAs where it controls FA dynamics and cell migration [106]. Taking an even more mechanistic approach, another group demonstrated the structural basis that allows talin to work as a mechanotransducer [107]. This work provided an idea on how cells could use proteins such as talin to accurately and rapidly respond to a noisy native environment. Talin and vinculin are also known to connect the FAs to the actin cytoskeleton to mediate mechanical signalling [22]. Vinculin, for instance, can bind to both FAs and the cytoskeleton and the structural domains by which this interaction occurs are well‐defined [108, 109]. Taking advantage of this knowledge, a group of authors developed a tool that allowed to directly measure the magnitude of forces that are transmitted from the substrate into the cell via vinculin [64]. One of the conclusions of this seminal work is that force‐dependent stabilization of FAs involves recruitment of vinculin to FAs, from where it can transmit force. Strikingly, they showed that these two steps can be independently controlled [64]. The role of vinculin in mechanotransduction and its interactors has been widely studied and discussed in the literature [110, 111, 112]. In vivo, the role of integrin, vinculin and talin in the collective migration of neural crest cells, a mechanosensitive cell population [102], revealed a physiological context in which these molecules can operate to potentially mediate force transmission in a native context. This offers an excellent platform to test the extent to which in vitro signalling mechanisms that have been shown to be regulated by the integrin pathway converge with those that may arise in more complex in vivo environments.
Finally, SACs such as Piezo1 allow calcium influx upon membrane stretch, activating an intriguing calcium‐mediated downstream signalling cascade [113, 114]. Interestingly, it has been shown that Piezo1‐mediated increase in cytosolic Ca2+ induces actomyosin contractility [115]. Also, a recent paper has shown that Piezo1 is involved in modulating the morphogenesis of zebrafish outflow tract tissue, which is constantly exposed to mechanical stress [116]. In this tissue, Piezo1 activity is essential for the activation of the transcription factor Yap1 [116], a well‐known mechanotransducer, which is an essential component of the Hippo pathway [117]. This study demonstrates how a mechanical stimulus that is received in the membrane of a cell can be internalized into the nucleus.
One of the most well‐studied long‐term modifications, driven by mechanical cues, is the activation and localization of the Yes‐associated protein (YAP) [118]. YAP is a family of transcriptional cofactors whose translocation to the nucleus is known to be controlled by mechanical cues of the environment, such as ECM rigidity, strain, shear stress, adhesive area or force [119]. Interestingly, YAP nuclear translocation is itself a short‐term modification, but the effects of this translocation have long‐term transcriptional effects. Once in the nucleus, YAP binds to TEAD transcription factors and induces the transcription of genes associated with proliferation and inhibition of differentiation or cell migration and invasion in cancer cells [120]. Accordingly, a study using optogenetic tools to modulate RhoA‐mediated cell contractility showed that cell contractility can induce YAP nuclear localization [121]. Furthermore, another group showed that Yap1 nuclear translocation is force‐dependent [122]. In this study, it was elegantly shown that upon AFM‐induced extrinsic stress, Yap1 can translocate into the nucleus via nuclear pores [122]. In this line, talin unfolding in response to increases in matrix rigidity leads to YAP nuclear translocation [123]. Finally, in vivo data revealed that YAP is required for CCM [124]. In this article, the authors showed that YAP is expressed predominantly in the dorsal neural tube, and its loss of function inhibits the migration of neural crest cells, while gain‐of‐function embryos exhibit an increase in neural crest migration from the neural tube [124]. Furthermore, recent studies show that Yap1 also mediates the migration of chicken neural crest cells and that this process is dependent on the metabolic state of these cells [124]. Although the authors did not take a mechanical approach, this system can offer a great platform where to study how the interplay of mechanics, and metabolism can modify gene transcription to allow cell migration in vivo.
Another molecule known to promote long‐term changes in response to mechanical cues is β‐catenin. β‐catenin is a subunit of the cadherin protein complex and thus is involved in several processes, such as maintaining cell‐to‐cell adhesion and promoting downstream signalling. This positions β‐catenin as one of the main intracellular transducers of the WNT pathway. Catenins are required for cell migration [125], and at the signalling level, it is known that Wnt/β‐catenin can be activated by mechanical cues to promote processes such as differentiation [126], patterning [127] and specific changes in gene expression [128]. These findings may suggest a role for β‐catenin in the response to mechanical cues during cell migration, although this possibility remains to be explored.
In this section, we have described the roles of only a few structural elements in the process of mechanotransduction during cell migration. However, several other structures and pathways have been suggested to work as mechanotransductive elements of cells, in a variety of cellular processes, aside from cell migration. We refer you to a number of excellent reviews, written on this topic [48, 129, 130, 131].
Durotaxis: mechanosensing and mechanotransduction in action
Cell migration is a dynamic process and when migrating on a chemical, electrical or stiffness gradient, cells are required to constantly monitor changes in their migratory substrate, that is, rigidity changes in the case of durotaxis (Fig. 4). We have described several cellular structures that have been proposed to work as mechanical sensors and transducers in cells adhering to substrates of isotropic stiffness (either soft or stiff). Nonetheless, during durotaxis, migration occurs in a rather anisotropic substrate, from which mechanical cues must be differentially ‘read’ and transmitted across the cell or the cluster. Hence, in this part of our work we will refer just to work performed by analysing cell behaviour in anisotropic surfaces.
Fig. 4.
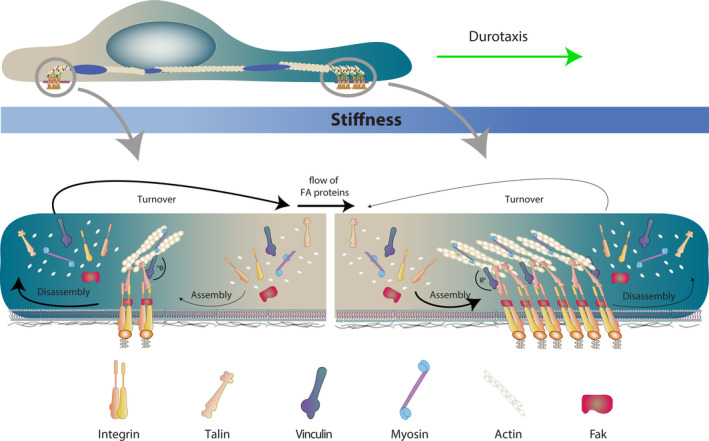
Durotaxis: mechanosensing and mechanotransduction in action. When cells follow stiffness gradients, different events can be observed at the front and the rear of the cells. At the front, due to higher rigidity, there is high recruitment of FA proteins, higher rate of assembly and thus larger FA size. On the other hand, in the rear of the cell, the softer substrate promotes lower recruitment of FA proteins, with an increased frequency of FA disassembly. This asymmetry between front and rear generates a net flow of FA proteins to the leading edge. Thus, mature FAs form mainly at the front to promote protrusion extension and establish the direction of migration to the stiffer regions of the substrate.
Once internalized, the output of this differential elasticity of the migratory substrate is translated into a cascade that stabilizes cell polarity to bias the direction of cell migration, transforming what is known as ‘random walk’ into ‘directed cell migration’ (Box 1. Here, we comment on advances that depict, in part, how the above‐described mechanisms of sensing and transduction of mechanical cues can work to direct cell migration via durotaxis.
As stated, most evidence points towards integrin‐based FAs and actomyosin‐based contractility as key components of the mechanism by which migrating cells sense substrate rigidity [50, 133]. For instance, it is known that polarized migratory cells sense rigidity at the leading edge in an integrin‐dependent manner [92, 95, 134]. Also, rigidity sensing and adaptation to substrates of varying stiffnesses are mediated by regulating integrin types [95]. These last studies were not performed on cells adhering to substrates with stiffness gradients, but more recent and direct evidence shows that FAK activity is required for durotaxis [47, 135, 136, 137]. In this study, FAK knockdown inhibited the directionality that fibroblast normally displays when undergoing durotaxis on a stiffness gradient [135]. In addition to FAK, vinculin and talin are involved in substrate sensing by enabling cells to generate traction, also known as ‘tugging’ [60, 138]. This process involves dynamic fluctuations of FA dynamics and in turn cell traction in response to changes in substrate rigidity via a mechanism that depends on ROCK activity [60]. Though we refer to few examples, there are plenty of articles that complement the studies described here and confirm the importance of FA‐associated proteins as part of the mechanosensing machineries that cells use during durotaxis.
Although FAs are known to work as a sensing hub for substrate rigidity, strong evidence also suggests that rigidity of the substrate is not only sensed at the FA. By using micropillars, the Ladoux group demonstrated that mechanosensing requires FAs. However, their observations suggested that mechanosensing requires a large‐scale feedback mechanism that involves reorganization of the actin cytoskeleton to align in the direction of the applied force and modify traction at the FAs [133]. Even before Ladoux's observations, the actomyosin cytoskeleton was proposed to be essential to transmit stiffness anisotropy from the migratory substrate to the cell interior, to allow cells to adapt to applied force [50, 52, 67, 69]. In one study, a tool to measure traction forces of isolated myoblasts in real time in response to varying levels of stiffness was generated. Using this tool, the authors demonstrated that the kinetics of myosin binding to actin is force‐dependent, meaning that contractile units of actomyosin themselves can work as mechanosensors [69]. It is important to note that these results were not performed in the presence of a stiffness gradient. However, the authors hypothesized that by using this mechanism, cells could translate substrate anisotropy into anisotropic cytoskeletal tension and, with that, locally adjust adhesion complexes to guide migration along stiffness gradients [69, 133]. Subsequently, Raab et al. demonstrated that myosin IIA (MIIA) and MIIB isoforms cooperate to promote cytoskeleton polarization in mesenchymal stem cells (MSCs) that ‘crawl’ from soft to stiff substrates. These authors found that while MSCs adhere to soft 2D or 3D substrates, MIIB remains in a nonpolarized configuration. Conversely, when MSCs are plated on stiffer substrates, MIIB acquires a polarized distribution by localizing at the centre and rear of the cell (see Box 2: cell rear definition). They also confirmed that this substrate‐induced polarized distribution of myosin II underpinned the formation of a nonprotrusive region, as it has been previously described in migrating cells [67]. The authors also showed that in their conditions there was not polarized distribution of MIIA; however, its assembly into fibrils increased as the cells moved towards stiffer substrates, in accordance with a reduction in phosphorylation levels. This work also revealed that both isoforms were required for durotaxis, but knockdown experiments revealed that MIIB was more sensitive to durotaxis than MIIA [67]. Thus, the mechanism described by these authors suggests that myosin II isoforms cooperate to work as mechanosensors. Also, a model was proposed where the accumulation of MIIB at the rear of the cell is primed by MIIA assembly, in a manner dependent on the stiffness of the migratory environment, supporting previous observations proposing the relevance of the actomyosin cytoskeleton for durotaxis [50, 69, 133]. More recently, a group of authors confirmed that sensing of matrix rigidity requires dynamic actin polymerization at the FA sites [139]. The examples discussed here confirm that there is a long‐range mechanism of cellular sensing of the substrate, where the actomyosin cytoskeleton globally senses changes in substrate stiffness to then locally ‘tug’ FA traction and polarize cells to perform durotaxis. In addition, several evidence confirms that FAs and actomyosin are also required for collective durotaxis; however, cellular clusters also rely on cell‐to‐cell contacts to transmit mechanical information from their front to the rear [85, 140, 141, 142].
Our knowledge of durotaxis does not solely derive from experimental work. In fact, much of what we know today about the mechanisms of durotaxis is the result of integrating experimental information into physical models. 2D models describe durotaxis as an elastic stability phenomenon [143]. Here, the authors consider the cytoskeleton as a planar system composed of prestressed elastic linear elements. These elements represent stress fibres, which are anchored to the substrates via dots that represent the FAs. Using this simple model, the authors proposed that although cytoskeleton and FAs are both important for migration, the elastic stability of the cytoskeleton was more important than the stability of FAs. As such, they suggested that the cytoskeleton may play a major role in migration and mechanosensing [143]. This and several other models nicely fit the experimental demonstrations discussed along this review [144, 145, 146, 147, 148]. More recently, other groups used cellular pots models where they analysed the morphology of migrating cells, migrated distance, spreading area and migration speed under five different configurations of durotaxis and found that their numerical results were also in agreement with experimental data [149]. Furthermore, a ‘molecular clutch’ model has been generated to explain mechanosensing and directional motion of durotacting cells. In this model, adhesion points engage when stiffness is sufficiently high, exposing binding sites of mechanosensitive proteins. Several publications that combine modelling and experimental approaches use this clutch model to explain the phenomenon of durotaxis in both single and collectively migrating cells [95, 123, 150]. A common feature of all the models mentioned so far is that they reproduce what has been shown with experimental systems: the migration from soft to stiff substrates. However, there is a very interesting case where two different groups proposed that cells can also migrate from stiff to soft substrate. The first evidence was provided in a theoretical work, in which the authors found that their model conditions revealed that cells would crawl not only from very soft to stiff regions but also from very stiff to soft surfaces [151]. Subsequently, in vitro data suggest that HT‐1080 cancer cells can also perform reverse durotaxis – as stated by the authors ‘at least in some matrix regimes’ [152]. Results from this work are in agreement with findings observed by groups studying directional motion of liquid nanodroplet [153]. In this case, these simple structures also undergo reverse durotaxis. Perhaps an explanation for this interesting variation of durotaxis may lay in advances made by the material sciences field. These examples show the relevance of studying durotaxis in different contexts and ranges of rigidity. Also, they suggest that it is important to combine experimental and modelling data, as this may lead to finding emergent properties of durotaxis that would otherwise be overlooked.
Conclusions and perspectives
A century ago, D'Arcy Thompson suggested that animal body shapes are conditioned by gravity [154] marking the beginning of an era in which researchers would set to understand the biophysical basis of life. Although advances in this field have been delayed due to the global interest in genetic studies, technical developments in the last decades allow us now to measure and challenge native mechanical environments in morphogenesis [102, 129, 155, 156] and cancer [156, 157, 158, 159, 160]. These discoveries have helped researchers to advance our knowledge in several biophysical aspects, particularly in elucidating a role for durotaxis in the guidance of cell migration in vivo.
Durotaxis in vivo
In addition to the 2D experimental and modelling approaches discussed here, 3D modelling and 3D in vitro and in vivo durotaxis approaches provide further support to the idea that mechanical guidance exists in physiologically relevant conditions. Using ex vivo systems, some groups have generated gradients with in vivo relevant stiffness values to study both blood vessels [161] and Schwann cells [162] durotactic responses. In these well‐controlled ex vivo systems, the authors observed that blood vessels and Schwann cells as well as stem cells directionally migrate by following physiologically relevant stiffness gradients [161, 162, 163], strongly suggesting that this may also be the case in vivo. Theoretical work using 3D modelling supports these experimental observations [146]. A group used a finite element model to describe a single‐cell migrating in a 3D stiffness gradient and not only found that cells follow stiffness gradients, but also found that filopodia may serve as an explorative or sensing structure for cells migrating across complex 3D environments [146]. In a more direct approach, Franze's Lab showed that nerves of the optic tract collectively follow a stiffness gradient in vivo [164, 165]. Remarkably, the authors observed that depleting observed stiffness gradients inhibited the direction of axon growth. In addition, this group provided an explanation for the origin of this stiffness gradient and suggested that the mechanically gated channel, Piezo1, is required to sense substrates of anisotropic stiffness [164, 165]. This, in our opinion, is the first and clearest example of the displacement of cellular structures from soft to stiff regions in their native environment. Furthermore, a more recent article clearly shows that a stiffness gradient arises during mouse limb bud formation and that limb cells migrate by following this stiffness gradient [166], yet assessing the migration of limb cells after modifying this gradient remains to be assessed. Building on these examples, an interesting question is whether the robustness of the mechanism described to support durotaxis in vitro would be maintained in complex native environments. Accordingly, Raab et al. demonstrated that all their results regarding MIIA/B‐based mechanosensing mechanisms are reproducible in cells performing durotaxis in 3D gels [167, 168]. This strongly suggests that mechanisms of durotaxis may operate in vivo, but further experiments are required to achieve such a conclusion. In addition, recent data revealed that cell stiffness is largely independent of substrate stiffness [169]. Although still hypothetical, this is an important point as most of the in vitro data generated by the field works under the assumption that cell stiffness matches that of its migratory substrate. In this context, we believe that it would be interesting to test these novel ideas in the context of durotaxis and to evaluate whether and how this new scenario could reshape our current understanding about the ‘mechanisms of durotaxis’.
Integrating chemotaxis and durotaxis
In spite of the establishment of a role for durotaxis in vivo, one of the challenges that the field is approaching is the integration of mechanical guidance with other types of guidance mechanisms such as chemotaxis. In a living organism, cells migrate in a convoluted environment where chemical and physical cues co‐exist; thus, it is likely that cells do not migrate by following just one or the other type of environmental cue. Instead, cells may perform a more integrative type of motion where they sense and translate multiple cues into directed migration by perhaps performing a sort of ‘mixotaxis’, as it is starting to be proposed [1]. In accordance with this, recently published evidence shows that the presence of a structured gradient of chemoattractant(s) seems dispensable to direct the migration of neural crest cells. The authors of this article propose that their data suggest that other types of nonhomogeneous environments, of so far unknown nature, may be guiding CCM of neural crest cells in vivo [170]. Considering that neural crest cells have been shown to be mechanosensitive [102], it is tempting to hypothesize that the nature of these putative nonhomogeneous environments may be in the form of stiffness gradients. In other systems but in line with these ideas, it has been proposed that durotaxis and chemotaxis interplay [171]. This is an interesting example as it shows that chemotaxis of hMSCs is more efficient in soft substrates, as stated by the authors, this highlights the synergic influence of chemical and physical cues in guiding cell migration [171]. Intriguingly, gradients of bound molecules can also guide cell migration in a process named as haptotaxis, a form of collective guidance where cells follow gradients of adhesive substrates, that is, ECM or chemokinetic molecules [172, 173]. As a concept, haptotaxis reinforces the idea that cells may migrate by integrating at least both chemical and mechanical cues. This idea of integrating chemotaxis and durotaxis in vivo has been recently shown. For instance, WNT5a was previously thought to promote chemotaxis during mouse limb bud formation, though recent data suggest that this molecule may not operate as a chemoattractant. Instead, the evidence suggests that a stiffness gradient may underlay WNT5a secretion in the shape of a gradient that will in turn modify the ECM to allow directional migration [166]. Together, these observations suggest that mechanical gradients may work in vivo by cooperating with chemical cues to promote directed cell migration. In our opinion, this is a clear demonstration that chemotaxis and durotaxis interact to enable directed cell motion by allowing haptotaxis in vivo. Nonetheless, in order to fully demonstrate this, the authors should confirm or rule out that a stiffness gradient underlies the graded deposition of matrix or the graded distribution of WNT5a. This would finally probe that haptotaxis operates during mouse limb development. A clearer example of in vivo haptotaxis has been shown in breast cancer where a gradient of fibronectin allows directional movement of cancer cells both, in vivo and in vitro [174]. In their in vivo approach, these authors used intravital imaging, a tool that is proving extremely useful in the visualization of biological processes in their native context [175]. Another important point that arises in this context stems from the fact that several cell types, that is dendritic cells of lymph nodes, migrate by responding to chemoattractant cues but closely interacting with their challenging biophysical environment by combining integrin‐dependent [176] and amoeboid‐like [177] migratory modes. In this context, we believe that an additional challenge faced by the field is to (a) explore whether cells can durotax by using amoeboid migratory strategies or a combination of amoeboid and integrin‐dependent migration; and (b) to understand how these potentially new modes of durotaxis can actually be integrated with known chemotactic mechanisms. Addressing this type of points is challenging as it would take the field to a new level of experimental complexity. However, these approaches can turn out to be highly rewarding in terms of our understanding of the complexity of directed cell motion in complex native environments.
Finally, we believe that the field requires deeper analysis when studying a potential role for durotaxis in vivo, since simplified assumptions can lead to misleading interpretations about the role of stiffness gradients in native contexts. In addition, we invite authors to depart from the prevalent ‘binary’ vision of directed cell motion in which results are analysed by considering the role of either chemical or biophysical cues and to consider more integrative approaches to directed cell migration.
Conflict of interest
The authors declare no conflict of interest.
Author contribution
JAE, CLM and EHB wrote the manuscript and prepared the figures.
Acknowledgements
Work at EHB Lab receives funding from the ‘European Research Council (ERC) under the European Union's Horizon 2020 Research and Innovation Programme (Grant Agreement No. 950254)’; ‘EMBO IG Project Number 4765’; and ‘la Caixa Junior Leader Incoming (94978)’. The authors would also like to thank Sofia Moreira for useful comments on the manuscript.
References
- 1. Barriga EH & Theveneau E (2020) In vivo neural crest cell migration is controlled by “mixotaxis”. Front Physiol 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shellard A & Mayor R (2020) All roads lead to directional cell migration. Trends Cell Biol 30, 852–868. [DOI] [PubMed] [Google Scholar]
- 3. Lo CM, Wang HB, Dembo M & Wang YL (2000) Cell movement is guided by the rigidity of the substrate. Biophys J 79, 144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sunyer R, Conte V, Escribano J, Elosegui‐Artola A, Labernadie A, Valon L, Navajas D, Garcia‐Aznar JM, Munoz JJ, Roca‐Cusachs P et al. (2016) Collective cell durotaxis emerges from long‐range intercellular force transmission. Science 353, 1157–1161. [DOI] [PubMed] [Google Scholar]
- 5. Sunyer R & Trepat X (2020) Durotaxis. Curr Biol 30, R383–R387. [DOI] [PubMed] [Google Scholar]
- 6. Lange JR & Fabry B (2013) Cell and tissue mechanics in cell migration. Exp Cell Res 319, 2418–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shellard A & Mayor R (2021) Durotaxis: the hard path from in vitro to in vivo . Dev Cell 56, 227–239. [DOI] [PubMed] [Google Scholar]
- 8. Carmona‐Fontaine C, Matthews H & Mayor R (2008) Directional cell migration in vivo: Wnt at the crest. Cell Adh Migr 2, 240–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Petrie RJ, Doyle AD & Yamada KM (2009) Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol 10, 538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Block J, Breitsprecher D, Kuhn S, Winterhoff M, Kage F, Geffers R, Duwe P, Rohn JL, Baum B, Brakebusch C et al. (2012) FMNL2 drives actin‐based protrusion and migration downstream of Cdc42. Curr Biol 22, 1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guilluy C, Garcia‐Mata R & Burridge K (2011) Rho protein crosstalk: another social network? Trends Cell Biol 21, 718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Itoh RE, Kurokawa K, Ohba Y, Yoshizaki H, Mochizuki N & Matsuda M (2002) Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer‐based single‐molecule probes in the membrane of living cells. Mol Cell Biol 22, 6582–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ridley AJ & Hall A (1992) The small GTP‐binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70, 389–399. [DOI] [PubMed] [Google Scholar]
- 14. Rohatgi R, Nollau P, Ho HY, Kirschner MW & Mayer BJ (2001) Nck and phosphatidylinositol 4,5‐bisphosphate synergistically activate actin polymerization through the N‐WASP‐Arp2/3 pathway. J Biol Chem 276, 26448–26452. [DOI] [PubMed] [Google Scholar]
- 15. Warner H, Wilson BJ & Caswell PT (2019) Control of adhesion and protrusion in cell migration by Rho GTPases. Curr Opin Cell Biol 56, 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burridge K & Wennerberg K (2004) Rho and Rac take center stage. Cell 116, 167–179. [DOI] [PubMed] [Google Scholar]
- 17. Caswell PT & Zech T (2018) Actin‐based cell protrusion in a 3D matrix. Trends Cell Biol 28, 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rottner K & Schaks M (2019) Assembling actin filaments for protrusion. Curr Opin Cell Biol 56, 53–63. [DOI] [PubMed] [Google Scholar]
- 19. Ridley AJ (2011) Life at the leading edge. Cell 145, 1012–1022. [DOI] [PubMed] [Google Scholar]
- 20. Chrzanowska‐Wodnicka M & Burridge K (1996) Rho‐stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol 133, 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Worthylake RA & Burridge K (2003) RhoA and ROCK promote migration by limiting membrane protrusions. J Biol Chem 278, 13578–13584. [DOI] [PubMed] [Google Scholar]
- 22. Burridge K & Guilluy C (2016) Focal adhesions, stress fibers and mechanical tension. Exp Cell Res 343, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lawson CD & Ridley AJ (2018) Rho GTPase signaling complexes in cell migration and invasion. J Cell Biol 217, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Z, Dong X, Wang Z, Liu W, Deng N, Ding Y, Tang L, Hla T, Zeng R, Li L et al. (2005) Regulation of PTEN by Rho small GTPases. Nat Cell Biol 7, 399–404. [DOI] [PubMed] [Google Scholar]
- 25. Vemula S, Shi J, Hanneman P, Wei L & Kapur R (2010) ROCK1 functions as a suppressor of inflammatory cell migration by regulating PTEN phosphorylation and stability. Blood 115, 1785–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang S & Kim HM (2012) The RhoA‐ROCK‐PTEN pathway as a molecular switch for anchorage dependent cell behavior. Biomaterials 33, 2902–2915. [DOI] [PubMed] [Google Scholar]
- 27. Bustos RI, Forget MA, Settleman JE & Hansen SH (2008) Coordination of Rho and Rac GTPase function via p190B RhoGAP. Curr Biol 18, 1606–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morishita Y, Tsutsumi K & Ohta Y (2015) Phosphorylation of serine 402 regulates RacGAP protein activity of FilGAP protein. J Biol Chem 290, 26328–26338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ohta Y, Hartwig JH & Stossel TP (2006) FilGAP, a Rho‐ and ROCK‐regulated GAP for Rac binds filamin A to control actin remodelling. Nature 8, 803–814. [DOI] [PubMed] [Google Scholar]
- 30. Heasman SJ, Carlin LM, Cox S, Ng T & Ridley AJ (2010) Coordinated RhoA signaling at the leading edge and uropod is required for T cell transendothelial migration. J Cell Biol 190, 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM & Danuser G (2009) Coordination of Rho GTPase activities during cell protrusion. Nature 461, 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lara Rodriguez L & Schneider IC (2013) Directed cell migration in multi‐cue environments. Integrative Biology 5, 1306–1323. [DOI] [PubMed] [Google Scholar]
- 33. Theveneau E, Marchant L, Kuriyama S, Gull M, Moepps B, Parsons M & Mayor R (2010) Collective chemotaxis requires contact‐dependent cell polarity. Dev Cell 19, 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pankov R, Endo Y, Even‐Ram S, Araki M, Clark K, Cukierman E, Matsumoto K & Yamada KM (2005) A Rac switch regulates random versus directionally persistent cell migration. J Cell Biol 170, 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bray D (1984) Axonal growth in response to experimentally applied mechanical tension. Dev Biol 102, 379–389. [DOI] [PubMed] [Google Scholar]
- 36. Chada S, Lamoureux P, Buxbaum RE & Heidemann SR (1997) Cytomechanics of neurite outgrowth from chick brain neurons. J Cell Sci 110 (Pt 10), 1179–1186. [DOI] [PubMed] [Google Scholar]
- 37. Lamoureux P, Buxbaum RE & Heidemann SR (1989) Direct evidence that growth cones pull. Nature 340, 159–162. [DOI] [PubMed] [Google Scholar]
- 38. Verkhovsky AB, Svitkina TM & Borisy GG (1999) Self‐polarization and directional motility of cytoplasm. Curr Biol 9, 11–20. [DOI] [PubMed] [Google Scholar]
- 39. Sunyer R, Jin AJ, Nossal R & Sackett DL (2012) Fabrication of hydrogels with steep stiffness gradients for studying cell mechanical response. PLoS One 7, e46107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Whang M & Kim J (2016) Synthetic hydrogels with stiffness gradients for durotaxis study and tissue engineering scaffolds. Tissue Eng Regen Med 13, 126–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ng MR, Besser A, Danuser G & Brugge JS (2012) Substrate stiffness regulates cadherin‐dependent collective migration through myosin‐II contractility. J Cell Biol 199, 545–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lagares D, Liu F, Auernheimer V, Adams DC, Ludovic V, Kapoor M, Suter MJ, Goldmann W, Engler AJ, Tschumperlin DJ et al. (2015) Feedback amplification of lung fibrosis through matrix stiffness gradient‐induced fibroblast durotaxis via alpha tat1‐mediated microtubule acetylation. Am J Resp Crit Care 191. [Google Scholar]
- 43. DuChez BJ, Doyle AD, Dimitriadis EK & Yamada KM (2019) Durotaxis by human cancer cells. Biophys J 116, 670–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McKenzie AJ, Hicks SR, Svec KV, Naughton H, Edmunds ZL & Howe AK (2018) The mechanical microenvironment regulates ovarian cancer cell morphology, migration, and spheroid disaggregation. Sci Rep 8, 7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wormer DB, Davis KA, Henderson JH & Turner CE (2014) The focal adhesion‐localized CdGAP regulates matrix rigidity sensing and durotaxis. PLoS One 9, e91815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lachowski D, Cortes E, Pink D, Chronopoulos A, Karim SA, Morton JP & Del Rio Hernandez AE (2017) Substrate rigidity controls activation and durotaxis in pancreatic stellate cells. Sci Rep 7, 2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lachowski D, Cortes E, Robinson B, Rice A, Rombouts K & Del Rio Hernandez AE (2018) FAK controls the mechanical activation of YAP, a transcriptional regulator required for durotaxis. FASEB J 32, 1099–1107. [DOI] [PubMed] [Google Scholar]
- 48. Petridou NI, Spiro Z & Heisenberg CP (2017) Multiscale force sensing in development. Nat Cell Biol 19, 581–588. [DOI] [PubMed] [Google Scholar]
- 49. van Helvert S, Storm C & Friedl P (2018) Mechanoreciprocity in cell migration. Nat Cell Biol 20, 8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Trichet L, Le Digabel J, Hawkins RJ, Vedula SR, Gupta M, Ribrault C, Hersen P, Voituriez R & Ladoux B (2012) Evidence of a large‐scale mechanosensing mechanism for cellular adaptation to substrate stiffness. Proc Natl Acad Sci USA 109, 6933–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stutchbury B, Atherton P, Tsang R, Wang DY & Ballestrem C (2017) Distinct focal adhesion protein modules control different aspects of mechanotransduction. J Cell Sci 130, 1612–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mitrossilis D, Fouchard J, Pereira D, Postic F, Richert A, Saint‐Jean M & Asnacios A (2010) Real‐time single‐cell response to stiffness. Proc Natl Acad Sci USA 107, 16518–16523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sawada Y, Tamada M, Dubin‐Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S & Sheetz MP (2006) Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell 127, 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kefauver JM, Ward AB & Patapoutian A (2020) Discoveries in structure and physiology of mechanically activated ion channels. Nature 587, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Murthy SE, Dubin AE & Patapoutian A (2017) Piezos thrive under pressure: mechanically activated ion channels in health and disease. Nat Rev Mol Cell Biol 18, 771–783. [DOI] [PubMed] [Google Scholar]
- 56. Jansen KA, Atherton P & Ballestrem C (2017) Mechanotransduction at the cell‐matrix interface. Semin Cell Dev Biol 71, 75–83. [DOI] [PubMed] [Google Scholar]
- 57. Luo T, Mohan K, Iglesias PA & Robinson DN (2013) Molecular mechanisms of cellular mechanosensing. Nat Materials 12, 1064–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Geiger B, Spatz JP & Bershadsky AD (2009) Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol 10, 21–33. [DOI] [PubMed] [Google Scholar]
- 59. Wu C (2007) Focal adhesion: a focal point in current cell biology and molecular medicine. Cell Adh Migr 1, 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Plotnikov SV, Pasapera AM, Sabass B & Waterman CM (2012) Force fluctuations within focal adhesions mediate ECM‐rigidity sensing to guide directed cell migration. Cell 151, 1513–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Austen K, Ringer P, Mehlich A, Chrostek‐Grashoff A, Kluger C, Klingner C, Sabass B, Zent R, Rief M & Grashoff C (2015) Extracellular rigidity sensing by talin isoform‐specific mechanical linkages. Nat Cell Biol 17, 1597–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nordenfelt P, Elliott HL & Springer TA (2016) Coordinated integrin activation by actin‐dependent force during T‐cell migration. Nat Commun 7, 13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Golji J, Lam J & Mofrad MR (2011) Vinculin activation is necessary for complete talin binding. Biophys J 100, 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T et al. (2010) Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Haining AW, Lieberthal TJ & Del Rio Hernandez A (2016) Talin: a mechanosensitive molecule in health and disease. FASEB J 30, 2073–2085. [DOI] [PubMed] [Google Scholar]
- 66. del Rio A, Perez‐Jimenez R, Liu R, Roca‐Cusachs P, Fernandez JM & Sheetz MP (2009) Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Raab M, Swift J, Dingal PC, Shah P, Shin JW & Discher DE (2012) Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin‐II heavy chain. J Cell Biol 199, 669–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Starke J, Wehrle‐Haller B & Friedl P (2014) Plasticity of the actin cytoskeleton in response to extracellular matrix nanostructure and dimensionality. Biochem Soc Trans 42, 1356–1366. [DOI] [PubMed] [Google Scholar]
- 69. Gupta M, Sarangi BR, Deschamps J, Nematbakhsh Y, Callan‐Jones A, Margadant F, Mege RM, Lim CT, Voituriez R & Ladoux B (2015) Adaptive rheology and ordering of cell cytoskeleton govern matrix rigidity sensing. Nat Commun 6, 7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rauzi M, Verant P, Lecuit T & Lenne PF (2008) Nature and anisotropy of cortical forces orienting Drosophila tissue morphogenesis. Nat Cell Biol 10, 1401–1410. [DOI] [PubMed] [Google Scholar]
- 71. Etienne J, Fouchard J, Mitrossilis D, Bufi N, Durand‐Smet P & Asnacios A (2015) Cells as liquid motors: mechanosensitivity emerges from collective dynamics of actomyosin cortex. Proc Natl Acad Sci USA 112, 2740–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gauthier NC, Masters TA & Sheetz MP (2012) Mechanical feedback between membrane tension and dynamics. Trends Cell Biol 22, 527–535. [DOI] [PubMed] [Google Scholar]
- 73. Morris CE & Homann U (2001) Cell surface area regulation and membrane tension. J Membr Biol 179, 79–102. [DOI] [PubMed] [Google Scholar]
- 74. Sinha B, Koster D, Ruez R, Gonnord P, Bastiani M, Abankwa D, Stan RV, Butler‐Browne G, Vedie B, Johannes L et al. (2011) Cells respond to mechanical stress by rapid disassembly of caveolae. Cell 144, 402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Le Roux AL, Quiroga X, Walani N, Arroyo M & Roca‐Cusachs P (2019) The plasma membrane as a mechanochemical transducer. Philos Trans R Soc Lond B Biol Sci 374, 20180221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ranade SS, Syeda R & Patapoutian A (2015) Mechanically activated ion channels. Neuron 87, 1162–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chachisvilis M, Zhang YL & Frangos JA (2006) G protein‐coupled receptors sense fluid shear stress in endothelial cells. Proc Natl Acad Sci USA 103, 15463–15468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jin P, Jan LY & Jan YN (2020) Mechanosensitive ion channels: structural features relevant to mechanotransduction mechanisms. Annu Rev Neurosci 43, 207–229. [DOI] [PubMed] [Google Scholar]
- 79. Kobayashi T & Sokabe M (2010) Sensing substrate rigidity by mechanosensitive ion channels with stress fibers and focal adhesions. Curr Opin Cell Biol 22, 669–676. [DOI] [PubMed] [Google Scholar]
- 80. Moroni M, Servin‐Vences MR, Fleischer R, Sanchez‐Carranza O & Lewin GR (2018) Voltage gating of mechanosensitive PIEZO channels. Nat Commun 9, 1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Volkers L, Mechioukhi Y & Coste B (2015) Piezo channels: from structure to function. Pflugers Arch 467, 95–99. [DOI] [PubMed] [Google Scholar]
- 82. Srivastava N, Traynor D, Piel M, Kabla AJ & Kay RR (2020) Pressure sensing through Piezo channels controls whether cells migrate with blebs or pseudopods. Proc Natl Acad Sci USA 117, 2506–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhao R, Afthinos A, Zhu T, Mistriotis P, Li Y, Serra SA, Zhang Y, Yankaskas CL, He S, Valverde MA et al. (2019) Cell sensing and decision‐making in confinement: the role of TRPM7 in a tug of war between hydraulic pressure and cross‐sectional area. Science Adv 5, eaaw7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Petho Z, Najder K, Bulk E & Schwab A (2019) Mechanosensitive ion channels push cancer progression. Cell Calcium 80, 79–90. [DOI] [PubMed] [Google Scholar]
- 85. Haeger A, Wolf K, Zegers MM & Friedl P (2015) Collective cell migration: guidance principles and hierarchies. Trends Cell Biol 25, 556–566. [DOI] [PubMed] [Google Scholar]
- 86. Hawkins PT & Stephens LR (2016) Emerging evidence of signalling roles for PI(3,4)P2 in Class I and II PI3K‐regulated pathways. Biochem Soc Trans 44, 307–314. [DOI] [PubMed] [Google Scholar]
- 87. Fukumoto M, Ijuin T & Takenawa T (2017) PI(3,4)P2 plays critical roles in the regulation of focal adhesion dynamics of MDA‐MB‐231 breast cancer cells. Cancer Sci 108, 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chin L, Theise ND, Loneker AE, Janmey PA & Wells RG (2020) Lipid droplets disrupt mechanosensing in human hepatocytes. bioRxiv [PREPRINT]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Price LS, Leng J, Schwartz MA & Bokoch GM (1998) Activation of Rac and Cdc42 by integrins mediates cell spreading. Mol Biol Cell 9, 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kreis NN, Louwen F & Yuan J (2019) The multifaceted p21 (Cip1/Waf1/CDKN1A) in cell differentiation, migration and cancer therapy. Cancers 11, 1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Berrier AL, Martinez R, Bokoch GM & LaFlamme SE (2002) The integrin beta tail is required and sufficient to regulate adhesion signaling to Rac1. J Cell Sci 115, 4285–4291. [DOI] [PubMed] [Google Scholar]
- 92. Schiller HB, Hermann MR, Polleux J, Vignaud T, Zanivan S, Friedel CC, Sun Z, Raducanu A, Gottschalk KE, Thery M et al. (2013) beta1‐ and alphav‐class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin‐based microenvironments. Nat Cell Biol 15, 625–636. [DOI] [PubMed] [Google Scholar]
- 93. Chen H, Qu J, Huang X, Kurundkar A, Zhu L, Yang N, Venado A, Ding Q, Liu G, Antony VB et al. (2016) Mechanosensing by the alpha6‐integrin confers an invasive fibroblast phenotype and mediates lung fibrosis. Nat Commun 7, 12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhou Y, Huang X, Hecker L, Kurundkar D, Kurundkar A, Liu H, Jin TH, Desai L, Bernard K & Thannickal VJ (2013) Inhibition of mechanosensitive signaling in myofibroblasts ameliorates experimental pulmonary fibrosis. J Clin Investig 123, 1096–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Elosegui‐Artola A, Bazellieres E, Allen MD, Andreu I, Oria R, Sunyer R, Gomm JJ, Marshall JF, Jones JL, Trepat X et al. (2014) Rigidity sensing and adaptation through regulation of integrin types. Nat Mater 13, 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sonnenberg A, Linders CJ, Daams JH & Kennel SJ (1990) The alpha 6 beta 1 (VLA‐6) and alpha 6 beta 4 protein complexes: tissue distribution and biochemical properties. J Cell Sci 96 (Pt 2), 207–217. [DOI] [PubMed] [Google Scholar]
- 97. Zhao X & Guan JL (2011) Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv Drug Deliv Rev 63, 610–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Cheresh DA, Leng J & Klemke RL (1999) Regulation of cell contraction and membrane ruffling by distinct signals in migratory cells. J Cell Biol 146, 1107–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Cho SY & Klemke RL (2000) Extracellular‐regulated kinase activation and CAS/Crk coupling regulate cell migration and suppress apoptosis during invasion of the extracellular matrix. J Cell Biol 149, 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Aquino JB, Lallemend F, Marmigere F, Adameyko II, Golemis EA & Ernfors P (2009) The retinoic acid inducible Cas‐family signaling protein Nedd9 regulates neural crest cell migration by modulating adhesion and actin dynamics. Neuroscience 162, 1106–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Liu JA, Rao Y, Cheung MPL, Hui MN, Wu MH, Chan LK, Ng IO, Niu B, Cheah KSE, Sharma R et al. (2017) Asymmetric localization of DLC1 defines avian trunk neural crest polarity for directional delamination and migration. Nat Commun 8, 1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Barriga EH, Franze K, Charras G & Mayor R (2018) Tissue stiffening coordinates morphogenesis by triggering collective cell migration in vivo . Nature 554, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Defilippi P, Di Stefano P & Cabodi S (2006) p130Cas: a versatile scaffold in signaling networks. Trends Cell Biol 16, 257–263. [DOI] [PubMed] [Google Scholar]
- 104. Dumbauld DW, Lee TT, Singh A, Scrimgeour J, Gersbach CA, Zamir EA, Fu J, Chen CS, Curtis JE, Craig SW et al. (2013) How vinculin regulates force transmission. Proc Natl Acad Sci USA 110, 9788–9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Rahikainen R, Ohman T, Turkki P, Varjosalo M & Hytonen VP (2019) Talin‐mediated force transmission and talin rod domain unfolding independently regulate adhesion signaling. J Cell Sci 132. [DOI] [PubMed] [Google Scholar]
- 106. Haining AWM, Rahikainen R, Cortes E, Lachowski D, Rice A, von Essen M, Hytonen VP & Del Rio Hernandez A (2018) Mechanotransduction in talin through the interaction of the R8 domain with DLC1. PLoS Biol 16, e2005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Tapia‐Rojo R, Alonso‐Caballero A & Fernandez JM (2020) Talin folding as the tuning fork of cellular mechanotransduction. Proc Natl Acad Sci USA 117, 21346–21353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GW, Jennings L, Critchley DR, Craig SW & Liddington RC (2004) Structural basis for vinculin activation at sites of cell adhesion. Nature 430, 583–586. [DOI] [PubMed] [Google Scholar]
- 109. Ziegler WH, Gingras AR, Critchley DR & Emsley J (2008) Integrin connections to the cytoskeleton through talin and vinculin. Biochem Soc Trans 36, 235–239. [DOI] [PubMed] [Google Scholar]
- 110. Goldmann WH (2016) Role of vinculin in cellular mechanotransduction. Cell Biol Int 40, 241–256. [DOI] [PubMed] [Google Scholar]
- 111. Wrighton KH (2014) Vinculin discrimination at adhesions. Nat Rev Mol Cell Biol 15, 367. [Google Scholar]
- 112. Ziegler WH, Liddington RC & Critchley DR (2006) The structure and regulation of vinculin. Trends Cell Biol 16, 453–460. [DOI] [PubMed] [Google Scholar]
- 113. Nourse JL & Pathak MM (2017) How cells channel their stress: Interplay between Piezo1 and the cytoskeleton. Semin Cell Dev Biol 71, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wu J, Lewis AH & Grandl J (2017) Touch, tension, and transduction ‐ the function and regulation of piezo ion channels. Trends Biochem Sci 42, 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT & Horwitz AR (2003) Cell migration: integrating signals from front to back. Science 302, 1704–1709. [DOI] [PubMed] [Google Scholar]
- 116. Pathak MM, Nourse JL, Tran T, Hwe J, Arulmoli J, Le DT, Bernardis E, Flanagan LA & Tombola F (2014) Stretch‐activated ion channel Piezo1 directs lineage choice in human neural stem cells. Proc Natl Acad Sci USA 111, 16148–16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Pan D (2010) The hippo signaling pathway in development and cancer. Dev Cell 19, 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Halder G, Dupont S & Piccolo S (2012) Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol 13, 591–600. [DOI] [PubMed] [Google Scholar]
- 119. Dasgupta I & McCollum D (2019) Control of cellular responses to mechanical cues through YAP/TAZ regulation. J Biol Chem 294, 17693–17706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Liu X, Li H, Rajurkar M, Li Q, Cotton JL, Ou J, Zhu LJ, Goel HL, Mercurio AM, Park JS et al. (2016) Tead and AP1 coordinate transcription and motility. Cell Rep 14, 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Valon L, Marin‐Llaurado A, Wyatt T, Charras G & Trepat X (2017) Optogenetic control of cellular forces and mechanotransduction. Nat Commun 8, 14396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Elosegui‐Artola A, Andreu I, Beedle AEM, Lezamiz A, Uroz M, Kosmalska AJ, Oria R, Kechagia JZ, Rico‐Lastres P, Le Roux AL et al. (2017) Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell 171, 1397–1410.e14. [DOI] [PubMed] [Google Scholar]
- 123. Elosegui‐Artola A, Oria R, Chen Y, Kosmalska A, Perez‐Gonzalez C, Castro N, Zhu C, Trepat X & Roca‐Cusachs P (2016) Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat Cell Biol 18, 540–548. [DOI] [PubMed] [Google Scholar]
- 124. Kumar D, Nitzan E & Kalcheim C (2019) YAP promotes neural crest emigration through interactions with BMP and Wnt activities. Cell Commun Signal 17, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Vassilev V, Platek A, Hiver S, Enomoto H & Takeichi M (2017) Catenins steer cell migration via stabilization of front‐rear polarity. Dev Cell 43, 463–479.e5. [DOI] [PubMed] [Google Scholar]
- 126. Norvell SM, Alvarez M, Bidwell JP & Pavalko FM (2004) Fluid shear stress induces beta‐catenin signaling in osteoblasts. Calcif Tissue Int 75, 396–404. [DOI] [PubMed] [Google Scholar]
- 127. Cha B, Geng X, Mahamud MR, Fu J, Mukherjee A, Kim Y, Jho EH, Kim TH, Kahn ML, Xia L et al. (2016) Mechanotransduction activates canonical Wnt/beta‐catenin signaling to promote lymphatic vascular patterning and the development of lymphatic and lymphovenous valves. Genes Dev 30, 1454–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Pukhlyakova E, Aman AJ, Elsayad K & Technau U (2018) beta‐Catenin‐dependent mechanotransduction dates back to the common ancestor of Cnidaria and Bilateria. Proc Natl Acad Sci USA 115, 6231–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Heisenberg CP & Bellaiche Y (2013) Forces in tissue morphogenesis and patterning. Cell 153, 948–962. [DOI] [PubMed] [Google Scholar]
- 130. Plusa B & Hadjantonakis AK (2016) Mammalian development: Mechanics drives cell differentiation. Nature 536, 281–282. [DOI] [PubMed] [Google Scholar]
- 131. Vogel V & Sheetz M (2006) Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol 7, 265–275. [DOI] [PubMed] [Google Scholar]
- 132. Oliveri H, Franze K & Goriely A (2020) An optic ray theory for durotactic axon guidance. bioRxiv [PREPRINT]. [DOI] [PubMed] [Google Scholar]
- 133. Gupta M, Doss B, Lim CT, Voituriez R & Ladoux B (2016) Single cell rigidity sensing: A complex relationship between focal adhesion dynamics and large‐scale actin cytoskeleton remodeling. Cell Adh Migr 10, 554–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Janostiak R, Pataki AC, Brabek J & Rosel D (2014) Mechanosensors in integrin signaling: the emerging role of p130Cas. Eur J Cell Biol 93, 445–454. [DOI] [PubMed] [Google Scholar]
- 135. Wang HB, Dembo M, Hanks SK & Wang Y (2001) Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc Natl Acad Sci USA 98, 11295–11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Plotnikov SV & Waterman CM (2013) Guiding cell migration by tugging. Curr Opin Cell Biol 25, 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Ringer P, Colo G, Fassler R & Grashoff C (2017) Sensing the mechano‐chemical properties of the extracellular matrix. Matrix Biol 64, 6–16. [DOI] [PubMed] [Google Scholar]
- 138. Wu Z, Plotnikov SV, Moalim AY, Waterman CM & Liu J (2017) Two distinct actin networks mediate traction oscillations to confer focal adhesion mechanosensing. Biophys J 112, 780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Puleo JI, Parker SS, Roman MR, Watson AW, Eliato KR, Peng L, Saboda K, Roe DJ, Ros R, Gertler FB et al. (2019) Mechanosensing during directed cell migration requires dynamic actin polymerization at focal adhesions. J Cell Biol 218, 4215–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Ladoux B, Mege RM & Trepat X (2016) Front‐rear polarization by mechanical cues: from single cells to tissues. Trends Cell Biol 26, 420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ladoux B & Mege RM (2017) Mechanobiology of collective cell behaviours. Nat Rev Mol Cell Biol 18, 743–757. [DOI] [PubMed] [Google Scholar]
- 142. Venhuizen JH & Zegers MM (2017) Making heads or tails of it: cell–cell adhesion in cellular and supracellular polarity in collective migration. Cold Spring Harb Perspect Biol 9, a027854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Lazopoulos KA & Stamenovic D (2008) Durotaxis as an elastic stability phenomenon. J Biomech 41, 1289–1294. [DOI] [PubMed] [Google Scholar]
- 144. Dokukina IV & Gracheva ME (2010) A model of fibroblast motility on substrates with different rigidities. Biophys J 98, 2794–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Escribano J, Sunyer R, Sanchez MT, Trepat X, Roca‐Cusachs P & Garcia‐Aznar JM (2018) A hybrid computational model for collective cell durotaxis. Biomech Model Mechanobiol 17, 1037–1052. [DOI] [PubMed] [Google Scholar]
- 146. Kim MC, Silberberg YR, Abeyaratne R, Kamm RD & Asada HH (2018) Computational modeling of three‐dimensional ECM‐rigidity sensing to guide directed cell migration. Proc Natl Acad Sci USA 115, E390–E399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Malik AA & Gerlee P (2019) Mathematical modelling of cell migration: stiffness dependent jump rates result in durotaxis. J Math Biol 78, 2289–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Marzban B, Yi X & Yuan H (2018) A minimal mechanics model for mechanosensing of substrate rigidity gradient in durotaxis. Biomech Model Mechanobiol 17, 915–922. [DOI] [PubMed] [Google Scholar]
- 149. Allena R (2018) The discriminant role of mechanics during cell migration. J Cell Immunother 4, 30–34. [Google Scholar]
- 150. Bennett M, Cantini M, Reboud J, Cooper JM, Roca‐Cusachs P & Salmeron‐Sanchez M (2018) Molecular clutch drives cell response to surface viscosity. Proc Natl Acad Sci USA 115, 1192–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Lober J, Ziebert F & Aranson IS (2014) Modeling crawling cell movement on soft engineered substrates. Soft Matter 10, 1365–1373. [DOI] [PubMed] [Google Scholar]
- 152. Singh SP, Schwartz MP, Lee JY, Fairbanks BD & Anseth KS (2014) A peptide functionalized poly(ethylene glycol) (PEG) hydrogel for investigating the influence of biochemical and biophysical matrix properties on tumor cell migration. Biomaterials Science 2, 1024–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Mahmood A, Chen S, Chen C, Weng D & Wang J (2018) Directional motion of water droplet on nanocone surface driven by curvature gradient: a molecular dynamics simulation study. J Phys Chem C 122, 14937–14944. [Google Scholar]
- 154. Thompson DAW (1961) On growth and form, An abridged edn, University Press, Cambridge Eng. [Google Scholar]
- 155. Bauer AL, Jackson TL & Jiang Y (2009) Topography of extracellular matrix mediates vascular morphogenesis and migration speeds in angiogenesis. PLoS Comput Biol 5, e1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Friedl P & Gilmour D (2009) Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol 10, 445–457. [DOI] [PubMed] [Google Scholar]
- 157. Baker EL, Lu J, Yu D, Bonnecaze RT & Zaman MH (2010) Cancer cell stiffness: integrated roles of three‐dimensional matrix stiffness and transforming potential. Biophys J 99, 2048–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Carter SB (1965) Principles of cell motility: the direction of cell movement and cancer invasion. Nature 208, 1183–1187. [DOI] [PubMed] [Google Scholar]
- 159. Li L, He Y, Zhao M & Jiang J (2013) Collective cell migration: Implications for wound healing and cancer invasion. Burns Trauma 1, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Wang M, Cheng B, Yang Y, Liu H, Huang G, Han L, Li F & Xu F (2019) Microchannel stiffness and confinement jointly induce the mesenchymal‐amoeboid transition of cancer cell migration. Nano Lett 19, 5949–5958. [DOI] [PubMed] [Google Scholar]
- 161. Hartman CD, Isenberg BC, Chua SG & Wong JY (2016) Vascular smooth muscle cell durotaxis depends on extracellular matrix composition. Proc Natl Acad Sci USA 113, 11190–11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Evans EB, Brady SW, Tripathi A & Hoffman‐Kim D (2018) Schwann cell durotaxis can be guided by physiologically relevant stiffness gradients. Biomater Res 22, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Smith L, Cho S & Discher DE (2017) Mechanosensing of matrix by stem cells: From matrix heterogeneity, contractility, and the nucleus in pore‐migration to cardiogenesis and muscle stem cells in vivo . Semin Cell Dev Biol 71, 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Koser DE, Thompson AJ, Foster SK, Dwivedy A, Pillai EK, Sheridan GK, Svoboda H, Viana M, Costa LD, Guck J et al. (2016) Mechanosensing is critical for axon growth in the developing brain. Nat Neurosci 19, 1592–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Thompson AJ, Pillai EK, Dimov IB, Foster SK, Holt CE & Franze K (2019) Rapid changes in tissue mechanics regulate cell behaviour in the developing embryonic brain. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Zhu M, Tao H, Samani M, Luo M, Wang X, Hopyan S & Sun Y (2020) Spatial mapping of tissue properties in vivo reveals a 3D stiffness gradient in the mouse limb bud. Proc Natl Acad Sci USA 117, 4781–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Rehfeldt F, Brown AE, Raab M, Cai S, Zajac AL, Zemel A & Discher DE (2012) Hyaluronic acid matrices show matrix stiffness in 2D and 3D dictates cytoskeletal order and myosin‐II phosphorylation within stem cells. Integr Biol 4, 422–430. [DOI] [PubMed] [Google Scholar]
- 168. Thiam HR, Vargas P, Carpi N, Crespo CL, Raab M, Terriac E, King MC, Jacobelli J, Alberts AS, Stradal T et al. (2016) Perinuclear Arp2/3‐driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat Commun 7, 10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Rheinlaender J, Dimitracopoulos A, Wallmeyer B, Kronenberg NM, Chalut KJ, Gather MC, Betz T, Charras G & Franze K (2020) Cortical cell stiffness is independent of substrate mechanics. Nat Mater 19, 1019–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Bajanca F, Gouignard N, Colle C, Parsons M, Mayor R & Theveneau E (2019) In vivo topology converts competition for cell‐matrix adhesion into directional migration. Nat Commun 10, 1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Saxena N, Mogha P, Dash S, Majumder A, Jadhav S & Sen S (2018) Matrix elasticity regulates mesenchymal stem cell chemotaxis. J Cell Sci 131. [DOI] [PubMed] [Google Scholar]
- 172. Carter SB (1967) Haptotaxis and the mechanism of cell motility. Nature 213, 256–260. [DOI] [PubMed] [Google Scholar]
- 173. Yamada KM & Sixt M (2019) Mechanisms of 3D cell migration. Nat Rev Mol Cell Biol 20, 738–752. [DOI] [PubMed] [Google Scholar]
- 174. Oudin MJ, Jonas O, Kosciuk T, Broye LC, Guido BC, Wyckoff J, Riquelme D, Lamar JM, Asokan SB, Whittaker C et al. (2016) Tumor cell‐driven extracellular matrix remodeling drives haptotaxis during metastatic progression. Cancer Discov 6, 516–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Scheele C, Maynard C & van Rheenen J (2016) Intravital Insights into heterogeneity, metastasis, and therapy responses. Trends Cancer 2, 205–216. [DOI] [PubMed] [Google Scholar]
- 176. Linder S, Nelson D, Weiss M & Aepfelbacher M (1999) Wiskott‐Aldrich syndrome protein regulates podosomes in primary human macrophages. Proc Natl Acad Sci USA 96, 9648–9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Renkawitz J, Kopf A, Stopp J, de Vries I, Driscoll MK, Merrin J, Hauschild R, Welf ES, Danuser G, Fiolka R et al. (2019) Nuclear positioning facilitates amoeboid migration along the path of least resistance. Nature 568, 546–550. [DOI] [PMC free article] [PubMed] [Google Scholar]


