Abstract
Polyploidization may precipitate dramatic changes to the genome, including chromosome rearrangements, gene loss, and changes in gene expression. In dioecious plants, the sex-determining mechanism may also be disrupted by polyploidization, with the potential evolution of hermaphroditism. However, while dioecy appears to have persisted through a ploidy transition in some species, it is unknown whether the newly formed polyploid maintained its sex-determining system uninterrupted, or whether dioecy re-evolved after a period of hermaphroditism. Here, we develop a bioinformatic pipeline using RNA-sequencing data from natural populations to demonstrate that the allopolyploid plant Mercurialis canariensis directly inherited its sex-determining region from one of its diploid progenitor species, M. annua, and likely remained dioecious through the transition. The sex-determining region of M. canariensis is smaller than that of its diploid progenitor, suggesting that the non-recombining region of M. annua expanded subsequent to the polyploid origin of M. canariensis. Homeologous pairs show partial sexual subfunctionalization. We discuss the possibility that gene duplicates created by polyploidization might contribute to resolving sexual antagonism.
Author summary
Polyploidization, or genome duplication, is widespread among plants, with consequences that include gene loss, genome reorganization, and large-scale changes in gene expression. These changes could also affect species with separate sexes, with implications for sex determination and the evolution of sexual dimorphism. In some plants, polyploidy may disrupt the mechanism of sex determination, potentially bringing about a transition to hermaphroditism. In others, such as the wind-pollinated annual plant Mercurialis canariensis, dioecy has been maintained through allopolyploidization (i.e. genome duplication associated with hybridization). Here, we show that M. canariensis arose via hybridization between the dioecious relative M. annua and an unidentified, possibly extinct, second species. We also demonstrate that M. canariensis inherited its sex-determining region from M. annua. Finally, a small number of gene duplicates created by polyploidization have diverged in gene expression between males and females of M. canariensis. Our finding suggests that expression of each of the duplicated loci may have been differentially optimized for each sex. This possibility of ‘sexual subfunctionalization’ further highlights the dynamic interplay between polyploidization, sex-determination, and the evolution of sexual dimorphism.
Introduction
It is estimated that ~30% of plant species also have recent polyploid ancestry [1,2]. These increases in ploidy can have wide-ranging consequences, including impacts on biogeography and ecology [3,4], rates of diversification [5–7], and genome evolution [8,9]. Whereas the importance and success of polyploidy in plants cannot be disputed, new polyploid lineages that persist eventually become re-diploidized, initially in terms of patterns of segregation [10,11], but ultimately also in terms of genome size [12,13], gene content, and gene expression [14]. For most genes, diploidization of polyploid genomes involves the loss of one of their duplicated copies through a process of ‘fractionation’ [14–16], which may be strongly influenced by differences in expression levels between homeologs [17,18]. Such fractionation is typically not random, so that one ‘dominant’ subgenome retains more genes than the other [19,20]. While gene loss appears to be the most likely fate of one or other of redundant copies in polyploid lineages, some genes may be retained via subfunctionalization [21], where each copy evolves a different pattern of expression (e.g., at different times during cellular or organ development, or in different cell or organ types; [22,23]).
The possibility of subfunctionalization is particularly interesting in dioecious plants, because it may contribute to the resolution of sexual antagonism [24,25], with one homeolog expressed more in males and the other more in females. This process of ‘sexual subfunctionalization’ has been detected in gene duplicates of diploid species across a broad phylogenetic scale, including Drosophila [26], brown algae [27], and mice [28], but it would seem to be particularly plausible in dioecious polyploid species, with each of the two homeologs expressed at a level closer to the phenotypic optimum of one sex or the other. Given that dioecious plants frequently display sexual dimorphism for a wide range of physiological, morphological, defense and life-history traits (summarized in [29]), the compartmentalization of the genome into two subgenomes in dioecious polyploids could overcome genetic correlations between the sexes that might otherwise limit the optimization of distinct male and female phenotypes. Because sexual subfunctionalization implies the retention of both gene copies after polyploidization, we might also expect the evolution of subgenome dominance to be less important in dioecious species.
Genome duplication in dioecious species also has implications for the evolution of sexual systems and sex determination. First, polyploidy can cause a minority cytotype disadvantage, where the lack of mates selects for self-fertile hermaphrodites [30,31]. And second, the substantial reorganization of the genome through polyploidization (including aneuploidies and chromosome rearrangements) can lead to a disruption of meiosis [30,32,33], which may cause the breakdown of the existing genetic sex-determination mechanisms to provoke a transition from dioecy to hermaphroditism [30,34], notably in species in which genome duplication interferes with sex determined by an X:A (or Z:A) balance, with the potential generation of ‘intersexes’ and hermaphroditism [32,34]. For instance, in species of both Silene and Rumex that have XY sex chromosomes, the progeny of synthetic polyploids are hermaphroditic [34]. Other species seem to maintain dioecy after polyploidization (e.g., Silene, Rumex, Bouteloua, and Atrichum; [30,35–38], but little is known about how this occurs. Outside of angiosperms, the maintenance of a sex-determining region through polyploidization has been suggested for sturgeon [39] and mosses [40]. While these studies illustrate the potential effect of polyploidy on the breakdown of dioecy, polyploidization can also precipitate the evolution of dioecy from hermaphroditism [30,41].
Only a few diploid/polyploid pairs have been thoroughly examined for their sex determination. The most well-studied taxa are wild strawberries of the genus Fragaria [42–49]. Dioecious species in Fragaria have octoploid genomes that consist of four distinct subgenomes, with little recombination between homeologous chromosomes [50]. The sex-determining region consists of a female-specific ‘cassette’ on the W chromosome that contains a candidate sex-determining gene [42]. The cassette is hypothesized to have evolved once, and then to have jumped between homeologous chromosomes in different species [42]. However, although sex determination in octoploid populations of Fragaria is well-characterized, it is unclear when separate sexes first arose. In persimmon, sex is determined in diploids by the autosomal gene MeGI and its Y-linked paralog OGI [51,52], but expression of OGI is disrupted in polyploids that may be responsible for reversion to hermaphroditism–although some polyploid lineages have secondarily re-established dioecy through the reactivation of OGI [53]. In contrast to these examples, the polyploid descendants of other diploid dioecious species appear to have maintained dioecy. In cases that involve allopolyploid hybridization between two diploid dioecious parents, it is pertinent to ask which of the two precursors contributed the sex determination system, and how the sex chromosomes might have evolved during the subsequent process of diploidization. The mainly European and north African clade of annual species of the genus Mercurialis presents one such case.
The genus Mercurialis shows remarkable variation in both its sexual system and ploidy [31]. Dioecy is ancestral in the genus, present in perennial species, but monoecy has evolved in polyploid annuals [31,54,55]. Polyploidization of diploid M. annua produced a monoecious tetraploid population, which subsequently hybridized with the M. huetii, the diploid sister species of M. annua, yielding hexaploid populations (still referred to as M. annua, or M. ambigua) with an androdioecious sexual system (the maintenance of males with hermaphrodites) [55]. Crosses among species [56] suggest that sex in hexaploid M. annua is determined by the same XY sex-chromosomal system as that characterized for diploid M. annua. Obbard et al [57] described a new dioecious species, M. canariensis (confined to the Canary Islands), which has a life history and morphology very similar to that of M. annua. However, while dioecious M. annua is diploid, M. canariensis is tetraploid [57]. The fact that M. canariensis is polyploid and dioecious is noteworthy because all other polyploid annuals in Mercurialis are monoecious. Obbard et al. [57] inferred that M. canariensis was an allotetraploid hybrid between diploid M. annua and a likely extinct progenitor that may also have been dioecious. It is not known which of the two putative progenitors of M. canariensis contributed the sex-determining locus.
Here, we use long-read RNA sequencing and expression data to re-examine the evolutionary origins of allotetraploid M. canariensis and to identify which of its two diploid progenitors contributed the sex chromosomes. We based our analysis on full-length transcripts from polyploid M. canariensis, thus overcoming the problem of assembling chimeric transcripts derived from different homeologs. As set out in Fig 1, our approach involved attributing the full-length transcripts to one of the two subgenomes of M. canariensis, M. can1 and M. can2, on the basis of differences in inferred times to their common ancestor, either with each other or with an outgroup, the putative progenitor M. annua. Our analysis confirmed that diploid M. annua was one of the two progenitors of M. canariensis, as inferred by Obbard et al. [55]. It also identified the M. can1 subgenome as the more closely related to present-day M. annua (P1 in Fig 1), and the M. can2 subgenome as derived from the unknown progenitor (P2). With the robust identification of the two subgenomes of M. canariensis, we then sought evidence for biased gene expression and gene retention in M. canariensis, including the possible evolution of subgenome dominance. We also asked whether such patterns are consistent with a model of sexual subfunctionalization.
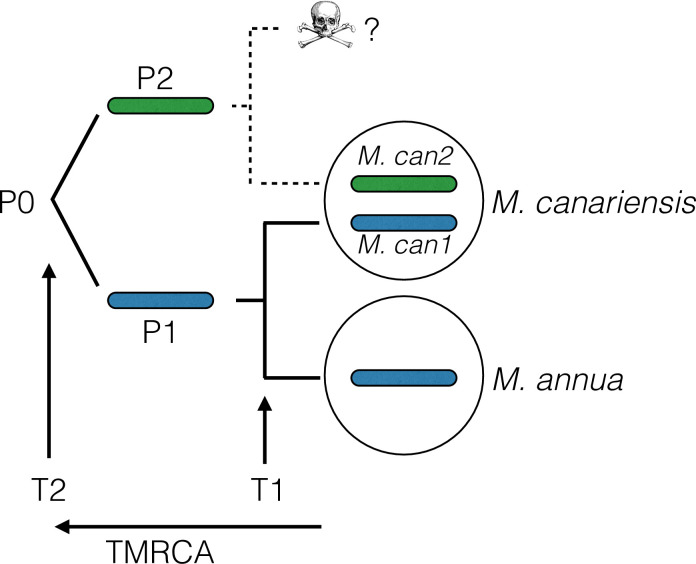
Fig 1. Conceptual approach for identifying the putative progenitors of each of the two subgenomes of M. canariensis, M. can1 and M. can2.
P0 is the most recent common ancestor (MRCA) of the two progenitors of allopolyploid M. canariensis, P1 is the progenitor of the first subgenome of M. canariensis, M. can1, and the MRCA of M. can1 and extant diploid M. annua. P2 is the progenitor of the second subgenome of M. canariensis, M. can2, and a second likely extinct lineage. T1 and T2 are the times to the most recent common ancestors (TMRCA), indicated by the coalescence time arrow. We assigned transcripts from M. canariensis to one or other of the two subgenomes on the basis of silent-site divergence, dS, from the homologous sequence in M. annua: we inferred that transcripts with lower dS were derived from P1, whereas those with a higher dS were derived from P2 (because they share the more ancient MRCA with M. annua, P0).
Results
Transcriptome assembly and homeolog identification
To obtain full-length transcripts of Mercurialis canariensis, we used PacBio long-read technology to sequence the RNA from the mature leaves of a female plant grown in the University of Lausanne glasshouses. The Isoseq pipeline identified 24,374 full-length transcripts (all datasets available in [58]). As this approach identifies and retains multiple isoforms per gene, we developed a pipeline to select only the longest isoform (See S1 Fig; all pipelines available in [59]). We used an all-vs-all blat to identify transcripts with high sequence identity to form transcript clusters, and we selected the longest isoform per cluster; this resulted in the identification of 12,918 transcripts for M. canariensis. We applied the same pipeline to select the longest isoform in the published M. annua transcriptome [60], from which we retained 21,230 of the 39,302 transcripts originally identified.
Next, we extracted homeologous sequence pairs derived from each of the two subgenomes from the reduced transcriptome of M. canariensis (i.e., consisting of only the longest isoform per gene cluster). To identify homeologs, we again used an all-vs-all blat within the reduced M. canariensis transcriptome and assigned reciprocal best blat hits to homeolog pairs. The median silent-site divergence, dS, between the putative homeologs was 0.14, consistent with a recent polyploidization event between two recently diverged parental lineages [61]. Each M. canariensis transcript was then mapped to the reduced M. annua transcriptome using blat, and dS was calculated between the M. canariensis transcript and the M. annua best hit (S1 Fig). We retained reciprocal best hit pairs as homeologs from our all-vs-all blat within M. canariensis if they shared a best hit in the M. canariensis-M. annua blat. This resulted in 3,135 putative homeolog pairs.
The M. annua lineage is one of the ancestors of M. canariensis
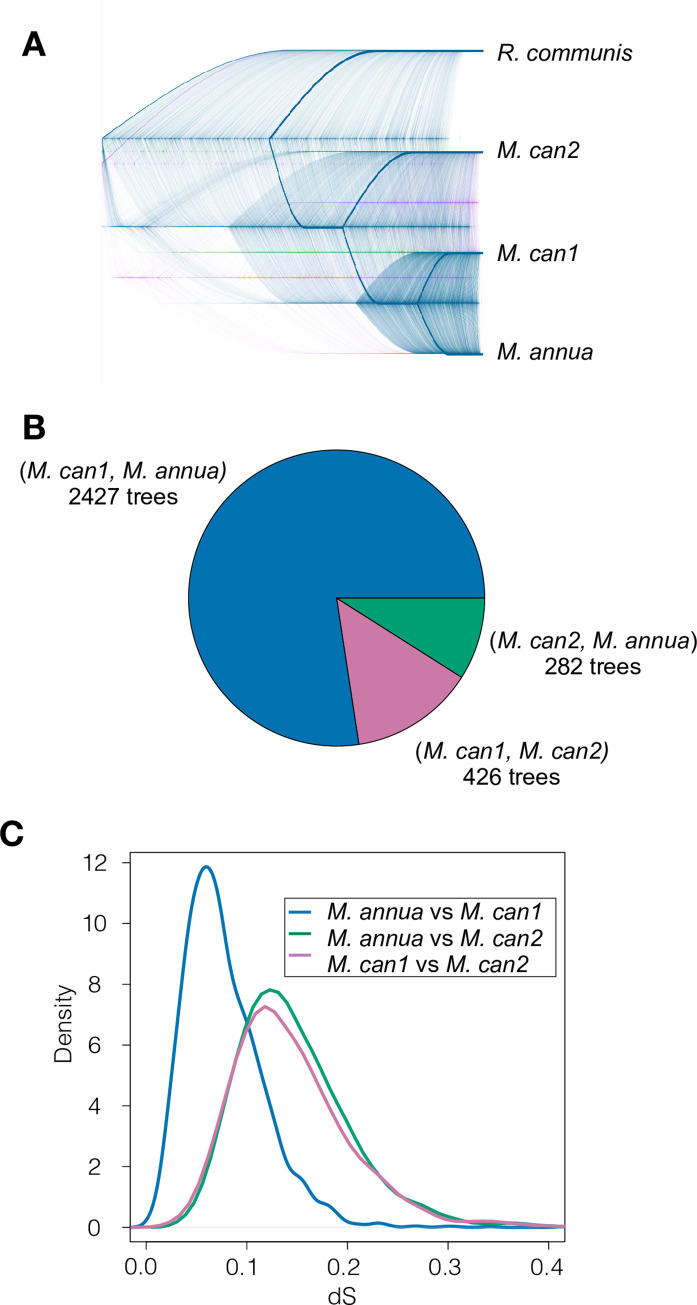
We used dS from orthologous sequences in M. annua to assign each homeolog to either subgenome M. can1 or M. can2 (Fig 1). We preliminarily assigned the homeolog with the lower dS to the M. can1 subgenome (Fig 1; derived from M. annua), and the homeolog with the higher dS to the M. can2 subgenome (derived from an unsampled and likely extinct progenitor; Fig 1). We constructed gene trees to test the hypothesis that M. annua was the progenitor of the M. can1 subgenome. For each homeolog pair, we included putative M. can1 and M. can2 homeologs, their ortholog in M. annua, and the corresponding 1:1 ortholog between M. annua and the more distant outgroup species Ricinus communis. The majority of trees (2427/3135 = 77%) had the expected topology of (((M. can1, M. annua), M. can2), R. communis) (Fig 2A and 2B).
Fig 2. The M. can1 subgenome is sister to M. annua.
(A) Gene trees constructed from 3,135 homeolog pairs in M. canariensis and their orthologs in M. annua and R. communis. Blue, pink and green gene trees indicate those in which M. can1 is sister to M. annua, M. can1 is sister to M. can2, and M. can2 is sister to M. annua, respectively. (B) Pie chart showing the proportion of trees in each category, and their respective topologies. (C). Density distribution of dS for M. annua vs. M. can1 subgenome (blue), M. annua vs. M. can2 subgenome (green), and M. can1 vs. M. can2 subgenomes (pink) among the 2,427 trees with the hypothesized topology.
We then examined the dS distributions of only those homeologs whose gene trees resulted in the expected topology. (Discordant topologies might be the result of assembly errors, or gene movement between subgenomes, e.g., due to recombination between homeologs.) First, we confirmed that the mean dS between M. annua and M. can2 was significantly larger than between M. annua and M. can1 (Fig 2C; Wilcoxon test, p < 2.2 x 10−16). Furthermore, if the M. can1 subgenome was derived from M. annua, and our subgenome assignments are correct, then the divergence between M. can1 and M. can2 should be approximately equal to the divergence between M. annua and M. can2 (Fig 1). Consistent with this hypothesis, we detected no difference between these two distributions (Fig 2C; Wilcoxon test, p = 0.067). Only homeologous pairs with the tree structure shown in Fig 2A were included in subsequent analyses.
To test the robustness of our results, we also employed an alternative method of homeolog subgenome assignment. Pairs were defined as above (having a shared a best hit in the M. canariensis-M. annua blat), but were filtered only on the basis of our hypothesized gene tree. The member of the pair that was inferred as sister to M. annua was assigned to the M. can1 subgenome, while other was assigned to the M. can2 subgenome. This more inclusive method resulted in 2709 pairs. Importantly, all of our results were consistent between the two methods (S2 and S3 Figs).
M. canariensis inherited the SDR from M. annua
To determine the location of the sex-determining region in M. canariensis, we used paired-end short-read RNA-sequencing from six males and six females sampled from six populations in Tenerife. Previous work by Veltsos et al. [60] used a combination of linkage mapping and data from natural populations to demonstrate that the sex-determining region of M. annua is on linkage group 1. We thus obtained linkage group assignments for M. annua transcripts from Veltsos et al. [60]. Of the 21,230 transcripts in the reduced transcriptome of M. annua, 8,181 could be assigned to linkage groups. We then used the M. annua ortholog position in the corresponding linkage group to assign M. canariensis homeologs to a genomic location in either the M. can1 or M. can2 subgenomes, as previously inferred. This resulted in 1,471 homeologous pairs assigned to linkage group positions in M. canariensis.
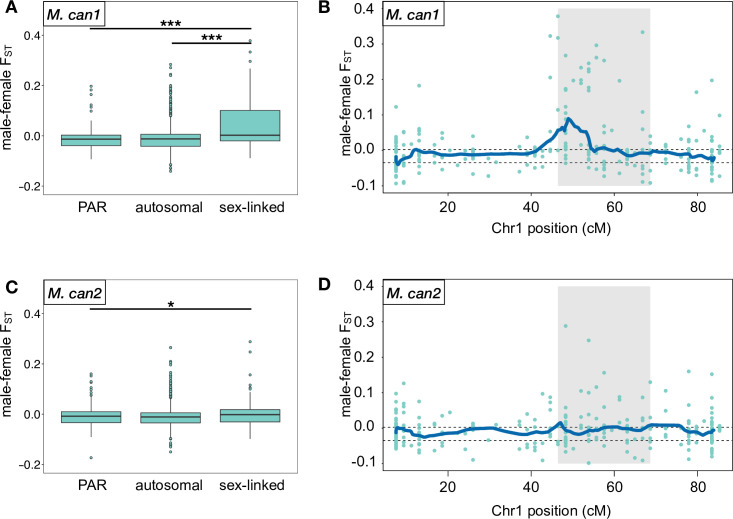
To detect sex-linked regions in the M. canariensis genome, we computed FST between males and females for each transcript. As a first step, we categorized each transcript as sex-linked, autosomal, or pseudoautosomal (i.e., in the recombining pseudoautosomal region, PAR), again based on the M. annua linkage map [60]. To test the hypothesis that M. canariensis inherited its sex-linked region from M. annua, we compared the sex-linked regions to the PAR and autosomes within each subgenome. We found that FST between males and females was elevated in the sex-linked region relative to the PAR and the autosomes in the M. can1 subgenome (Fig 3A; sex-linked vs. PAR, Wilcoxon test: p = 4.80 x 10−6; sex-linked vs. autosomes: p = 1.70 x 10−7). In the M. can2 subgenome, by contrast, there was no difference between the sex-linked region and the PAR (Fig 3C; Wilcoxon test: p = 0.2351), and FST in the sex-linked region was elevated relative to the autosomes, but the effect was much weaker than in the M. can1 subgenome and likely due to residual mapping of reads derived from M. can1 (Fig 3C; Wilcoxon test: p = 0.02998). Moreover, there was no significant difference between the autosomes or PAR in either subgenome (Fig 3A; Wilcoxon test: p = 0.9191, Fig 3C; Wilcoxon test: p = 0.2917), and no regions of elevated FST were detected on the autosomes (S4 Fig). This pattern implies that the sex-determining region is in the M. can1 subgenome and thus derived directly from a recent ancestor of extant M. annua.
Fig 3. FST between males and females in M. canariensis.
(A) Boxplot of FST between males and females in the M. can1 subgenome, with autosomal, pseudoautosomal, and sex-linked categories defined according to the linkage map for M. annua [60]. (B). Rolling average of 20 transcripts across linkage group 1 in the M. can1 subgenome. Grey shaded regions indicate the sex-linked region inferred for M. annua [60]. Horizontal dashed lines show 95% CI based on comparison with autosomes. (C). Boxplot of FST between males and females in autosomal, pseudoautosomal, and sex-linked regions in M. can2 subgenome. (D) Rolling average of 20 transcripts across linkage group 1 in M. can2 subgenome.
To locate the sex-determining region more precisely, we computed FST in a sliding window. We calculated FST for each transcript and computed a rolling average of FST for 20 consecutive transcripts. To compute 95% confidence intervals, we similarly computed the rolling average for 20 consecutive transcripts on autosomes 1,000 times (linkage groups 2 to 8 in M. can1, i.e., excluding the putative sex chromosome; all linkage groups of M. can2). This identified a single region of elevated FST corresponding to linkage group 1 of the M. can1 subgenome (Fig 3B). This region largely overlapped with the sex-determining region identified in M. annua [60]. We detected no other regions of elevated FST elsewhere in the M. can1 or M. can2 subgenomes (Figs 3D and S4).
No evidence of subgenome dominance
To evaluate the possibility of preferential gene loss or inactivation in one subgenome over the other, we aligned our short reads from M. canariensis to the M. annua transcriptome. Both members of the homeologous pair should align to their ortholog in M. annua, resulting in increased within-individual heterozygosity when both members of the pair are present relative to when only one homeolog is present or expressed. It is worth noting that reads derived from the M. can2 subgenome are expected to map less efficiently than reads derived from the M. can1 subgenome, potentially leading to an underestimate of heterozygosity. The distribution of heterozygosity (measured in terms of within-individual SNP density, S5 Fig) shows that the majority of genes are highly heterozygous, as expected for a polyploid genome. A secondary smaller peak is found at low heterozygosity (consisting almost exclusively of genes for which only one transcript was found in the assembly), arguing against widespread gene loss. Since M. can1 is less diverged from M. annua than M. can2, divergence with M. annua should be lower when M. can1 is present than when M. can2 is maintained instead. We therefore computed the average SNP density relative to M. annua as a proxy for divergence. Putative single-copy genes (heterozygosity<0.01) had a wide range of divergence levels, suggesting that gene loss has not occurred primarily in one of the two subgenomes (S5 Fig).
We then asked whether the strength of purifying selection differed between M. can1 and M. can2. We computed Tajima’s D and πN/πS for each transcript in both subgenomes. Tajima’s D is lower in M. can2 than M. can1 for all eight linkage groups (S6 Fig; paired Wilcoxon test, p = 0.007813). The fact that Tajima’s D is low across the whole genome points to demographic fluctuations rather than selection as responsible for distorting the coalescent branch lengths. Furthermore, πN/πS did not differ between the two subgenomes (S7 Fig; paired Wilcoxon test, p = 0.3828). All measures of π (π, πN, and πS) are elevated in M. can1 compared to M. can2 (S8–S10 Figs; paired Wilcoxon test: p = 0.00781 for all three measures). M. can1 is thus generally more polymorphic than M. can2, likely because there was a greater level of ancestral polymorphism in the M. annua-like ancestor of M. can1 than the (unknown) ancestor of M. can2. We therefore conclude that there is no evidence of different levels of purifying selection between the two subgenomes.
Finally, we characterized expression for each transcript in the reduced M. canariensis transcriptome. In our analyses, we considered all genes assigned to a subgenome, regardless of whether or not they can be assigned to a linkage group location based on the M. annua linkage map. Of the 2,427 pairs examined, nearly half (1,018) had significantly different expression between the two subgenomes in females, and approximately 1/3 (880 genes) were expressed at different levels in males. However, in both sexes the direction in subgenome bias was approximately equal, with 515 expressed more highly in M. can1 and 503 expressed more highly in M. can2 in females and 447 expressed more highly in M. can1 and 433 expressed more highly in M. can2 in males (S11 Fig). We therefore find no evidence for subgenome dominance in M. canariensis.
Low levels of sex-biased gene expression and sexual subfunctionalization
We detected 12,918 genes expressed in mature leaves; of these, 80 (0.6%) were sex-biased, with 46 and 34 male- and female-biased, respectively (S12 Fig). We could only assign 17 of the 80 sex-biased genes to linkage groups, and only 10 to linkage groups in the M. can1 subgenome. However, four of the ten sex-biased genes (40%) in the M. can1 subgenome were located on linkage group 1, a proportion that is substantially greater than the 17% (262/1,500) of all expressed genes on linkage group 1 in the M. can1 subgenome. However, although this trend is similar to that found for diploid M. annua [62], it falls short of statistical significance (Fisher’s exact test, p = 0.0807).
We asked whether homeologous pairs show evidence of sexual subfunctionalization. Sexual subfunctionalization could consist of individual homeologous pairs, with one member being male-biased and the other being female-biased. Alternatively, only one member of a homeologous pair may evolve sex-bias, while the other may retain a sex-independent function [26]. Finally, it is also possible that both members of the homeologous pair are sex-biased in the same direction, with one more so than the other.
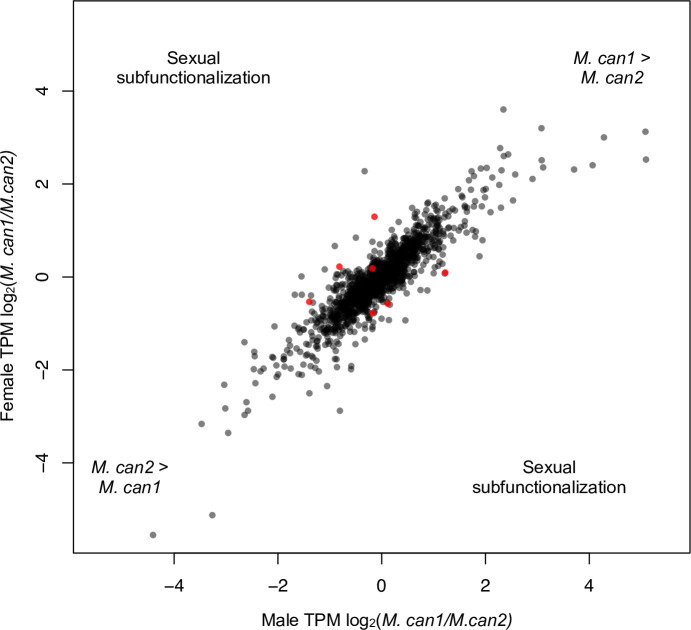
To identify genes with the most divergent sex bias between the homeolog pairs, we computed the log2(TPM M. can1/TPM M. can2) for each individual. We then compared these ratios between males and females using the “robust” option in the R package Limma [63]. We identified seven genes showing evidence of sexual subfunctionalization (Fig 4). We found a similar number of genes that were female-biased in M. can1 and male-biased in M. can2 (3) as the reverse (3), suggesting that sexual subfunctionalization evolves independently within a given homeologous pair. Additionally, we detected one gene that was more male-biased in M. can1 than M. can2. While we only identify seven genes with evidence of sexual subfunctionalization, two of these genes were located in the SDR and were more female-biased in M. can1 than M. can2. While this result suggests that the SDR of M. can1 is enriched for sexually subfunctionalized genes, it falls short of statistical significance (Fisher’s exact test, p = 0.05742). Importantly, these results suggests that genes on the Y chromosome may be undergoing degeneration on a gene-by-gene basis, but we do not detect a large-scale pattern of Y degeneration.
Fig 4. Potential subfunctionalization of M. can1 and M. can2 subgenomes.
Expression is shown in TPM (transcripts per million). Several homeolog pairs are subfunctionalized (expressed higher in one subgenome). Seven pairs of homeologs showed evidence of sexual subfunctionalization (shown in red).
Discussion
Origin of allopolyploid M. canariensis and its sex-determining region
Our results indicate that the diploid M. annua lineage was one of the two progenitors of allopolyploid M. canariensis, confirming the conclusions of Obbard et al. [55]. Further, M. canariensis inherited its sex-determining region (SDR) directly from M. annua (see S13 Fig for a more detailed hypothesis of M. canariensis formation). The SDR of M. canariensis spans a region from approximately 40 cM to 55 cM on linkage group 1 of the M. annua linkage map [60]. As in the sex-determining region of M. annua [60], divergence between the X and Y chromosomes in this region in M. canariensis is low, with no deficit of heterozygosity in males (S14 Fig). Interestingly, the inferred SDR for M. canariensis only partially overlaps with that inferred for M. annua ([60]; Fig 3B), suggesting that the M. annua SDR may have expanded since the allopolyploid origin of M. canariensis. This interpretation is consistent with the previous identification of two strata on the sex chromosomes of M. annua, the younger of which emerged after M. annua diverged from its sister species, M. huetii, approximately a million years ago [60], though that study was unable to locate these strata precisely using the M. annua linkage map. Given that the SDR of M. canariensis overlaps with that of M. annua in the region of 46 cM to 55 cM (based on the corresponding female linkage map of M. annua; [60]), this region might harbor the sex-determining locus of all three species and have stopped recombination earliest. Finally, we note that the SDR of M. canariensis might have expanded in the opposite direction to that of M. annua after its allopolyploid origin (i.e., towards ~ 40 cM), although evidence for this possibility is much weaker.
Subgenome dominance and sexual subfunctionalization
We could detect no evidence for subgenome dominance in gene loss, purifying selection, or gene expression in mature leaf tissues of M. canariensis, in contrast to findings for other polyploid species (e.g.,[64–66]. It is possible that genome dominance is absent in M. canariensis because the two progenitor species were not sufficiently highly diverged, with little difference in the content and distribution of transposable elements at the time of polyploidization, which is thought to be its basis [65,67,68]. It is also possible that M. canariensis originated so recently that gene loss and silencing have simply not had time to establish a dominant subgenome. To better understand the timing of these events, we estimated the TMRCA of M. can1 and M. annua, and M. can1 and M. can2. Using a mutation rate of 7.5 x 10−9 [60], we estimated that the two subgenomes diverged ~9.4 million generations ago, and that M. can1 and M. annua diverged ~4.7 million generations ago. Finally, subgenome dominance may indeed have evolved in M. canariensis, though in tissues other than the mature leaves that we sampled, as has been found, for instance, in allotetraploid blueberry [69] and cotton [65].
The small proportion of sex-biased genes that we observed in mature leaves of M. canariensis was similar to that found for diploid M. annua (~2%; [62]) as well as the vegetative tissues of several other plant species including Populus tremula [70], Populus balsamifera [71], and Salix viminalis [72]. However, in diploid M. annua, sex-biased genes were three times more likely to be female-biased than male-biased, whereas in M. canariensis we found slightly more male-biased genes. Sex-biased genes detected in vegetative tissues are overrepresented on the sex chromosomes of M. annua [62]. Similarly, examination of gene expression reproductive tissues has detected an enrichment of sex-biased genes in the sex-linked region in Asparagus officinalis [73], Silene latifolia [74], and Cannabis sativa [75], but not in Populus balsamifera [71] or Salix viminalis [72]. We also detected an overrepresentation of sex-biased genes on the sex chromosomes, though the difference was not statistically significant.
The differences in sex-biased gene expression between two closely related species that share a genome contribute to an emerging picture of rapid change in gene expression over time, and between different expression contexts [76]. In the case of Mercurialis, Cossard et al. [62] found little correspondence between the identity of male- or female-biased genes even between different stages of leaf development in diploid M. annua separated by a few days in early seedling growth. Thus, while sex-biased gene expression in plants is of substantial interest for understanding the evolution of sexual dimorphism [71–74,77,78], and indeed the evolution of gene expression itself in different genetic and phenotypic contexts, interpretations of its importance and meaning ultimately need to be articulated in the context of these apparent vicissitudes in expression.
We found that seven gene pairs that showed evidence of sexual subfunctionalization in M. canariensis, i.e., a divergence in expression between the two homeologous gene pairs that corresponds to male versus female functions. Thus, in our analysis, sexual subfunctionalization in M. canariensis was limited to a few genes in vegetative tissues. However, we examined only a small proportion of the genome (1503 homeologous pairs out of an estimated 21,000), so our power to detect sexual subfunctionalization was limited. Nevertheless, sexual dimorphism in gene expression tends to be greater in reproductive tissues than vegetative tissues in plants [71,72], so it seems possible that sexual subfunctionalization may have evolved for more genes in M. canariensis that are expressed in sexually dimorphic reproductive tissue, e.g., flower buds.
Evolutionary history of dioecy in Mercurialis
Our study throws further light on the role of allopolyploidy in the diversification of Mercurialis. Obbard et al.[55] hypothesized that the diploid M. annua was a progenitor of M. canariensis on the basis of phylogenetic inference using ITS and chloroplast DNA sequences. In addition, both tetraploid M. annua, a monoecious lineage currently distributed in central western Morocco, and the two hexaploid lineages currently distributed in the Iberian Peninsula [79] also have ITS and chloroplast sequences that point to an ancestor of diploid M. annua as a progenitor [55]. It is now clear that an ancestor of diploid M. annua has been a progenitor for more than one polyploid species, with two or more independent origins. Polyploidy is thought to be polyphyletic in other plant clades too, with ploidal races comprising populations with more than one independent origin, involving the same progenitor or progenitors [79–81]. In other cases, the same diploid progenitor has contributed to the polyploid origin of more than one species by hybridizing with more than one other diploid species [81,82]. For instance, the diploid plant Tragopogon dubius was inferred to have been a progenitor of both T. mirus and T. miscellus via allopolyploid hybridization with T. porrifolius and T. pratensis, respectively [83]. M. annua provides a further such example, with no fewer than three distinct lineages now carrying and expressing its genome.
As just noted, the allotetraploid and allohexaploid lineage of M. annua to which diploid M. annua has contributed its genome are composed largely of monoecious individuals. Perhaps significantly, the early descendants of artificial neo-polyploids of M. annua, as reported by Durand [54], are also monoecious, suggesting that polyploidization might have played a role in the breakdown of dioecy in polyploid M. annua, at least initially. If this is a general feature of the effect of genome duplication in the genus [30,31], then the fact that M. canariensis is dioecious would indicate that dioecy re-evolved subsequent to its loss. In part of the range of hexaploid M. annua, androdioecy appears to have evolved back to subdioecy, with the monoecious individuals producing almost no male flowers (thus being functionally female), and population sex ratios being close to 1:1 [84]. Such a process might have given rise to dioecy in M. canariensis, too. Obbard et al. [55] hypothesized that the male-determining allele in androdioecious hexaploid M. annua might have been donated by its other putative diploid progenitor, M. huetii, because its plastid genome evidently came from M. annua. Verification of that scenario awaits comparison of the hexaploid M. annua Y chromosome with the Y chromosomes of M. huetii. In contrast, because our study here involved characterization of the sex-determining region of M. canariensis, it is immediately possible to infer not only that M. annua was one of its two diploid progenitors, but also that M. annua is the origin of the M. canariensis sex-determining locus. Thus, whether or not dioecy in M. canariensis evolved via androdioecy, as might be the case for the subdioecious populations of hexaploid M. annua in Morocco, it seems that its dioecious sexual system did not originate in either lineage with the evolution of a completely new sex-determining locus.
Our results also suggest that the same sex-determining system is likely shared across all the annual Mercuries lineages with separate sexes. Previous work by Russell and Pannell [56] used interspecific crosses to show that the underlying architecture of sex determination has been maintained throughout the M. annua and M. huetii species complex. Interspecific crosses of males from diploid M. annua, M. huetii, or hexaploid androdioecious M. annua with hermaphrodites or females of any of these species (including tetraploid M. annua) produced ~ 50% male offspring, suggesting that the Y chromosome in males of Mercurialis retain their dominance over the recessive X chromosome locus in females and hermaphrodites [56]. Although Russell and Pannell [56] did not include M. canariensis in their crossing scheme, our study suggests that they would have had similar results with it, too.
Implications for our understanding of ploidy and sexual-system evolution
Polyploidization is a process that can involve a disruption of meiotic segregation, gene expression, the proliferation of transposable elements, and epigenetic marking [8,30,85]. When it involves dioecious species, polyploidization might also interfere with sex determination, precipitating the breakdown of dioecy and the reversion to hermaphroditism [30], as was found in artificially generated tetraploids of M. annua cited above (Durand 1963). However, our study demonstrates the inheritance of an intact sex-determining region in an allopolyploid from a diploid progenitor. Although this a possibility was implied by the results Obbard et al [55] and Russell and Pannell [56], our results here represent the first more direct indication on the basis of genome analysis for the retention of dioecy through allopolyploidization.
Gene duplication has been shown to resolve sexual antagonism in a number of diploid taxa by breaking sexual correlations due to pleiotropic constraints [26–28]. Our study extends this idea to homeologous pairs in a polyploid plant. While we only found evidence for sexual subfunctionalization for a few genes, it may also have played a role in the evolution of gene expression in reproductive tissues in M. canariensis, which we did not sample, and may be important in other polyploid species. In this sense, we believe that sexual subfunctionalization may be an underappreciated possibility in evolution of dioecy in polyploids, including those derived from hermaphroditic diploids. It is of course also possible that sexual subfunctionalization may be an alternative route to subgenome dominance in dioecious polyploids, though this does not yet appear to have been examined in any other species.
Finally, we note that M. canariensis is the only known polyploid Mercurialis species extant on an island. Dioecy is more common in island taxa than their relatives on the mainland [86–91], a fact that may appear surprising, given that self-compatible hermaphrodites are thought to be able to colonize islands more easily than self-incompatible hermaphrodites or dioecious species [92,93]. It is possible that the higher frequency of dioecy on oceanic islands is due to the evolution of separate sexes after colonization [92]. It is often difficult to know whether a dioecious island species evolved separate sexes prior to colonization or afterwards, although phylogenetic information can be helpful [88]. In the case of M. canariensis, it seems likely that dioecy evolved prior to colonization, given that at least one of its progenitors, M. annua, is a widespread continental species. If so, either a male and a female colonized together or, more likely perhaps, the island was colonized by a ‘leaky’ male that produced one or a few female flowers. Such leakiness in sex expression is common in diploid M. annua [94,95] and may have been present in the colonizing ancestors of extant M. canariensis.
Materials & methods
Study material
We sampled seeds of M. canariensis from six populations on the island of Tenerife. Six males and seven females were grown together in the greenhouse at the University of Lausanne in the spring of 2017. For each individual, we extracted RNA from 10 mature leaves using the Qiagen plant RNAeasy kit. For six males and six females, individual libraries were prepared using the TruSeq Stranded mRNA Sample Prep Kit and sequenced on the Illumina HiSeq2000. The remaining female was sequenced on a PacBio SMRT Cell on the Sequel I platform. All samples were sequenced at the Center of Integrative Genomics at the University of Lausanne.
Transcriptome and quality control
The Center of Integrative Genomics processed the PacBio sequencing data using the long-read isoform sequencing pipeline, IsoSeq3. This sidesteps the problem of assembling transcripts from short reads by producing full-length transcripts. This resulted in a transcriptome assembly consisting of 24,375 transcripts. ORFs were extracted using Transdecoder v.5.5 [96].
We assessed sequence quality of our short reads using FASTQC v.0.11.6 (www.bioinformatics.babraham.ac.uk/projects/fastqc). Adapter removal and trimming used Trimmomatic v.0.36 [97], with options ILLUMINACLIP: TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW: 4:15 MINLEN:36. After filtering, we retained an average of 44.7 million paired-end sequences per sample.
Homeolog identification and genomic location
To identify homeologs with confidence, we first removed all redundant transcripts and isoforms from our IsoSeq assembly using a custom perl script. We first ran an all-vs-all blat within the IsoSeq3 assembly and identified transcripts with high sequence identity. We grouped transcripts together that had a match score > 50 and divergence of < 1%, and retained only the longest transcript. We also implemented this pipeline on the published transcriptome of M. annua.
We then used our reduced assemblies to identify homeologs with M. canariensis. We again implemented an all-vs-all blat within the M. canariensis transcriptome. We selected transcript pairs in which the second-best hits were reciprocal (as the best hit is the ‘self’). These were preliminarily assigned as homeolog pairs. In parallel, for each M. canariensis transcript, we used blat to identify the best hit in the nonredundant M. annua transcriptome. We required putative homeolog pairs to have the same best blat hit in the M. annua transcriptome.
For each homeolog pair, we calculated dS with the best-hit M. annua transcript using KaKs_calculator (http://code.google.com/p/kaks-calculator/). Within each pair, the transcript with lower dS was preliminarily assigned to the M. can1 subgenome (Fi 1A; putatively derived from M. annua) and the higher dS was assigned to M. can2 (putatively derived from an unknown progenitor species). Furthermore, we used Ka/Ks calculator (http://code.google.com/p/kaks-calculator/) to compute dS between homeologs.
To ensure that our homeolog pairs were indeed homeologs, as opposed to isoforms or alleles of the same gene, we constructed gene trees. We included both members of the homeolog pair, their ortholog in M. annua, and the reciprocal best-hit match between M. annua and Ricinus communis. We constructed gene trees with default parameters and the GTRGAMMA model of nucleotide evolution in RAxML [98], and rooted trees using Treebender in the Phylommand software package [99]. To visualize the resulting gene trees, we constructed a cloudogram using Densitree v.2.2.27 [100]. Homeologs were considered correctly assigned if the gene trees had the expected structure of (((M. can1, M. annua), M. can2), R. communis).
We used linkage map information from M. annua [60] to assign each homeolog pair to linkage groups. Veltsos et al. [60] assigned a total of 8490 transcripts to linkage groups, however some of these transcripts were removed in the nonredundant transcriptome. In total, 8181 nonredundant transcripts were assigned linkage group positions in M. annua. We parsed each pair into the M. can1 and M. can2 subgenomes based on their dS values and gene tree structure.
Identifying the sex-determining region in M. canariensis
To identify the sex-determining region, we evaluated allele frequency differences between males and females by computing FST [101–103]. In order to maximize the number of reads that map to each homeolog, we kept only the longest isoform per gene in all subsequent analyses, and used the exon-junction aware software STAR v.2.6 [104] for the alignment. This allows us to incorporate all read information when identifying the sex-determining region of M. canariensis. Further, it allows us to compare sex-biased gene expression, homeolog-biased gene expression, and sexual subfunctionalization of genes with confidence. We then processed the alignments with Picard Tools v.1.141 (http://broadinstitute.github.io/picard/). We ran Samtools v.1.11 mpileup [105] with the probabilistic alignment disabled, and called SNPs using Varscan v2.4.3 [106], with a minimum variant allele frequency of 0.15, and a minimum threshold for homozygotes of 0.85. We used VCFtools v.0.1.16 [107] to require a minimum of 10 reads per site for all individuals and a Phred quality score > 20. We then computed FST for each transcript over 100 bp using VCFtools v.0.1.16 [107].
As a second measure, we computed sex-biased heterozygosity for each transcript following the method outlined in Toups et al. [108]. We defined heterozygosity as the fraction of heterozygous sites per transcript, and we then computed the log10 ratio of male heterozygosity:female heterozygosity. This measure of sex-biased heterozygosity should be ~ 0 for autosomal transcripts. For sex chromosomes, sex-biased heterozygosity should be positive in less differentiated XY systems where males have an excess of heterozygous sites, and negative in highly differentiated XY systems due to hemizygosity in males.
We defined sex-linked, autosomal, and pseudoautosomal (PAR) regions of the two subgenomes of M. canariensis according to the M. annua linkage map [60] and assessed the significance between these regions using Wilcoxon signed rank tests. We computed moving averages in sliding windows of 20 transcripts on each linkage group using the rollmean function from the package zoo in R v.4.0.3. To identify regions of elevated FST and sex-biased heterozygosity, we computed 95% confidence intervals by sampling rolling averages of 20 consecutive autosomal transcripts 1000 times.
Expression analysis
To assess gene expression, we generated pseudoalignments of our reads to the reduced M. canariensis transcriptome using Kallisto v.0.43.1 [109]. We used the tximport package [110] to import transcript-level abundance, estimated counts, and transcript lengths for downstream analyses in DEseq2 [111]. As a first step, we analysed sex-biased gene expression for each M. canariensis transcript using count matrices. As a second step, we assessed differential expression of homeologs between subgenomes. In both analyses, we corrected p-values for multiple tests using Benjamini and Hochberg’s algorithm [112].
For our sexual subfunctionalization analysis, we converted our count information to TPM, and then computed the ratio of log2(M. can1 TPM/M. can2 TPM) for each sample. We then constructed a linear model for each gene pair in the R package limma [63] using the “robust” option. We corrected for multiple tests using the Benjamini and Hochberg algorithm [112], and identified significant gene pairs using an adjusted p-value of <0.05.
Computing measures of selection
To determine if one subgenome was under stronger purifying selection than the other, we computed two measures of selection, Tajima’s D and πN/πS, for each transcript. In addition, to better understand the differing demographic effects on the two subgenomes, we also calculated π, πN, and πS. Tajima’s D and pi were calculated in VCFTools v.0.1.16 [107] and πN, πA, and πN/πS were calculated using the PopPhyl pipeline [113]. To eliminate the effects of linkage, we computed the mean metric for each chromosome and compared the two subgenomes using paired Wilcoxon tests.
Supporting information
After assembly of long-read PacBio sequencing using the Isoseq3 pipeline, an all-vs-all blat was used to form clusters of highly similar sequences and only the longest isoform per cluster was retained in the transcriptome. A second all-vs-all blat was performed on the transcriptome consisting of the longest isoforms, and this was used to locate reciprocal best hits. These reciprocal best hits were treated as potential homeolog pairs. We then used blat to identify the closest sequence for each homeolog in the M. annua transcriptome, and filtered only for homeolog pairs that blat to the same M. annua transcript. Finally, we calculated dS between each member of a retained homeolog pair and M. annua, and the homeolog with lower dS was assigned to the M. can1 subgenome (putatively derived from P1 –the same lineage as M. annua) and the other homeolog was assigned to the M. can2 subgenome (putatively derived from P2, the unknown progenitor).
(TIF)
(A) Boxplot of FST between males and females in the M. can1 subgenome, with autosomal, pseudoautosomal, and sex-linked categories defined according to the linkage map for M. annua. (B) Rolling average of 20 transcripts across linkage group 1 in the M. can1 subgenome. Grey shaded regions indicate the sex-linked region inferred for M. annua. Horizontal dashed lines show 95% CI based on comparison with autosomes. (C) Boxplot of FST between males and females in autosomal, pseudoautosomal, and sex-linked regions in the M. can2 subgenome. (D) Rolling average of 20 transcripts across linkage group 1 in the M. can2 subgenome.
(TIF)
Expression is shown in TPM (transcripts per million). Several homeolog pairs are subfunctionalized (expressed higher in one subgenome). Six pairs of homeologs showed evidence of sexual subfunctionalization (shown in red).
(TIF)
FST in rolling windows of 20 transcripts across linkage groups 2–8 in both the M. can1 and M. can2 subgenomes. Horizontal dashed lines show 95% CI.
(TIF)
Blue triangles show values for transcripts in which both homeologs are present, whereas green circles show values when no homeolog is identified for a transcript. We hypothesized that when only the M. can1 homeolog was present, within-individual heterozygosity and divergence from M. annua should be low, while when only the M. can2 homeolog was present, within-individual heterozygosity should be low while divergence with M. annua should be high. However, because there are no two distinct distributions in the divergence from M. annua, it is not possible ¨to assign single homeologs to subgenomes.
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Genes that are significantly differently expressed between subgenomes are shown in red.
(TIF)
Genes that are significantly differently expressed between the sexes are shown in red.
(TIF)
M. canariensis most likely formed via a triploid bridge, which requires at least two hybridization events: one resulting in males and one resulting in females. Males most likely evolved when an unreduced gamete from diploid, male M. canariensis fused with a haploid gamete containing the X chromosome of the unknown progenitor species. This resulted in a triploid offspring, which backcrossed with a haploid gamete containing the X chromosome from the unknown progenitor species. Females also likely formed via a triploid bridge pathway, though there are at least three possible crossing scenarios that result in the XXXX genotype. Figure adapted from [114].
(TIF)
(A) Boxplot of SBH between males and females in the M. can1 subgenome, with autosomal, pseudoautosomal, and sex-linked categories defined according to the linkage map for M. annua [60]. (B) Rolling average of 20 transcripts across linkage group 1 in the M. can1 subgenome. The grey rectangle indicate the sex-linked region in M. annua [60]. Horizontal dashed lines show 95% CI based on comparison with autosomes. (C) Boxplot of SBH between males and females in autosomal, pseudoautosomal, and sex-linked regions in the M. can2 subgenome. (D) Rolling average of 20 transcripts across linkage group 1 in the M. can2 subgenome.
(TIF)
Acknowledgments
Plants were grown in Lausanne by Aline Revel, and RNA extraction and library preparation were performed by Dessislava Savova Bianchi. All sequencing and the IsoSeq3 analysis were carried out by Center for Integrative Genomics at the University of Lausanne. All other computational analyses were performed on the server at IST Austria.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. Sequencing data from this project are archived on the SRA under accession: BioProject PRJNA835047. The long-read assembly, bioinformatic pipeline, and all scripts are made available on Dryad at https://doi.org/10.5061/dryad.gf1vhhmsj.
Funding Statement
JRP was supported by the Swiss National Science Foundation (https://www.snf.ch/en), Sinergia grant 26073998. BV was supported by the European Research Council (https://erc.europa.eu/) under the European Union’s Horizon 2020 research and innovation program, grant number 715257. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci U S A. 2009;106(33): 13875–13879. doi: 10.1073/pnas.0811575106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA. On the relative abundance of autopolyploids and allopolyploids. New Phytol. 2016;210(2): 391–398. doi: 10.1111/nph.13698 [DOI] [PubMed] [Google Scholar]
- 3.Rice A, Šmarda P, Novosolov M, Drori M, Glick L, Sabath N, et al. The global biogeography of polyploid plants. Nat Ecol Evol. 2019;3(2): 265–273. doi: 10.1038/s41559-018-0787-9 [DOI] [PubMed] [Google Scholar]
- 4.Van de Peer Y, Ashman TL, Soltis PS, Soltis DE. Polyploidy: an evolutionary and ecological force in stressful times. Plant Cell. 2021;33(1): 11–26. doi: 10.1093/plcell/koaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landis JB, Soltis DE, Li Z, Marx HE, Barker MS, Tank DC, et al. Impact of whole-genome duplication events on diversification rates in angiosperms. Am J Bot. 2018;105(3): 348–363. doi: 10.1002/ajb2.1060 [DOI] [PubMed] [Google Scholar]
- 6.Tank DC, Eastman JM, Pennell MW, Soltis PS, Soltis DE, Hinchliff CE, et al. Nested radiations and the pulse of angiosperm diversification: Increased diversification rates often follow whole genome duplications. New Phytol. 2015;207(2): 454–467. doi: 10.1111/nph.13491 [DOI] [PubMed] [Google Scholar]
- 7.Soltis PS, Soltis DE. Ancient WGD events as drivers of key innovations in angiosperms. Curr Opin Plant Biol. 2016;30: 159–165. doi: 10.1016/j.pbi.2016.03.015 [DOI] [PubMed] [Google Scholar]
- 8.Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8(2): 135–141. doi: 10.1016/j.pbi.2005.01.001 [DOI] [PubMed] [Google Scholar]
- 9.Soltis PS, Marchant DB, Peer Y Van De, Soltis DE. Polyploidy and genome evolution in plants. Curr Opin Genet Dev. 2015;35: 119–125. doi: 10.1016/j.gde.2015.11.003 [DOI] [PubMed] [Google Scholar]
- 10.Wolfe KH. Yesterday’s polyploids and the mystery of diploidization. Nat Rev. 2001;2(5): 333–341. doi: 10.1038/35072009 [DOI] [PubMed] [Google Scholar]
- 11.Le Comber SC, Ainouche ML, Kovarik A, Leitch AR. Making a functional diploid: From polysomic to disomic inheritance. New Phytol. 2010;186(1): 113–122. doi: 10.1111/j.1469-8137.2009.03117.x [DOI] [PubMed] [Google Scholar]
- 12.Wendel JF. The wondrous cycles of polyploidy in plants. Am J Bot. 2015;102(11): 1753–1756. doi: 10.3732/ajb.1500320 [DOI] [PubMed] [Google Scholar]
- 13.Thomas BC, Pedersen B, Freeling M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006;16(7): 934–946. doi: 10.1101/gr.4708406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freeling M, Scanlon MJ, Fowler JF. Fractionation and subfunctionalization following genome duplications: Mechanisms that drive gene content and their consequences. Curr Opin Genet Dev. 2015;35: 110–118. doi: 10.1016/j.gde.2015.11.002 [DOI] [PubMed] [Google Scholar]
- 15.Langham RJ, Walsh J, Dunn M, Ko C, Goff SA, Freeling M. Genomic duplication, fractionation and the origin of regulatory novelty. Genetics. 2004;166(2): 935–945. doi: 10.1534/genetics.166.2.935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng F, Wu J, Cai X, Liang J, Freeling M, Wang X. Gene retention, fractionation and subgenome differences in polyploid plants. Nat Plants. 2018;4(5): 258–268. doi: 10.1038/s41477-018-0136-7 [DOI] [PubMed] [Google Scholar]
- 17.Schnable JC, Springer NM, Freeling M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci U S A. 2011;108(10): 4069–4074. doi: 10.1073/pnas.1101368108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freeling M, Woodhouse MR, Subramaniam S, Turco G, Lisch D, Schnable JC. Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr Opin Plant Biol. 2012;15(2): 131–139. doi: 10.1016/j.pbi.2012.01.015 [DOI] [PubMed] [Google Scholar]
- 19.Wendel JF, Lisch D, Hu G, Mason AS. The long and short of doubling down: polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr Opin Genet Dev. 2018;49: 1–7. doi: 10.1016/j.gde.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 20.Yoo MJ, Liu X, Pires JC, Soltis PS, Soltis DE. Nonadditive gene expression in polyploids. Annu Rev Genet. 2014;48: 485–517. doi: 10.1146/annurev-genet-120213-092159 [DOI] [PubMed] [Google Scholar]
- 21.Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4): 1531–1545. doi: 10.1093/genetics/151.4.1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones DM, Wells R, Pullen N, Trick M, Irwin JA, Morris RJ. Spatio-temporal expression dynamics differ between homologues of flowering time genes in the allopolyploid Brassica napus. Plant J. 2018;96(1): 103–118. doi: 10.1111/tpj.14020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramírez-González RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, et al. The transcriptional landscape of polyploid wheat. Science (80-). 2018;361(6403). doi: 10.1126/science.aar6089 [DOI] [PubMed] [Google Scholar]
- 24.Connallon T, Clark AG. The resolution of sexual antagonism by gene duplication. Genetics. 2011;187(3): 919–937. doi: 10.1534/genetics.110.123729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallach M, Betrán E. Intralocus sexual conflict resolved through gene duplication. Trends Ecol Evol. 2011;26(5): 222–228. doi: 10.1016/j.tree.2011.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyman MJ, Cutter AD, Rowe L. Gene duplication in the evolution of sexual dimorphism. Evolution (N Y). 2011;66(5): 1556–1566. [DOI] [PubMed] [Google Scholar]
- 27.Lipinska A, Cormier A, Luthringer R, Peters AF, Corre E, Gachon CMM, et al. Sexual dimorphism and the evolution of sex-biased gene expression in the brown alga ectocarpus. Mol Biol Evol. 2015;32(6): 1581–1597. doi: 10.1093/molbev/msv049 [DOI] [PubMed] [Google Scholar]
- 28.Cocquet J, Ellis PJI, Mahadevaiah SK, Affara NA, Vaiman D, Burgoyne PS. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet. 2012;8(9). doi: 10.1371/journal.pgen.1002900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett SCH, Hough J. Sexual dimorphism in flowering plants. J Exp Bot. 2013;64(1): 67–82. doi: 10.1093/jxb/ers308 [DOI] [PubMed] [Google Scholar]
- 30.Ashman T-L, Kwok A, Husband BC. Revisiting the dioecy-polyploidy association: alternate pathways and research opportunities. Cytogenet Genome Res. 2013;140: 241–55. doi: 10.1159/000353306 [DOI] [PubMed] [Google Scholar]
- 31.Pannell JR, Obbard DJ, Buggs RJA. Polyploidy and the sexual system: What can we learn from Mercurialis annua? Biol J Linn Soc. 2004;82(4): 547–560. doi: 10.1111/j.1095-8312.2004.00340.x [DOI] [Google Scholar]
- 32.Wertheim B, Beukeboom LW, Van De Zande L. Polyploidy in animals: Effects of gene expression on sex determination, evolution and ecology. Cytogenet Genome Res. 2013;140: 256–269. doi: 10.1159/000351998 [DOI] [PubMed] [Google Scholar]
- 33.Bomblies K, Higgins JD, Yant L. Meiosis evolves: Adaptation to external and internal environments. New Phytol. 2015;208: 306–323. doi: 10.1111/nph.13499 [DOI] [PubMed] [Google Scholar]
- 34.Westergaard M. The mechanism of sex determination in dioecious flowering plants. Adv Genet. 1958;9: 217–281. doi: 10.1016/s0065-2660(08)60163-7 [DOI] [PubMed] [Google Scholar]
- 35.Kinney M, Columbus T, Friar E. Dicliny in Bouteloua (Poaceae: Chloridoideae): Implications for the Evolution of Dioecy. Aliso. 2007;23(1): 605–614. doi: 10.5642/aliso.20072301.45 [DOI] [Google Scholar]
- 36.Singh RB, Smith BW. The mechanism of sex determination in Rumex acetosella. Theor Appl Genet. 1971;41(8): 360–364. doi: 10.1007/BF00277336 [DOI] [PubMed] [Google Scholar]
- 37.Smith BENW. Cytogeography and Cytotaxonomic Relationships of Rumex paucifolius. Am J Bot. 1968;55(6): 673–683. [Google Scholar]
- 38.Perley DS, Jesson LK. Hybridization is associated with changes in sexual system in the bryophyte genus Atrichum. Am J Bot. 2015;102: 555–565. doi: 10.3732/ajb.1400494 [DOI] [PubMed] [Google Scholar]
- 39.Kuhl H, Guiguen Y, Höhne C, Kreuz E, Du K, Klopp C, et al. A 180 Myr-old female-specific genome region in sturgeon reveals the oldest known vertebrate sex determining system with undifferentiated sex chromosomes. Proc R Soc B Biol Sci. 2021;376: 20200089. doi: 10.1098/rstb.2020.0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carey SB, Jenkins J, Lovell JT, Maumus F, Sreedasyam A, Payton AC, et al. Gene-rich UV sex chromosomes harbor conserved regulators of sexual development. Sci Adv. 2021;7: eabh2488. doi: 10.1126/sciadv.abh2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller JS, Venable DL. Polyploidy and the evolution of gender dimorphism in plants. Science (80-). 2000;289(5488): 2335–2338. doi: 10.1126/science.289.5488.2335 [DOI] [PubMed] [Google Scholar]
- 42.Tennessen JA, Wei N, Straub SCK, Govindarajulu R, Liston A, Ashman TL. Repeated translocation of a gene cassette drives sex-chromosome turnover in strawberries. PLoS Biol. 2018;16(8): 1–22. doi: 10.1371/journal.pbio.2006062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei N, Tennessen JA, Liston A, Ashman TL. Present-day sympatry belies the evolutionary origin of a high-order polyploid. New Phytol. 2017;216(1): 279–290. doi: 10.1111/nph.14711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liston A, Cronn R, Ashman TL. Fragaria: A genus with deep historical roots and ripe for evolutionary and ecological insights. Am J Bot. 2014;101(10_: 1686–1699. doi: 10.3732/ajb.1400140 [DOI] [PubMed] [Google Scholar]
- 45.Tennessen JA, Govindarajulu R, Liston A, Ashman TL. Homomorphic ZW chromosomes in a wild strawberry show distinctive recombination heterogeneity but a small sex-determining region. New Phytol. 2016;211(4): 1412–1423. doi: 10.1111/nph.13983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spigler RB, Lewers KS, Ashman TL. Genetic architecture of sexual dimorphism in a subdioecious plant with a proto-sex chromosome. Evolution (N Y). 2011;65(4): 1114–1126. doi: 10.1111/j.1558-5646.2010.01189.x [DOI] [PubMed] [Google Scholar]
- 47.Wei N, Govindarajulu R, Tennessen JA, Liston A, Ashman TL, Sayres MW. Genetic mapping and phylogenetic analysis reveal intraspecific variation in sex chromosomes of the Virginian strawberry. J Hered. 2017;108(7): 731–739. doi: 10.1093/jhered/esx077 [DOI] [PubMed] [Google Scholar]
- 48.Spigler RB, Lewers KS, Johnson AL, Ashman TL. Comparative mapping reveals autosomal origin of sex chromosome in octoploid Fragaria virginiana. J Hered. 2010;101: 107–117. doi: 10.1093/jhered/esp081 [DOI] [PubMed] [Google Scholar]
- 49.Spigler RB, Lewers KS, Main DS, Ashman TL. Genetic mapping of sex determination in a wild strawberry, Fragaria virginiana, reveals earliest form of sex chromosome. Heredity (Edinb). 2008;101(6): 507–517. doi: 10.1038/hdy.2008.100 [DOI] [PubMed] [Google Scholar]
- 50.Tennessen JA, Govindarajulu R, Ashman TL, Liston A. Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biol Evol. 2014;6(12): 3295–3313. doi: 10.1093/gbe/evu261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akagi T, Shirasawa K, Nagasaki H, Hirakawa H, Tao R, Comai L, et al. The persimmon genome reveals clues to the evolution of a lineage-specific sex determination system in plants. PLoS Genet. 2020;16(2): 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akagi T, Henry IM, Tao R, Comai L. A Y-chromosome–encoded small RNA acts as a sex determinant in persimmons. Science (80-). 2014;346(6209): 646–650. [DOI] [PubMed] [Google Scholar]
- 53.Masuda K, Fujita N, Yang HW, Ushijima K, Kubo Y, Tao R, et al. Molecular mechanism underlying derepressed male production in hexaploid persimmon. Front Plant Sci. 2020;11(12): 1–12. doi: 10.3389/fpls.2020.567249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Durand B. Le complexe Mercurialis annua L. s.l. une étude biosystématique. Ann des Sci Nat Bot Paris. 1963;12: 579–736. [Google Scholar]
- 55.Obbard DJ, Harris SA, Buggs RJA, Pannell JR. Hybridization, polyploidy, and the evolution of sexual systems in Mercurialis (Euphorbiaceae). Evolution (N Y). 2006;60(9): 1801–1815. [PubMed] [Google Scholar]
- 56.Russell JRW, Pannell JR. Sex determination in dioecious Mercurialis annua and its close diploid and polyploid relatives. Heredity (Edinb). 2015;114(3): 262–271. doi: 10.1038/hdy.2014.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Obbard DJ, Pannell JR, Harris SA. Mercurialis canariensis (Euphorbiaceae), a new endemic to the Canary Islands. Kew Bull. 2006;61(1): 99–106. doi: 10.2307/20443250 [DOI] [Google Scholar]
- 58.Toups MA, Vicoso B, Pannell JR. The maintenance of dioecy and chromosomal sex determination through allopolyploid speciation in the plant genus Mercurialis. Dryad, Dataset. 2022. 10.5061/dryad.gf1vhhmsj [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toups MA, Vicoso B, Pannell JR. The maintenance of dioecy and chromosomal sex determination through allopolyploid speciation in the plant genus Mercurialis. Zenodo, Softw. 2022. 10.5281/zenodo.6623367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Veltsos P, Ridout KE, Toups MA, Gonzalez-Martinez SC, Muyle A, Emery O, et al. Early sex-chromosome evolution in the diploid dioecious plant Mercurialis annua. Genetics. 2019;212(7): 815–835. doi: 10.1534/genetics.119.302045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cui L, Wall PK, Leebens-Mack JH, Lindsay BG, Soltis DE, Doyle JJ, et al. Widespread genome duplications throughout the history of flowering plants. Genome Res. 2006;16(6): 738–749. doi: 10.1101/gr.4825606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cossard GG, Toups MA, Pannell JR. Sexual dimorphism and rapid turnover in gene expression in pre-reproductive seedlings of a dioecious herb. Ann Bot. 2019;123(7): 1119–1131. doi: 10.1093/aob/mcy183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. 2015;43. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flagel LE, Wendel JF. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 2010;186(1): 184–193. doi: 10.1111/j.1469-8137.2009.03107.x [DOI] [PubMed] [Google Scholar]
- 65.Alger EI, Edger PP. One subgenome to rule them all: underlying mechanisms of subgenome dominance. Curr Opin Plant Biol. 2020;54: 108–113. doi: 10.1016/j.pbi.2020.03.004 [DOI] [PubMed] [Google Scholar]
- 66.Xu P, Xu J, Liu G, Chen L, Zhou Z, Peng W, et al. The allotetraploid origin and asymmetrical genome evolution of the common carp Cyprinus carpio. Nat Commun. 2019;10(1): 1–11. doi: 10.1038/s41467-019-12644-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edger PP, Smith R, McKain MR, Cooley AM, Vallejo-Marin M, Yuan Y, et al. Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, and a 140-year-old naturally established neo-allopolyploid monkeyflower. Plant Cell. 2017;29(9): 2150–2167. doi: 10.1105/tpc.17.00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun H, Wu S, Zhang G, Jiao C, Guo S, Ren Y, et al. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol Plant. 2017;10(19): 1293–1306. doi: 10.1016/j.molp.2017.09.003 [DOI] [PubMed] [Google Scholar]
- 69.Colle M, Leisner CP, Wai CM, Ou S, Bird KA, Wang J, et al. Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. Gigascience. 2019;8(3): 1–15. doi: 10.1093/gigascience/giz012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson KM, Delhomme N, Mähler N, Schiffthaler B, Önskog J, Albrectsen BR, et al. Populus tremula (European aspen) shows no evidence of sexual dimorphism. BMC Plant Biol. 2014;14: 1–14. doi: 10.1186/s12870-014-0276-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanderson BJ, Wang L, Tiffin P, Wu Z, Olson MS. Sex-biased gene expression in flowers, but not leaves, reveals secondary sexual dimorphism in Populus balsamifera. New Phytol. 2019;221(1): 527–539. doi: 10.1111/nph.15421 [DOI] [PubMed] [Google Scholar]
- 72.Darolti I, Wright AE, Pucholt P, Berlin S, Mank JE. Slow evolution of sex-biased genes in the reproductive tissue of the dioecious plant Salix viminalis. Mol Ecol. 2018;27(3): 694–708. doi: 10.1111/mec.14466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harkess A, Mercati F, Shan HY, Sunseri F, Falavigna A, Leebens-Mack J. Sex-biased gene expression in dioecious garden asparagus (Asparagus officinalis). New Phytol. 2015;207(3): 883–892. doi: 10.1111/nph.13389 [DOI] [PubMed] [Google Scholar]
- 74.Zemp N, Tavares R, Muyle A, Charlesworth D, Marais GAB, Widmer A. Evolution of sex-biased gene expression in a dioecious plant. Nat Plants. 2016;2(11). doi: 10.1038/nplants.2016.168 [DOI] [PubMed] [Google Scholar]
- 75.Prentout D, Razumova O, Rhoné B, Badouin H, Henri H, Feng C, et al. An efficient RNA-seq-based segregation analysis identifies the sex chromosomes of Cannabis sativa. Genome Res. 2020;30: 164–172. doi: 10.1101/gr.251207.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grath S, Parsch J. Sex-Biased Gene Expression. Annu Rev Genet. 2016;50: 29–44. doi: 10.1146/annurev-genet-120215-035429 [DOI] [PubMed] [Google Scholar]
- 77.Muyle A, Shearn R, Marais GAB. The evolution of sex chromosomes and dosage compensation in plants. Genome Biol Evol. 2017;9(3): 627–645. doi: 10.1093/gbe/evw282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muyle A. How different is the evolution of sex-biased gene expression between plants and animals? A commentary on: “Sexual dimorphism and rapid turnover in gene expression in pre-reproductive seedlings of a dioecious herb.” Ann Bot. 2019;123(7): IV–V. doi: 10.1093/aob/mcz081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma WJ, Santos Del Blanco L, Pannell JR. A new biological species in the Mercurialis annua polyploid complex: Functional divergence in inflorescence morphology and hybrid sterility. Ann Bot. 2019;124(1): 165–178. doi: 10.1093/aob/mcz058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gordon SP, Contreras-Moreira B, Levy JJ, Djamei A, Czedik-Eysenberg A, Tartaglio VS, et al. Gradual polyploid genome evolution revealed by pan-genomic analysis of Brachypodium hybridum and its diploid progenitors. Nat Commun. 2020;11(1): 1–16. doi: 10.1038/s41467-020-17302-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soltis DE, Soltis PS. Polyploidy: Recurrent formation and genome evolution. Trends Ecol Evol. 1999;14(9): 348–352. doi: 10.1016/s0169-5347(99)01638-9 [DOI] [PubMed] [Google Scholar]
- 82.Guo X, Mandáková T, Trachtová K, Özüdoğru B, Liu J, Lysak MA. Linked by ancestral bonds: Multiple whole-genome duplications and reticulate evolution in a Brassicaceae tribe. Mol Biol Evol. 2021;38(5): 1695–1714. doi: 10.1093/molbev/msaa327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Soltis DE, Soltis PS, Pires JC, Kovarik A, Tate JA, Mavrodiev E. Recent and recurrent polyploidy in Tragopogon (Asteraceae): Cytogenetic, genomic and genetic comparisons. Biol J Linn Soc. 2004;82(4): 485–501. doi: 10.1111/j.1095-8312.2004.00335.x [DOI] [Google Scholar]
- 84.Pannell JR, Eppley SM, Dorken ME, Berjano R. Regional variation in sex ratios and sex allocation in androdioecious Mercurialis annua. J Evol Biol. 2014;27(7): 1467–1477. doi: 10.1111/jeb.12352 [DOI] [PubMed] [Google Scholar]
- 85.Baduel P, Bray S, Vallejo-Marin M, Kolář F, Yant L. The “Polyploid Hop”: Shifting challenges and opportunities over the evolutionary lifespan of genome duplications. Front Ecol Evol. 2018;6: 1–19. doi: 10.3389/fevo.2018.00117 [DOI] [Google Scholar]
- 86.Pannell JR, Auld JR, Brandvain Y, Burd M, Busch JW, Cheptou P-O, et al. The scope of Baker’s law. New Phytol. 2015;208: 656–667. doi: 10.1111/nph.13539 [DOI] [PubMed] [Google Scholar]
- 87.Baker HG, Cox PA. Further thoughts on dioecism and islands. Ann Missouri Bot Gard. 1984;71(1): 244–253. [Google Scholar]
- 88.Sakai AK., Wagner WL, Ferguson DM, Herbst DR. Origins of dioecy in the Hawaiian flora. Ecology. 1995;76(8): 2517–2529. [Google Scholar]
- 89.Webb CJ, Lloyd DG, Delph LF. Gender dimorphism in indigenous New Zealand seed plants. New Zeal J Bot. 1999;37(1): 119–130. doi: 10.1080/0028825X.1999.9512618 [DOI] [Google Scholar]
- 90.Barrett SCH. The reproductive biology and genetics of island plants. Philos Trans R Soc B Biol Sci. 1996;351: 725–733. doi: 10.1098/rstb.1996.0067 [DOI] [Google Scholar]
- 91.Carlquist S. The biota of long-distance dispersal. III. Loss of dispersibility in the Hawaiian flora. Brittonia. 1966;18(4): 310–335. doi: 10.2307/2805148 [DOI] [Google Scholar]
- 92.Baker HG. Self-compatibility and establishment after “long-distance” dispersal. Evolution (N Y). 1955;9(3): 347–349. [Google Scholar]
- 93.Baker HG. Support for Baker’s Law—as a rule. Evolution (N Y). 1967;21(4): 853–856. doi: 10.1111/j.1558-5646.1967.tb03440.x [DOI] [PubMed] [Google Scholar]
- 94.Cossard GG, Pannell JR. Enhanced leaky sex expression in response to pollen limitation in the dioecious plant Mercurialis annua. J Evol Biol. 2021;34(2): 416–422. doi: 10.1111/jeb.13720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cossard GG, Pannell JR. A functional decomposition of sex inconstancy in the dioecious, colonizing plant Mercurialis annua. Am J Bot. 2019;106: 722–732. doi: 10.1002/ajb2.1277 [DOI] [PubMed] [Google Scholar]
- 96.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Philip D, Bowden J, et al. De novo transcript sequence recostruction from RNA-Seq: reference generation and analysis with Trinity. Nat Protoc. 2013;8(8): 1–43. doi: 10.1038/nprot.2013.084.De [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15): 2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9): 1312–1313. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Phylommand Ryberg M. -a command line software package for phylogenetics. F1000Research. 2019;2903(5): 1–8. [Google Scholar]
- 100.Bouckaert RR. DensiTree: Making sense of sets of phylogenetic trees. Bioinformatics. 2010;26(10): 1372–1373. doi: 10.1093/bioinformatics/btq110 [DOI] [PubMed] [Google Scholar]
- 101.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution (N Y). 1984;38(6): 1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x [DOI] [PubMed] [Google Scholar]
- 102.Gammerdinger WJ, Toups MA, Vicoso B. Disagreement in FST estimators: A case study from sex chromosomes. Mol Ecol Resour. 2020;20(6): 1517–1525. doi: 10.1111/1755-0998.13210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rodrigues N, Dufresnes C. Using conventional F-statistics to study unconventional sex-chromosome differentiation. PeerJ. 2017;4. doi: 10.7717/peerj.3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1): 15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16): 2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, et al. VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25(17): 2283–2285. doi: 10.1093/bioinformatics/btp373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15): 2156–2158. doi: 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Toups MA, Rodrigues N, Perrin N, Kirkpatrick M. A reciprocal translocation radically reshapes sex-linked inheritance in the common frog. Mol Ecol. 2019;28(8): 1877–1889. doi: 10.1111/mec.14990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34(5): 525–527. doi: 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- 110.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2015;4(2): 1521. doi: 10.12688/f1000research.7563.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12): 1–21. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57(1): 289–300. [Google Scholar]
- 113.Gayral P, Melo-Ferreira J, Glémin S, Bierne N, Carneiro M, Nabholz B, et al. Reference-free population genomics from next-generation transcriptome data and the vertebrate-invertebrate gap. PLoS Genet. 2013;9(4). doi: 10.1371/journal.pgen.1003457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Svačina R, Sourdille P, Kopecký D, Bartoš J. Chromosome Pairing in Polyploid Grasses. Front Plant Sci. 2020;11. doi: 10.3389/fpls.2020.01056 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
After assembly of long-read PacBio sequencing using the Isoseq3 pipeline, an all-vs-all blat was used to form clusters of highly similar sequences and only the longest isoform per cluster was retained in the transcriptome. A second all-vs-all blat was performed on the transcriptome consisting of the longest isoforms, and this was used to locate reciprocal best hits. These reciprocal best hits were treated as potential homeolog pairs. We then used blat to identify the closest sequence for each homeolog in the M. annua transcriptome, and filtered only for homeolog pairs that blat to the same M. annua transcript. Finally, we calculated dS between each member of a retained homeolog pair and M. annua, and the homeolog with lower dS was assigned to the M. can1 subgenome (putatively derived from P1 –the same lineage as M. annua) and the other homeolog was assigned to the M. can2 subgenome (putatively derived from P2, the unknown progenitor).
(TIF)
(A) Boxplot of FST between males and females in the M. can1 subgenome, with autosomal, pseudoautosomal, and sex-linked categories defined according to the linkage map for M. annua. (B) Rolling average of 20 transcripts across linkage group 1 in the M. can1 subgenome. Grey shaded regions indicate the sex-linked region inferred for M. annua. Horizontal dashed lines show 95% CI based on comparison with autosomes. (C) Boxplot of FST between males and females in autosomal, pseudoautosomal, and sex-linked regions in the M. can2 subgenome. (D) Rolling average of 20 transcripts across linkage group 1 in the M. can2 subgenome.
(TIF)
Expression is shown in TPM (transcripts per million). Several homeolog pairs are subfunctionalized (expressed higher in one subgenome). Six pairs of homeologs showed evidence of sexual subfunctionalization (shown in red).
(TIF)
FST in rolling windows of 20 transcripts across linkage groups 2–8 in both the M. can1 and M. can2 subgenomes. Horizontal dashed lines show 95% CI.
(TIF)
Blue triangles show values for transcripts in which both homeologs are present, whereas green circles show values when no homeolog is identified for a transcript. We hypothesized that when only the M. can1 homeolog was present, within-individual heterozygosity and divergence from M. annua should be low, while when only the M. can2 homeolog was present, within-individual heterozygosity should be low while divergence with M. annua should be high. However, because there are no two distinct distributions in the divergence from M. annua, it is not possible ¨to assign single homeologs to subgenomes.
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Genes that are significantly differently expressed between subgenomes are shown in red.
(TIF)
Genes that are significantly differently expressed between the sexes are shown in red.
(TIF)
M. canariensis most likely formed via a triploid bridge, which requires at least two hybridization events: one resulting in males and one resulting in females. Males most likely evolved when an unreduced gamete from diploid, male M. canariensis fused with a haploid gamete containing the X chromosome of the unknown progenitor species. This resulted in a triploid offspring, which backcrossed with a haploid gamete containing the X chromosome from the unknown progenitor species. Females also likely formed via a triploid bridge pathway, though there are at least three possible crossing scenarios that result in the XXXX genotype. Figure adapted from [114].
(TIF)
(A) Boxplot of SBH between males and females in the M. can1 subgenome, with autosomal, pseudoautosomal, and sex-linked categories defined according to the linkage map for M. annua [60]. (B) Rolling average of 20 transcripts across linkage group 1 in the M. can1 subgenome. The grey rectangle indicate the sex-linked region in M. annua [60]. Horizontal dashed lines show 95% CI based on comparison with autosomes. (C) Boxplot of SBH between males and females in autosomal, pseudoautosomal, and sex-linked regions in the M. can2 subgenome. (D) Rolling average of 20 transcripts across linkage group 1 in the M. can2 subgenome.
(TIF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. Sequencing data from this project are archived on the SRA under accession: BioProject PRJNA835047. The long-read assembly, bioinformatic pipeline, and all scripts are made available on Dryad at https://doi.org/10.5061/dryad.gf1vhhmsj.