Abstract
Concurrent with recent advances seen with Cryptosporidium parvum detection in both treated and untreated water is the need to properly evaluate these advances. A micromanipulation method by which known numbers of C. parvum oocysts, even a single oocyst, can be delivered to a test matrix for detection sensitivity is presented. Using newly developed nested PCR-restriction fragment length polymorphism primers, PCR sensitivity was evaluated with 1, 2, 3, 4, 5, 7, or 10 oocysts. PCR detection rates (50 samples for each number of oocysts) ranged from 38% for single oocysts to 92% for 5 oocysts, while 10 oocysts were needed to achieve 100% detection. The nested PCR conditions amplified products from C. parvum, Cryptosporidium baileyi, and Cryptosporidium serpentis but no other Cryptosporidium sp. or protozoan tested. Restriction enzyme digestion with VspI distinguished between C. parvum genotypes 1 and 2. Restriction enzyme digestion with DraII distinguished C. parvum from C. baileyi and C. serpentis. Use of known numbers of whole oocysts encompasses the difficulty of liberating DNA from the oocyst and eliminates the standard deviation inherent within a dilution series. To our knowledge this is the first report in which singly isolated C. parvum oocysts were used to evaluate PCR sensitivity. This achievement illustrates that PCR amplification of a single oocyst is feasible, yet sensitivity remains an issue, thereby illustrating the difficulty of dealing with low oocyst numbers when working with environmental water samples.
Cryptosporidiosis is a self-limiting diarrheal infection in the immunocompetent individual, and with proper oral hydration therapy a full recovery is expected (6). The disease is caused by apicomplexan parasite Cryptosporidium parvum and to date is still without effective chemotherapy (19). The lack of drug therapy is of major concern in immunocompromised individuals in which the parasite's unique self-perpetuating life cycle can cause long-duration diarrhea resulting in major fluid loss (6).
The infectious stage of C. parvum, the oocyst, is shed in the feces of an infected individual. The oocysts have three major features that contribute greatly to the survival and spread of the organism. First, when shed in the feces, oocysts are immediately infectious to the next host (8). This readily leads to direct fecal/oral transmission. Second, oocysts are environmentally resistant and can remain infectious for 2 to 3 months or longer under proper conditions (17). Last, oocysts are small in size (4 to 6 μm in diameter) and resistant to the normal chlorine disinfection level used in water treatment plants and distribution systems, consequently allowing for the spread of C. parvum via drinking water (10, 17). This was illustrated in 1993, when an estimated 400,000 persons in Milwaukee, Wis., succumbed to a waterborne disease outbreak in which Cryptosporidium played a major contributing role (13).
The 1993 Milwaukee outbreak has led to continued monitoring of C. parvum in both untreated (lakes, reservoirs, rivers, and groundwater) and treated water supplies. Also, since 1993, detection methods have progressively improved. Initial techniques included the Information Collection Rule (EPA-600-R-95–178) and membrane filter dissolution (1) and calcium carbonate flocculation protocols (16). The latest oocyst recovery technique from water samples is immunomagnetic separation (IMS) as described by Method 1623: Cryptosporidium and Giardia in Water by Filtration/IMS/FA (EPA-821-R-99–006). This protocol is becoming widely accepted, with recovery rates ranging from 62 to 100% (4, 9).
A limiting factor of all these detection techniques is that none is C. parvum specific. The protocols all follow the basic flow chart of concentrating a volume of water containing oocysts, followed by a purification step and detection with fluorescein isothiocyanate (FITC)-labeled monoclonal antibodies that lack C. parvum specificity. This is important, because not all species of Cryptosporidium are infectious for mammals but many do have the same size and shape of C. parvum oocysts. With the current detection methods lacking the ability to distinguish among species, some scientists have turned to molecular techniques to identify which species have been isolated from a water sample. These techniques include the PCR, restriction fragment length polymorphism (RFLP), and direct DNA sequencing (3, 11, 20). The need for improved oocyst isolation from water samples, followed by identification of the species of isolated Cryptosporidium oocysts, has led to the combination of IMS followed by PCR detection and species determination with RFLP (7, 12). This combination has been reported to detect small numbers of oocysts (12).
In addition, recent molecular studies have classified C. parvum into genotypes 1 and 2 (5, 18, 21). Genotype 1 is found in humans, while genotype 2 can be found in both infected humans and animals. These findings are of major concern in understanding the epidemiology of C. parvum infections and have resulted in the hypothesis of two separate transmission cycles, one anthroponotic and one zoonotic (14).
These current methods for detecting low numbers of oocysts utilize the dilution of either oocysts or C. parvum DNA to achieve the nominal value of a single oocyst. Using the dilution method with oocysts results in a number plus or minus the standard deviation, and therefore exact numbers of oocysts are not known. Using the dilution method with C. parvum DNA does not take into account the difficulty of liberating DNA from the oocyst.
This report outlines a protocol that assesses the efficiency of low DNA template detection via PCR. By using micromanipulation techniques, low numbers of C. parvum oocysts, even a single oocyst, can be confidently and accurately delivered directly into PCR tubes for subsequent DNA liberation and PCR detection. Using this approach, the current techniques for detection and species determination of C. parvum can be evaluated.
MATERIALS AND METHODS
C. parvum oocyst stocks.
The Iowa C. parvum genotype 2 isolate was propagated at the University of Arizona. A C. parvum genotype 1 isolate was collected by Asociacion Benefica PRISMA field workers (Lima, Peru) engaged in long-term collaborative efforts and shipped to the University of Arizona. Oocysts were purified using discontinuous sucrose gradients and cesium chloride (2) and enumerated by hemocytometer, and aliquots of a single genotype were stored in antibiotic solution (0.01% Tween 20, 100 U of penicillin, 0.1 mg of streptomycin/ml, and 0.1 mg of gentamicin/ml) at 4°C. Genotype 2 oocysts used in this study were 2 months old. The genotype 1 oocysts used were less than 6 months old.
FITC labeling of C. parvum oocysts.
Purified oocysts were FITC labeled in solution using the indirect Hydrofluor Combo detection kit for Cryptosporidium and Giardia (Ensys Inc., Research Triangle Park, N.C.). Briefly, primary and secondary labeling reagents and bovine serum albumin (1:10 [vol/vol] each) were added to a 100-μl water matrix suspension containing approximately 105 oocysts (Iowa isolate). The labeling mixture was incubated in the dark for 1 h at room temperature with brief vortexing every 10 min. FITC-labeled suspensions were then washed three times in 1× PCR buffer (10 mM Tris-HCl [pH 8.0], 50 mM KCl) (PE Applied Biosystems, Branchburg, N.J.). Washing the oocysts in 1× PCR buffer consisted of centrifugation at 14,000 × g for 3 min followed by resuspension of the packed pellet in 500 μl of 1× PCR buffer. After the final centrifuge step, the pellet was resuspended in 100 μl of 1× PCR buffer.
Microscopic isolation of single oocysts.
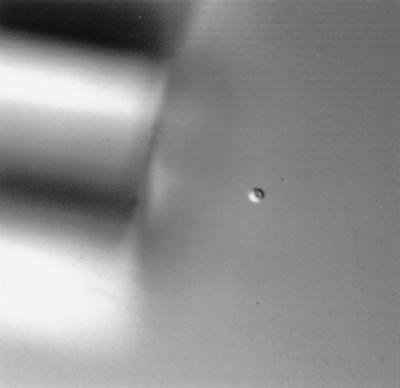
An aliquot of approximately 200 FITC-labeled oocysts was dispensed onto a microscope slide and diluted further with 1× PCR buffer to achieve two to three oocysts per field of view at ×200. Single, intact, fluorescing, FITC-labeled oocysts were located using epifluorescence microscopy. For oocyst isolation, the single oocyst was observed with 100× phase microscopy and subsequently isolated using a manually pulled glass micropipette (≈20-μm-inner-diameter tip opening) together with an UltraMicroPump II (World Precision Instruments [WPI], Sarasota, Fla.) and Micro4 controller (microprocessor-based controller) (WPI) (Fig. 1). A joystick micromanipulator (WPI) stabilized the micropipette ultramicropump. Single oocysts were then transferred and dispensed to the test matrix of interest, a microscope slide, or 10 μl of 1× PCR buffer within a thin-walled PCR tube.
FIG. 1.
Differential interference contrast microscopy of a single C. parvum oocyst during micromanipulation. Magnification, ×400. FITC-labeled oocysts were located using epifluorescence microscopy and isolated with 100× phase microscopy using a manually pulled glass micropipette (≈20-μm-inner-diameter tip opening) together with a micropump stabilized by a micromanipulator.
To validate the transfer of single oocysts, 75 single oocysts were isolated, as described above, and transferred to 75 microscope slides. Single oocysts were dispensed into 3 μl of double-distilled H2O on a microscope slide followed by the addition of 3 μl of mounting medium (Merifluor Giardia and Cryptosporidium kit; Meridian Diagnostics, Cincinnati, Ohio). The suspension containing the oocyst was covered with a 15-mm-diameter coverslip and sealed with clear fingernail polish. The entire coverslip area was scanned using 200× fluorescence microscopy to confirm the presence of singly isolated oocysts.
PCR.
The nested PCR primers designed for this study amplify a region within the 18S rRNA gene. External primers ExCry1 (GCC AGT AGT CAT ATG CTT GTC TC) (bp 16 to 38) and ExCry2 (ACT GTT AAA TAG AAA TGC CCC C) (bp 838 to 859) amplify an 844-bp fragment from genotype 1 and an 840-bp fragment from genotype 2. Nested primers NesCry3 (GCG AAA AAA CTC GAC TTT ATG GAA GGG) (bp 173 to 199) and NesCry4 (GGA GTA TTC AAG GCA TAT GCC TGC) (bp 739 to 765) amplify a 593-bp fragment from genotype 1 and a 590-bp fragment from genotype 2. Base pair positions are relative to the entire 18S rRNA gene of C. parvum isolates carrying genotype 1 (GenBank accession no. AF093491) and genotype 2 (GenBank accession no. AF164102).
The external PCR master mixture incorporated a 200 nM concentration of each primer (ExCry1 and ExCry2), 1× PCR Gold buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl) (PE Applied Biosystems), 2 mM MgCl2 (PE Applied Biosystems), 200 μM (each) dATP, dCTP, dGTP, and dTTP (Promega, Madison, Wis.), 1 U of Taq polymerase (Promega), and purified, sterile water. Ten microliters (approximately 50 ng) of purified C. parvum template DNA, genotype 1 or 2, was added to give a total volume of 50 μl. The nested PCR master mixture was essentially the same as the external PCR master mixture with the exception of nested primers (NesCry3 and NesCry4) and 1.5 mM MgCl2. Two microliters of the amplified external reaction mixture was transferred to 48 μl of the nested master mixture.
PCR parameters used in the external reaction included an initial denaturation at 95°C for 5 min and a 10-min hold at 80°C (Taq polymerase was added at this step), followed by 40 cycles of 94°C for 45 s, 53°C for 75 s, and 72°C for 45 s. Final extension was carried out at 72°C for 7 min. The nested-reaction parameters were the same except that 35 cycles were performed at an annealing temperature of 65°C and dehybridization, annealing, and extension time periods were 25 s each. PCRs were performed in an Mastercycler gradient thermal cycler (Eppendorf Scientific, Inc., Westbury, N.Y.). PCR amplicons were visualized and photographed on 1.2% agarose gels stained with ethidium bromide (0.5 μg/μl) following UV transillumination. Two negative-control tubes, containing 10 μl of sterile double-distilled H2O instead of DNA, were the first and last samples completed with each PCR round.
Primer evaluation.
The nested primer set was evaluated for C. parvum specificity. Isolates of C. parvum (KSU-1), Cryptosporidium serpentis (KSU-2), Cryptosporidium andersoni (KSU-3), C. parvum (KSU-4), Cryptosporidium muris (108735), and Hammondia heydorni were provided by Steve Upton (Kansas State University). Cryptosporidium baileyi was provided by Byron Blagburn (Auburn University). An isolate of Neospora caninum was provided by David Lindsay (Virginia Polytechnic and State University) via S. Upton. Isolates of Cyclospora cayetanensis (Robert Gilman, Johns Hopkins), Eimeria neischulzi (Don Duszynski, University of New Mexico), Encephalitozoon (septata) intestinalis (ATCC 50603), Giardia duodenalis (CDC0284), Bacillus subtilis (ATCC 27370), and Escherichia coli (ATCC 15224) were also tested. DNA from previously purified organisms was isolated using freezing and thawing and phenol-chloroform-ethanol extraction. DNA from organisms still in fecal material was isolated using the QIAamp DNA stool minikit (Qiagen Inc., Valencia, Calif.) in accordance with the manufacturer's instructions.
PCR sensitivity.
Testing nested-PCR sensitivity with low numbers of oocysts consisted of transferring 1, 2, 3, 4, 5, 7, or 10 oocysts into PCR tubes containing 10 μl of 1× PCR buffer. In total, 50 samples for each number of oocysts were subjected to PCR.
The 10-μl 1× PCR buffer solution containing the single oocyst or multiple oocysts was subjected to six freeze/thaw cycles (2 min in liquid nitrogen followed by 2 min in a 98°C water bath). A 30-s centrifugation (14,000 × g) was added between the third and forth cycles. PCR conditions were those described above and consisted of adding 40 μl of the external master mixture directly to the 10 μl of 1× PCR buffer containing the ruptured single or multiple oocysts. Again, 2 μl of the amplified external reaction mixture was added to 48 μl of the nested master mixture.
RFLP analysis.
Restriction sites within the nested amplicons allow for differentiation between C. parvum genotypes 1 and 2 (VspI) (23) and between C. parvum isolates from C. baileyi and C. serpentis (DraII). In 20-μl reaction volumes, 15 μl of the nested PCR amplicon was digested with 6 U of VspI (Promega) or 4 U of DraII (Hoffmann-La Roche Inc., Nutley, N.J.) in the supplied buffer for 6 h at 37°C. Digested products were visualized on 2% agarose gels stained with ethidium bromide (0.5 μg/μl) following UV transillumination.
RESULTS AND DISCUSSION
Technology for the detection of C. parvum has seen advances related to concentration of oocysts in filtered-water samples, and IMS has replaced differential-centrifugation techniques to enhance oocyst isolation and concentration when screening environmental samples (4, 15). Subsequent detection with advanced molecular techniques has been used to detect and determine the species of low numbers of oocysts (12). Along with development of these sensitive techniques for isolation and detection needs to be proper evaluation with known numbers of oocysts. To recapitulate, the two main methods for evaluating PCR sensitivity for detecting C. parvum are (i) dilution of intact oocysts from a stock suspension enumerated with a hemocytometer and (ii) dilution of DNA extracted from a large population of oocysts.
Using a dilution series from an enumerated stock of oocysts innately contains a standard deviation. Due to this standard deviation, the actual number of oocysts used to evaluate a detection limit is not known. Flow cytometry and other studies in which there are microscopically confirmed numbers of oocysts are beginning to be used to ensure that evaluation is being performed with known numbers of oocysts (22, 24). Dilution of DNA to the single-oocyst level does not take into account the difficulty of liberating genomic DNA from individual oocysts or the loss of DNA during purification procedures such as phenol-chloroform extraction or those associated with commercial kits to rid the DNA preparation of PCR inhibitors encountered in fecal and environmental samples.
In view of the need to determine the true limit of PCR for detecting whole oocysts, the combination of an ultramicropump attached to a micromanipulator was used to deliver the desired number of oocysts (1 or 10 oocysts) into PCR tubes (Fig. 1). To validate oocyst isolation and delivery, 75 individual FITC-labeled oocysts were transferred to microscope slides and confirmation of single-oocyst transfer was performed by epifluorescence microscopy. Sixty-seven of the 75 (89.3%) singly isolated oocysts were detected. In no case were additional oocysts detected in a single transfer. These data indicate that isolation and delivery of single oocysts can be performed accurately and confidently.
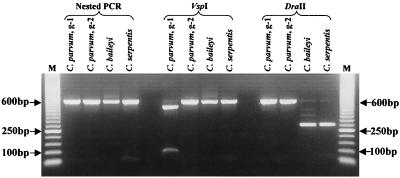
The newly designed nested primer set described in this report was evaluated for C. parvum specificity and detection sensitivity with low numbers of oocysts. PCR conditions were optimized using purified DNA from C. parvum genotypes 1 and 2. As expected, the external primer set amplified 844- and 840-bp fragments from genotype 1 and genotype 2, respectively. Similarly, the nested primer set amplified 593- and 590-bp fragments from genotype 1 and genotype 2, respectively (Fig. 2). Isolated DNA from test organisms C. parvum (KSU-1), C. parvum (KSU-4), C. andersoni, C. baileyi, C. muris, C. serpentis, H. heydorni, N. caninum, Cyclospora cayetanensis, E. neischulzi, E. (septata) intestinalis, G. duodenalis, B. subtilis, and E. coli was also subjected to PCR and RFLP digestion. PCR with the external primers amplified products from C. parvum (KSU-1), C. parvum (KSU-4), C. baileyi, C. serpentis, Cyclospora cayetanensis, E. neischulzi, and N. caninum DNA (Table 1). With the exception of that from C. parvum, C. baileyi, and C. serpentis, amplified DNA from the external-primer reaction was not amplified with the nested-primer set (Table 1). Differentiation between C. parvum genotypes 1 and 2 was accomplished with restriction enzyme VspI (Fig. 2). Genotype 1 contains the restriction site producing 503- and 90-bp fragments, while genotype 2 does not contain the restriction cut site. Differentiation of C. parvum from C. baileyi and C. serpentis was accomplished with restriction enzyme DraII (Fig. 2). The DraII digestion site within C. baileyi produced 295- and 284-bp fragments (indistinguishable within the agarose gel) (Fig. 2). The DraII digestion site within C. serpentis produced 298- and 284-bp fragments (indistinguishable within the agarose gel) (Fig. 2).
FIG. 2.
Nested PCR amplification and restriction enzyme digestions with VspI and DraII of a segment within the 18S rRNA of Cryptosporidium species; g-1 and g-2, C. parvum genotypes 1 and 2, respectively. Lanes M, 50-bp molecular marker (Pharmacia, Piscataway, N.J.).
TABLE 1.
Evaluation of nested PCR amplification and restriction enzyme digestion of a segment within the 18S rRNA of Cryptosporidium for species differentiationa
| Test organism (strain, genotype) | PCR product (sizeb[bp]) obtained from:
|
Results of RFLP digestion with:
|
||
|---|---|---|---|---|
| External primers | Nested primers | VspI | DraII | |
| C. parvum (Peru, 1) | + (844) | + (593) | + | − |
| C. parvum (Iowa, 2) | + (840) | + (590) | − | − |
| C. parvum (KSU-1, 2) | + (840) | + (590) | − | ND |
| C. serpentis (KSU-2) | + (836) | + (583) | − | + |
| C. andersoni (KSU-3) | − | − | ND | ND |
| C. parvum (KSU-4, 2) | + (840) | + (590) | − | ND |
| C. baileyi | + (831) | + (579) | − | + |
| C. muris (108735) | − | − | ND | ND |
| Cyclospora cayetanensis | + (883) | − | ND | ND |
| Eimeria neischulzi | + (885) | − | ND | ND |
| Encephalitozoon intestinalis | − | − | ND | ND |
| Giardia duodenalis | − | − | ND | ND |
| Neospora caninum | +c | − | ND | ND |
| Bacillus subtilis | − | − | ND | ND |
| Escherichia coli | − | − | ND | ND |
+, amplification or digestion; −, no amplification or digestion; ND, not done.
Amplicon size based on the following representative GenBank accession numbers: C. parvum genotypes 1 and 2, AF093491 and AF164102, respectively; C. baileyi, L19068; C. serpentis, AF151376; C. cayetanensis, AF111183; E. neischulzi, U40263.
Multiple PCR fragments observed.
The amplification of DNA from C. baileyi and C. serpentis lowers the specificity of this primer set at the species level, but the VspI and DraII restriction digestion sites readily differentiate between C. parvum genotypes and other Cryptosporidium species, respectively. Unfortunately, due to the difficulty in acquiring oocysts, DNA from Cryptosporidium meleagridis could not be evaluated using these primers.
Finally, nested PCR sensitivity was evaluated with single and multiple C. parvum genotype 2 oocysts. PCR amplification rates were as follows: 100% for PCR tubes with 10 oocysts, 94% for tubes containing 7 oocysts, 92% for tubes containing 5 oocysts, 88% for tubes containing 4 oocysts, 76% for tubes containing 3 oocysts, 56% for tubes with 2 oocysts, and 38% for tubes containing a single oocyst.
To our knowledge, this is the first report evaluating C. parvum detection by PCR at the confirmed single-oocyst level. Under these conditions, single oocysts were detected by PCR with a 38% PCR amplification rate and five oocysts were required to achieve the 90% level. These results demonstrate the difficulty of PCR amplification with intact oocysts and are a reflection of the inherent problems when applying PCR detection to environmental samples.
REFERENCES
- 1.Aldom J E, Chagla A H. Recovery of Cryptosporidium oocysts from water by a membrane filter dissolution method. Lett Appl Microbiol. 1995;20:186–187. doi: 10.1111/j.1472-765x.1995.tb00423.x. [DOI] [PubMed] [Google Scholar]
- 2.Arrowood M J, Donaldson K. Improved purification methods for calf-derived Cryptosporidium parvum oocysts using discontinuous sucrose and cesium chloride gradients. J Eukaryot Microbiol. 1996;43:89S. doi: 10.1111/j.1550-7408.1996.tb05015.x. [DOI] [PubMed] [Google Scholar]
- 3.Awad-El-Kariem F M, Warhurst D C, McDonald V. Detection and species identification of Cryptosporidium oocysts using a system based on PCR and endonuclease restriction. Parasitology. 1993;109:19–22. doi: 10.1017/s0031182000077714. [DOI] [PubMed] [Google Scholar]
- 4.Bukhari Z, McCuin R M, Fricker C R, Clancy J L. Immunomagnetic separation of Cryptosporidium parvum from source water samples of various turbidities. Appl Environ Microbiol. 1998;64:4495–4499. doi: 10.1128/aem.64.11.4495-4499.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carraway M, Tzipori S, Widmer G. A new restriction fragment length polymorphism from Cryptosporidium parvum identifies genetically heterogeneous parasite populations and genotypic changes following transmission from bovine to human hosts. Infect Immun. 1997;65:3958–3960. doi: 10.1128/iai.65.9.3958-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark D P. New insights into human cryptosporidiosis. Clin Microbiol Rev. 1999;12:554–563. doi: 10.1128/cmr.12.4.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng M Q, Cliver D O, Mariam T W. Immunomagnetic capture PCR to detect viable Cryptosporidium parvum oocysts from environmental samples. Appl Environ Microbiol. 1997;63:3134–3138. doi: 10.1128/aem.63.8.3134-3138.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fayer R, Speer C A, Dubey J P. The general biology of Cryptosporidium. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. New York, N.Y: CRC Press, Inc.; 1997. pp. 1–41. [Google Scholar]
- 9.Hallier-Soulier S, Guillot E. Detection of cryptosporidia and Cryptosporidium parvum oocysts in environmental water samples by immunomagnetic separation-polymerase chain reaction. J Appl Microbiol. 2000;89:5–10. doi: 10.1046/j.1365-2672.2000.01029.x. [DOI] [PubMed] [Google Scholar]
- 10.Korich D G, Mead J R, Madore M S, Sinclair N A, Sterling C R. Effects of ozone, chlorine dioxide, chlorine, and monochloramine on Cryptosporidium parvum oocyst viability. Appl Environ Microbiol. 1990;56:1423–1428. doi: 10.1128/aem.56.5.1423-1428.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laxer M A, Timblin B K, Patel R J. DNA sequences for the specific detection of Cryptosporidium parvum by the polymerase chain reaction. Am J Trop Med Hyg. 1991;45:688–694. doi: 10.4269/ajtmh.1991.45.688. [DOI] [PubMed] [Google Scholar]
- 12.Lowery C J, Moore J E, Millar B C, Burke D P, McCorry K A, Crothers E, Dooley J S. Detection and speciation of Cryptosporidium spp. in environmental water samples by immunomagnetic separation, PCR and endonuclease restriction. J Med Microbiol. 2000;49:779–785. doi: 10.1099/0022-1317-49-9-779. [DOI] [PubMed] [Google Scholar]
- 13.MacKenzie W R, Hoxie N J, Proctor M E, Gradus M S, Blair K A, Peterson D E, Kazmierczak J J, Addis D G, Fox K R, Rose J B, Davis J P. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N Engl J Med. 1994;331:161–167. doi: 10.1056/NEJM199407213310304. [DOI] [PubMed] [Google Scholar]
- 14.Peng M M, Xiao L, Freeman A R, Arrowood M J, Escalante A A, Weltman A C, Ong C S, MacKenzie W R, Lal A A, Beard C B. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–573. doi: 10.3201/eid0304.970423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rochelle P A, De Leon R, Johnson A, Stewart M H, Wolfe R L. Evaluation of immunomagnetic separation for recovery of infectious Cryptosporidium parvum oocysts from environmental samples. Appl Environ Microbiol. 1999;65:841–845. doi: 10.1128/aem.65.2.841-845.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shepherd K M, Wyn-Jones A P. An evaluation of methods for the simultaneous detection of Cryptosporidium oocysts and Giardia cysts from water. Appl Environ Microbiol. 1996;62:1317–1322. doi: 10.1128/aem.62.4.1317-1322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith H V, Rose J B. Waterborne cryptosporidiosis: current status. Parasitol Today. 1998;14:14–22. doi: 10.1016/s0169-4758(97)01150-2. [DOI] [PubMed] [Google Scholar]
- 18.Spano F, Putignani L, Guida S, Crisanti A. Cryptosporidium parvum: PCR-RFLP analysis of the TRAP-C1 (thrombospondin-related adhesive protein of Cryptosporidium-1) gene discriminates between two alleles differentially associated with parasite isolates of animal and human origin. Exp Parasitol. 1998;90:195–198. doi: 10.1006/expr.1998.4324. [DOI] [PubMed] [Google Scholar]
- 19.Sterling C R. Cryptosporidiosis: the treatment dilemma. J Med Microbiol. 2000;49:207–208. doi: 10.1099/0022-1317-49-3-207. [DOI] [PubMed] [Google Scholar]
- 20.Sulaiman I M, Xiao L, Lal A A. Evaluation of Cryptosporidium parvum genotyping techniques. Appl Environ Microbiol. 1999;65:4431–4435. doi: 10.1128/aem.65.10.4431-4435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sulaiman I M, Xiao L, Yang C, Escalante L, Moore A, Beard C B, Arrowood M J, Lal A A. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg Infect Dis. 1998;4:681–685. doi: 10.3201/eid0404.980424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiedenmann A, Steuer S, Kruger P, Botzenhart K. A simple procedure for an exact evaluation of the sensitivity of the selective detection of viable Cryptosporidium oocysts by in vitro excystation and PCR. In: Fricker C, Rochelle P, editors. Proceedings of the 1997 International Symposium on Waterborne Cryptosporidium. Denver, Colo: American Water Works Association; 1997. pp. 109–113. [Google Scholar]
- 23.Xiao L, Escalante L, Yang C, Sulaiman I, Escalante A, Montali R, Fayer R, Lal A. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang S, Benson S K, Du C, Healey M C. Infection of immunosuppressed C57BL/6N adult mice with a single oocyst of Cryptosporidium parvum. J Parasitol. 2000;86:884–887. doi: 10.1645/0022-3395(2000)086[0884:IOICAM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]