Abstract
Recently, advances in genomic technology such as RNA sequencing and genome‐wide profiling have enabled the identification of considerable numbers of non‐coding RNAs (ncRNAs). MicroRNAs have been studied for decades, leading to the identification of those with disease‐causing and/or protective effects in vascular disease. Although other ncRNAs such as long ncRNAs have not been fully described yet, recent studies have indicated their important functions in the development of vascular diseases. Here, we summarize the current understanding of the mechanisms and functions of ncRNAs, focusing on microRNAs, circular RNAs and long ncRNAs in vascular diseases.
Keywords: atherosclerosis, circRNA, endothelial cells, lncRNA, miRNA, vascular smooth muscle cells
Considerable numbers of non‐coding RNAs have been identified by RNA sequencing and genome‐wide profiling. These include microRNAs studied for decades and other non‐coding RNAs (ncRNAs) such as long ncRNAs. Recent studies have indicated their important functions in the development of vascular diseases. Here, we summarize the current understanding of the mechanisms and functions of these ncRNAs.
Abbreviations
- ABCA1
ATP‐binding cassette transporter subfamily A member 1
- CAD
coronary artery disease
- CCL2
C‐C motif chemokine ligand 2
- ChIRP
chromatin isolation by RNA purification
- ceRNA
competing endogenous RNA
- CHART
capture hybridization analysis of RNA targets
- CLASH
cross‐linking, ligation and sequencing of hybrids
- CLIP‐seq
cross‐linking immunoprecipitation sequencing
- CRISPR
clustered regularly interspaced short palindromic repeat
- CTGF
connective tissue growth factor
- ENCODE
Encyclopedia of DNA Elements
- EC
enhancer cell
- eNOS
endothelial nitric oxide synthase
- EZH2
enhancer of zeste homolog 2
- FANTOM
Functional Annotation of the Mammalian Genome
- FISH
fluorescence in situ hybridization
- GENCODE
Encyclopedia of Genes and Gene Variants
- HDL
high‐density lipoprotein
- LOXL2
lysyl oxidase homolog 2
- LXR
liver X receptors
- MDM2
murine double minute 2
- MI
myocardial infarction
- PPAR
peroxisome proliferator‐activated receptor
- RAP
RNA antisense purification
- RIP
RNA immunoprecipitation
- SHAPE
selective 2'‐hydroxyl acylation and primer extension
- SPAAR
small regulatory polypeptide of amino acid response
- SREBF
sterol regulatory element binding factor
- VCAM1
vascular cell adhesion molecule 1
- VSMC
vascular smooth muscle cell
Introduction
Although only approximately 2% of the human genome encodes mRNAs, it is well known that a large portion of the human genome (approximately 70%) is transcribed and that the majority of the transcripts are non‐coding RNAs (ncRNAs) (Encyclopedia of DNA Elements – ENCODE, https://www.encodeproject.org; Encyclopedia of Genes and Gene Variants – GENCODE, https://www.gencodegenes.org; or Functional Annotation of the Mammalian Genome – FANTOM, https://fantom.gsc.riken.jp). The group of ncRNAs can be divided into small ncRNAs [e.g. microRNAs (miRNAs)] and transcripts > 200 nucleotides, named long ncRNAs (lncRNAs). Among the 228 000 known transcripts, approximately 7500 are classified as small ncRNAs and 48 000 are grouped as lncRNAs (GENCODE, version 34).
miRNAs constitute the vast majority of the studied group of ncRNAs. Extensive research has led to strategies to target disease‐relevant miRNAs as potential therapies. On the other hand, it has already been reported that almost 20 000 lncRNAs are functional in humans (FANTOM5) [1]. Thus, many researchers are now trying to identify lncRNAs that are related to vascular biology and diseases. However, lncRNAs have a wide variety of functions, and even their classification has not yet been fully determined.
In this review, we describe the current understanding and functions of miRNAs and lncRNAs, which include circular RNAs (circRNAs), in vascular biology and diseases. In particular, we try to classify lncRNAs in terms of their molecular functions and describe the current understanding and experimental methods of ncRNA research in vascular diseases (Fig. 1).
Fig. 1.
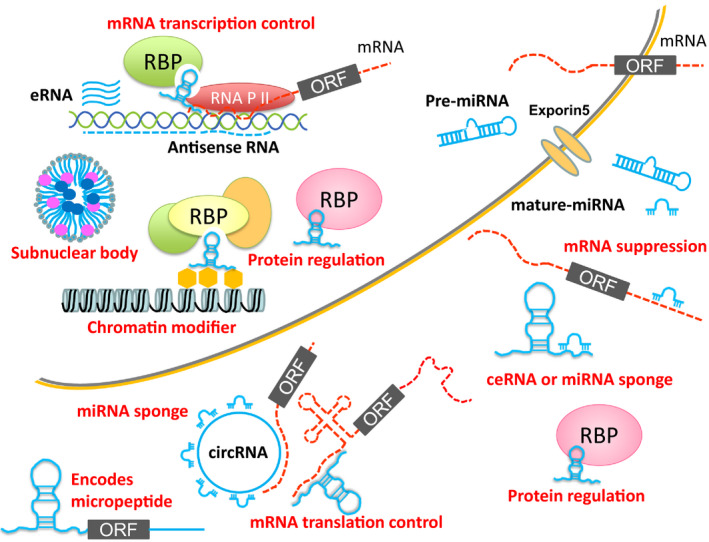
Schematic classification of ncRNAs based on the function of each ncRNA. MicroRNAs are known to suppress target mRNA functions by degradation or inhibition of translation. Some circRNAs and lncRNAs work as miRNA sponges or ceRNAs to inhibit the functions of miRNAs. lncRNAs can enhance mRNA transcription working as enhancer RNAs or by recruiting transcription factor complexes. Moreover, they can suppress mRNA transcription as antisense RNAs. Some ncRNAs may encode micropeptides at the same time. Chromatin structure can also be modified by lncRNAs. Because lncRNAs can bind to nucleic acids and proteins, there may be many other functions that are not illustrated. eRNA, enhancer RNA; RBP, RNA binding protein; RNA P II, RNA polymerase II.
miRNAs
miRNAs are the best studied family of ncRNAs. miRNAs are 20–25 nucleotides in length and are known to regulate protein‐coding genes through translational repression or degradation of mRNAs by binding to sequences in the 3'‐UTR of specific mRNAs. The mechanisms and the functions of miRNA are well known and strategies to target miRNAs have already been developed, including miRNA mimics to augment their functions, miRNA inhibitors (antisense against miRNA) and miRNA sponges to suppress their functions. miRNAs are stable in plasma and they have been proposed as biomarkers of myocardial infarction (MI) and other vascular diseases.
There is already extensive evidence demonstrating that miRNAs are involved in many pathological processes in vascular diseases and atherosclerosis. Recent reviews have already provided insights into the mechanisms of how miRNAs exert an influence on atherosclerosis, their potential use in diagnostics and strategies for improving RNA therapeutics [2, 3, 4]. Thus, we briefly summarize their regulation in lipid handling, inflammation and cellular mechanisms, which are involved in endothelial cell (EC) and vascular smooth muscle cell (VSMC) proliferation, migration and phenotypic switching.
Functions of specific miRNAs in vascular diseases
Several studies have demonstrated an important role for miR‐21 with respect to negatively regulating inflammation and suppressing pro‐inflammatory signaling cascades [5, 6]. In addition to its role in regulating pro‐inflammatory responses, many studies have already reported that miR‐21 has essential functions in ECs and VSMCs [7, 8, 9]. miR‐21 targets peroxisome proliferator‐activated receptor‐α (PPAR‐α), which modulates flow‐induced endothelial inflammation. The role of miR‐21 with respect to VSMC functions and vascular remodeling is well established [10, 11]. The expression of miR‐21 is increased following balloon angioplasty or vascular injury induced by carotid artery ligation, as well as in human atherosclerotic lesions. [10, 12]. Moreover, miR 21 in hematopoietic cells is important for the progression of atherosclerosis. The lack of miR‐21 in hematopoietic cells enhances atherosclerosis formation [13]. In addition to its effects on vascular biology, miR‐21 has been implicated in the pathogenesis of myocardial fibrosis and hypertrophy. Currently, the investigation of oligonucleotide‐based therapeutics against miR‐21 to prevent cardiac fibrosis is underway [14].
Recent studies have indicated that miR‐33a and miR‐33b control lipid metabolism in vivo [15, 16, 17, 18, 19]. These miRNAs are encoded in the introns of sterol regulatory element binding factor (SREBF)2 and SREBF1, respectively, in larger mammals, including humans [16, 20]. However, there is a deletion in part of miR‐33b in rodents and miR‐33b cannot be expressed. Several groups, including our own, have reported that Abca1 and Abcg1 are the targets of miR‐33a in vivo through the use of either antisense oligonucleotides or by generating miR‐33a‐deficient mice [15, 16, 17, 18]. ATP‐binding cassette transporter subfamily A member 1 (ABCA1) promotes cholesterol efflux to lipid‐poor apolipoprotein A‐I and forms nascent high‐density lipoprotein (HDL) particles in the liver and intestine, whereas ABCG1 transports cellular cholesterol to HDL2 and HDL3. Therefore, upregulation of ABCA1 and ABCG1 results in a 35–50% increase in plasma HDL without affecting other lipoproteins in mice treated with anti‐miR‐33a‐oligonucleotides [15, 16, 17]. Similarly, miR‐33a‐deficient mice demonstrated a 25–40% increase in HDL [18] and showed reduced atherosclerosis formation in an Apoe‐deficient background [21]. On the other hand, miR‐33b knock‐in mice, in which miR‐33b is inserted in the same intron as in humans, have levels of HDL‐cholesterol that are reduced by almost 35%, in addition to severe atherosclerosis, when they are crossed with Apoe‐deficient mice [22, 23]. Moreover, miR‐33a deficiency ameliorates aortic aneurysm both in Ca2+‐ and angiotensin II‐induced mice models, which suggested that inhibition of miR‐33 may be a novel therapeutic strategy for abdominal aortic aneurysm [24]. There are several articles summarizing the role of miRNAs in lipid and lipoprotein metabolism [25, 26].
The miR‐17‐92 cluster is an important regulator of angiogenesis. It consists of six mature miRNAs including miR‐17, ‐18a, ‐19a, ‐19b, ‐20a and ‐92a, which are transcribed from a polycistronic transcription unit C13orf25 [27]. This region is closely related to the transcription factor c‐Myc, and some of these miRNAs promote angiogenesis in response to Myc. In particular, miR‐18 and miR‐19 target thrombospondin‐1 and connective tissue growth factor to promote tumor angiogenesis via the stimulation of Myc [28]. On the other hand, miR‐92a in the same cluster inhibits angiogenesis [29]. Overexpression of miR‐92a in ECs blocks the growth of new blood vessels, and inhibition of miR‐92a leads to enhanced angiogenesis and shows functional recovery from limb ischemia and MI in mice. Thus, miR‐92a may serve as an important target for the treatment of ischemic disease. The miR‐17‐92 cluster also has effects on neurological disorders [30].
miR‐126 is an endothelial‐specific miRNA encoded in intron 7 of epidermal growth factor‐like domain multiple 7 (Egfl7). Mechanosensitive transcription factor Krüppel‐like factor 2a induces miR‐126 expression for the activation of vascular endothelial growth factor signaling [31]. Thus, miR‐126 facilitates the integration of physiological stimuli with growth factor signaling in ECs to promote angiogenesis. miR‐126 also has anti‐inflammatory effects. Indeed, transfection of ECs with an oligonucleotide that decreases miR‐126 levels results in an increase in tumor necrosis factor‐α‐stimulated vascular cell adhesion molecule 1 (VCAM1) expression and increased leukocyte adherence to ECs [32]. Thus, miR‐126 overexpression can be utilized as a therapeutic approach and miR‐126‐conjugated stents have been developed to inhibit neointimal hyperplasia in rabbits. [33]. Bubble liposome‐mediated systemic delivery of miR‐126 also improved blood flow in a hind‐limb ischemia model [34].
The miR‐143/‐145 encoding genes are located in close proximity to each other on murine chromosome 18 and human chromosome 5. miR‐145 is essential for VSMC differentiation. It has been shown that miR‐145 is necessary and sufficient for directing the fate of VSMCs from multipotent neural crest stem cells [35]. miR‐145 is selectively expressed in VSMCs of the vascular wall in adult rats, and it is downregulated during the formation of neointimal lesions [36]. The target of miR‐145 is KLF5, and a corresponding increase in myocardin expression is observed by the induction of miR‐145. Overexpression of miR‐145 suppressed neointimal formation in balloon‐injured arteries and might be utilized for the treatment of vascular diseases. The lncRNA miR143HG is located in a similar locus to miR‐143/145, and it was recently implicated in cardiac specification and smooth muscle differentiation [37, 38].
miR‐221 and miR‐222 expression levels are elevated in rat carotid arteries after balloon injury [10, 39]. p27 (Kip1) and p57 (Kip2) are the target genes of miR‐221‐ and miR‐222, which mediate the effects on VSMC growth. Thus, knockdown of miR‐221 and miR‐222 results in decreased VSMC proliferation both in vitro and in vivo. Moreover, endothelial progenitor cells, quiescent ECs and umbilical vein ECs highly express miR‐221/222, which suggests an essential role for this miRNA cluster in endothelial physiology [40].
These examples only highlight some of the miRNAs related to vascular diseases (Table 1). There are many miRNAs that have similar actions and a single miRNA can have a variety of functions. Thus, any targeting therapy against miRNAs requires a specificity of action to reduce off‐target problems [41].
Table 1.
Examples of miRNAs that have functions in vascular diseases.
| miRNA | Target genes that are important for vascular diseases | Disease | References |
|---|---|---|---|
| miR‐21 | PPARα, PTEN, SPRY1, SMAD7, BCL‐2 | Inflammation resolution, atherosclerosis inhibition, fibrosis progression | [5, 6, 7, 8, 9, 10, 11, 12, 13, 14] |
| miR‐33a/b | ABCA1 | Atherosclerosis inhibition, aortic aneurysm inhibition | [15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26] |
| miR‐18 (miR‐17‐92 cluster) | CTGF | Angiogenesis promotion | [28] |
| miR‐19 (miR‐17‐92 cluster) | TSP1, PPARα, PTEN | Angiogenesis promotion | [28] |
| miR‐92a (miR‐17‐92 cluster) | ITGA5 | Angiogenesis inhibition | [29] |
| miR‐126 | SPRED1, PIK3R2, VCAM1, and ALCAM1 | Angiogenesis promotion | [31, 32, 33, 34] |
| miR‐143 | ELK1 | VSMC differentiation | [35] |
| miR‐145 | MYOCD, KLF5 | VSMC differentiation | [35, 36] |
| miR‐221/222 | KIP1, KIP2 | VSMC growth | [37] |
lncRNAs
Recent work has shown that many lncRNAs are functional and represent a large class of potential therapeutic targets and agents [42]. Although clear functional classification of lncRNAs is not established yet, we will attempt to classify vascular disease‐related lncRNAs according to their molecular functions, such as genomic scaffolds, enhancers, miRNA sponges, regulators of proteins and so on. However, some lncRNAs have been shown to exhibit several different mechanisms of action concurrently. Thus, a better classification may be created in the future.
circRNAs
In 2012, the ubiquitous expression of circRNA from genes traditionally assumed to express only mRNAs or lncRNAs was found and reported [43]. These molecules derive from a noncanonical type of splicing defined as tail‐to‐head because it goes from a downstream 5′ splice site to an upstream 3′ splice site, a process suggested to be guided by specific repetitive sequences [44]. circRNAs are enriched in the brain and it has been shown that cerebellar degeneration‐related protein 1 antisense RNA has over 70 binding sites for miR‐7, acting as a sponge for this miRNA and being able to modulate its activity on miR‐7 target genes. Several studies have addressed circRNAs expression in the cardiovascular system. The circRNA derived from alternative splicing of lipoprotein receptor 6 is named circ_Lrp6. It has a role as an miR‐145 sponge. Silencing of circ_Lrp6 increases the levels of miR‐145 and thereby reduces the expression of miR‐145 target genes, such as integrin‐β8, fascin actin‐bundling protein 1, KLF‐4, YES proto‐oncogene 1 (Yes1) and lysyl oxidase (Lox). Short hairpin RNA against circ_Lrp6 reduces the neointima formation in a model of stenosis induced by perivascular carotid collar placement in ApoE−/− mice [45].
Exons of the lncRNA anti‐sense ncRNA in the INK4 locus (ANRIL), which is described below, can form circANRIL. circANRIL binds to pescadillo homologue 1, an essential 60S‐preribosomal assembly factor, thereby impairing exonuclease‐mediated pre‐ribosomal RNA processing and ribosome biogenesis in VSMCs and macrophages. As a consequence, circANRIL induces nucleolar stress and p53 activation, resulting in the induction of apoptosis and inhibition of proliferation, which are key cell functions in atherosclerosis [46].
The Nrg‐1‐ICD‐induced circular alpha‐actin‐2 (circACTA2) acts as a sponge, which binds miR‐548f‐5p. circACTA2 upregulates α‐smmoth muscle actin expression by the suppression of miR‐548f‐5p, thereby facilitating stress fiber formation and cell contraction in human arterial smooth muscle cells [47].
lncRNAs that modulate chromatin architecture
Many lncRNAs are known to affect the transcription of genes. They often impact chromatin readers and writers to control the gene promoter [48]. Mechanistically, lncRNAs can recruit chromatin remodelers to target genes via binding to DNA, or control their enzymatic activity or function as negative decoys. Such lncRNAs with relevance to atherosclerosis include ANRIL, lincRNA‐p21, MALAT1, MEG3, TUG1, GAS5 and MANTIS.
ANRIL is transcribed from the well‐known 9p21.3 cardiovascular disease locus, which has been strongly implicated in coronary artery disease (CAD) through genome‐wide association studies [49, 50, 51, 52]. Although important tumor suppressor genes CDK2A and CDKN2B are found near this region, these genes are not dysregulated in animal models of atherosclerosis or human samples. Indeed, the approximately 60‐kb risk haplotype is human‐specific and lacks coding genes, hindering efforts to clarify its function. Recently, induced pluripotent stem cells from risk and non‐risk individuals were generated. After differentiation into VSMCs, risk VSMCs exhibit globally altered transcriptional networks that resemble the previously identified CAD risk genetic pathways. Of note, deleting the risk haplotype rescues VSMCs, whereas expressing ANRIL induces cardiovascular risk phenotypes in non‐risk VSMCs [53]. Some part of the functions of ANRIL is mediated by interactions among ANRIL, enhancer of zeste homolog 2 (EZH2; as part of the polycomb repressive complex 2 complex) and the histone acetyltransferase p300 [54].
lincRNA‐p21 is downregulated in a mice model of atherosclerosis and in patients with CAD. Silencing of lincRNA‐p21 was shown to induce cell proliferation and inhibit apoptosis in VSMCs and macrophages in vitro. Moreover, inhibition of lincRNA‐p21 increased neointimal hyperplasia in a carotid artery injury model in vivo. Mechanistically, lincRNA‐p21 enhances p53 transcriptional activity via binding to the E3 ubiquitin ligase mouse double minute 2 (MDM2). The association of lincRNA‐p21 and MDM2 releases MDM2 repression of p53, thereby enabling p53 to interact with p300, which enhances p53 transcriptional activity [55].
Metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1) was originally identified as a nuclear‐enriched prognostic lung cancer metastasis marker. Genetic deletion or silencing of MALAT1 in vivo inhibits EC proliferation, postnatal retina vascularization and ischemia‐induced neovascularization [56]. In humans, reduced MALAT1 expression levels are associated with a worse prognosis [57]. A recent study suggests that the action of MALAT1 is mediated by interaction with polycomb repressive complex 2, binding of the transactivation domain of TEAD proteins, activities of competing endogenous RNAs (ceRNAs) and the regulation of various signaling pathways, including phosphatidylinositol‐3‐kinase‐AKT, mitogen‐activated protein kinase, WNT and nuclear factor‐kappa B [58].
Maternally expressed gene 3 (MEG3) was previously shown to regulate tumor suppressor genes through stimulating p53 accumulation [59]. GapmeR‐mediated silencing of MEG3 in aged mice promotes neovascularization after hindlimb ischemia in vivo [60]. In humans, MEG3 is significantly downregulated in the lung tissue of patients with pulmonary arterial hypertension [61]. Mechanistically, two distal motifs interact by base pairing to form alternative, mutually exclusive pseudoknot structures, which are called ‘kissing loops’, in an evolutionarily conserved region of MEG3. Mutations that destroy these interactions impair MEG3‐dependent p53 stimulation in vivo [62]. In addition to the MEG3‐p53 interaction, several studies have suggested that it functions as a sponge for miR‐9, ‐21, ‐26 and ‐328 [63, 64, 65, 66].
Taurine up‐regulated gene 1 (TUG1) is expressed in rat VSMCs and its level is increased in synthetic VSMCs [67]. Because EZH2‐mediated methylation of α‐actin is dependent on TUG1, F‐actin polymerization is promoted by TUG1 in synthetic VSMCs.
Growth arrest‐specific transcript 5 (GAS5), which is located antisense to another lncRNA, GAS5 antisense, was identified and named as a result of its elevation upon cell growth arrest. Overexpressed GAS5 increases lipid accumulation in THP‐1 macrophages. GAS5 inhibits the expression of ABCA1 by binding to EZH2. Knockdown of GAS5 promotes reverse‐transportation of cholesterol and inhibits intracellular lipid accumulation, ultimately preventing atherosclerosis progression [68]. GAS5 was also shown to have multiple molecular mechanisms, such as binding to DNA sequences and forming an RNA–DNA triplex complex, which results in the triggering or suppression of the expression of genes in human cancer [69].
Recently, epigenetically controlled lncRNAs in human umbilical vein ECs were searched using an exon‐array technique after silencing histone demethylase JARID1B. MANTIS was identified as the most strongly regulated lncRNA [70]. Deletion or silencing of MANTIS inhibits angiogenic sprouting and alignment of ECs in response to shear stress. Mechanistically, the nuclear MANTIS interacts with BRG1, a subunit of the switch/sucrose nonfermentable (SWI/SNF) chromatin remodeling complex. This interaction is required for nucleosome remodeling and the regulation of key endothelial genes such as SMAD6, SOX18 and COUP‐TFII by facilitating the recruitment of RNA polymerase II to their promoter regions.
lncRNAs work as enhancer RNAs or regulate neighboring mRNAs
Many ncRNAs were found to be transcribed as active enhancers for gene promoters and they are called enhancer lncRNAs, which confer enhancer activity by capturing the promoter‐contacting mediator protein complex. There are also ncRNAs that act in cis to regulate their neighboring mRNAs. Examples of such important ncRNAs in the field of atherosclerosis include HOTTIP, LEENE, SMILR and lncRNA‐CCL2.
HOXA transcript at the distal tip (HOTTIP) expression level is higher in CAD tissues than in normal arterial tissues. Ectopic expression of HOTTIP promotes EC proliferation and increases the expression of cyclin D1 and PCNA. On the other hand, downregulated expression of HOTTIP suppresses EC proliferation and migration [71].
lncRNA that enhances endothelial nitric oxide synthase (eNOS) expression (LEENE) was identified by combining RNA‐sequencing (RNA‐seq) techniques and chromatin conformation capture methods [72]. LEENE facilitates the recruitment of RNA Pol II to the eNOS promoter region to enhance eNOS mRNA transcription.
Smooth muscle‐induced lncRNA enhances replication (SMILR) was identified as an lncRNA for which the expression was altered in human saphenous vein VSMCs following stimulation with interleukin‐1α and platelet‐derived growth factor. Mechanistically, the expression of genes proximal to SMILR was also altered by treatment with these cytokines. In addition, HAS2, which is one of the proximal transcripts of SMILR, was also reduced by SMILR knockdown. Increased expression of SMILR is observed in unstable atherosclerotic plaques and in plasma from patients with high plasma C‐reactive protein [73].
lncRNA‐CCL2 is transcribed divergently to C‐C motif chemokine ligand 2 (CCL2), a pro‐atherosclerotic chemokine. lncRNA‐CCL2 and CCL2 are up‐regulated in response to inflammatory stimuli, and their expression is elevated in unstable human atherosclerotic plaques [74]. Knockdown experiments showed the positive regulation of CCL2 by lncRNA‐CCL2. This regulation involves the interaction of lncRNA‐CCL2 with RNA binding proteins such as HNRNPU and IGF2BP2.
lncRNA localized in the subnuclear body
Some lncRNAs can affect other genes through their architectural roles in subnuclear territories. A previous study reported that the nuclear enriched abundant transcript 1 (NEAT1) is critical for the structural constituent of paraspeckles and tumorigenesis by promoting cell migration and proliferation [75]. Moreover, another study provided evidence demonstrating a critical role for NEAT1 with respect to promoting VSMC proliferation, migration and dedifferentiation during phenotypic switching. A loss‐of‐function study of NEAT1 in VSMCs resulted in enhanced expression of smooth muscle‐specific genes at the same time as attenuated VSMC proliferation and migration [76]. Mechanistically, NEAT1 sequesters the key chromatin modifier WD Repeat Domain 5 (WDR5) from smooth muscle‐specific gene loci and initiates an epigenetic off state, which consequently impairs SRF accessibility to the CArG boxes, resulting in down‐regulation of smooth muscle‐contractile gene expression.
lncRNAs that work as ceRNAs to absorb miRNAs
There are several lncRNAs that work as ceRNAs to absorb miRNAs. However, most of them are not sufficiently highly expressed compared to the number of corresponding target miRNAs per cell.
H19 is among the first discovered eukaryotic lncRNAs and is known to be transcribed as intergenic RNA from the imprinted H19/IGF2 gene locus [77]. Although it is downregulated after birth, some vascular diseases are accompanied by re‐expression of this lncRNA. Increased expression of H19 is reported in aortic aneurysms and in calcific aortic valves [78, 79, 80]. Because of a high degree of secondary structure conservation, H19 is assumed to function as a structure‐dependent lncRNA. However, miR‐675‐3p and ‐5p are also encoded in H19, which may play a role in disease progression [81]. Moreover, H19 has a potential binding site for the let‐7 miRNA family and may also work as a molecular sponge [82].
Cholesterol homeostasis regulator of miRNA expression (CHROME) was identified as an important regulator of cellular and systemic cholesterol homeostasis [83]. CHROME levels are elevated in the plasma and atherosclerotic plaques of patients with CAD. It has been shown that CHROME promotes cholesterol efflux and HDL biogenesis by changing the levels of miRNAs that repress genes in these pathways. Indeed, CHROME binds specifically to miR‐27b, miR‐33a, miR‐33b and miR‐128, which are miRNAs that repress genes mediating cholesterol transport.
Through a genome‐wide association study using single nucleotide polymorphisms, chromosome 22q12.1 was identified as a susceptible locus for MI. Within this locus, a novel ncRNA was isolated and designated as MI‐associated transcript (MIAT). MIAT consists of five exons and does not encode any translational product. Several studies have reported that MIAT functions as a sponge for many miRNAs to regulate tumorigenesis and progression. These include miRNA‐155‐5p, miR‐29c, miR‐141 and miR‐212. MIAT also functions as a ceRNA against miR‐150‐5p to form a feedback loop with vascular endothelial growth factor [84]. Moreover, MIAT also acts to sponge miR‐204‐5p in the MIAT/miR‐204‐5p/HMGB1 axis in cerebral ischemia [85].
lncRNAs that bind to and regulate proteins
Liver X receptors (LXRs) are transcription factors that regulate cellular and systemic cholesterol homeostasis. Liver‐expressed LXR‐induced sequence (LeXis) was shown to control LXRs [86]. Hepatic LeXis expression is strongly induced in response to a Western diet or pharmacological LXR activation. Changes in LeXis levels in the liver affect the levels of cholesterol biosynthesis‐related genes. This effect is mediated by the interaction of LeXis with the heterogeneous ribonucleoprotein RALY, which acts as a transcriptional cofactor for cholesterol biosynthetic genes in the mouse liver.
Macrophage‐expressed LXR‐induced sequence (MeXis) was identified as an amplifier of LXR‐dependent transcription of Abca1, which is an important regulator of cholesterol efflux [87]. Mice lacking the MeXis show reduced Abca1 expression in a tissue‐selective manner. Moreover, MeXis‐deficient bone marrow cells altered chromosome architecture at the Abca1 locus and accelerated the development of atherosclerosis in mice. Mechanistically, MeXis interacts with and guides transcriptional coactivator DDX17 to the promoter region of Abca1 in a context‐specific manner.
lncRNAs that are transcribed as antisense RNAs
Some lncRNAs reside within protein‐coding gene units and overlap coding exons in antisense. Because the effects on host genes can be positive, negative or neutral, this classification is not directly related to their functions. Examples of such ncRNAs include ANRIL, MALAT1, HOXC‐AS1, SENCR, GATA6‐AS, STEEL and NEXN‐AS1. The functions of ANRIL and MALAT1 have already been described above.
HOXC cluster antisense RNA 1 (HOXC‐AS1) and homeobox C6 (HOXC6) were shown to be downregulated in carotid atherosclerosis via microarray analysis. Lentivirus‐mediated overexpression of HOXC‐AS1 induces HOXC6 expression at mRNA and protein levels in THP‐1 macrophages [88]. However, the precise functions of HOXC‐AS1 in atherosclerosis need to be clarified in further experiments.
Smooth muscle and EC‐enriched migration/differentiation‐associated lncRNA (SENCR) was revealed by RNAseq) of human coronary artery SMCs. SENCR is transcribed antisense from the 5'‐end of the FLI1 gene and two splice variants exist [89]. Knockdown studies revealed little to no cis‐acting effect of SENCR on FLI1 or neighboring gene expression. Loss‐of‐function studies indicated that SENCR inhibits SMC migration and maintains EC membrane integrity. Mechanistically, SENCR physically associates with cytoskeleton‐associated protein 4, thereby stabilizing cell membrane‐bound cadherin‐5 to promote EC adherens junction integrity [90].
GATA transcription factors are involved in variety of processes in development and diseases. The GATA locus expresses a noncoding antisense transcript of GATA6, named GATA6‐AS [91]. GATA6‐AS is upregulated in ECs during hypoxia. Silencing of GATA6‐AS diminished transforming growth factor‐β2‐induced endothelial–mesenchymal transition and promoted blood vessel formation in mice. Lysyl oxidase homolog 2 (LOXL2), which is known to remove activating H3K4me3 chromatin marks, is identified as a direct binding partner of GATA6‐AS. Moreover, a set of angiogenesis‐related genes were inversely regulated by LOXL2 and GATA6‐AS negatively regulated nuclear LOXL2 function. Thus, GATA6‐AS controls EC function as a negative regulator of nuclear LOXL2 function and activates angiogenesis‐related genes by increasing H3K4me3 methylation.
Spliced‐transcript endothelial‐enriched lncRNA (STEEL) is expressed from the homeobox D locus and is transcribed as antisense to homeobox D transcription factors [92]. STEEL promotes blood vessel formation in vivo. STEEL up‐regulates both eNOS and KLF2 and is inhibited by both of them in a feedback manner. Mechanistically, up‐regulation of eNOS and KLF2 is mediated via the recruitment of poly‐ADP ribosylase, PARP1 by STEEL. Feedback inhibition of STEEL expression may modulate angiogenic behavior in a position‐ and shear‐dependent fashion.
Nexilin F‐actin binding protein antisense RNA 1 (NEXN‐AS1) interacts with the chromatin remodeler BAZ1A and upregulates the expression of the actin‐binding protein NEXN. NEXN deficiency results in enhanced atherosclerosis, whereas NEXN overexpression reduces atherosclerosis in mice model of atherosclerosis. Both NEXN‐AS1 and NEXN are reduced in human atherosclerotic plaques, and patients with CAD have lower plasma NEXN levels [93].
lncRNAs that encode micropeptides
Emerging evidence indicates that several lncRNA molecules have short ORFs that encode functional peptides. LINC00961 was first annotated as a lncRNA but reassigned as a protein coding gene for the small regulatory polypeptide of amino acid response (SPAAR) micropeptide. LINC00961 was increased during the differentiation in ECs. Silencing of LINC00961 via siRNA or a GapmeR strategy significantly reduced EC adhesion, tube formation, migration, proliferation and endothelial membrane barrier integrity. On the other hand, overexpression of the SPAAR ORF increased tubule formation. Furthermore, overexpression of an ATG mutant of the full length LINC00961 transcript reduced network formation, which suggests that a ncRNA function of the transcript is opposed to the effects of SPAAR. Mechanistically, LINC00961 RNA binds the G‐actin sequestering protein thymosin beta‐4 (Tβ4) and Tβ4 depletion acts similarly to the overexpression of the ATG mutant. SPAAR binding partners includes the actin binding protein, spectrin repeat containing nuclear envelope protein 1 [94].
Currently, many lncRNAs related to vascular diseases are being identified and a summary of the lncRNAs is provided in Table 2.
Table 2.
Examples of lncRNAs that have functions in vascular disease.
| lncRNA | Function | Disease | References |
|---|---|---|---|
| circ_Lrp6 | CeRNA/miRNA sponge | Atherosclerosis progression | [45] |
| CircANRIL | Inhibiting rRNA processing | Atherosclerosis inhibition | [46] |
| circACTA2 | CeRNA/miRNA sponge | VSMC differentiation | [47] |
| ANRIL |
Transcription Guiding chromatin regulators |
CAD progression | [53, 54] |
| lincRNA‐p21 |
Transcription Protein regulation |
CAD progression | [55] |
| MALAT1 |
Transcription Binding chromatin remodelers |
EC proliferation | [56, 57, 58] |
| MEG3 |
Tethering of chromatin modifier CeRNA/miRNA sponge |
EC and VSMC proliferation inhibition Pulmonary hypertension inhibition |
[59, 60, 61, 62, 63, 64, 65, 66] |
| TUG1 | 3D chromatin positioning | VSMC proliferation | [67] |
| GAS5 | Transcription | Macrophage lipid accumulation | [68, 69] |
| MANTIS | Scaffold of chromatin modifier | EC angiogenesis promotion | [70] |
| HOTTIP |
eRNA Transcription |
EC proliferation | [71] |
| LEENE |
eRNA Transcription |
EC inflammation inhibition | [72] |
| SMILR |
eRNA Transcription |
VSMC proliferation Atherosclerosis promotion |
[73] |
| lncRNA‐CCL2 |
eRNA Transcription |
Macrophage chemotaxis promotion | [74] |
| NEAT1 | Decoy for chromatin regulator | VSMC proliferation | [75, 76] |
| H19 |
CeRNA/miRNA sponge mRNA decay |
Aortic aneurysm and aortic valve calcification promotion | [77, 78, 79, 80, 81, 82] |
| CHROME | CeRNA/miRNA sponge | Cholesterol efflux promotion | [83] |
| MIAT | CeRNA/miRNA sponge | EC proliferation | [84, 85] |
| LeXis | Transcription | Cholesterol biosynthesis promotion | [86] |
| MeXis | Transcription | Cholesterol efflux promotion | [87] |
| HOXC‐AS1 | Transcription | Atherosclerosis inhibition | [88] |
| SENCR | Transcription | VSMC migration inhibition | [89, 90] |
| GATA6‐AS |
Transcription Binding chromatin modifier |
Angiogenesis inhibition | [91] |
| STEEL | Transcription | Angiogenesis promotion | [92] |
| NEXN‐AS1 | Binding chromatin modifier | Atherosclerosis inhibition | [93] |
| LINC00961 | Encoding micropeptide | Angiogenesis inhibition | [94] |
Strategies for the identification of functional ncRNAs
There are already many databases of published miRNA sequences, annotations and predicted targets (e.g. http://www.mirbase.org and http://www.targetscan.org/vert_72). On the other hand, genome‐wide transcriptomic approaches such as RNA‐seq, microarray and cap analysis of gene expression technology are currently being used for the detection of lncRNAs [95]. Recently, a computational approach was also applied for the identification of lncRNAs as a result of an improvement in lncRNA annotations from RNA‐seq data [1]. RNA‐seq is superior with respect to detecting low‐abundance transcripts, although arrays have the advantage of rapid analysis.
After the identification, validation of the transcripts is necessary for the next step of the analysis. Quantitative‐PCR is commonly utilized for the validation of the candidate ncRNAs expression and the RACE technique is applied for the identification of the full sequence of the lncRNA. Moreover, identification of the localization of the lncRNA is useful and important with respect to hypothesizing its potential biological functions. Precise information of its subcellular localization can be obtained by RNA fluorescence in situ hybridization (FISH) [96]. Determination of tissue expression is also important.
As mentioned earlier above regarding lncRNAs that encode micropeptides, it is necessary to test the coding potential of novel lncRNAs. For example, a dwarf ORF encoded by an annotated lncRNA was reported to encode a peptide of 34 amino acids with unknown function [97].
Several techniques have also been developed for the identification of the interactions between lncRNAs and the genome, RNAs and proteins. Just as chromatin immunoprecipitation followed by microarray or deep sequencing has greatly improved our understanding of protein–DNA interactions on a genomic scale, chromatin isolation by RNA purification (ChIRP) was first developed by Chu et al. [98] to map long RNA occupancy genome‐wide at high resolution. This method is based on the affinity of antisense DNA oligonucleotides to capture the target lncRNA:chromatin complexes, which then generates a map of genomic binding sites. Other high‐throughput experimental technologies include capture hybridization analysis of RNA targets (CHART), RNA antisense purification (RAP), RNA immunoprecipitation (RIP), cross‐linking immunoprecipitation sequencing (CLIP‐seq), ChIRP‐mass spectrometry (MS) and CHART‐MS. These techniques have led to a rapid expansion of lncRNA research and also resulted in many publicly available databases [99, 100, 101, 102, 103]. RNA–RNA interactions can also be assessed by ChIRP easily because the design of affinity‐probes is straightforward; however, it cannot differentiate direct RNA–RNA binding from possible interactions with other intermediate proteins. By contrast, cross‐linking, ligation and sequencing of hybrids (CLASH) can be utilized to detect only direct hybridization between RNA molecules [104].
The unique secondary and tertiary structure of each lncRNA may contribute to its biological function. Thus, several techniques have been developed to obtain the RNA structure. Recent advances in probing the RNA structurome, including the use of RNA‐selective 2′‐hydroxyl acylation and primer extension (SHAPE) or kethoxal reagents or dimethyl sulfate, can provide unprecedented insights into the architecture of RNA molecules in living cells [105, 106, 107]. However, it is still unclear what controls lncRNA folding in the complex nuclear environment and to what extent secondary and tertiary structures are important to mediate lncRNA function.
Finally, loss‐of‐function strategies are required to determine the physiological functions of lncRNAs in animal models. Typical knockdown assays make use of short hairpin RNAs, siRNAs or locked nucleic acid GapmeRs. Their results can be assigned with higher confidence when oligonucleotide‐based strategies are complemented by the recent development of clustered regularly interspaced short palindromic repeat (CRISPR) technology [108, 109]. A summary of lncRNA analysis is provided in Table 3.
Table 3.
Summary of ncRNA research techniques.
| Stage | Technique | References |
|---|---|---|
| Identification | Microarray, RNA‐seq, Cap‐assisted gene expression sequencing, and nuclear run‐on assay | [95] |
| Validation | Database, quantitative PCR, RACE and RNA‐FISH | [96] |
| Assessment of the coding potential | Bioinformatic tool, and in vitro transcription assays | [97] |
| Genome‐wide mapping of binding sites | ChIRP, Chart, RAP, RIP and CLIP‐seq | [98, 99, 100, 101] |
| Identification of binding proteome | ChIRP‐MS and Chart‐MS | [102, 103] |
| Identification of RNA‐RNA interaction | CLASH | [104] |
| Analysis of RNA secondary structure | SHAPE and use of kethoxal reagents or dimethyl sulfate | [105, 106, 107] |
| Identification of function in vivo | Genetic KO, promoter insertion, polyA insertion, RNA interference, CRISPR interference and transgenics | [108, 109] |
Future perspectives
In summary, recent studies have provided considerable evidence about the functions of miRNAs and lncRNAs on various stages of cardiovascular diseases. They have indicated that these ncRNAs participate especially in the regulation of a broad spectrum of atherosclerosis (Fig. 2). Most of the ncRNAs have their own promoter and are regulated by distinct transcriptional factors in the disease condition. However, some of miRNAs are located within an intron of its host gene and the regulation totally depends on the levels of their host genes.
Fig. 2.
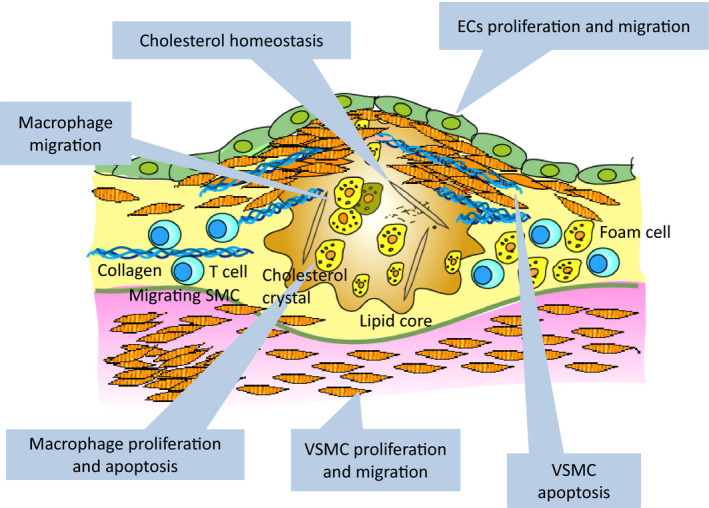
Possible targets of ncRNAs in atherosclerosis formation. EC proliferation and migration is affected by miR‐21, miR‐126, miR‐221/222, ANRIL, MIAT, MEG3, MALAT1, MANTIS, HOTTIP and LINC00961. VSMC proliferation and migration is mediated by miR‐21, miR‐143/145, H19, lncRNA‐p21, circ‐Lrp6, TUG1, SMILR, NEAT1 and SENCR. VSMC apoptosis is affected by miR‐125b, lncRNA‐p21 and circ‐ANRIL. Macrophage proliferation and apoptosis is enhanced by miR‐19 and lncRNA‐p21 and macrophage migration is mediated by lncRNA‐CCL2. Cholesterol homeostasis is also important for atherosclerosis formation, which is regulated by miR‐33a/b, GAS5, CHROME, MeXis and LeXis (liver).
Currently, RNA therapeutics are diverse and include antisense oligonucleotides, siRNAs, miRNAs, mRNAs, RNA aptamers, short activating RNAs and single guide RNAs for CRISPR/Cas9 systems. Molecular functions for miRNAs are relatively clear, and anti‐miRs and miRNA mimics have been developed. Anti‐miRs bind directly to the target miRNA and inhibit its function. Locked nucleic acids enhance the function of anti‐miRs by increasing their affinity and stability [110]. miRNA mimics are synthetic, double‐stranded RNAs that resemble a naturally generated miRNA. Several miRNAs that are currently being investigated in clinical trials are summarized in Table 4.
Table 4.
miRNAs that are currently being investigated in clinical trials.
| miRNA | Inhibition or augmentation | Name of drug | Target disease | Clinical stage | Year and clinical trial number |
|---|---|---|---|---|---|
| miR‐122 | Inhibition | Miravirsen | Hepatitis C | Phase II |
2010/2015 |
| miR‐103/107 | Inhibition | RG‐125 (AZD4076) | Nonalcoholic steatohepatitis | Phase I |
2015 |
| miR‐21 | Inhibition | SAR339375 | Alport’s syndrome | Phase II |
2016 |
| miR‐155 | Inhibition | MRG‐106 (Cobomarsen) |
Cutaneous T cell lymphoma Mycosis fungoides/lymphoma and leukemia |
Phase II/ Phase I |
2018/2015 |
| miR‐92a | Inhibition | MRG‐110 | Wound | Phase I |
2018 |
| miR‐16 | Augmentation | TargomiRs |
Malignant pleural mesothelioma Non‐small cell lung cancer |
Phase I |
2015 |
| miR‐29 | Augmentation | MRG‐201 | Keloid | Phase II |
2018 |
On the other hand, for most of the lncRNAs, the molecular mode of action remains elusive. Thus, further investigations are required aiming to understand the functions of lncRNAs in vivo. In any case, inhibition or activation of lncRNAs also leads to beneficial effects on disease conditions; therefore, the development of techniques that enable the fine regulation of lncRNAs is awaited.
Conflicts of interest
The authors declare no conflict of interest.
Author contributions
KO prepared the manuscript. TK, OB, MK, ST, RRR, SM, and TK analyzed current data and discussed the contents.
Acknowledgements
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant Numbers 17H05599, 20H03675 and 20K21600 to K. Ono, and 20K08904 to T. Horie) and by AMED under Grant Number JP19fk0210112 to K. Ono. This work was also supported by grants from THE VEHICLE RACING COMMEMORATIVE FOUNDATION to K. Ono.
References
- 1. Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J et al. (2017) An atlas of human long non‐coding RNAs with accurate 5' ends. Nature 543, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Feinberg MW & Moore KJ (2016) MicroRNA regulation of atherosclerosis. Circ Res 118, 703–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lu Y, Thavarajah T, Gu W, Cai J & Xu Q (2018) Impact of miRNA in atherosclerosis. Arterioscler Thromb Vasc Biol 38, e159–e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lucas T, Bonauer A & Dimmeler S (2018) RNA therapeutics in cardiovascular disease. Circ Res 123, 205–220. [DOI] [PubMed] [Google Scholar]
- 5. Sheedy FJ, Palsson‐McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, Johnson DS, Chen Y & O'Neill LA (2010) Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR‐21. Nat Immunol 11, 141–147. [DOI] [PubMed] [Google Scholar]
- 6. Sheedy FJ (2015) Turning 21: Induction of miR‐21 as a Key Switch in the Inflammatory Response. Front Immunol 6, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu LZ, Li C, Chen Q, Jing Y, Carpenter R, Jiang Y, Kung HF, Lai L & Jiang BH (2011) MiR‐21 induced angiogenesis through AKT and ERK activation and HIF‐1alpha expression. PLoS One 6, e19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou J, Wang KC, Wu W, Subramaniam S, Shyy JY, Chiu JJ, Li JY & Chien S (2011) MicroRNA‐21 targets peroxisome proliferators‐activated receptor‐alpha in an autoregulatory loop to modulate flow‐induced endothelial inflammation. Proc Natl Acad Sci USA 108, 10355–10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maegdefessel L, Azuma J, Toh R, Deng A, Merk DR, Raiesdana A, Leeper NJ, Raaz U, Schoelmerich AM, McConnell MV et al. (2012) MicroRNA‐21 blocks abdominal aortic aneurysm development and nicotine‐augmented expansion. Sci Transl Med 4, 122ra22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB & Zhang C (2007) MicroRNA expression signature and antisense‐mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res 100, 1579–1588. [DOI] [PubMed] [Google Scholar]
- 11. Davis‐Dusenbery BN, Wu C & Hata A (2011) Micromanaging vascular smooth muscle cell differentiation and phenotypic modulation. Arterioscler Thromb Vasc Biol 31, 2370–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng Y & Zhang C (2010) MicroRNA‐21 in cardiovascular disease. J Cardiovasc Transl Res 3, 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Canfran‐Duque A, Rotllan N, Zhang X, Fernandez‐Fuertes M, Ramirez‐Hidalgo C, Araldi E, Daimiel L, Busto R, Fernandez‐Hernando C & Suarez Y (2017) Macrophage deficiency of miR‐21 promotes apoptosis, plaque necrosis, and vascular inflammation during atherogenesis. EMBO Mol Med 9, 1244–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ben‐Nun D, Buja LM & Fuentes F (2020) Prevention of heart failure with preserved ejection fraction (HFpEF): reexamining microRNA‐21 inhibition in the era of oligonucleotide‐based therapeutics. Cardiovasc Pathol 49, 107243. [DOI] [PubMed] [Google Scholar]
- 15. Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ & Fernandez‐Hernando C (2010) MiR‐33 contributes to the regulation of cholesterol homeostasis. Science 328, 1570–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Najafi‐Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE & Naar AM (2010) MicroRNA‐33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 328, 1566–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marquart TJ, Allen RM, Ory DS & Baldan A (2010) miR‐33 links SREBP‐2 induction to repression of sterol transporters. Proc Natl Acad Sci USA 107, 12228–12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y et al. (2010) MicroRNA‐33 encoded by an intron of sterol regulatory element‐binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci USA 107, 17321–17326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA & Bommer GT (2010) Expression of miR‐33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem 285, 33652–33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brown MS, Ye J & Goldstein JL (2010) Medicine. HDL miR‐ed down by SREBP introns. Science 328, 1495–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horie T, Baba O, Kuwabara Y, Chujo Y, Watanabe S, Kinoshita M, Horiguchi M, Nakamura T, Chonabayashi K, Hishizawa M et al. (2012) MicroRNA‐33 deficiency reduces the progression of atherosclerotic plaque in ApoE(‐/‐) mice. J Am Heart Assoc 1, e003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Horie T, Nishino T, Baba O, Kuwabara Y, Nakao T, Nishiga M, Usami S, Izuhara M, Nakazeki F, Ide Y et al. (2014) MicroRNA‐33b knock‐in mice for an intron of sterol regulatory element‐binding factor 1 (Srebf1) exhibit reduced HDL‐C in vivo. Sci Rep 4, 5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nishino T, Horie T, Baba O, Sowa N, Hanada R, Kuwabara Y, Nakao T, Nishiga M, Nishi H, Nakashima Y et al. (2018) SREBF1/microRNA‐33b axis exhibits potent effect on unstable atherosclerotic plaque formation in vivo. Arterioscler Thromb Vasc Biol 38, 2460–2473. [DOI] [PubMed] [Google Scholar]
- 24. Nakao T, Horie T, Baba O, Nishiga M, Nishino T, Izuhara M, Kuwabara Y, Nishi H, Usami S, Nakazeki F et al. (2017) Genetic ablation of microRNA‐33 attenuates inflammation and abdominal aortic aneurysm formation via several anti‐inflammatory pathways. Arterioscler Thromb Vasc Biol 37, 2161–2170. [DOI] [PubMed] [Google Scholar]
- 25. Zhang X, Price NL & Fernandez‐Hernando C (2019) Non‐coding RNAs in lipid metabolism. Vascul Pharmacol 114, 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sedgeman LR, Michell DL & Vickers KC (2019) Integrative roles of microRNAs in lipid metabolism and dyslipidemia. Curr Opin Lipidol 30, 165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mendell JT (2008) miRiad roles for the miR‐17‐92 cluster in development and disease. Cell 133, 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT et al. (2006) Augmentation of tumor angiogenesis by a Myc‐activated microRNA cluster. Nat Genet 38, 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K et al. (2009) MicroRNA‐92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 324, 1710–1713. [DOI] [PubMed] [Google Scholar]
- 30. Li X, Teng C, Ma J, Fu N, Wang L, Wen J & Wang TY (2019) miR‐19 family: a promising biomarker and therapeutic target in heart, vessels and neurons. Life Sci 232, 116651. [DOI] [PubMed] [Google Scholar]
- 31. Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE & Lawson ND (2010) MicroRNA‐mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature 464, 1196–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harris TA, Yamakuchi M, Ferlito M, Mendell JT & Lowenstein CJ (2008) MicroRNA‐126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA 105, 1516–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Izuhara M, Kuwabara Y, Saito N, Yamamoto E, Hakuno D, Nakashima Y, Horie T, Baba O, Nishiga M, Nakao T et al. (2017) Prevention of neointimal formation using miRNA‐126‐containing nanoparticle‐conjugated stents in a rabbit model. PLoS One 12, e0172798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Endo‐Takahashi Y, Negishi Y, Nakamura A, Ukai S, Ooaku K, Oda Y, Sugimoto K, Moriyasu F, Takagi N, Suzuki R et al. (2014) Systemic delivery of miR‐126 by miRNA‐loaded Bubble liposomes for the treatment of hindlimb ischemia. Sci Rep 4, 3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN & Srivastava D (2009) miR‐145 and miR‐143 regulate smooth muscle cell fate and plasticity. Nature 460, 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q, Deitch EA, Huo Y, Delphin ES & Zhang C (2009) MicroRNA‐145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res 105, 158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, Pezzuto I, Crippa S, Nemir M, Sarre A, Johnson R et al. (2015) Genome‐wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart‐specific long non‐coding RNAs. Eur Heart J 36, 353–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Plaisance I, Perruchoud S, Fernandez‐Tenorio M, Gonzales C, Ounzain S, Ruchat P, Nemir M, Niggli E & Pedrazzini T (2016) Cardiomyocyte lineage specification in adult human cardiac precursor cells via modulation of enhancer‐associated long noncoding RNA expression. JACC Basic Transl Sci 1, 472–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu X, Cheng Y, Zhang S, Lin Y, Yang J & Zhang C (2009) A necessary role of miR‐221 and miR‐222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res 104, 476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Celic T, Metzinger‐Le Meuth V, Six I, Massy ZA & Metzinger L (2017) The mir‐221/222 cluster is a key player in vascular biology via the fine‐tuning of endothelial cell physiology. Curr Vasc Pharmacol 15, 40–46. [DOI] [PubMed] [Google Scholar]
- 41. Wang L, Liu C, Li C, Xue J, Zhao S, Zhan P, Lin Y, Zhang P, Jiang A & Chen W (2015) Effects of microRNA‐221/222 on cell proliferation and apoptosis in prostate cancer cells. Gene 572, 252–258. [DOI] [PubMed] [Google Scholar]
- 42. Kuwabara Y, Tsuji S, Nishiga M, Izuhara M, Ito S, Nagao K, Horie T, Watanabe S, Koyama S, Kiryu H et al. (2020) Lionheart LincRNA alleviates cardiac systolic dysfunction under pressure overload. Commun Biol 3, 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Salzman J, Gawad C, Wang PL, Lacayo N & Brown PO (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7, e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M et al. (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. [DOI] [PubMed] [Google Scholar]
- 45. Hall IF, Climent M, Quintavalle M, Farina FM, Schorn T, Zani S, Carullo P, Kunderfranco P, Civilini E, Condorelli G et al. (2019) Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA‐145 function. Circ Res 124, 498–510. [DOI] [PubMed] [Google Scholar]
- 46. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A et al. (2016) Circular non‐coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 7, 12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sun Y, Yang Z, Zheng B, Zhang XH, Zhang ML, Zhao XS, Zhao HY, Suzuki T & Wen JK (2017) A novel regulatory mechanism of smooth muscle alpha‐actin expression by NRG‐1/circACTA2/miR‐548f‐5p axis. Circ Res 121, 628–635. [DOI] [PubMed] [Google Scholar]
- 48. Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M & Lander ES (2016) Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Holdt LM, Beutner F, Scholz M, Gielen S, Gabel G, Bergert H, Schuler G, Thiery J & Teupser D (2010) ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol 30, 620–627. [DOI] [PubMed] [Google Scholar]
- 50. Congrains A, Kamide K, Oguro R, Yasuda O, Miyata K, Yamamoto E, Kawai T, Kusunoki H, Yamamoto H, Takeya Y et al. (2012) Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis 220, 449–455. [DOI] [PubMed] [Google Scholar]
- 51. Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE et al. (2007) Genomewide association analysis of coronary artery disease. N Engl J Med 357, 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wellcome Trust Case Control, C. (2007) Genome‐wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lo Sardo V, Chubukov P, Ferguson W, Kumar A, Teng EL, Duran M, Zhang L, Cost G, Engler AJ, Urnov F et al. (2018) Unveiling the role of the most impactful cardiovascular risk locus through haplotype editing. Cell 175, 1796–1810 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M & Xiong Y (2011) Long non‐coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30, 1956–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y et al. (2014) LincRNA‐p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation 130, 1452–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S et al. (2014) Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 114, 1389–1397. [DOI] [PubMed] [Google Scholar]
- 57. Cremer S, Michalik KM, Fischer A, Pfisterer L, Jae N, Winter C, Boon RA, Muhly‐Reinholz M, John D, Uchida S et al. (2019) Hematopoietic deficiency of the long noncoding RNA MALAT1 promotes atherosclerosis and plaque inflammation. Circulation 139, 1320–1334. [DOI] [PubMed] [Google Scholar]
- 58. Sun Y & Ma L (2019) New insights into long non‐coding RNA MALAT1 in cancer and metastasis. Cancers (Basel) 11, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Boon RA, Hofmann P, Michalik KM, Lozano‐Vidal N, Berghauser D, Fischer A, Knau A, Jae N, Schurmann C & Dimmeler S (2016) Long noncoding RNA Meg3 controls endothelial cell aging and function: implications for regenerative angiogenesis. J Am Coll Cardiol 68, 2589–2591. [DOI] [PubMed] [Google Scholar]
- 60. Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C & Klibanski A (2007) Activation of p53 by MEG3 non‐coding RNA. J Biol Chem 282, 24731–24742. [DOI] [PubMed] [Google Scholar]
- 61. Sun Z, Nie X, Sun S, Dong S, Yuan C, Li Y, Xiao B, Jie D & Liu Y (2017) Long non‐coding RNA MEG3 downregulation triggers human pulmonary artery smooth muscle cell proliferation and migration via the p53 signaling pathway. Cell Physiol Biochem 42, 2569–2581. [DOI] [PubMed] [Google Scholar]
- 62. Uroda T, Anastasakou E, Rossi A, Teulon JM, Pellequer JL, Annibale P, Pessey O, Inga A, Chillon I & Marcia M (2019) Conserved pseudoknots in lncRNA MEG3 are essential for stimulation of the p53 pathway. Mol Cell 75, 982–995.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. He C, Yang W, Yang J, Ding J, Li S, Wu H, Zhou F, Jiang Y, Teng L & Yang J (2017) Long noncoding RNA MEG3 negatively regulates proliferation and angiogenesis in vascular endothelial cells. DNA Cell Biol 36, 475–481. [DOI] [PubMed] [Google Scholar]
- 64. Wu Z, He Y, Li D, Fang X, Shang T, Zhang H & Zheng X (2017) Long noncoding RNA MEG3 suppressed endothelial cell proliferation and migration through regulating miR‐21. Am J Transl Res 9, 3326–3335. [PMC free article] [PubMed] [Google Scholar]
- 65. Bai Y, Zhang Q, Su Y, Pu Z & Li K (2019) Modulation of the proliferation/apoptosis balance of vascular smooth muscle cells in atherosclerosis by lncRNA‐MEG3 via regulation of miR‐26a/Smad1 axis. Int Heart J 60, 444–450. [DOI] [PubMed] [Google Scholar]
- 66. Xing Y, Zheng X, Fu Y, Qi J, Li M, Ma M, Wang S, Li S & Zhu D (2019) Long noncoding RNA‐maternally expressed gene 3 contributes to hypoxic pulmonary hypertension. Mol Ther 27, 2166–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen R, Kong P, Zhang F, Shu YN, Nie X, Dong LH, Lin YL, Xie XL, Zhao LL, Zhang XJ et al. (2017) EZH2‐mediated alpha‐actin methylation needs lncRNA TUG1, and promotes the cortex cytoskeleton formation in VSMCs. Gene 616, 52–57. [DOI] [PubMed] [Google Scholar]
- 68. Meng XD, Yao HH, Wang LM, Yu M, Shi S, Yuan ZX & Liu J (2020) Knockdown of GAS5 inhibits atherosclerosis progression via reducing EZH2‐mediated ABCA1 transcription in ApoE(‐/‐) mice. Mol Ther Nucleic Acids 19, 84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yu Y & Hann SS (2019) Novel tumor suppressor lncRNA growth arrest‐specific 5 (GAS5) in human cancer. Onco Targets Ther 12, 8421–8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Leisegang MS, Fork C, Josipovic I, Richter FM, Preussner J, Hu J, Miller MJ, Epah J, Hofmann P, Gunther S et al. (2017) Long noncoding RNA MANTIS facilitates endothelial angiogenic function. Circulation 136, 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liao B, Chen R, Lin F, Mai A, Chen J, Li H, Xu Z & Dong S (2018) Long noncoding RNA HOTTIP promotes endothelial cell proliferation and migration via activation of the Wnt/beta‐catenin pathway. J Cell Biochem 119, 2797–2805. [DOI] [PubMed] [Google Scholar]
- 72. Miao Y, Ajami NE, Huang TS, Lin FM, Lou CH, Wang YT, Li S, Kang J, Munkacsi H, Maurya MR et al. (2018) Enhancer‐associated long non‐coding RNA LEENE regulates endothelial nitric oxide synthase and endothelial function. Nat Commun 9, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ballantyne MD, Pinel K, Dakin R, Vesey AT, Diver L, Mackenzie R, Garcia R, Welsh P, Sattar N, Hamilton G et al. (2016) Smooth muscle enriched long noncoding RNA (SMILR) regulates cell proliferation. Circulation 133, 2050–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Khyzha N, Khor M, DiStefano PV, Wang L, Matic L, Hedin U, Wilson MD, Maegdefessel L & Fish JE (2019) Regulation of CCL2 expression in human vascular endothelial cells by a neighboring divergently transcribed long noncoding RNA. Proc Natl Acad Sci USA 116, 16410–16419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Naganuma T & Hirose T (2013) Paraspeckle formation during the biogenesis of long non‐coding RNAs. RNA Biol 10, 456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ahmed ASI, Dong K, Liu J, Wen T, Yu L, Xu F, Kang X, Osman I, Hu G, Bunting KM et al. (2018) Long noncoding RNA NEAT1 (nuclear paraspeckle assembly transcript 1) is critical for phenotypic switching of vascular smooth muscle cells. Proc Natl Acad Sci USA 115, E8660–E8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bartolomei MS, Zemel S & Tilghman SM (1991) Parental imprinting of the mouse H19 gene. Nature 351, 153–155. [DOI] [PubMed] [Google Scholar]
- 78. Li DY, Busch A, Jin H, Chernogubova E, Pelisek J, Karlsson J, Sennblad B, Liu S, Lao S, Hofmann P et al. (2018) H19 induces abdominal aortic aneurysm development and progression. Circulation 138, 1551–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hadji F, Boulanger MC, Guay SP, Gaudreault N, Amellah S, Mkannez G, Bouchareb R, Marchand JT, Nsaibia MJ, Guauque‐Olarte S et al. (2016) Altered DNA methylation of long noncoding RNA H19 in calcific aortic valve disease promotes mineralization by silencing NOTCH1. Circulation 134, 1848–1862. [DOI] [PubMed] [Google Scholar]
- 80. Sun Y, Zhong L, He X, Wang S, Lai Y, Wu W, Song H, Chen Y, Yang Y, Liao W et al. (2019) LncRNA H19 promotes vascular inflammation and abdominal aortic aneurysm formation by functioning as a competing endogenous RNA. J Mol Cell Cardiol 131, 66–81. [DOI] [PubMed] [Google Scholar]
- 81. Dey BK, Pfeifer K & Dutta A (2014) The H19 long noncoding RNA gives rise to microRNAs miR‐675‐3p and miR‐675‐5p to promote skeletal muscle differentiation and regeneration. Genes Dev 28, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H et al. (2013) The imprinted H19 lncRNA antagonizes let‐7 microRNAs. Mol Cell 52, 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hennessy EJ, van Solingen C, Scacalossi KR, Ouimet M, Afonso MS, Prins J, Koelwyn GJ, Sharma M, Ramkhelawon B, Carpenter S et al. (2019) The long noncoding RNA CHROME regulates cholesterol homeostasis in primate. Nat Metab 1, 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q & Jiang Q (2015) lncRNA‐MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res 116, 1143–1156. [DOI] [PubMed] [Google Scholar]
- 85. Deng W, Fan C, Shen R, Wu Y, Du R & Teng J (2020) Long noncoding MIAT acting as a ceRNA to sponge microRNA‐204‐5p to participate in cerebral microvascular endothelial cell injury after cerebral ischemia through regulating HMGB1. J Cell Physiol 235, 4571–4586. [DOI] [PubMed] [Google Scholar]
- 86. Sallam T, Jones MC, Gilliland T, Zhang L, Wu X, Eskin A, Sandhu J, Casero D, Vallim TQ, Hong C et al. (2016) Feedback modulation of cholesterol metabolism by the lipid‐responsive non‐coding RNA LeXis. Nature 534, 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sallam T, Jones M, Thomas BJ, Wu X, Gilliland T, Qian K, Eskin A, Casero D, Zhang Z, Sandhu J et al. (2018) Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat Med 24, 304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huang C, Hu YW, Zhao JJ, Ma X, Zhang Y, Guo FX, Kang CM, Lu JB, Xiu JC, Sha YH et al. (2016) Long noncoding RNA HOXC‐AS1 suppresses Ox‐LDL‐induced cholesterol accumulation through promoting HOXC6 expression in THP‐1 macrophages. DNA Cell Biol 35, 722–729. [DOI] [PubMed] [Google Scholar]
- 89. Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, Zhou Q, Han Y, Spector DL, Zheng D et al. (2014) Identification and initial functional characterization of a human vascular cell‐enriched long noncoding RNA. Arterioscler Thromb Vasc Biol 34, 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lyu Q, Xu S, Lyu Y, Choi M, Christie CK, Slivano OJ, Rahman A, Jin ZG, Long X, Xu Y et al. (2019) SENCR stabilizes vascular endothelial cell adherens junctions through interaction with CKAP4. Proc Natl Acad Sci USA 116, 546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Neumann P, Jae N, Knau A, Glaser SF, Fouani Y, Rossbach O, Kruger M, John D, Bindereif A, Grote P et al. (2018) The lncRNA GATA6‐AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nat Commun 9, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Man HSJ, Sukumar AN, Lam GC, Turgeon PJ, Yan MS, Ku KH, Dubinsky MK, Ho JJD, Wang JJ, Das S et al. (2018) Angiogenic patterning by STEEL, an endothelial‐enriched long noncoding RNA. Proc Natl Acad Sci USA 115, 2401–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hu YW, Guo FX, Xu YJ, Li P, Lu ZF, McVey DG, Zheng L, Wang Q, Ye JH, Kang CM et al. (2019) Long noncoding RNA NEXN‐AS1 mitigates atherosclerosis by regulating the actin‐binding protein NEXN. J Clin Invest 129, 1115–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Spencer HL, Sanders R, Boulberdaa M, Meloni M, Cochrane A, Spiroski AM, Mountford J, Emanueli C, Caporali A, Brittan M et al. (2020) The LINC00961 transcript and its encoded micropeptide SPAAR regulate endothelial cell function. Cardiovasc Res 116, 1981–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lizio M, Abugessaisa I, Noguchi S, Kondo A, Hasegawa A, Hon CC, de Hoon M, Severin J, Oki S, Hayashizaki Y et al. (2019) Update of the FANTOM web resource: expansion to provide additional transcriptome atlases. Nucleic Acids Res 47, D752–D758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Isobe M, Toya H, Mito M, Chiba T, Asahara H, Hirose T & Nakagawa S (2020) Forced isoform switching of Neat1_1 to Neat1_2 leads to the loss of Neat1_1 and the hyperformation of paraspeckles but does not affect the development and growth of mice. RNA 26, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET et al. (2016) A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chu C, Qu K, Zhong FL, Artandi SE & Chang HY (2011) Genomic maps of long noncoding RNA occupancy reveal principles of RNA‐chromatin interactions. Mol Cell 44, 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI & Kingston RE (2011) The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci USA 108, 20497–20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Engreitz JM, Pandya‐Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES et al. (2013) The Xist lncRNA exploits three‐dimensional genome architecture to spread across the X chromosome. Science 341, 1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Cao M, Zhao J & Hu G (2019) Genome‐wide methods for investigating long noncoding RNAs. Biomed Pharmacother 111, 395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E & Chang HY (2015) Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY & Kingston RE (2014) The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell 55, 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Helwak A & Tollervey D (2014) Mapping the miRNA interactome by cross‐linking ligation and sequencing of hybrids (CLASH). Nat Protoc 9, 711–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Spitale RC, Crisalli P, Flynn RA, Torre EA, Kool ET & Chang HY (2013) RNA SHAPE analysis in living cells. Nat Chem Biol 9, 18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Weng X, Gong J, Chen Y, Wu T, Wang F, Yang S, Yuan Y, Luo G, Chen K, Hu L et al. (2020) Keth‐seq for transcriptome‐wide RNA structure mapping. Nat Chem Biol 16, 489–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Lempereur L, Nicoloso M, Riehl N, Ehresmann C, Ehresmann B & Bachellerie JP (1985) Conformation of yeast 18S rRNA. Direct chemical probing of the 5' domain in ribosomal subunits and in deproteinized RNA by reverse transcriptase mapping of dimethyl sulfate‐accessible. Nucleic Acids Res 13, 8339–8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hsu PD, Lander ES & Zhang F (2014) Development and applications of CRISPR‐Cas9 for genome engineering. Cell 157, 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Pickar‐Oliver A & Gersbach CA (2019) The next generation of CRISPR‐Cas technologies and applications. Nat Rev Mol Cell Biol 20, 490–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rupaimoole R & Slack FJ (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16, 203–222. [DOI] [PubMed] [Google Scholar]


