Abstract
Objectives
To assess the feasibility and uptake of a community‐based prostate cancer (PCa) screening programme selecting men according to their genetic risk of PCa. To assess the uptake of PCa screening investigations by men invited for screening. The uptake of the pilot study would guide the opening of the larger BARCODE1 study recruiting 5000 men.
Subjects and Methods
Healthy males aged 55–69 years were invited to participate via their general practitioners (GPs). Saliva samples were collected via mailed collection kits. After DNA extraction, genotyping was conducted using a study specific assay. Genetic risk was based on genotyping 130 germline PCa risk single nucleotide polymorphisms (SNPs). A polygenic risk score (PRS) was calculated for each participant using the sum of weighted alleles for 130 SNPs. Study participants with a PRS lying above the 90th centile value were invited for PCa screening by prostate magnetic resonance imaging (MRI) and biopsy.
Results
Invitation letters were sent to 1434 men. The overall study uptake was 26% (375/1436) and 87% of responders were eligible for study entry. DNA genotyping data were available for 297 men and 25 were invited for screening. After exclusions due to medical comorbidity/invitations declined, 18 of 25 men (72%) underwent MRI and biopsy of the prostate. There were seven diagnoses of PCa (38.9%). All cancers were low‐risk and were managed with active surveillance.
Conclusion
The BARCODE1 Pilot has shown this community study in the UK to be feasible, with an overall uptake of 26%. The main BARCODE1 study is now open and will recruit 5000 men. The results of BARCODE1 will be important in defining the role of genetic profiling in targeted PCa population screening.
Patient Summary
What is the paper about?
Very few prostate cancer screening programmes currently exist anywhere in the world. Our pilot study investigated if men in the UK would find it acceptable to have a genetic test based on a saliva sample to examine their risk of prostate cancer development. This test would guide whether men are offered prostate cancer screening tests.
What does it mean for patients?
We found that the study design was acceptable: 26% of men invited to take part agreed to have the test. The majority of men who were found to have an increased genetic risk of prostate cancer underwent further tests offered (prostate MRI scan and biopsy). We have now expanded the study to enrol 5000 men. The BARCODE1 study will be important in examining whether this approach could be used for large‐scale population prostate cancer screening.
Keywords: prostate cancer, SNPs, screening, precision medicine, GWAS
Introduction
In common with other complex diseases, the genetic heritability of prostate cancer (PCa) is composed of both rare, high to moderately penetrant variants conferring a high risk of disease (e.g. BRCA2 DNA repair associated [BRCA2] gene mutations), and commonly occurring single‐nucleotide variants (SNVs) that confer risks of lower magnitude. With the advent of the genome‐wide association study (GWAS) and the increasing numbers of cases and controls included in such studies, PCa GWAS and meta‐analyses have identified more than 200 loci associated with PCa development [1, 2, 3]. Most of these SNVs are commonly occurring single nucleotide polymorphisms (SNPs; i.e. minor allele frequency [MAF] ≥5%) and although each locus is associated with a low to moderate per allele odds ratio (OR), the genetic risk increases with increasing number of risk alleles in a person’s germline DNA [4]. The currently known PCa susceptibility loci explain an estimated 34–43% of the familial relative risk (RR) of PCa [2, 3, 5, 6].
There are very few population PCa screening programmes due to the lack of a test with adequate sensitivity and specificity required for large‐scale population screening. The use of the PSA blood test was investigated in two large, randomised screening studies: the Prostate, Lung, Colorectal and Ovary (PLCO) study and the European Randomised Study of Screening for Prostate Cancer (ERSPC) [7, 8]. Although long‐term follow‐up in the ERSPC studies has shown a survival benefit (absolute reduction of 13 PCa deaths per 10 000 men), the complications of prostate biopsies, high rate of false‐positive results (10% in PLCO and 18% in ERSPC), as well as over‐diagnosis of indolent PCa has led to caution around the use of PSA testing.
Guidelines from national screening programmes such as the United States Preventive Services Task Force (USPSTF) have fluctuated from advising against PSA screening (in 2012) to recommending that men make an individualised decision regarding PSA testing in conjunction with their clinician (for those aged 55–69 years) [9, 10]. In the UK, the National Screening Committee recommends against universal PSA screening.
To our knowledge, no prospective studies utilising genetic profiling to stratify men in the population according to their PCa risk have been performed. The use of a genetic test utilising the known PCa risk SNPs could allow PCa screening to be targeted to men at high risk of PCa development. Germline DNA for genotyping requires a one‐off collection, usually in the form of a blood draw or saliva sample. Using the genotyping data, a polygenic risk score (PRS) can be calculated for an individual to estimate their risk of PCa development relative to the average population risk. When used at scale, a genotyping test is relatively cheap and can be carried out ahead of likely disease onset to identify men with a high PRS who may benefit from screening starting at a pre‐determined time‐point. In parallel, men with a low PRS may avoid the potential complications of invasive tests.
Utilising the known PCa risk loci, the RR of PCa for European ancestry men in the top 1% of the PRS distribution is 5.7 compared with the average risk of men in the general population; this level of risk is in the range of that associated with BRCA2 mutations [11]. For men in the top 10% of PRS distribution the RR is 2.7 [2].
The use of PRS stratification within a PCa screening programme has been shown through modelling studies to reduce the level of over‐diagnosis of indolent disease through screening [12]. In the breast cancer setting, using genetics to take a risk‐stratified approach to refine screening also reduces the rate of over‐diagnosis and improves cost‐effectiveness of screening [13].
The BARCODE1 study (NCT03857477) is enrolling men of European ancestry from the community via their GPs to undergo a germline genetic profile test utilising 130 PCa risk SNPs. Men in the top 10% of the study population’s genetic risk distribution are offered screening with MRI of the prostate followed by systematic and targeted biopsy. As the PRS used in BARCODE1 is based on SNPs discovered in European populations, there is a portability problem across different populations. Although multi‐ethnic GWAS have demonstrated that many PCa risk SNPs are shared between populations, the risk associated with a variant may vary according to ethnicity [3]. Additionally, some PCa risk SNPs will be population specific. Studies are underway to investigate the use of genetic profiling in PCa screening in other ethnic groups [14].
The present study is the first prospective study to assess the utility of genetic profiling in a community setting to guide PCa screening and aims to recruit a total of 5000 men. In the present study, we report the results of the BARCODE1 Pilot, which was set up as a feasibility study to examine the acceptability of the study to men in the UK community prior to opening the main BARCODE1 study.
Subjects and Methods
Trial Design
The BARCODE1 Pilot was a feasibility study aiming to recruit 300 participants prior to the launch of a larger trial with 5000 participants. The study was co‐ordinated and managed by the Oncogenetics Team at The Institute of Cancer Research, UK. Recruitment was carried out via seven GP centres in London, and participants were invited into the study by letter.
Eligible responders were sent a consent form to sign and return to the study team. They were also sent a saliva collection kit to provide a saliva sample for DNA extraction and genotyping (Fig. 1). Participants found to lie in the top 10% of the PRS distribution were offered a MRI of the prostate followed by prostate biopsy. Participants with negative biopsies were followed up with annual PSA measurement and repeat biopsies were performed if there was a significant PSA rise (study protocol in Appendix S1).
Fig. 1.
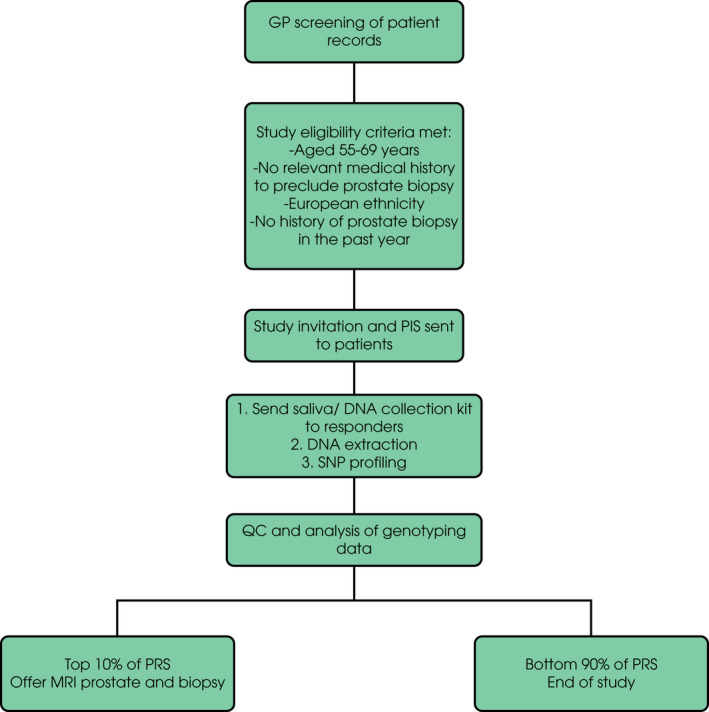
BARCODE1 study outline.
Trial Registration
This study was reviewed by the London‐Chelsea Research Ethics Committee under reference number: 15/LO/1992 and was approved to be conducted in NHS England by the Health Research Authority under reference number: 147536.
Participants
The GP sites acted as recruiting centres. Practice records were reviewed to identify men aged 55–69 years meeting study eligibility criteria. A participant information sheet (PIS) and health questionnaire was sent to all suitable men and responses screened to ensure eligibility. Participants were required to have no history of PCa, no prostate biopsy within 12 months, no contra‐indications to MRI or prostate biopsy, and to be of European ethnicity. Responders meeting the criteria were requested to complete a postal saliva (DNA) sample.
Sample Size
For the main BARCODE1 study, a sample size of 5000 men will be required to identify 500 men within the top 10% of the PRS distribution, who will undergo screening. The pilot study was planned with the aim of recruiting 300 men, with ∼30 in the top 10% of PRS distribution. This sample size was chosen pragmatically assuming recruitment of 100 men from each of the (initial) three GP centres. A further four centres were added during the course of the pilot study to complete recruitment due to a lower than anticipated recruitment rate. All initial study procedures including invitations to participate, informed consent and saliva collection were carried out using the postal system.
DNA Extraction and Genotyping
DNA extraction from saliva was carried out externally by Tepnel Pharma Services, Livingston, UK. Extracted DNA was sent to Affymetrix® (part of Thermo Fisher Scientific Inc., Waltham, MA, USA) for genotyping using a custom high throughput ‘genotyping by sequencing’ assay (Eureka myDesign Genotyping Panel; https://tools.thermofisher.com/content/sfs/brochures/Eureka_Genotyping_Solution_datasheet.pdf) [15]. At the time of BARCODE1 study setup, 147 index PCa susceptibility SNPs had been reported in European ancestry populations [2]. Our assay design aimed to capture all of these known variants; however, 17 index SNPs could not be successfully designed or failed quality control (QC) using this technology, whilst a further three index variants needed to be substituted for a suitable proxy variant to achieve inclusion. Our final genotyping panel therefore consisted of 130 European ancestry PCa risk SNPs (Appendix S2). Additional QC was performed on the raw computer assigned genotype calls, through manual examination of cluster plots using Eureka™ Analysis Suite (https://www.thermofisher.com/uk/en/home/life‐science/microarray‐analysis/microarray‐analysis‐instruments‐software‐services/microarray‐analysis‐software/eureka‐analysis‐suite.html).
Statistical Methods
We calculated a PRS for each participant based on the sum of risk alleles for the 130 PCa risk loci, weighted by their per‐allele log OR (formula in Appendix S2). In instances where a variant failed genotype QC within a specific individual, to reflect the probability of their carrying the risk allele for that variant, missing genotype(s) were assigned a risk allele count corresponding to the population risk allele frequency for the variant, doubled for autosomal variants.
The PRS mean and standard deviation (SD) for the BARCODE1 Pilot cohort were used to calculate the PRS threshold at the 90th centile based on a normal distribution (Appendix S2). Using this method to calculate the 90th centile accounts for the potential large rise in PRS value at the extremes of the distribution in the small sample size. BARCODE1 Pilot participants with a PRS >90th centile were identified for PCa screening.
Screening Procedures
Participants received a letter informing them of the outcome of genotyping and whether their PRS fell in the top 10% of the study PRS distribution. Those found to have a PRS >90th centile were invited to undergo PCa screening at The Royal Marsden Hospital (RMH). Screening involved a multi‐parametric MRI scan of the prostate and a prostate biopsy. Baseline PSA level was measured but did not alter management. The MRI included T2‐weighted, diffusion‐weighted (DW) and dynamic contrast (gadolinium)‐enhanced images. Scans were performed either at 3‐T with endorectal coil or at 1.5‐T with an external phased array coil and reported by a specialist uro‐radiologist. Prostate lesions identified were scored 1–5 according to Prostate Imaging ‐ Reporting and Data System (PI‐RADS) as developed by the European Society of Urogenital Radiology [16]. A score of 1 indicated clinically significant disease is highly unlikely to be present, while a score of 5 indicated clinically significant cancer is highly likely to be present.
The MRI was followed by prostate biopsy. A systematic 12‐core TRUS biopsy was carried out by the study urologist. If a lesion/lesions were identified on DW‐MRI, additional targeted sampling was undertaken (Koeils Urostation™). Prostate biopsy samples were examined by a specialist uro‐pathologist at RMH and reported as per the 2014 International Society of Urological Pathologists guidelines.
Outcomes
Response rates across all recruiting centres were examined, as well as uptake of screening tests (MRI and biopsy) in men who were invited for screening based on their PRS. Outcomes from screening include the number of cancers diagnosed (clinically significant and insignificant), MRI lesions detected and biopsy‐related complications.
An additional sub‐study is ongoing to examine the psychosocial impact of screening in this group of high‐risk individuals, the results of which will be reported separately.
Results
Study Recruitment
Recruitment to the BARCODE1 Pilot commenced in April 2016 and completed in April 2018. Initially, three GP centres were involved. Four more sites were added to the study in April 2017 to increase recruitment rate and complete the pilot study.
The GPs screened the medical records of 1802 men registered at their practices; 1436 potentially eligible men (80% of those screened) were sent a study invitation letter, which included the study PIS and a health questionnaire. The health questionnaire was used to exclude men who did not meet study eligibility criteria, as well as those with any comorbidities that would make the risk of prostate biopsy unacceptable.
Of the 1436 men invited to the study, 375 men responded to the invitation letter giving a study uptake rate of 26% (range 13–47%, Table 1) of whom 329 (88%) were eligible for study entry (Fig. 2). Reasons for exclusion from study entry included medical comorbidities and non‐European ethnicities.
Table 1.
BARCODE1 pilot study screening, response rates and saliva returns.
| Site | Screened, n | Mail‐outs, n | Total responders, n (%) | Eligible, n | Returned saliva samples, n (% of eligible responders) |
|---|---|---|---|---|---|
| GP 1 | 148 | 148 | 45 (30) | 45 | 42 (93) |
| GP 2 | 350 | 326 | 78 (24) | 66 | 62 (94) |
| GP 3 | 277 | 175 | 23 (13) | 12 | 12 (100) |
| GP 4 | 267 | 232 | 46 (20) | 44 | 41 (93) |
| GP 5 | 223 | 211 | 51 (24) | 49 | 44 (90) |
| GP 6 | 390 | 200 | 93 (47) | 77 | 74 (96) |
| GP 7 | 145 | 142 | 37 (26) | 34 | 30 (88) |
| RMH* | 2 | 2 | 2 | 2 | 2 |
| Total | 1802 | 1436 | 375 (26) | 329 | 307 (93) |
GP, General Practice site.
Two participants recruited via Oncogenetics Research clinic in RMH.
Fig. 2.
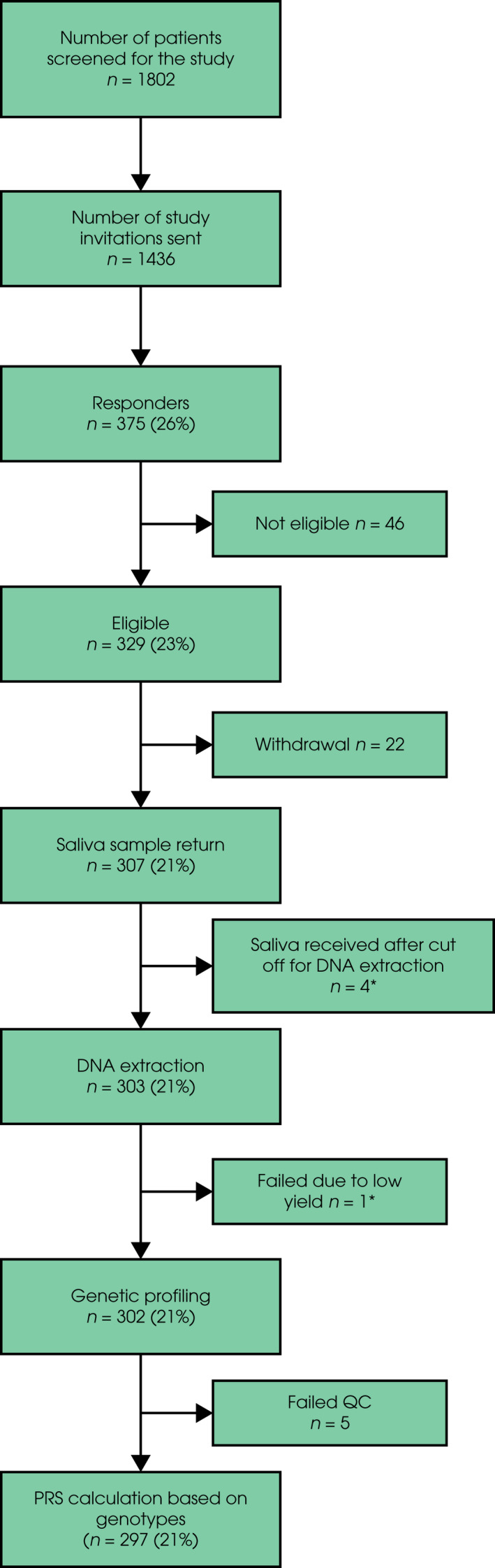
BARCODE1 pilot study recruitment. Percentages relate to the proportion of invitations to the study. *Saliva samples from four participants were received after 15 April 2018, which was the cut‐off date for DNA extraction in the pilot study. These participants will have DNA extracted and analysed in the main BARCODE1 study. One saliva sample did not yield sufficient DNA at extraction and a new saliva sample was requested. This participant will also be added to the main BARCODE1 study.
Saliva collection kits were posted to 328 eligible participants with a study consent form. A total of 307 saliva samples were returned, giving a return rate of 94%; 21 participants withdrew from the study after providing a saliva sample and one withdrew prior to providing a saliva sample, giving an overall withdrawal rate of 6.7% of eligible responders.
Participant Characteristics and Genotyping
The mean (range) age of study participants at the time of consent was 61 (55–69) years. DNA was extracted for 303 participants whose saliva sample was returned before the cut‐off date (15 April 2018) for the pilot study. Of 303 saliva samples that underwent DNA extraction, one sample had a low DNA yield and could not be genotyped (further saliva was collected later from this participant). Genotyping was therefore carried out for 302 samples. 17 samples (5.6%) failed QC processes due to a SNP call rate of <90%; for 12 of these, a repeat saliva sample was subsequently returned by the participant and passed genotyping QC. Therefore, 297 samples underwent successful genotyping. The mean genotype call rate across the 130 SNPs for all study participants was 99.6% and median 100% (in total 162 of 38 610 genotypes across all 297 samples failed genotyping QC).
PRS Results
The PRS for 297 men in the BARCODE1 Pilot ranged from 8.42 to 12.21 (median 10.34). The mean (10.33) and SD (0.64) were used to calculate the 90th centile value, which was 11.15 (Fig. 3). Using this threshold value, 26 participants were eligible for screening based on their PRS. The 271 men whose PRS fell in the bottom 90% were not eligible for screening and received standard care through their GP.
Fig. 3.
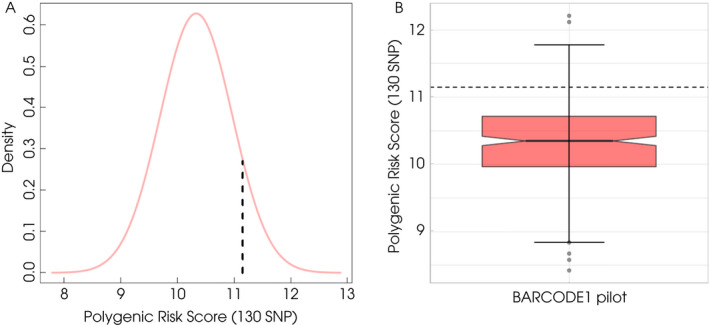
PRS distribution in BARCODE1 Pilot. (A) The dashed line denotes the 90th centile PRS value (11.15). (B) Boxplot of the PRS distribution in BARCODE1 pilot displaying minimum and maximum values of PRS in whole study cohort as well as outliers. The dashed line denotes the 90th centile PRS value (11.15).
To ensure that participants in the BARCODE1 Pilot were representative of the general population, we compared the PRS distribution to one calculated in another UK genotyping dataset taken from the Prostate Testing for Cancer Treatment (ProtecT) trial [17]. There was no significant difference in the PRS distribution (Fig. S2; P = 0.92).
Uptake of Screening by Men in the Top 10% of PRS Distribution
Of the 26 participants identified to be in the top 10% of the PRS distribution, seven men did not proceed with PCa screening: one patient died of a non‐prostate‐related cause after entering the study, two were lost to follow‐up, and four withdrew when offered PCa screening. An eighth participant underwent MRI but did not proceed to biopsy due to persistent sterile pyuria. Therefore, the overall uptake of PCa screening was 72% (18 of 25 living eligible participants).
Outcomes of PCa Screening
The MRI of the prostate (18 with endorectal coil at 3‐T, one with phased array coil at 1.5‐T) followed by systematic (±targeted sampling) biopsies was carried out for 18 participants. The PSA was measured prior to MRI. The mean (range) PSA for the 19 men undergoing MRI was 1.60 (0.3–5.8) ng/mL.
Assigned PI‐RADS scores were 1–2 in 10 participants (53%), PI‐RADS 3 in five (26%) and PI‐RADS 4 in four (21%). There were no PIRADS 5 lesions detected (Table 2).
Table 2.
Distribution of PI‐RADS score in 19 men who underwent MRI prostate in BARCODE1 pilot.
| PI‐RADS Score | MRI, n (%) | Number of repeat MRI |
|---|---|---|
| 1 | 2 (11) | 0 |
| 2 | 8 (42) | 2 |
| 3 | 5 (26) | 0 |
| 4 | 4 (21) | 0 |
| 5 | 0 | 0 |
| Total | 19 | 2 |
Two scans repeated due to initial findings of ASAP and HGPIN on first biopsy. PI‐RADS score was unchanged as both had a score of 2 on first and second scans.
Biopsy results were available for 18 men. One man did not proceed to biopsy due to persistent sterile pyuria for which he was referred for urological investigations. Table 3 displays biopsy outcomes. All initial screening biopsies were performed via the transrectal route except for one (Participant 2 in Table 3), which was carried out via the transperineal route to allow adequate sampling of small bilateral lesions detected on MRI.
Table 3.
Outcomes of screening for 19 men in the BARCODE1 pilot study.
| Age, years | Pre‐biopsy PSA level, ng/mL | PI‐RADS Score pre‐biopsy MRI | Histology (Gleason score) | Management | Repeat MRI P‐IRADS Score | Repeat biopsy histology | Management | PRS |
|---|---|---|---|---|---|---|---|---|
| 62 | 0.78 | 4 | 3 + 3 | AS | 12.12 | |||
| 66 | 5.8 | 4 | 3 + 3 | AS | 11.19 | |||
| 66 | 2.3 | 3 | 3 + 3 | AS | 11.32 | |||
| 55 | 2.4 | 1 | 3 + 4 | AS | 2 | 3 + 3 | AS | 11.17 |
| 56 | 1 | 2 | 3 + 3 | AS | 11.53 | |||
| 64 | 3.6 | 4 | 3 + 3 | AS | 11.31 | |||
| 64 | 0.64 | 2 | ASAP | Repeat MRI and Bx at 6 months | 2 | 3 + 4 | AS | 11.42 |
| 55 | 0.35 | 2 | HGPIN | Repeat MRI and Bx at 6 months | 2 | Benign | Trial f/u | 11.32 |
| 56 | 2.6 | 2 | Benign | Trial f/u | 11.49 | |||
| 59 | 0.89 | 4 | Benign | Trial f/u | 11.38 | |||
| 60 | 0.46 | 3 | Benign | Trial f/u | 11.17 | |||
| 56 | 0.89 | 2 | Benign | Trial f/u | 11.69 | |||
| 56 | 0.3 | 2 | Benign | Trial f/u | 11.32 | |||
| 59 | 1.5 | 1 | Benign | Trial f/u | 11.53 | |||
| 60 | 0.93 | 3 | Benign | Trial f/u | 11.73 | |||
| 55 | 0.41 | 2 | Benign | Trial f/u | 12.21 | |||
| 66 | 1.7 | 2 | Benign | Trial f/u | 11.49 | |||
| 59 | 1.8 | 3 | Benign | Trial f/u | 11.44 |
Bx, biopsy; f/u, follow‐up.
Participant 4 underwent an MRI prostate and transperineal template biopsy as part of the local AS protocol and the Gleason score was downgraded on the second biopsy to 3 + 3. Patients 7 and 8 underwent a repeat MRI and biopsy as part of the pilot study, 6 months after initial biopsy due to findings of ASAP and HGPIN, respectively.
Prostate cancer was diagnosed in six of the 18 participants (33%) who underwent an initial screening biopsy; four of these had a target lesion identified on MRI (three PI‐RADS 4 lesions and one PI‐RADS 3 lesion), while four participants with identified target lesions (three PI‐RADS 3 and one PI‐RADS 4) were negative on biopsy. The mean maximum cancer core length (MCCL) in the four MRI‐identified lesions was 1.2 mm with a mean pre‐biopsy PSA level of 1.6 ng/mL, and in the two cancers not identified on MRI, the mean MCCL was 1 mm with a mean pre‐biopsy PSA level of 1.1 ng/mL.
In the remaining 12 participants, two were diagnosed with atypical small acinar proliferation (ASAP) and high‐grade prostatic intraepithelial neoplasia (HGPIN); a repeat MRI and biopsy was carried out for both at 6 months as per trial protocol. The participant with ASAP was diagnosed with Gleason 3 + 4 PCa (MCCL 1.5 mm, 1%) and the participant with HGPIN had benign findings. All cancers were discussed in the RMH uro‐oncology multidisciplinary team meeting and management by active surveillance (AS) was recommended for all seven cases (Table 4).
Table 4.
Features of prostate cancers diagnosed in pilot study.
| Number of biopsies carried out | Gleason score on final biopsy | MCCL, mm | TCCL, mm | |
|---|---|---|---|---|
| Participant 1 | 1 | 3 + 3 | 1 | 2 |
| Participant 2 | 1 | 3 + 3 | 2 | 2 |
| Participant 3 | 1 | 3 + 3 | 1 | 1 |
| Participant 4 | 2 | 3 + 3 | 1 | 1 |
| Participant 5 | 1 | 3 + 3 | 1 | 1 |
| Participant 6 | 1 | 3 + 3 | 0.5 | 0.5 |
| Participant 7 | 2 | 3 + 4 | 1.5 | 2 |
TCCL, total cancer core length.
Incidental Findings and Post‐Biopsy Complications
There was one case of an incidental finding reported on MRI of the prostate that required further investigation, resulting in a diagnosis of low‐grade B‐cell non‐Hodgkin lymphoma. An uncomplicated lower urinary tract infection was diagnosed in two out of 18 men (11%) who underwent a prostate biopsy. Both cases were managed successfully with oral antibiotics. There were no other post‐biopsy complications.
Post‐Biopsy Follow‐Up
Men diagnosed with PCa in this pilot study (seven; six initially and one at follow‐up) are currently undergoing follow‐up in the RMH uro‐oncology clinic. For the 11 participants who had a benign biopsy result, study follow‐up will continue with annual PSA measurement for at least 5 years with repeat MRI and biopsy if the PSA rises above the protocol set threshold for repeat biopsy. Cancer registries will be reviewed, and data will be collected for all men enrolled in the study for a minimum of 10 years to determine cancer incidence and mortality.
Discussion
The BARCODE1 Pilot was successful in demonstrating the acceptability and feasibility of community‐based PCa screening guided by germline genetics in the UK. There was good uptake by men invited by their GPs, and a high level of interest from primary care centres wishing to participate after the pilot transitioned to the larger main study. Uptake of PCa screening by men offered it was 72%, which is comparable to the uptake of national screening programmes such as the bowel and breast cancer screening programmes in England (59–72%) [18, 19].
The BARCODE1 main study has now commenced recruitment with the aim of genotyping 5000 men in the community and in turn, offer screening to those lying in the top 10% of the PRS distribution. The PRS threshold for screening within BARCODE1 was chosen pragmatically so as to lead to a feasible number of men that could be offered screening procedures within the study. It was also guided by the latest scientific estimates that men of European ancestry with a PRS between the 90th and 99th centiles have a 2.7‐fold higher risk of developing PCa compared with the population average. This is similar to the PCa risk of a man with a first‐degree family history and for whom guidance advises PCa screening is considered [20].
A 26% study uptake rate by invitees was comparable to the PROFILE pilot screening study reported by our team in 2016 [21], which had an uptake of 12.8% in men with a family history of PCa. The uptake in the BARCODE1 Pilot was lower than that reported in the ProtecT [17] study that invited participants via their GPs for PSA‐based screening and had an uptake of 36%. This may partly be explained by one of the GP sites in our present study having a large proportion of patients of non‐European ethnicities (in keeping with the local population demographic) who were not eligible for study entry; unfortunately, patient ethnicity is not recorded routinely at some practices therefore some study invitations would have been sent to men who were not eligible and therefore would not have responded to the study invitation. To offset the lower than expected uptake, an additional four GP sites opened recruitment, and for the main study a large network of over 70 GP sites will be recruiting. By expanding the main study to a large number of GPs and with modifications in response to participants’ feedback, such as reducing the size of the information pack sent out with study invitations, we may find that uptake increases in the main BARCODE1 study.
In the present pilot, 39% (seven of 18) of men screened were diagnosed with low Gleason grade (Gleason 3 + 3 or 3 + 4), low‐stage (no nodal involvement on imaging) PCa amenable to management by AS. The rate of progression to disease that requires treatment could not be evaluated because of the short follow‐up in the pilot phase of the study (mean [range] 4.5 [1–11] months). Management with AS (regular PSA tests, interval MRI scans and repeat biopsies), although resource intensive, potentially offers early curative treatment options in this high‐risk relatively young population.
The PRS used in BARCODE1 includes 130 PCa risk SNPs that associate with both aggressive and non‐aggressive PCa. There have been no risk loci identified to date that are specific to aggressive PCa. Despite the lack of predictive biomarkers for aggressive disease, a recent pan‐ancestry meta‐analysis of PCa GWAS data found that approximately a third of aggressive PCa cases occurred in men whose genetic risk lay in the top 10% of the risk distribution [3]. Therefore, using a PRS‐based approach to target screening could identify a subset of men in the population in which a substantial proportion of aggressive PCa cases will develop. The results of the larger BARCODE1 study and long‐term follow‐up will examine this further.
Apart from identifying men at risk of PCa, some of whom will develop aggressive disease, utilising a PRS for PCa screening may reduce the level of over‐diagnosis of indolent disease that has prevented the adoption of PSA‐based population screening [22]. Over‐diagnosis is defined as the ‘detection by screening of tumours that would not have presented clinically in a person’s lifetime in the absence of screening’ [12]. In the context of PSA‐based PCa screening, the proportion of over‐diagnosed cases has been shown to be inversely proportional to PRS [12]. In a retrospective analysis of a PCa PRS in 17 000 men in the UK, there was a 56% reduction in over‐diagnosis between the lowest and highest PRS quartiles when using PSA for screening. Therefore, utilising a PRS as part of a programme to target screening to men in the top quartile or upper decile of the genetic risk distribution has the potential to reduce over‐diagnosis compared with unselected PSA screening. This in turn would spare men at low genetic risk from undergoing screening procedures [12, 23, 24].
Although over‐diagnosis is unavoidable in most screening programmes, attempts to minimise this by utilising a targeted approach based on genetic risk while still balancing the identification of high‐risk cancers will be investigated in the BARCODE1 study. Although all the cancers diagnosed in the pilot study were low grade, data from the full BARCODE1 study are needed to draw conclusions on the effect of PRS‐based screening on over‐diagnosis. With the increased acceptability and safety of using AS, study participants whose low‐grade PCa does not progress can avoid the complications of radical treatment while those who experience disease progression can be treated promptly.
As the BARCODE1 study PRS is based on SNPs identified in European populations, study recruitment was restricted to men of European ethnicity. Dedicated studies in populations of other ethnicities are in progress to establish population‐specific genotyping panels and thresholds for PCa screening. In a recent multi‐ancestry PCa GWAS meta‐analysis reported by Conti et al. [3], case‐control data was analysed from European, African and East Asian ancestry men. That analysis confirmed that many PCa loci are shared across populations, although the risk associated with PRS varied between populations. The absolute risk of PCa in men lying in the top decile of the genetic risk distribution was highest in men of European and African ancestry at 38%, while the top decile in East Asian ancestry men had the lowest risk of 26%. Identifying this ancestry‐related variation in genetic risk will be useful in the development of tailored approaches to PCa risk assessment in different populations [3]. In this vein, one of the screening arms in the ongoing PROFILE screening study (NCT02543905) is recruiting men of African and Afro‐Caribbean ancestry to investigate the relationship between a genetic profile and outcome at prostate biopsy during screening. Several international efforts are also trying to identify ethnic‐specific risk SNPs and assess generally the utility of the polygenic risk scores currently in use [3, 25]. These will help guide the development of a cross‐ethnic PCa screening programme in diverse populations [26, 27, 28].
The BARCODE1 study is the first prospective study to utilise a germline SNP profile to target PCa screening in a community‐based setting. The STOCKHOLM3 (STHLM3) study [29], reported in 2015, was the first large prospective and population‐based PCa screening study that assessed a targeted approach to screening. The study utilised a screening model combining plasma protein biomarkers (including PSA), 232 risk SNPs and a set of defined clinical variables (e.g. age, family history) and compared this with PSA measurement alone (≥3 ng/mL) [29]. Although the sensitivity for the detection of high‐risk PCa was significantly improved with the STHLM3 model (area under the curve 0.74 vs 0.56), the value of the SNP profile within the larger model was difficult to ascertain. Since that study was reported, the STHLM3 model has been combined with MRI targeted biopsies in a prospective study comparing it with PSA screening and reported a 69% reduction in over‐diagnosis while maintaining the sensitivity for the detection of significant PCa [30]. By examining the role of targeting screening using only a germline SNP profile in BARCODE1, the contribution of a PRS in such multi‐factor models can be evaluated and guide the future design of screening programmes that may use a SNP profile alongside other biomarkers and/or imaging. By carrying out a biopsy for all men who undergo a MRI of the prostate (as well as PSA test), the utility of MRI as a screening tool for men at increased genetic risk of PCa will be investigated in BARCODE1. To date, the usefulness of pre‐biopsy MRI has been reported in men with an elevated PSA in studies such as the PROstate MRI Imaging Study (PROMIS) and PRECISION [31, 32]. By correlating the results of MRI and biopsy in the ∼500 men screened in BARCODE1, we aim to define the role of MRI in guiding the decision to undergo a prostate biopsy in men with an increased genetic risk of PCa.
Additional studies are required to establish the role of MRI outside and within the context of genetic risk. The ReIMAGINE Prostate Cancer Screening study (NCT04063566) is currently inviting PSA naïve men aged 50–75 years via their GP to undergo MRI of the prostate. Those with lesions assigned a PI‐RADS score of 3–5 (or with a suspicious PSA density >0.12 ng/ml2) will be referred for prostate biopsy. In the sister study, ReIMAGINE Prostate Cancer Risk (NCT04060589), participants accepting a biopsy based on their study MRI will be invited to donate samples including germline DNA, which will be genotyped to retrospectively assess polygenic risk. The BARCODE1 study and complimentary studies such as ReIMAGINE will allow a robust assessment of different screening modalities and provide valuable information to guide the application of less invasive screening methods, and potentially plan for precision PCa screening in the population.
Funding
The BARCODE1 study is funded by the European Research Council Seventh Framework Programme under grant 339208 (ERC‐2013‐AdG‐339208). We acknowledge support from the National Institute for Health Research (NIHR) to the Biomedical Research Centre at The Institute of Cancer Research and Royal Marsden Foundation NHS Trust. The Institute of Cancer Research is the Sponsor of the BARCODE1 study.
Conflict of Interest
Prof. Rosalind Eeles reports grants from European Research Council funding the submitted work. Prof Eeles reports personal fees from AstraZeneca UK Limited for her role as a member of an external Expert Committee as part of the Prostate Cancer Diagnosis Advisory Panel, and University of Chicago for an invited lecture, both of which are outside the submitted work. Dr Sarah Benafif, Dr Elizabeth Bancroft, Mrs Sarah Wakerell, Mrs Natalie Taylor, Mr Edward Saunders, Dr Holly Ni Raghallaigh, Ms Kathryn Myhill, Dr Eva McGrowder, Dr Zsofia Kote‐Jarai, Mr Denzil James, Mr Matthew Hogben, Mrs Lucia D’Mello, Mrs Barbara Benton, Dr Mark Brook, Mr Anthony Chamberlain, Mrs Reshma Rageevakuma and Dr Sibel Saya report funding for the submitted work from a European Research Council grant for this study. Dr Imran Rafi, Dr Tamsin Sevenoaks, Dr Andre Beattie, Dr Hywel Bowen‐Perkins, Dr Juliet Bower, Dr Shophia Kuganolipava report funding of research costs for the submitted work from The Institute of Cancer Research and from National Institute for Health Research: Clinical Research Network. Dr Michelle Ferris reports funding of research costs for the submitted work from The Institute of Cancer Research and from National Institute for Health Research: Clinical Research Network. Dr Ferris reports financial activities outside the submitted work related to the Befriend Your Boobs project for which she also has a patent, as well as a research grant from Noclor. Prof Antonis Antoniou is listed as creator of the BOADICEA algorithm that has been licensed by Cambridge Enterprise (outside the submitted work). The following authors have no conflict of interest to disclose: Dr Steve Hazell, Dr Christos Mikropoulos, Mr Pardeep Kumar, Prof Nandita De Souza.
Abbreviations
- AS
active surveillance
- ASAP
atypical small acinar proliferation
- BRCA2
BReast CAncer type 2 susceptibility protein
- DW
diffusion‐weighted
- ERSPC
European Randomised Study of Screening for Prostate Cancer
- GWAS
genome‐wide association study
- HGPIN
high‐grade prostatic intraepithelial neoplasia
- MCCL
maximum cancer core length
- OR
odds ratio
- PCa
prostate cancer
- PI‐RADS
Prostate Imaging ‐ Reporting and Data System
- PIS
participant information sheet
- PLCO
Prostate, Lung, Colorectal and Ovary
- ProtecT
Prostate Testing for Cancer Treatment
- PRS
polygenic risk score
- QC
quality control
- RMH
Royal Marsden Hospital
- RR
relative risk
- SNP
single‐nucleotide polymorphisms
- SNV
single‐nucleotide variant
- STHLM3
STOCKHOLM3 (study)
Supporting information
Appendix S1. The BARCODE1 Pilot Study protocol.
Appendix S2. Supplementary material.
Acknowledgements
We thank all the participants who took part in this research and collaborators: The Kent, Surrey and Sussex Clinical Research Network (CRN) led by facilitators Anne Oliver and Natalie Billington who provided six GP practices as participant identification centres within this network; The North Thames CRN for providing one GP practice to support this work. The BARCODE1 Pilot Study Steering Committee: J. Offman, King’s College London; A. Antoniou, University of Cambridge; I. Rafi, GP CRN Kent, Surrey & Sussex; M. Ferris, GP CRN North Thames; H. Bowen‐Perkins, GP CRN Kent, Surrey & Sussex; K. Muir, University of Manchester; N. Pashayan, University College London; P. Kumar, Royal Marsden NHS Foundation Trust; N. van As, Royal Marsden NHS Foundation Trust; D. Dearnaley, Royal Marsden NHS Foundation Trust and The Institute of Cancer Research; V. Khoo, Royal Marsden NHS Foundation Trust; A. Arden‐Jones, Royal Marsden NHS Foundation Trust; C. Moynihan, The Institute of Cancer Research; J. Taylor, Patient Representative; D. Nicol, Royal Marsden NHS Foundation Trust; C. Ogden, Royal Marsden NHS Foundation Trust; A. Thompson, Royal Marsden NHS Foundation Trust; C. Woodhouse, Royal Marsden NHS Foundation Trust. The BARCODE 1 Pilot Study Independent Data Monitoring Committee: S. Duffy (Chair), P. White (UK NEQAS representative), J. McGrath (BAUS representative), and S. Wallace (Ethics Advisor).
References
- 1. Benafif S, Kote‐Jarai Z, Eeles RA. A review of prostate cancer genome wide association studies (GWAS). Cancer Epidemiol Biomarkers Prev 2018; 27: 845–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schumacher FR, Al Olama AA, Berndt SI et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet 2018; 50: 928–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Conti DV, Darst BF, Moss LC et al. Trans‐ancestry genome‐wide association meta‐analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat Genet 2021; 53: 65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Khera AV, Chaffin M, Aragam KG et al. Genome‐wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018; 50: 1219–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matejcic M, Saunders EJ, Dadaev T et al. Germline variation at 8q24 and prostate cancer risk in men of European ancestry. Nat Commun 2018; 9: 4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dadaev T, Saunders EJ, Newcombe PJ et al. Fine‐mapping of prostate cancer susceptibility loci in a large meta‐analysis identifies candidate causal variants. Nat Commun 2018; 9: 2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schröder FH, Hugosson J, Roobol MJ et al. Screening and prostate‐cancer mortality in a randomized European study. N Engl J Med 2009; 360: 1320–8 [DOI] [PubMed] [Google Scholar]
- 8. Pinsky PF, Prorok PC, Yu K et al. Extended mortality results for prostate cancer screening in the PLCO trial with median follow‐up of 15 years. Cancer 2017; 123: 592–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moyer VA, United States Preventive Services Task Force. Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 2012; 157: 120–34 [DOI] [PubMed] [Google Scholar]
- 10. Carroll PR. USPTF prostate cancer screening recommendations‐A step in the right direction. JAMA Surg. 2018; 153: 701–2 [DOI] [PubMed] [Google Scholar]
- 11. Nyberg T, Frost D, Barrowdale D et al. Prostate cancer risks for male BRCA1 and BRCA2 mutation carriers: a prospective cohort study. Eur Urol 2020; 77: 24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pashayan N, Duffy SW, Neal DE et al. Implications of polygenic risk‐stratified screening for prostate cancer on overdiagnosis. Genet Med 2015; 17: 789–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pashayan N, Morris S, Gilbert FJ, Pharoah PDP. Cost‐effectiveness and benefit‐to‐harm ratio of risk‐stratified screening for breast cancer: a life‐table model. JAMA Oncol. 2018; 4: 1504–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Darst BF, Wan P, Sheng X et al. A Germline variant at 8q24 contributes to familial clustering of prostate cancer in men of African ancestry. Eur Urol 2020; 78: 316–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Missirian V, Curry JD, Patil MA, Piraniermo A. Ultra High Throughput, High Quality Genotypes using the Applied Biosystems™ Eureka Genotyping Platform. Available at: http://assets.thermofisher.com/TFS‐Assets/GSD/posters/high‐quality‐genotypes‐using‐eureka‐genotyping‐poster.pdf. Accessed July 2021
- 16. Barentsz JO, Richenberg J, Clements R et al. ESUR prostate MR guidelines 2012. Eur Radiol 2012; 22: 746–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lane JA, Donovan JL, Davis M et al. Active monitoring, radical prostatectomy, or radiotherapy for localised prostate cancer: study design and diagnostic and baseline results of the ProtecT randomised phase 3 trial. Lancet Oncol 2014; 15: 1109–18 [DOI] [PubMed] [Google Scholar]
- 18. Public Health England . NHS Screening Programmes in England, 1 April 2016 to 31 March 2017. Available at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/661677/NHS_Screening_Programmes_in_England_2016_to_2017_web_version_final.pdf. Accessed July 2021
- 19. Public Health England . Cancer Services. Available at: https://fingertips.phe.org.uk/profile/cancerservices. Accessed July 2021
- 20. National Comprehensive Cancer Network (NCCN) . NCCN Guideline for Prostate Cancer Early Detection, 2021. Available at: https://www.nccn.org/professionals/physician_gls/PDF/prostate_detection.pdf
- 21. Castro E, Mikropoulos C, Bancroft EK et al. The PROFILE feasibility study: Targeted screening of men with a family history of prostate cancer. Oncologist 2016; 21: 716–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Draisma G, Boer R, Otto SJ et al. Lead times and overdetection due to prostate‐Specific antigen screening: Estimates from the European randomized study of screening for prostate cancer. J Natl Cancer Inst 2003; 95: 868–78 [DOI] [PubMed] [Google Scholar]
- 23. Callender T, Emberton M, Morris S et al. Polygenic risk‐tailored screening for prostate cancer: A cost‐effectiveness analysis. Cancer Res 2019; 79: Abstract 2415. Available at: https://cancerres.aacrjournals.org/content/79/13_Supplement/2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liss MA, Xu J, Chen H, Kader AK. Prostate genetic score (PGS‐33) is independently associated with risk of prostate cancer in the PLCO trial. Prostate 2015; 75: 1322–8 [DOI] [PubMed] [Google Scholar]
- 25. Han Y, Rand KA, Hazelett DJ et al. Prostate cancer susceptibility in men of African Ancestry at 8q24. J Natl Cancer Inst 2016; 108: djv431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Darst BF, Wan P, Sheng X et al. A Germline variant at 8q24 contributes to familial clustering of prostate cancer in men of african ancestry. Eur Urol 2020; 78: 316–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Conti DV, Wang K, Sheng X et al. Two novel susceptibility loci for prostate cancer in men of African ancestry. JNCI: Journal of the National Cancer Institute 2017; 109: djx084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harlemon M, Ajayi O, Kachambwa P et al. A custom genotyping array reveals population‐level heterogeneity for the genetic risks of prostate cancer and other cancers in Africa. Can Res 2020; 80:2956–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gronberg H. Prostate cancer screening in men aged 50–69 years (STHLM3): a prospective population‐based diagnostic study. Lancet Oncol 2015; 16: 1667–76 [DOI] [PubMed] [Google Scholar]
- 30. Nordstrom T, Discacciati A, Bergman M, et al. Prostate cancer screening using a combination of risk‐prediction, magnetic resonance imaging and targeted prostate biopsies: A randomised trial, 2021. Available at SSRN: https://ssrncom/abstract=3788918 or http://dxdoiorg/102139/ssrn3788918
- 31. Ahmed HU, El‐Shater Bosaily A, Brown LC et al. Diagnostic accuracy of multi‐parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet 2017; 389: 815–22 [DOI] [PubMed] [Google Scholar]
- 32. Kasivisvanathan V, Rannikko AS, Borghi M et al. MRI‐targeted or standard biopsy for prostate‐cancer diagnosis. N Engl J Med 2018; 378: 1767–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. The BARCODE1 Pilot Study protocol.
Appendix S2. Supplementary material.


