Abstract
Human papillomavirus (HPV)‐induced anal intraepithelial neoplasia (AIN, graded 1‐3) is highly prevalent in HIV‐positive (HIV+) men who have sex with men (MSM), but only a minority of lesions progresses to cancer. Our study aimed to characterise comprehensively anal tissue samples from a cross‐sectional series (n = 104) of HIV+ MSM and longitudinal series (n = 40) of AIN2/3 progressing to cancer using different biomarkers. The cross‐sectional series consisted of 8 normal, 26 AIN1, 45 AIN2, 15 AIN3 and 10 anal squamous cell carcinoma. Tissue sections were immunohistochemically (IHC) stained for p16 (viral transformation marker), Ki‐67 (cellular proliferation marker) and HPV‐E4 (viral production marker). We evaluated the expression of IHC markers and compared it with DNA methylation, a marker for malignant transformation. E4 positivity decreased, whereas p16 and Ki‐67 scores and methylation marker positivity increased (P values < .001) with increasing severity of anal lesions. Within AIN2, a heterogeneous biomarker pattern was observed concerning E4, p16 and methylation status, reflecting the biological heterogeneity of these lesions. In the longitudinal series, all AIN2/3 and carcinomas showed high p16 and Ki‐67 expression, strong methylation positivity and occasional E4 positivity. We earlier showed that high methylation levels are associated with progression to cancer. The observed E4 expression in some AIN2/3 during the course of progression to cancer and absence of E4 in a considerable number of AIN1 lesions make the potential clinical significance of E4 expression difficult to interpret. Our data show that IHC biomarkers can help to characterise AIN; however, their prognostic value for cancer risk stratification, next to objective methylation analysis, appears to be limited.
Keywords: anal high‐grade squamous intraepithelial lesion, host cell DNA methylation markers, human immunodeficiency virus, human papillomavirus, immunohistochemistry
Short abstract
What's new?
Anal high‐grade squamous intraepithelial lesions (HSILs) constitute a heterogeneous group of precancerous lesions. Understanding which HSILs progress to cancer could facilitate early detection and treatment of anal cancer. Here, the prognostic value of expression of the immunohistochemical (IHC) markers p16, Ki‐67, and HPV‐E4 was evaluated in anal tissues with evidence of precancerous lesions. While analyses revealed positive associations between p16 and Ki‐67 expression and severity of anal dysplasia, HSILs overall exhibited complex biomarker patterns, reflecting their ambiguous clinical behaviour. Thus, while IHC markers are useful for molecular characterisation of HSIL, their prognostic value, particularly compared to methylation analysis, is limited.
Abbreviations
- AIN
anal intraepithelial neoplasia
- CIN
cervical intraepithelial neoplasia
- CpG
cytosine located 5′ of a guanine
- Cq
quantification cycle
- DEIA
DNA enzyme immunoassay
- DNA
deoxyribonucleic acid
- H&E
haematoxylin and eosin
- HIV+
human immunodeficiency virus positive
- HPV
human papillomavirus
- hrHPV
high‐risk human papillomavirus
- HSIL
high‐grade squamous intraepithelial lesion
- IHC
immunohistochemistry
- J
Youden's index
- LAST project
Lower Anogenital Squamous Terminology Standardization Project
- LiPA
line probe assay
- LSIL
low‐grade squamous intraepithelial lesion
- MSM
men who have sex with men
- Norm(al)
normal control sample
- Pos
positive
- PP
predicted probability
- qMSP
quantitative methylation‐specific polymerase chain reaction
- SCC
squamous cell carcinoma
- SPANC
Study of the Prevention of Anal Cancer
- UMC
university medical centre
- Und
undetermined
1. INTRODUCTION
Anal cancer is an increasing problem with the highest risk for human immunodeficiency virus‐positive (HIV+) men who have sex with men (MSM). 1 , 2 Most anal cancers are squamous cell carcinoma (SCC), for the large majority caused by a persistent infection with high‐risk (hr) human papillomavirus (HPV). 3 Anal intraepithelial neoplasia (AIN) is considered the precursor of anal SCC. 4 Historically, AIN is histopathologically graded as AIN1‐3, with AIN2‐3 also being referred to as high‐grade AIN and AIN1 as low‐grade AIN. 5 The Lower Anogenital Squamous Terminology Standardization (LAST) Project formulated recommendations for histopathological grading incorporating HPV biology. 6 In this two‐tier system, a distinction is made between low‐grade squamous intraepithelial lesions (LSILs) and high‐grade squamous intraepithelial lesions (HSILs). LSILs, often caused by low‐risk HPV types, are associated with productive HPV infection and lesion regression. HSILs are associated with HPV persistence and a transforming HPV infection that poses a risk of progression to cancer. 4 , 6
HSILs consist of a heterogeneous group of lesions: only a minority eventually progresses to cancer at an estimated progression rate of 1 per 377 per person‐year (for HIV+ MSM), 7 while 22% to 28% of HSILs spontaneously regresses. 8 , 9 The current lack of capability to predict the clinical course of HSIL has directed clinical practice in some countries to treat all HSILs, resulting in substantial overtreatment. Hence, new biomarkers for cancer risk stratification of HSILs are needed to identify lesions for treatment and reduce current overtreatment.
The immunohistochemical (IHC) marker p16INK4a (further referred to as p16) is an important characteristic for the transformation of lesions. Its expression is the result of cell cycle deregulation induced by hrHPV E7 viral oncogene activity. 10 The LAST project recommends the use of p16 as adjunct to conventional haematoxylin and eosin (H&E) to support an HSIL diagnosis in case of uncertainty between HSIL and a benign mimic or to confirm HSIL when a lesion looks like AIN2 on H&E. It recommends against the use of p16 as a routine adjunct to lesions negative for AIN, AIN1 or AIN3 on H&E. 6 Another IHC marker is Ki‐67, indicating cell proliferation by identifying all cells with cell cycle activity. 11 In comparable cervical lesions increasing Ki‐67 staining is found to be associated with increasing cervical intraepithelial neoplasia (CIN) grade. 12 Although p16 13 , 14 , 15 and the combination of p16/Ki‐67 16 , 17 , 18 have been reported to support AIN grading and to improve reproducibility, their prognostic value for cancer risk stratification has thus far not be established. Recently, a new IHC marker, the panHPV‐E4 antibody (further referred to as E4), detecting the HPV viral gene E4, has been developed. 19 , 20 , 21 E4 is expressed at initiation of the productive phase of the HPV life cycle and is involved in viral genome amplification and virion assembly. 22 As a marker for productive lesions, E4 could be used to identify low‐grade lesions and subsets of high‐grade lesions with productive characteristics in both cervical lesions 23 , 24 , 25 , 26 , 27 and anal lesions, 19 , 20 as was recently described.
DNA methylation of host cell gene promoters can lead to inactivation of tumour suppressor genes and is considered an epigenetic hallmark of HPV‐induced carcinogenesis. 28 Recently, we identified and validated several methylation markers for accurate detection of HSIL and anal cancer in HIV+ MSM. 29 , 30 Methylation analysis showed that the group of histopathologically similar HSILs is heterogeneous on a molecular level, displaying either low or high/‘cancer‐like’ methylation levels. The presence of the latter ‘cancer‐like’ pattern was found to be associated with progression towards cancer. 30
In this study, we aimed to characterise anal lesions using both IHC markers and host cell DNA methylation markers to increase our insight into changes of biomarker patterns during anal carcinogenesis and to investigate their potential as a cancer risk stratification tool. Hereto, we evaluated methylation results and the expression of IHC markers p16, Ki‐67 and E4 in both a cross‐sectional series representing the complete spectrum of anal lesions and a unique longitudinal series of HSIL progressing to cancer.
2. MATERIALS AND METHODS
2.1. Study specimen series and ethics
For this study, clinical specimens were included from a cross‐sectional and a longitudinal series of anal tissue samples that were previously analysed for HPV and DNA methylation. 29 , 30 In the present study, we further characterised these samples using staining with IHC markers p16, Ki‐67 and HPV‐E4. Figure S1 provides a schematic overview of the study procedures.
From the cross‐sectional series, a large subset of in total 108 samples from 93 HIV+ MSM were included in the present study, representing the full spectrum of anal carcinogenesis. This subset (selection based on local pathology diagnosis) consisted of all (n = 88) AIN biopsies as well as a random selection of 10 anal SCC and 10 normal control samples as a positive and negative reference, respectively. These samples were obtained between 1999 and 2016 in HIV+ MSM during screening for anal cancer and retrieved from the Pathology archives of the Amsterdam University Medical Centers (Amsterdam UMC) and OLVG, Amsterdam, The Netherlands, as described previously in more detail. 29 Normal control samples were taken from clinically non‐suspect anal epithelium during high‐resolution anoscopy in HIV+ men, and histopathologically graded as non‐dysplastic or reactive anal epithelium.
From the longitudinal series, we included 40 tissue samples of 10 cases (eight HIV+ men, one HIV‐negative woman and one HIV‐negative man), who over time developed SCC (n = 5) or were suspected to have developed (n = 5) (superficially invasive) anal SCC (ie, clinical strong suspicion and/or continuous suspicion in subsequent biopsies). Each case comprised multiple consecutive tissue samples taken from the same anatomic location with corresponding HPV types, including one or more samples of histopathologically confirmed SCC or ‘HSIL/AIN3, highly suspicious for infiltrative growth’ (endpoint; in total 15 samples), and all suitable and available biopsies obtained months up to years preceding the endpoint diagnosis (in total 25 HSIL biopsies). These samples, obtained between 2009 and 2019, were retrospectively identified and retrieved from several European pathology archives (see Supplementary Methods), as described previously in more detail. 30
We adhered to the Declaration of Helsinki and Code of Conduct for Responsible Use of Left‐over Material of the Dutch Federation of Biomedical Scientific Societies. Ethical approval was granted or waived (see Supplementary Methods) as reported previously. 29 , 30
2.2. Histological sample processing and HPV testing
Formalin‐fixed paraffin‐embedded anal tissue samples were cut into serial sections of 3 μm using the ‘sandwich’ sectioning method. In short, the first and last sections were H&E stained for histopathological review, for confirmation of lesion presence throughout the sections. Three in‐between sections were mounted on slides for IHC staining of p16, Ki‐67 and E4. Additional in‐between sections were used for DNA isolation and subsequent HPV testing and methylation analysis. HPV detection and genotyping was performed at DDL Diagnostic Laboratory, Rijswijk, The Netherlands, using the SPF10 DEIA/LiPA25 version 1 system (see Supplementary Methods) and was reported previously. 29 , 30
2.3. Immunohistochemistry
IHC staining of p16, Ki‐67 and E4 for all biopsies was performed on in‐between sections using the Ventana BenchMark ULTRA automated slide stainer (Ventana Medical Systems, Roche, Tucson, AZ) at DDL Diagnostic Laboratory, Rijswijk, The Netherlands. 19 , 20 IHC staining was performed after heat‐induced epitope retrieval with ULTRA Cell Conditioning Solution 1 (ULTRA CC1) and protease 3 (Roche, Basel, Switzerland) using mouse monoclonal antibodies against p16INK4a antigen (clone E6H4, CINtec, p16 Histology, Roche, Basel, Switzerland), rabbit monoclonal antibody against Ki‐67 [CONFIRM anti‐Ki‐67 (30‐9), Roche, Basel, Switzerland] and mouse monoclonal antibody against panHPV‐E4 antigen (SILgrade‐E4‐1 kit containing XR‐E4‐1 monoclonal antibody, Labo Biomedical Products B.V., Rijswijk, The Netherlands). The panHPV‐E4 antigen is at least reactive to HPV6, 11, 16, 18, 31, 33, 35, 39, 42, 43, 44, 45, 51, 52, 53, 56, 58, 59, 66, 67, 70 and 74; and skin HPV genotypes 27 and 57 (in part unpublished data). 19 , 21 , 26 Reactivity was visualised using the OptiView DAB IHC Detection Kit for p16 and Ki‐67 detection, and the OptiView DAB IHC Detection Kit with OptiView Amplification Kit for HPV‐E4 detection (Roche, Tucson, AZ).
2.4. Methylation analysis
DNA methylation analysis on these series was performed using quantitative methylation‐specific polymerase chain reaction (qMSP) on sample DNA (see Supplementary Methods) and reported previously. 29 , 30 In the cross‐sectional and longitudinal series, methylation markers involved in HPV‐induced carcinogenesis were evaluated using multiplex qMSP assays, each targeting multiple host cell genes and the reference gene, β‐actin. 31 Using multivariable logistic regression analysis optimal methylation marker panels were identified for the detection of [AIN3+] (AIN3 and anal SCC). The methylation result (ie, the outcome of the multivariable logistic regression model for the panels) is expressed as predicted probabilities (PPs). The PP values range from 0 to 1 and represent the risk for [AIN3+]; 0 indicates no risk and 1 indicates high risk. For samples with PPs above the Youden's index (J)‐threshold (threshold that maximises the sum of sensitivity and specificity), the methylation result was considered methylation ‘positive’. Although the methylation marker panels and J‐threshold slightly differed between the series, the cross‐validated diagnostic performance for [AIN3+] detection of the panels was similar. 29 , 30
2.5. Study diagnosis and scoring of immunohistochemical marker staining in anal tissue samples
First, an expert pathologist confirmed the presence of a dysplastic anal lesion in the studied biopsies by reviewing the first and last H&E slide (Figure S1). The review of H&E and IHC stained slides was blinded for the previous local diagnosis, HPV genotyping and methylation results. In case of multiple distinct regions/lesions in the specimen, the region with the most dysplastic features was selected for grading and IHC scoring. Four samples with one or more IHC staining slides being uninterpretable due to a technical staining issue or no more lesion being present in the slide were scored as ‘non‐diagnostic samples’ and excluded from the analyses (Figure S1).
The pathologist made a diagnosis according to the LAST project recommendations by using conventional AIN grading (normal, AIN1, AIN2, AIN3 or SCC) based on morphologic characteristics on H&E, 5 together with interpretation of the p16 staining (see p16 scoring) only when indicated according to LAST (ie, to distinguish with benign mimics of AIN or in case of an AIN2 lesion on H&E to differentiate between low‐ and high‐grade lesions). 6 Because a two‐tier (LSIL‐HSIL) classification is unable to sufficiently cover heterogeneity within high‐grade lesions to accurately correlate biomarker patterns, a three‐tier study diagnosis was used with extended annotation (AIN1‐3) by considering LSIL as AIN1 and reporting HSIL as (p16 positive) AIN2 or AIN3. Finally, the remaining p16, all Ki‐67 (see Ki‐67 scoring) and all E4 (see E4 scoring) stained sections were reviewed and scored.
2.5.1. p16 scoring
Staining of p16 was scored [score 0‐4] as described by Leeman et al. No staining was scored as [0]; focally scattered (non‐diffuse) stained cells or small cell clusters (ie, ‘patchy’) as [1]; and diffuse or ‘block’ staining of the cell cytoplasm and/or nucleus in squamous epithelium restricted to the lower one‐third of the epithelium, the lower two‐thirds of the epithelium and the full thickness of the epithelium as [2–4], respectively. 19 , 20 Any diffuse or ‘block’ staining [score 2‐4] was considered ‘p16 positive’ and no staining or patchy staining [score 0‐1] as ‘p16 negative’. SCCs were only scored as positive or negative for p16.
2.5.2. Ki‐67 scoring
Ki‐67 IHC staining was scored [score 0‐3] as described by Leeman et al. A normal staining pattern (ie, scattered staining of nuclei in the basal layers) was scored as [0]; score [1‐3] were defined as increased nuclear staining predominantly found in the lower one‐third, lower two‐third and more than two‐thirds of the epithelium, respectively. 12 , 20 , 24 , 27 In SCC, Ki‐67 was scored by estimating the proportion of Ki‐67 positive stained nuclei.
2.5.3. E4 scoring
Membranous and/or cytoplasmic E4 IHC staining was scored [score 0‐2] as: ‘no staining’ [0]; ‘focal’: focally stained cells (ie, limited staining of some [≤5] cells restricted to the upper quarter of the epithelium) [1] or ‘extensive’: (ie, widespread) staining in the upper one‐third of the epithelium and/or below [2]. 24 , 25 , 27
2.6. Statistical analysis
IHC staining results of p16, Ki‐67 and E4 are presented stratified by the study diagnosis. Due to the currently unknown clinical relevance of ‘focal’ E4 staining, for the analyses we only considered ‘extensive’ E4 staining (score [2]) as status ‘E4 positive’, while ‘focal’ (score [1]) and ‘no staining’ (score [0]) were both considered as status ‘E4 negative’. Differences in proportions of p16 scores, Ki‐67 scores, E4 status (negative or positive), methylation result (ie, PPs), methylation status (negative or positive) and HPV16 positivity, and trends with increasing severity of anal lesions were assessed using the χ2 or Fisher's exact test and χ2 tests for trends (P trend), when appropriate. Statistical analyses were performed using IBM SPSS Statistics software (version 26; IBM Corporation, Armonk, NY). Reported P values are two‐sided, with .05 as significance threshold.
3. RESULTS
3.1. Cross‐sectional series
From the cross‐sectional series, a total of 104 samples with adequate IHC staining were included: 8 normal control samples (7.7%), 26 AIN1 (25.0%), 45 AIN2 (43.3%), 15 AIN3 (14.4%) and 10 SCC (9.6%) (Figure S1; study diagnosis).
3.1.1. Results of immunohistochemical markers p16, Ki‐67 and E4 in the cross‐sectional series of anal tissue samples
Figure 1 provides an overview of the p16, Ki‐67 and E4 expression patterns, together with HPV genotyping and methylation results in the individual samples of the cross‐sectional series.
FIGURE 1.
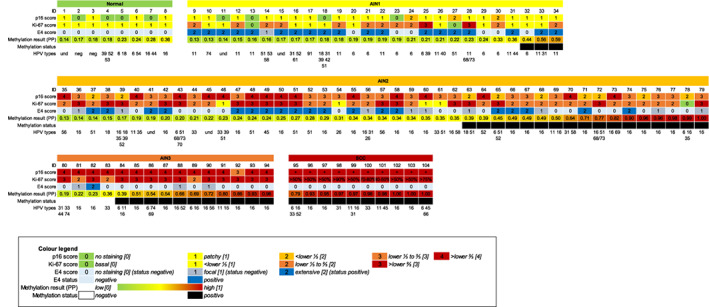
Overview of the p16, Ki‐67 and E4 expression pattern, HPV genotyping and methylation results in the individual samples of the cross‐sectional series of anal biopsies from HIV+ MSM. Immunohistochemical (IHC) marker staining scores (see Colour legend) for p16 [0‐4], Ki‐67 [0‐3] and E4 [0‐2], E4 status (‘negative’: no staining [0] or focal [1]; ‘positive’: extensive [2]), methylation results expressed in predicted probability (PP; values ranging from 0 to 1, representing the risk for AIN3+), methylation status (‘negative’ or ‘positive’) and HPV genotyping, across histological subgroups. The colours refer to the extent of IHC staining and methylation, as indicated in the Colour legend. For SCC, p16 positivity (+) and percentage of Ki‐67 positive nuclei are reported. In each subgroup, samples are consecutively arranged low [(PP = 0); green] to high [(PP = 1); red] based on their methylation result. Each column within a subgroup represents one anal tissue sample: 8 normal, 26 AIN1, 45 AIN2, 15 AIN3 and 10 SCC. AIN, anal intraepithelial neoplasia (grades 1‐3); HPV, human papillomavirus; ID, identification number; MSM, men who have sex with men; neg., negative; Norm(al), normal control samples; SCC, anal squamous cell carcinoma; und., HPV type undetermined [Color figure can be viewed at wileyonlinelibrary.com]
Table 1 shows the results of p16, Ki‐67 and E4 scoring stratified by study diagnosis. Overall, an increase in p16 and Ki‐67 scores (P trend < .001 and P trend < .001, respectively) was observed with increasing severity of anal lesions.
TABLE 1.
Results of p16 (A), Ki‐67 (B) and E4 (C) scoring, by study diagnosis
| A. | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study diagnosis | Total | p16 score | |||||||||||
| No staining (score 0) | Patchy (score 1) | ≤Lower 1/3 (score 2) | ≤Lower 2/3 (score 3) | >Lower 2/3 (score 4) | |||||||||
| Normal | 8 | 4 | 50.0% | 4 | 50.0% | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | ||
| AIN1 | 26 | 4 | 15.4% | 19 | 73.1% | 3 | 11.5% | 0 | 0.0% | 0 | 0.0% | ||
| AIN2 | 45 | 0 | 0.0% | 0 | 0.0% | 9 | 20.0% | 21 | 46.7% | 15 | 33.3% | ||
| AIN3 | 15 | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | 1 | 6.7% | 14 | 93.3% | ||
| Total | 94 | 8 | 8.5% | 23 | 24.5% | 12 | 12.8% | 22 | 23.4% | 29 | 30.9% | ||
| B. | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Study diagnosis | Total | Ki‐67 score | |||||||||
| Normal basal (score 0) | ≤Lower 1/3 (score 1) | ≤Lower 2/3 (score 2) | >Lower 2/3 (score 3) | ||||||||
| Normal | 8 | 0 | 0.0% | 8 | 100.0% | 0 | 0.0% | 0 | 0.0% | ||
| AIN1 | 26 | 2 | 7.7% | 11 | 42.3% | 11 | 42.3% | 2 | 7.7% | ||
| AIN2 | 45 | 1 | 2.2% | 4 | 8.9% | 23 | 51.1% | 17 | 37.8% | ||
| AIN3 | 15 | 0 | 0.0% | 0 | 0.0% | 3 | 20.0% | 12 | 80.0% | ||
| Total | 94 | 3 | 3.2% | 23 | 24.5% | 37 | 39.4% | 31 | 33.0% | ||
| C. | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Study diagnosis | Total | E4 score | |||||||
| No staining (score 0) | Focal (score 1) | Extensive (score 2) | |||||||
| Normal | 8 | 8 | 100.0% | 0 | 0.0% | 0 | 0.0% | ||
| AIN1 | 26 | 6 | 23.1% | 1 | 3.8% | 19 | 73.1% | ||
| AIN2 | 45 | 17 | 37.8% | 9 | 20.0% | 19 | 42.2% | ||
| AIN3 | 15 | 11 | 73.3% | 3 | 20.0% | 1 | 6.7% | ||
| Total | 104 | 52 | 50.0% | 13 | 12.5% | 39 | 37.5% | ||
Note: Data are numbers, percentage. SCCs are not reported here since all SCCs were p16 and Ki‐67 positive and E4 negative (Figure 1).
Abbreviations: AIN, anal intraepithelial neoplasia (grades 1‐3); Normal, normal control samples; SCC, anal squamous cell carcinoma.
E4 staining was mostly observed in AIN1. In 73.1% of AIN1, E4 was scored as ‘extensive’ (Table 1). In AIN2, extensive E4 staining was observed in 42.2% of lesions and in AIN3 in only one lesion (6.7%). In an additional 20% of AIN2 and AIN3, only focal E4 staining was observed. No E4 staining was found in normal control samples or in SCC. Table 2A shows E4 positivity stratified by study diagnosis. With increasing severity of AIN grade (AIN1‐3), E4 positivity decreased significantly (P trend < .001), from 73.1% in AIN1 to 6.7% in AIN3. Figure 2A‐E shows examples of AIN lesions with extensive E4 expression and an AIN2 (mixed) lesion without E4 staining (in the most dysplastic region).
TABLE 2.
E4 (A) and methylation (B) status by study diagnosis and the relationship between E4 and methylation status (C)
| A. | B. | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| E4 status | Methylation status | ||||||||
| Study diagnosis | Negative | Positive | Negative | Positive | |||||
| Normal | 8 | 100.0% | 0 | 0.0% | 8 | 100.0% | 0 | 0.0% | 8 |
| AIN1 | 7 | 26.9% | 19 | 73.1% | 23 | 88.5% | 3 | 11.5% | 26 |
| AIN2 | 26 | 57.8% | 19 | 42.2% | 28 | 62.2% | 17 | 37.8% | 45 |
| AIN3 | 14 | 93.3% | 1 | 6.7% | 4 | 26.7% | 11 | 73.3% | 15 |
| SCC | 10 | 100.0% | 0 | 0.0% | 0 | 0.0% | 10 | 100.0% | 10 |
| Total | 65 | 62.5% | 39 | 37.5% | 63 | 60.6% | 41 | 39.4% | 104 |
| C. | E4 status | Total | ||||||
|---|---|---|---|---|---|---|---|---|
| Study diagnosis | Negative | Positive | ||||||
| Methylation negative (n = 63) | Normal | 8 | 100.0% | 0 | 0.0% | 8 | 12.7% | |
| AIN1 | 6 | 26.1% | 17 | 73.9% | 23 | 36.5% | ||
| AIN2 | 14 | 50.0% | 14 | 50.0% | 28 | 44.4% | ||
| AIN3 | 3 | 75.0% | 1 | 25.0% | 4 | 6.3% | ||
| SCC | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | ||
| Subtotal | 31 | 49.2% | 32 | 50.8% | 63 | 60.6% | ||
| Methylation positive (n = 41) | Normal | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | |
| AIN1 | 1 | 33.3% | 2 | 66.7% | 3 | 7.3% | ||
| AIN2 | 12 | 70.6% | 5 | 29.4% | 17 | 41.5% | ||
| AIN3 | 11 | 100.0% | 0 | 0.0% | 11 | 26.8% | ||
| SCC | 10 | 100.0% | 0 | 0.0% | 10 | 24.4% | ||
| Subtotal | 34 | 82.9% | 7 | 17.1% | 41 | 39.4% | ||
| Total | 65 | 62.5% | 39 | 37.5% | 104 | 100.0% | ||
Note: Data are numbers, percentage.
Abbreviations: AIN, anal intraepithelial neoplasia (grades 1‐3); Normal, normal control samples; SCC, anal squamous cell carcinoma.
FIGURE 2.
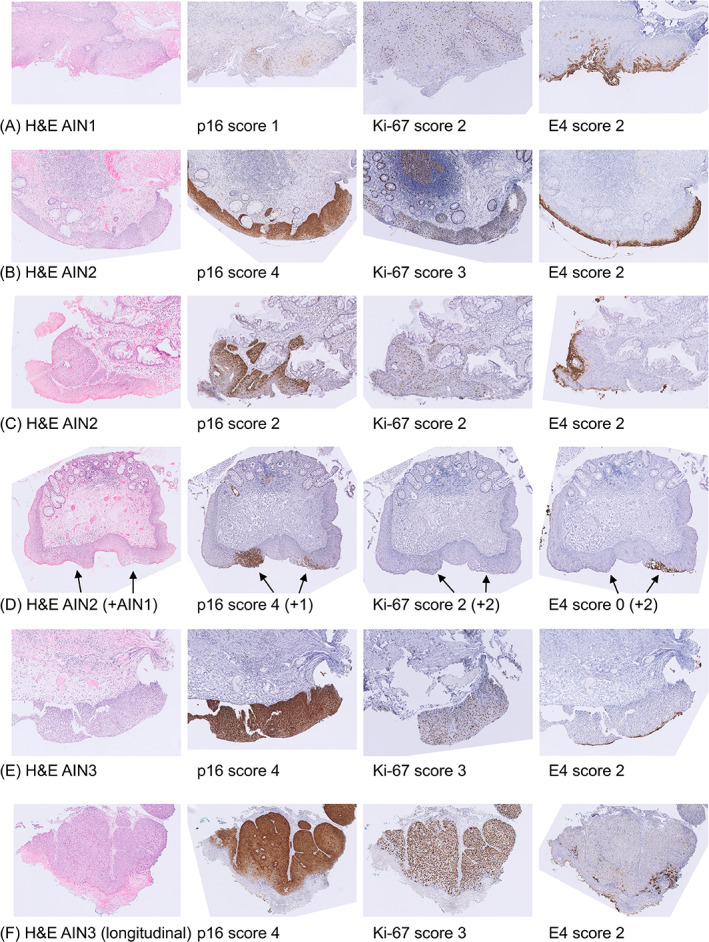
Examples of immunohistochemical stainings. A, AIN1 lesion (ID19; methylation negative) with patchy p16 staining (score 1), Ki‐67 staining up to the lower two‐third of the epithelium (score 2) and extensive membranous and cytoplasmic E4 staining (score 2); B, AIN2 lesion (ID49; methylation negative) with diffuse p16 staining (score 4), Ki‐67 staining reaching more than two‐thirds of the epithelium (score 3) and extensive E4 staining (score 2); C, AIN2 ‘composite’ (both transforming and productive characteristics) lesion (ID71; methylation positive) with diffuse p16 staining up to the lower one‐third of the epithelium (score 2), Ki‐67 staining up to the lower two‐third of the epithelium (score 2) and extensive E4 staining (score 2); D, AIN2 lesion (ID73; methylation positive) with adjacent AIN1 lesion (see arrows; mixed lesion) with diffuse full‐thickness p16 block staining (score 4), Ki‐67 staining up to the lower two‐third of the epithelium (score 2), and no E4 staining (score 0) in the AIN2 region and patchy p16 staining (score 1), Ki‐67 staining up to the lower two‐third of the epithelium (score 2) and extensive E4 staining (score 2) in the AIN1 region; E, AIN3 lesion (ID82; methylation negative) with diffuse full‐thickness p16 staining (score 4), Ki‐67 staining reaching more than two‐thirds of the epithelium (score 3) and extensive E4 staining (score 2); F, AIN3 (longitudinal series, case 10, sample number 2; methylation positive) with diffuse up to full‐thickness p16 staining (score 4), Ki‐67 staining reaching more than two‐thirds of the epithelium (score 3) and extensive E4 staining (score 2). AIN, anal intraepithelial neoplasia (grades 1‐3); H&E, haematoxylin and eosin; ID, identification number (matches with Figure 1); Normal, normal control samples; SCC, anal squamous cell carcinoma [Color figure can be viewed at wileyonlinelibrary.com]
Simultaneous high p16 and Ki‐67 scores were often observed in AIN2/3. Several AIN lesions showed simultaneous E4 positivity and p16 positivity, particularly within AIN2 (42.2%, 19/45). In E4 positive lesions, high Ki‐67 scores were frequently observed. Comparison of p16 and Ki‐67 staining scores between E4 negative and positive samples, stratified by study diagnosis (Table S1), revealed no significant differences (P values > .05).
3.1.2. Correlation of immunohistochemical marker expression with methylation positivity in the cross‐sectional series
Methylation positivity (ie, methylation result above threshold) increased significantly (P trend < .001) from 0% in normal, 11.5% in AIN1, 37.8% in AIN2 up to 73.3% in AIN3 and 100% in SCC. Table 2B shows methylation positivity stratified by study diagnosis.
Besides higher p16 scores in methylation negative AIN2 (P = .01), no clear relation (P values > .05) between p16 and Ki‐67 and methylation positivity was found by comparing p16 and Ki‐67 staining scores between methylation negative and positive samples stratified by study diagnosis (Table S2).
Table 2C shows the relationship between E4 and methylation positivity stratified by study diagnosis. Overall, the proportion of E4 positive lesions was higher in methylation negative samples compared to methylation positive samples (50.8%, 32/63, vs 17.1%, 7/41; P < .001; Table 2C). Moreover, the proportion of methylation positive lesions was higher in E4 negative samples compared to E4 positive samples (52.3%, 34/65 vs 17.9%, 7/39; P < .001). Figure S2 shows the methylation results (ie, PP) per sample stratified by E4 status and study diagnosis. We found no statistical differences (P values > .1) in methylation results comparing E4 negative and positive lesions per AIN grade; however, numbers were low.
The overview presented in Figure 1 highlights the heterogeneity in anal precancerous lesions regarding IHC and methylation patterns. Still, three main biomarker patterns were observed: (a) E4 positive, methylation negative lesions [particularly in AIN1 (65.4%; 17/26), a subset of AIN2 (31.1%; 14/45) and only one AIN3 (1/15) lesion]; (b) E4 negative, methylation positive lesions [particularly in AIN3 (73.3%; 11/15) and a subset of AIN2 (26.7%; 12/45)] and (c) E4 positive, methylation positive lesions (so‐called composite lesions) present in 7.7% (2/26) of AIN1 and 11.1% (5/45) of AIN2 (Figure 2C).
3.2. Longitudinal series
An overview of the full characterisation of the 10 cases in the longitudinal series using IHC marker staining patterns, HPV genotyping and methylation results is shown in Figure 3. In this series, all anal cancer samples and all AIN2‐3 during progression towards cancer were methylation positive and exhibited high p16 and Ki‐67 expression. Interestingly, although lesions exhibiting productive characteristics being less common in this series, three composite HSILs (showing simultaneous extensive E4 staining and methylation positivity) were found in two HIV+ patients: two AIN3 (case 9; sample no. 1 and case 10; sample no. 2; Figure 2F) and an AIN2 (case 10; sample no. 3). In both cases, E4 positivity was not detected at the endpoint diagnosis.
FIGURE 3.
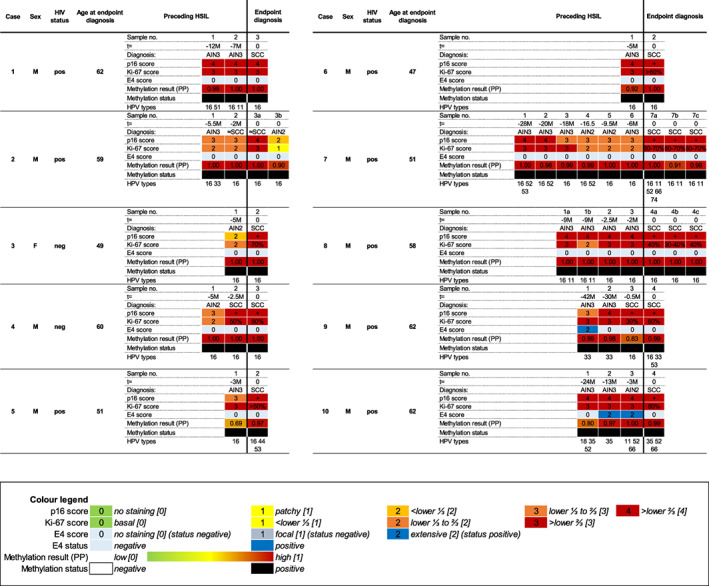
Overview of characterisation of anal tissue samples from 10 cases in the longitudinal series with development of anal SCC over time. Each sample number within a case represents one anal tissue sample of the endpoint diagnosis (SCC or suspected SCC (≈SCC; HSIL/AIN3 lesion with high suspicion for infiltrative growth) or preceding HSIL and was taken from the same anatomic location [t = time in months (M) before endpoint diagnosis]. In some cases, multiple biopsies were taken at the same time point, indicated with a letter (eg, 1a, 1b, etc). Per sample immunohistochemical (IHC) marker staining scores (see Colour legend) for p16 [0‐4], Ki‐67 [0‐3] and E4 [0‐2], E4 status (‘negative’: no staining [0] or focal [1]; ‘positive’: extensive [2]), methylation results expressed in predicted probability [PP; values ranging from low [(PP = 0); green] to high [(PP = 1); red], representing the risk for AIN3+], methylation status (‘negative’ or ‘positive’) and HPV genotyping is provided. The colours refer to the extent of IHC staining and methylation, as indicated in the Colour legend. For SCC, p16 positivity (+) and percentage of Ki‐67 positive nuclei are reported. AIN, anal intraepithelial neoplasia (grades 1‐3); F, female; HPV, human papillomavirus; HSIL, high‐grade squamous intraepithelial lesion; M, male; NDS, non‐diagnostic sample (staining uninterpretable due to technical staining issue or no more lesion being present in the slide); neg, negative; pos, positive; SCC, anal squamous cell carcinoma [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
In this study, we provided a comprehensive characterisation using IHC and DNA methylation markers of a cross‐sectional series of anal lesions representing the full spectrum of anal carcinogenesis in HIV+ MSM and of a longitudinal series of lesions with a known course of progression to cancer. In the cross‐sectional series, increasing p16 and Ki‐67 expression was found to be associated with increasing severity of anal dysplasia. Conversely, a decrease in E4 expression, from 73.1% in AIN1 to 42.2% in AIN2 and 6.7% in AIN3, was seen. E4 positivity was also inversely associated with methylation positivity. Interestingly, AIN lesions, in particular AIN2, were commonly found (42.2%) to exhibit both E4 and p16 expression. In a few AIN1 (7.7%) and AIN2 (11.1%) lesions, both E4 and methylation positivity were found. These so‐called composite lesions were also found in three samples from two HIV+ patients in the longitudinal series with a known course of progression towards cancer.
Previously, Leeman et al found similar IHC expression patterns in anal lesions, although in general we observed more E4 positivity in our cross‐sectional series for all histological grades, most prominently in AIN1 (73.1% vs 43%‐49%). 19 , 20 A likely explanation are differences in cohort composition. Our cross‐sectional series consisted exclusively of HIV+ MSM who were biopsied during screening, whereas their cohort was in part constituted of referral patients and also included HIV‐negative men. 19 , 20
The present study builds on this previous evidence 19 , 20 by showing that biomarker patterns in HSIL are complex, reflecting the heterogeneity of these lesions. Biomarker heterogeneity was most prominent in AIN2 and may explain the ambiguous clinical course of these lesions, ranging from regressive to progressive disease. This is supported by recent findings from the Study of the Prevention of Anal Cancer (SPANC) AIN natural history study: AIN2 had a higher probability of regression compared to AIN3 (hazard ratio 1.79). 8 A two‐tier classification (LSIL‐HSIL) would therefore insufficiently cover the heterogeneity within high‐grade lesions. For this reason, we chose to report our study diagnosis with a three‐tier annotation of AIN1‐3 extended from the LSIL‐HSIL classification to allow further characterisation of lesions while incorporating the LAST recommendations. 6 A similar approach was also recently advocated for cervical lesions given the clinical heterogeneity within high‐grade CIN. 32
The proportion of methylation positivity increased from AIN1 to cancer and we have shown that high methylation levels are associated with progression to cancer. 30 We therefore consider methylation markers as a promising and objective cancer risk stratification tool to identify advanced HSIL with a presumed high short‐term cancer progression risk, for which treatment seems appropriate, in contrast to early HSIL with a presumed low short‐term progression risk for which treatment can be withheld. 30 Our study supports the value of p16 to improve grading of AIN. 13 , 14 , 15 , 16 , 17 , 18 Ki‐67 seems less informative, as increased Ki‐67 expression was also observed in E4 positive, methylation negative, so‐called productive AIN lesions, especially in AIN1. So far, a prognostic value of p16 and Ki‐67 for cancer risk stratification of HSIL has not been established. Our findings and previous studies support that E4 expression, as a marker for viral production, can help to further characterise AIN lesions. 19 , 20
The presence of a methylation positive, E4 negative biomarker pattern, mainly seen in anal cancer, the majority (73.3%) of AIN3 and a proportion (26.7%) of AIN2 lesions is supportive of a high short‐term progression risk to cancer of AIN lesions with this biomarker pattern. Conversely, the presence of an E4 positive, methylation negative biomarker pattern in a high proportion of AIN1 lesions (65.4%) and almost one‐third of AIN2 lesions (31.1%) and the near, respectively, complete absence of this biomarker pattern in AIN3 and SCC support our assumption that these lesions have a low risk of progression to cancer. However, the observed E4 expression in some HSILs during the course of progression to cancer and absence of E4 in a considerable number of AIN1 lesions make the potential clinical significance of absence or presence of E4 expression difficult to interpret and argue that prognostic information is mainly derived from the methylation results.
Of interest are the composite lesions with simultaneous E4 expression and methylation positivity as observed in both the cross‐sectional (n = 7) and longitudinal (n = 3) series. These results indicate that production of viral particles and cellular transformation by HPV can overlap. Although gradual progression from low‐grade to high‐grade lesions remains under debate, 33 it may be hypothesised that these composite lesions reflect a temporal evolution from a lower grade lesion to a high‐grade lesion in which the productive lesion is replaced by transformed cells migrating up. Their occurrence in HIV+ MSM may relate to the impaired HPV clearance induced by the HIV infection and a higher susceptibility to HPV‐induced transformation. 34 Indeed, an upregulating effect of the HIV protein ‘tat’ on HPV oncogenes E6 and E7 expression has been described. 35 , 36 In addition, HPV and HIV proteins were reported to upregulate DNA methyl transferases resulting in methylation‐mediated silencing of tumour suppressor genes, thereby contributing to faster HPV‐induced carcinogenesis. 37 , 38 Similar composite lesions were also reported in the cervix of HIV+ women with more abundant E4 expression and higher methylation levels compared to HIV‐negative women. 25 , 27
Collectively, our data suggest limited prognostic value for E4 as sole biomarker. E4 expression seems to provide at best some supportive information next to methylation analysis results. Figure 4 depicts the conceptual relationship of E4 expression and methylation status in relation to AIN grade and their presumed anal cancer risk in terms of early HSIL and advanced HSIL. In this conceptual scheme, the composite lesions, with a currently uncertain clinical behaviour, are positioned in the middle.
FIGURE 4.
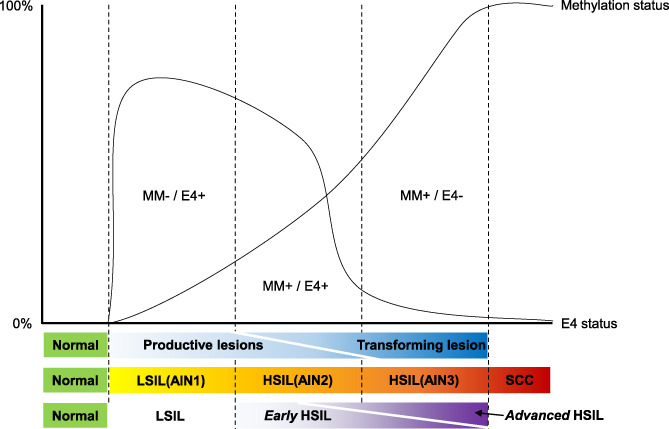
Concept of anal carcinogenesis in HIV+ men: association with E4 expression and host cell DNA methylation. Patterns of productive (E4 immunohistochemical marker expression) and transforming (host cell DNA methylation status) characteristics during anal carcinogenesis in HIV+ men. With progression to cancer, E4 positivity decreases, whereas methylation positivity increases, but they show considerable overlap, corresponding with biological heterogeneity in HSIL. Methylation analysis might aid a clinically relevant subdivision of HSIL into early and advanced HSIL, representing low and high short‐term risk of progression to cancer, respectively. +, positive; −, negative; AIN, anal intraepithelial neoplasia (grades 1‐3); HSIL, high‐grade squamous intraepithelial lesion; MM, methylation marker status; Normal, normal control samples; SCC, anal squamous cell carcinoma [Color figure can be viewed at wileyonlinelibrary.com]
To the best of our knowledge, this is the first study that comprehensively characterised anal lesions from a large cross‐sectional series as well as a well‐documented longitudinal series of patients who developed anal cancer over time, providing in‐depth information on anal carcinogenesis and potential prognostic value of IHC markers. Another strength of the study is the identification of the most dysplastic region within tissue samples for scoring of the IHC markers. The latter is of importance since tissue samples can include a mix of different types of lesions (Figure 2D). Although whole tissue sections were used for methylation analysis, qMSP assays are very sensitive and have been shown to represent the most dysplastic region or highest grade of dysplasia present. 39
We acknowledge several limitations of our study. First, the retrospective collection in the longitudinal series. 30 Although detailed information of the location of the biopsies was given and they had matching HPV genotypes, we cannot completely exclude that consecutive biopsies were not from exactly the same location. Second, although this is a unique longitudinal series of rare well‐documented cases, its size remains relatively small. Third, slightly different methylation marker panels were used for the cross‐sectional and longitudinal series and although their diagnostic performance have previously been proven similar, this may limit a direct comparison. 29 , 30 Our findings warrant confirmation in a larger prospective study. Moreover, our study is limited to HIV+ patients and while our recent data show that methylation patterns are similar between HIV+ and HIV‐negative individuals, it is currently unknown whether this is also the case regarding E4 expression. 40
In conclusion, we showed that anal lesions in the spectrum of carcinogenesis in HIV+ MSM harboured complex heterogeneous biomarker patterns. A decrease in productive (E4 expression) characteristics and increase in transforming characteristics (p16) and methylation was observed with increasing severity of disease. In particular in AIN2, we found a heterogeneous pattern displaying either transforming or productive characteristics or both, which supports the ambiguous clinical course of these lesions. IHC biomarkers can help to characterise anal precancerous lesions; however, their prognostic value for cancer risk stratification, next to objective methylation analysis, appears to be limited.
CONFLICT OF INTEREST
Renske D. M. Steenbergen and Chris J. L. M. Meijer are minority stockholders of Self‐screen BV, a spin‐off company of VUmc, which owns patents on methylation markers and HPV detection. Chris J. L. M. Meijer is part‐time director of Self‐screen BV. He has been on the speakers bureau and served occasionally on the scientific advisory board (expert meeting) of GSK, Qiagen, SPMSD/Merck. He has been co‐investigator on a SPMSD sponsored trial, of which his institute received research funding; has a very small number of Qiagen and MDxHealth shares. Henry J. C. de Vries received financial compensation or goods for research from Medigene, Gilead and MSD; financial compensation for presentations from Abbott and Janssen and financial compensation for advice to Medigene and Novartis. All other authors report no potential conflicts. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript or in the decision to publish the results.
ETHICS STATEMENT
We adhered to the Declaration of Helsinki and Code of Conduct for Responsible Use of Left‐over Material of the Dutch Federation of Biomedical Scientific Societies. Ethical approval was granted under reference numbers 07/318 (AIN biopsies) and 05/031 (normal control samples) and waived under reference number 17/151 (SCC) and 17/234 (longitudinal series) by the Institutional Review Board of the Amsterdam UMC. For the longitudinal series, additional local ethical approval was granted by the NHS Health Research Authority, United Kingdom (IRAS ID 226196), and the Ethical Committee of the University Witten/Herdecke, Germany (reference no. 166/2017).
Supporting information
Appendix S1: Supporting Information
ACKNOWLEDGEMENTS
We thank Sylvia Duin, Annina P. van Splunter, Timo J. ter Braak (Amsterdam UMC; Pathology) and Henk A. M. van den Munckhof (DDL Diagnostic Laboratory, Rijswijk) for excellent technical assistance; and Mayura Nathan (Homerton University Hospital; Anal Neoplasia Service, London) and Michael Sheaff (Barts Health NHS Trust; Cellular Pathology, London) for providing clinical data and samples for the longitudinal series.
van der Zee RP, Meijer CJLM, Cuming T, et al. Characterisation of anal intraepithelial neoplasia and anal cancer in HIV‐positive men by immunohistochemical markers p16, Ki‐67, HPV‐E4 and DNA methylation markers. Int. J. Cancer. 2021;149(10):1833‐1844. 10.1002/ijc.33748
Funding information KWF Kankerbestrijding (Dutch Cancer Society), Grant/Award Number: 2016‐10781
Contributor Information
Ramon P. van der Zee, Email: r.p.vanderzee@amsterdamumc.nl.
Renske D. M. Steenbergen, Email: r.steenbergen@amsterdamumc.nl.
DATA AVAILABILITY STATEMENT
Study data that underlie the results reported in this article (text, tables, figures and appendices) and a data dictionary will be available for 5 years after publication after de‐identification and at request to the corresponding author. Data will be securely transferred to researchers, who provide a methodologically sound research proposal and only to achieve aims in the approved proposal, after: approval of the ethics review board; additional approval of study participants (if applicable); and signing a data access agreement.
REFERENCES
- 1. Islami F, Ferlay J, Lortet‐Tieulent J, Bray F, Jemal A. International trends in anal cancer incidence rates. Int J Epidemiol. 2017;46:924‐938. [DOI] [PubMed] [Google Scholar]
- 2. Clifford GM, Georges D, Shiels MS, et al. A meta‐analysis of anal cancer incidence by risk group: toward a unified anal cancer risk scale. Int J Cancer. 2021;148:38‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hoots BE, Palefsky JM, Pimenta JM, Smith JS. Human papillomavirus type distribution in anal cancer and anal intraepithelial lesions. Int J Cancer. 2009;124:2375‐2383. [DOI] [PubMed] [Google Scholar]
- 4. Berry JM, Jay N, Cranston RD, et al. Progression of anal high‐grade squamous intraepithelial lesions to invasive anal cancer among HIV‐infected men who have sex with men. Int J Cancer. 2014;134:1147‐1155. [DOI] [PubMed] [Google Scholar]
- 5. Fenger C, Nielsen VT. Intraepithelial neoplasia in the anal canal. The appearance and relation to genital neoplasia. Acta Pathol Microbiol Immunol Scand A. 1986;94:343‐349. [DOI] [PubMed] [Google Scholar]
- 6. Darragh TM, Colgan TJ, Cox JT, et al. The Lower Anogenital Squamous Terminology Standardization Project for HPV‐associated lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. J Low Genit Tract Dis. 2012;16:205‐242. [DOI] [PubMed] [Google Scholar]
- 7. Machalek DA, Poynten M, Jin F, et al. Anal human papillomavirus infection and associated neoplastic lesions in men who have sex with men: a systematic review and meta‐analysis. Lancet Oncol. 2012;13:487‐500. [DOI] [PubMed] [Google Scholar]
- 8. Poynten IM, Jin F, Roberts JM, et al. The natural history of anal high‐grade squamous intraepithelial lesions in gay and bisexual men. Clin Infect Dis. 2021;72(5):853‐861. [DOI] [PubMed] [Google Scholar]
- 9. Goldstone SE, Lensing SY, Stier EA, et al. A randomized clinical trial of infrared coagulation ablation versus active monitoring of intra‐anal high‐grade dysplasia in adults with human immunodeficiency virus infection: an AIDS malignancy consortium trial. Clin Infect Dis. 2019;68:1204‐1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klaes R, Friedrich T, Spitkovsky D, et al. Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer. 2001;92:276‐284. [DOI] [PubMed] [Google Scholar]
- 11. Sasaki K, Murakami T, Kawasaki M, Takahashi M. The cell cycle associated change of the Ki‐67 reactive nuclear antigen expression. J Cell Physiol. 1987;133:579‐584. [DOI] [PubMed] [Google Scholar]
- 12. van Zummeren M, Leeman A, Kremer WW, et al. Three‐tiered score for Ki‐67 and p16(ink4a) improves accuracy and reproducibility of grading CIN lesions. J Clin Pathol. 2018;71:981‐988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Albuquerque A, Rios E, Dias CC, Nathan M. p16 immunostaining in histological grading of anal squamous intraepithelial lesions: a systematic review and meta‐analysis. Mod Pathol. 2018;31:1026‐1035. [DOI] [PubMed] [Google Scholar]
- 14. Krishnamurti U, Mohammad M, Monsrud A, et al. Diagnosing anal squamous intraepithelial lesions with and without p16: an interobserver variability study. J Low Genit Tract Dis. 2020;24:69‐74. [DOI] [PubMed] [Google Scholar]
- 15. Liu Y, McCluggage WG, Darragh TM, et al. Classifying anal intraepithelial neoplasia 2 based on LAST recommendations. Am J Clin Pathol. 2021;155(6):845‐852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bean SM, Eltoum I, Horton DK, Whitlow L, Chhieng DC. Immunohistochemical expression of p16 and Ki‐67 correlates with degree of anal intraepithelial neoplasia. Am J Surg Pathol. 2007;31:555‐561. [DOI] [PubMed] [Google Scholar]
- 17. Walts AE, Lechago J, Bose S. P16 and Ki67 immunostaining is a useful adjunct in the assessment of biopsies for HPV‐associated anal intraepithelial neoplasia. Am J Surg Pathol. 2006;30:795‐801. [DOI] [PubMed] [Google Scholar]
- 18. Pirog EC, Quint KD, Yantiss RK. P16/CDKN2A and Ki‐67 enhance the detection of anal intraepithelial neoplasia and condyloma and correlate with human papillomavirus detection by polymerase chain reaction. Am J Surg Pathol. 2010;34:1449‐1455. [DOI] [PubMed] [Google Scholar]
- 19. Leeman A, Jenkins D, Marra E, et al. Grading immunohistochemical markers p16(INK4a) and HPV E4 identifies productive and transforming lesions caused by low‐ and high‐risk HPV within high‐grade anal squamous intraepithelial lesions. Br J Dermatol. 2020;182:1026‐1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leeman A, Pirog EC, Doorbar J, et al. Presence or absence of significant HPVE4 expression in high‐grade anal intraepithelial neoplasia with p16/Ki‐67 positivity indicates distinct patterns of neoplasia: a study combining immunohistochemistry and laser capture microdissection PCR. Am J Surg Pathol. 2018;42:463‐471. [DOI] [PubMed] [Google Scholar]
- 21. Griffin H, Soneji Y, Van Baars R, et al. Stratification of HPV‐induced cervical pathology using the virally encoded molecular marker E4 in combination with p16 or MCM. Mod Pathol. 2015;28:977‐993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Doorbar J. The E4 protein; structure, function and patterns of expression. Virology. 2013;445:80‐98. [DOI] [PubMed] [Google Scholar]
- 23. Griffin H, Wu Z, Marnane R, et al. E4 antibodies facilitate detection and type‐assignment of active HPV infection in cervical disease. PLoS One. 2012;7:e49974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zummeren MV, Kremer WW, Leeman A, et al. HPV E4 expression and DNA hypermethylation of CADM1, MAL, and miR124‐2 genes in cervical cancer and precursor lesions. Mod Pathol. 2018;31:1842‐1850. [DOI] [PubMed] [Google Scholar]
- 25. Kremer WW, Vink FJ, van Zummeren M, et al. Characterization of cervical biopsies of women with HIV and HPV co‐infection using p16(ink4a), ki‐67 and HPV E4 immunohistochemistry and DNA methylation. Mod Pathol. 2020;33:1968‐1978. [DOI] [PubMed] [Google Scholar]
- 26. van Baars R, Griffin H, Wu Z, et al. Investigating diagnostic problems of CIN1 and CIN2 associated with high‐risk HPV by combining the novel molecular biomarker PanHPVE4 with P16INK4a. Am J Surg Pathol. 2015;39:1518‐1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vink FJ, Dick S, Heideman DAM, et al. Classification of high‐grade CIN by p16ink4a, Ki‐67, HPV E4 and FAM19A4/miR124‐2 methylation status demonstrates considerable heterogeneity with potential consequences for management. Int J Cancer. 2021;149(3):707‐716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV‐induced cervical precancerous lesions. Nat Rev Cancer. 2014;14:395‐405. [DOI] [PubMed] [Google Scholar]
- 29. van der Zee RP, Richel O, van Noesel CJM, et al. Host cell deoxyribonucleic acid methylation markers for the detection of high‐grade anal intraepithelial neoplasia and anal cancer. Clin Infect Dis. 2019;68:1110‐1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van der Zee RP, Richel O, van Noesel CJM, et al. Cancer risk stratification of anal intraepithelial neoplasia in HIV‐positive men by validated methylation markers associated with progression to cancer. Clin Infect Dis. 2021;72(12):2154‐2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmittgen TD, Livak KJ. Analyzing real‐time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101‐1108. [DOI] [PubMed] [Google Scholar]
- 32. Castle PE, Adcock R, Cuzick J, et al. Relationships of p16 immunohistochemistry and other biomarkers with diagnoses of cervical abnormalities: implications for LAST terminology. Arch Pathol Lab Med. 2020;144:725‐734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jongen VW, Steenbergen RDM, van der Loeff MFS. Reply to Fang & Buchwald. J Infect Dis. 2021;jiab080. https://doi.org10.1093/infdis/jiab080. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 34. Brickman C, Palefsky JM. Human papillomavirus in the HIV‐infected host: epidemiology and pathogenesis in the antiretroviral era. Curr HIV/AIDS Rep. 2015;12:6‐15. [DOI] [PubMed] [Google Scholar]
- 35. Barillari G, Palladino C, Bacigalupo I, Leone P, Falchi M, Ensoli B. Entrance of the Tat protein of HIV‐1 into human uterine cervical carcinoma cells causes upregulation of HPV‐E6 expression and a decrease in p53 protein levels. Oncol Lett. 2016;12:2389‐2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tornesello ML, Buonaguro FM, Beth‐Giraldo E, Giraldo G. Human immunodeficiency virus type 1 tat gene enhances human papillomavirus early gene expression. Intervirology. 1993;36:57‐64. [DOI] [PubMed] [Google Scholar]
- 37. Youngblood B, Reich NO. The early expressed HIV‐1 genes regulate DNMT1 expression. Epigenetics. 2008;3:149‐156. [DOI] [PubMed] [Google Scholar]
- 38. Sen P, Ganguly P, Ganguly N. Modulation of DNA methylation by human papillomavirus E6 and E7 oncoproteins in cervical cancer. Oncol Lett. 2018;15:11‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Baars R, van der Marel J, Snijders PJ, et al. CADM1 and MAL methylation status in cervical scrapes is representative of the most severe underlying lesion in women with multiple cervical biopsies. Int J Cancer. 2016;138:463‐471. [DOI] [PubMed] [Google Scholar]
- 40. van der Zee RP, van Noesel CJM, Martin I, et al. DNA methylation markers have universal prognostic value for anal cancer risk in HIV‐negative and HIV‐positive individuals. Mol Oncol. 2021. 10.1002/1878-0261.12926. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information
Data Availability Statement
Study data that underlie the results reported in this article (text, tables, figures and appendices) and a data dictionary will be available for 5 years after publication after de‐identification and at request to the corresponding author. Data will be securely transferred to researchers, who provide a methodologically sound research proposal and only to achieve aims in the approved proposal, after: approval of the ethics review board; additional approval of study participants (if applicable); and signing a data access agreement.


