Abstract
We investigated induction of biphenyl dioxygenase in the psychrotolerant polychlorinated biphenyl (PCB) degrader Pseudomonas strain Cam-1 and in the mesophilic PCB degrader Burkholderia strain LB400. Using a counterselectable gene replacement vector, we inserted a lacZ-Gmr fusion cassette between chromosomal genes encoding the large subunit (bphA) and small subunit (bphE) of biphenyl dioxygenase in Cam-1 and LB400, generating Cam-10 and LB400-1, respectively. Potential inducers of bphA were added to cell suspensions of Cam-10 and LB400-1 incubated at 30°C, and then beta-galactosidase activity was measured. Biphenyl induced beta-galactosidase activity in Cam-10 to a level approximately six times greater than the basal level in cells incubated with pyruvate. In contrast, the beta-galactosidase activities in LB400-1 incubated with biphenyl and in LB400-1 incubated with pyruvate were indistinguishable. At a concentration of 1 mM, most of the 40 potential inducers tested were inhibitory to induction by biphenyl of beta-galactosidase activity in Cam-10. The exceptions were naphthalene, salicylate, 2-chlorobiphenyl, and 4-chlorobiphenyl, which induced beta-galactosidase activity in Cam-10, although at levels that were no more than 30% of the levels induced by biphenyl. After incubation for 24 h at 7°C, biphenyl induced beta-galactosidase activity in Cam-10 to a level approximately four times greater than the basal level in cells incubated with pyruvate. The constitutive level of beta-galactosidase activity in LB400-1 grown at 15°C was approximately five times less than the level in LB400-1 grown at 30°C. Thus, there are substantial differences in the effects of physical and chemical environmental conditions on genetic regulation of PCB degradation in different bacteria.
Bioremediation of soil contaminated with polychlorinated biphenyls (PCBs) is an attractive clean-up strategy due to its potential to mineralize pollutants and to be inexpensive. Many PCB-degrading bacteria have been isolated and characterized (2, 3, 6, 8, 10, 22, 39). Some of these bacteria can grown on monochlorinated and dichlorinated biphenyls, and most cometabolize more highly chlorinated biphenyls while using biphenyl as a growth substrate (1, 7, 11). In some cases, the presence of biphenyl as a potential growth substrate and inducer of PCB metabolism (14) is important for maintaining PCB biodegradation activity in soil (5, 17). However, adding biphenyl to soil to stimulate PCB degradation activity is problematic due to the low water solubility of biphenyl and its possible adverse health effects (1, 19). Biphenyl is rare in natural environments, and it is possible that other, more common compounds also induce genes encoding biphenyl-degrading enzymes, termed bph genes (23). Such inducers may be less toxic and more water soluble than biphenyl, so that they could be added to soil to stimulate PCB degradation activity in bioremediation projects.
Several studies have investigated induction of PCB removal in cell suspensions of PCB-degrading bacteria by compounds other than biphenyl. Notably, cell suspensions of Arthrobacter sp. strain B1B grown on fructose medium supplemented with l-carvone, limonene, p-cymene, or isoprene remove Aroclor 1242 (27). Alcaligenes eutrophus H850 and Corynebacterium sp. strain MB1 grown on plant phenolic compounds and Pseudomonas sp. strain LB400 (10) (now a member of the genus Burkholderia [47]) grown on plant phenolic compounds, glucose, or glycerol degrade certain PCB congeners (9, 15). Also, other workers have amplified mRNA transcripts of 2,3-dihydroxybiphenyl dioxygenase (bphC) in A. eutrophus H850 grown on fructose plus l-carvone; however, these transcripts were not quantified to determine if there is a significant difference between the levels of bphC mRNA in cells grown on fructose alone and the levels of bphC mRNA in cells grown with carvone (38). Finally, Cellulomonas sp. strain T109 and Rhodococcus rhodochrous T100 grown on cymene and limonene, respectively, remove over 80% more Aroclor 1242 than these organisms grown on glucose (28). These studies support the hypothesis that certain compounds other than biphenyl may be used to stimulate PCB biodegradation. However, investigations so far have not shown that bacteria grown on substrates other than biphenyl remove PCBs as a result of induction of bph genes at levels above constitutive levels. Moreover, it is possible that the compounds used to induce bacterial PCB degradation activity did not induce bph genes but instead induced genes that encode other enzymes that also degrade PCBs or stimulated PCB degradation via mechanisms other than genetic regulation.
To determine if compounds other than biphenyl induce bph genes (Fig. 1), we constructed a chromosomal bphA-lacZ reporter in the psychrotolerant PCB-degrading bacterium Pseudomonas sp. strain Cam-1 (34) to generate strain Cam-10. We also constructed a chromosomal bphA-lacZ reporter in the mesophilic PCB-degrading bacterium Burkholderia sp. strain LB400 to generate strain LB400-1. Construction of Cam-10 and LB400-1 allowed us to study the regulation of bph genes in a chromosomal context. We incubated Cam-10 and LB400-1 with compounds that previously have been shown to stimulate PCB degradation in other bacteria or that are structurally similar to biphenyl. Then we performed beta-galactosidase assays to determine if the lacZ reporter gene was induced. Induction of beta-galactosidase activity was correlated to induction of bphA. Our results suggest that regulation of bphA in Cam-1 is highly specific. In contrast, the beta-galactosidase activities were indistinguishable in LB400-1 cells incubated with biphenyl and LB400-1 cells incubated with pyruvate, suggesting that in LB400 bphA is expressed constitutively.
FIG. 1.
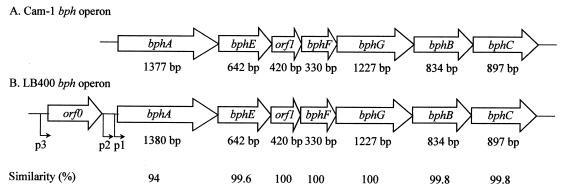
Organization and similarity of the bph gene clusters in Pseudomonas sp. strain Cam-1 (A) and Burkholderia sp. strain LB400 (B). bphA, gene encoding the terminal dioxygenase large subunit; bphE, gene encoding the terminal dioxygenase small subunit; bphF, gene encoding ferredoxin; bphG, gene encoding ferredoxin reductase; bphB, gene encoding dihydrodiol dehydrogenase; bphC, gene encoding 2,3-dihydroxybiphenyl dioxygenase. In the LB400 operon the locations of promoter regions are indicated by p1, p2, and p3.
Few studies thus far have compared how different PCB-degrading bacteria regulate genes that encode enzymes involved in the biphenyl degradative pathway. We investigated induction of bphA in two PCB-degrading bacteria and found that bph genes in these organisms are regulated differently. This result has implications for PCB bioremediation strategies, as it suggests that the optimal methods for stimulating PCB degradation activity may depend on which PCB-degrading bacteria are present.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise specified, Escherichia coli was cultured at 37°C in Luria-Bertani (LB) medium, and Pseudomonas sp. strain Cam-1 and Burkholderia sp. strain LB400 were grown at 15 or 30°C in tryptic soy broth or mineral medium (6) containing 1.3 or 9 mM pyruvate as the growth substrate.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| Pseudomonas sp. strain Cam-1 | Wild type | 34 |
| Pseudomonas sp. strain Cam-10 | bphA-lacZ-Gmr | This study |
| Pseudomonas sp. strain Cam-20 | bphA-xylE-Gmr | This study |
| Burkholderia sp. strain LB400 | Wild type | 10 |
| Burkholderia sp. strain LB400-1 | bphA-lacZ-Gmr | This study |
| Escherichia coli XL1-Blue MR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac | Stratagene |
| Escherichia coli DH5α | endA1 hsdR17(rk− mk−) supE44 thi-1 recA1 gyrA(Nalr) relA1 Δ(lacZYA-argF) U169 deoR[φ80dlac (lacZ)M15] | Gibco BRL |
| Escherichia coli S17-1 | recA pro thi hsdR with integrated RP4-2-tc::Mu-kan::Tn7; Tra+ Trr Smr | 44 |
| Plasmids | ||
| pUC19 | Cloning vector; Ampr | 49 |
| pT7-7 | Cloning vector; T7 promoter; Ampr | 46 |
| pT7-6a | bphAEFG from Burkholderia sp. strain LB400 inserted into multiple cloning site of pT7-6 | 30 |
| pEM1 | SuperCosl cosmid library clone containing bphAEFGBC gene cluster from Cam-1 | This study |
| pEM10 | bphAEFGBC containing 8-kb SacI fragment from pEM1 inserted into SacI site of pUC19 | This study |
| pT7-7a | 4-kb BamHI fragment PCR amplified from pEM10 inserted into BamHI site of pT7-7 | This study |
| pEX100T | sacB conjugable plasmid for gene replacement; Ampr | 43 |
| pX1918GT | Plasmid containing the xylE-gentamicin resistance cassette; Ampr Gmr | 43 |
| pUCGm | Plasmid containing the gentamicin resistance cassette; Ampr Gmr | 42 |
| pIND/lacZ | Plasmid containing lacZ, Ampr, Neor | Invitrogen |
| pEM2 | 4-kb BamHI fragment from pT7-7a inserted into SmaI site of pEX100T | This study |
| pEM20 | 877-bp XbaI fragment from pUCGm inserted into XbaI site of pIND/lacZ | This study |
| pEM21 | 4-kb PmeI fragment from pEM20 inserted into EcoRI site of pEM2 | This study |
Chemicals.
Biphenyl (99%), (±)-camphor (96%), (s)-(+)-carvone (96%), beta-citronellol (95%), cumene (99%), p-cymene (99%), anthracene (99%), benzoate (99%), fluorene (99%), naphthalene (99%), 2-methylnaphthalene (97%), 1,4-dimethylnaphthalene (95%), and phenanthrene (99.5%) were obtained from Aldrich Chemical Co. (+)-Limonene (97%), linoleic acid (60%), myricetin (85%), naringenin (95%), (+)-(α)-pinene (99%), salicylic acid (99%), and o-nitrophenyl-beta-d-galactopyranoside were obtained from Sigma. Benzene (99.9%) and toluene (99.8%) were obtained from Fisher Scientific. 2-Chlorobiphenyl (99%), 3-chlorobiphenyl (99%), 4-chlorobiphenyl (99%), 2,2′-dichlorobiphenyl (99%), 4,4′-dichlorobiphenyl (99%), and Aroclor 1242 (99%) were obtained from AccuStandard.
Cloning bph genes from strain Cam-1.
Total genomic DNA was isolated from strain Cam-1 by using hexadecyltrimethylammonium bromide (4) and was partially digested with Sau3A. The partially digested DNA was size fractionated with a 10 to 40% linear sucrose gradient. DNA fragments approximately 20 kb long were cloned into SuperCos by following the instructions of the manufacturer (Stratagene). In vitro packaging of the recombinant molecules was performed with GigapackII Gold packaging extract (Stratagene), and packaging reactions were used to infect E. coli XL1-Blue MR. The resulting cosmid library was amplified and screened for production of 2-hydroxy-6-oxo-6-phenyl-hexa-2,4-dienoic acid (a yellow meta-cleavage product) from biphenyl (32). Removal of biphenyl by yellow colonies was confirmed by adding 25 mg of biphenyl per liter to cell suspensions of the clones and then extracting the remaining biphenyl with hexane after incubation and analyzing the extracts by gas chromatography-mass spectrometry (34). Cosmid pEM1 was obtained from constructs that transformed biphenyl. Restriction fragments of cosmid pEM1 were separated on an agarose gel, transferred to a maximum-strength Nytran Plus nylon membrane (S&S Nytran Plus), and then hybridized with 32P-labeled bphA, bphF, and bphG from pT7-6a (Table 1). A nick translation system from Life Technologies was used to label bphA and bphG with [α-32P]dCTP. An Oligolabelling Kit from Pharmacia Biotech (Uppsala, Sweden) was used to label bphF with [α-32P]dCTP. A SacI restriction fragment that hybridized to all three probes was subcloned into pUC19, giving pEM10. The sequence of the cloned DNA from Cam-1 was obtained by generating successive unidirectional deletions of pEM10 with the double-stranded nested-deletion system from Pharmacia Biotech. Oligonucleotide primers synthesized at the Nucleic Acid and Protein Services Unit of the University of British Columbia were used to sequence DNA regions not covered by the deletions. DNA sequences were determined by the Nucleic Acid and Protein Services Unit by using AmpliTaq FS dyedeoxy terminator cycle sequencing chemistry (Applied Biosystems) and Centri-Sep columns (Princeton Separation, Adelphia, N.J.) to purify the extension products. ClustalX was used to align the cloned Cam-1 DNA sequence with the bph operon sequence from LB400 (Fig. 1). Vent polymerase (New England Biolabs) and PCR primers with 5′ extensions containing BamHI recognition sites were used to subclone bphAEFG from pEM10 into pT7-7, which yielded pT7-7a.
Insertion of a lacZ-Gmr cassette into the bph operon.
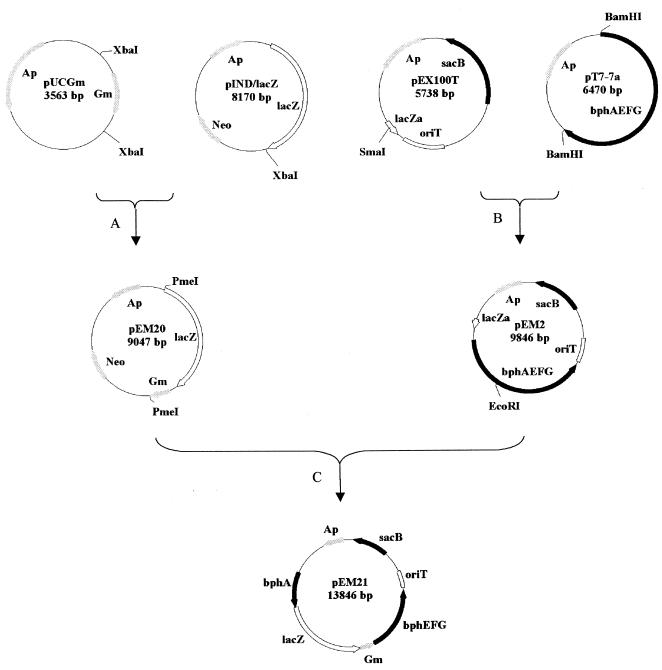
The pEX100T gene replacement vector (42, 43) was used to insert a selectable lacZ reporter gene cassette between the bphA and bphE genes in Cam-1 and LB400 (Fig. 2). Plasmids pEM2, pEM20, and pEM21 were selected in E. coli DH5α grown on LB medium containing appropriate antibiotics and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Plasmid pEM21 was transformed into the mobilizer strain E. coli S17-1 and conjugally transferred into Cam-1 and LB400 (24). Cam-1 and LB400 grow on pyruvate; however, E. coli S17-1 does not. Therefore, exconjugants were plated on minimal medium containing 9 mM pyruvate and 10 μg of gentamicin per ml, and colonies which appeared on this medium after 48 h of incubation at 30°C were streaked onto LB agar containing 10 μg of gentamicin per ml. Colonies of Cam-1 and LB400 in which double homologous recombination had occurred were selected on LB medium containing 10 μg of gentamicin per ml and 5% sucrose and were designated Cam-10 and LB400-1, respectively. Sucrose-resistant colonies were also sensitive to ampicillin, indicating that these colonies had lost the pEX100T vector-associated sequences. Gene insertions in Cam-10 and LB400-1 were also confirmed by performing colony PCR with 20-mer primers annealing to the 3′ region of bphA (5′-GACCTGGCAGAACAGCGACT) and the 5′ regions of bphE (5′-TCTGCACATGCACGTCCAGC-3′) and the lacZ reporter gene (5′-GTATCGCTCGCCACTTCAAC-3′) (50).
FIG. 2.
Construction of a selectable lacZ reporter cassette and pEM21. Step A was construction of pEM20. The 877-bp XbaI fragment from pUCGm was gel purified and ligated into the XbaI site of pIND/lacZ. Restriction digests obtained with EcoRV were used to isolate plasmids containing lacZ and the gene encoding gentamicin acetyltransferase 3-1 in the same transcriptional orientation. Step B was construction of pEM2. The 4-kb BamHI fragment from pT7-7a was treated with the large fragment of DNA polymerase I (PolK) and then ligated into the SmaI site of pEX100T. Restriction digests obtained with SacI and SacII were used to isolate plasmids containing lacZα and bphAEFG in the same transcriptional orientation. Step C was construction of pEM21. The 4-kb PmeI fragment of pEM20 was gel purified and ligated into the PolK-treated EcoRI site of pEM2. Colonies containing bphAEFG and lacZ-Gmr in the same transcriptional orientation were detected by formation of a blue color when the organisms were grown on LB medium supplemented with gentamicin, ampicillin, and X-Gal. The locations of restriction sites and genes and their transcriptional orientations are shown. Ap, β-lactamase-encoding gene; Gm, gentamicin acetyltransferase 3-1-encoding gene; Neo, neomycin resistance gene; oriT, origin of transfer.
To verify that Cam-1 requires the bph genes for biphenyl degradation, we inserted the xylE-Gmr cassette from pX1918GT between the chromosomal bphA and bphE genes in Cam-1 to form Cam-20. The xylE-Gmr cassette contains a transcriptional termination sequence downstream of the gentamicin resistance gene. Consequently, transcription of genes downstream of the cassette is inhibited. The method used to generate Cam-20 was similar to that used to generate Cam-10, except that pEM20 was replaced by pX1918GT and the xylE-Gmr cassette was ligated as an EcoRI fragment to the EcoRI site in pEM2. The resulting plasmid was transformed into the mobilizer strain E. coli S17-1 and conjugally transferred into Cam-1 (24). Exconjugants were selected as described above. Gene insertions in Cam-20 were confirmed by performing colony PCR with primers annealing to the 3′ region of bphA (5′-GCCGGCACAACATCC) and the 5′ region of bphB (5′-CCAGCTCTGCAAGGCGC-3′) (50).
Beta-galactosidase assays.
Unless otherwise specified, Cam-10 and LB400-1 were grown at 30°C on 9 mM pyruvate in the presence of 10 μg of gentamicin per ml to the mid-log phase and then cooled on ice for 15 min. Cultures were centrifuged at 5,000 × g for 15 min at 4°C and washed with mineral buffer. Washed cells were suspended in mineral medium with 1 mM pyruvate and adjusted to a final optical density at 600 nm of 0.6. Cell suspensions (20 ml) were prepared in 125-ml Erlenmeyer flasks and then were inoculated with potential inducers of the bphA gene at concentrations of 0.001 to 1 mM. Unless otherwise specified, triplicate cell suspensions were incubated with potential inducers for 3 h at 30°C on a rotary shaker at 200 rpm. Beta-galactosidase assays were performed as described by Miller (35). Precise volumes of chloroform (20 μl) and 0.1% sodium dodecyl sulfate (10 μl) were used to permeabilize cells (25). Test samples without o-nitrophenyl-β-d-galactopyranoside were used as negative controls. Protein concentrations of cell suspensions were determined by a bicinchoninic acid protein assay (4).
Biphenyl removal by Cam-10 and LB400-1.
Cell suspensions of Cam-10 and LB400-1 were prepared as described above, and then 2.5-ml aliquots were transferred to Teflon-lined screw-cap tubes. Duplicate cell preparations were inoculated with 0.1 mM biphenyl and then incubated on a tube roller at 30°C for 3 or 6 h. Boiled cells and mineral medium containing 0.1 mM biphenyl were used as negative controls. The remaining biphenyl was extracted from cell suspensions with hexane, and extracts were analyzed by gas chromatography as described previously (34).
Nucleotide sequence accession number.
The Cam-1 nucleotide sequence determined in this study has been deposited in the GenBank database under accession no. AY027651.
RESULTS AND DISCUSSION
Optimization of lacZ reporter gene expression in Cam-10 and LB400-1.
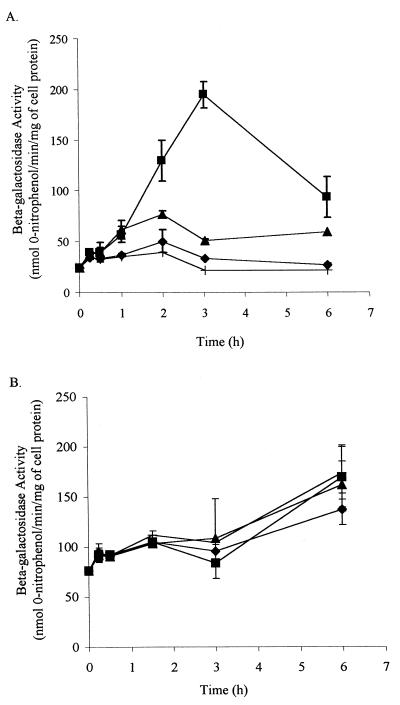
Maximum expression of the lacZ reporter gene in Pseudomonas sp. strain Cam-10 was observed after 3 h of incubation with 1 mM biphenyl (Fig. 3A). Other compounds that were studied to determine their abilities to induce beta-galactosidase activity in Cam-10 were tested under these conditions. Expression of the lacZ reporter gene in Cam-10 did not increase as the amount of biphenyl was increased above 1 mM. At concentrations of biphenyl less than 0.1 mM, beta-galactosidase activity was consistently higher when 1 mM pyruvate was also supplied. Pyruvate may provide cells with energy that allows greater beta-galactosidase production. Thus, unless otherwise stated, 1 mM pyruvate was added to all subsequent preparations in which potential inducers of bphA were tested.
FIG. 3.
Induction of beta-galactosidase activity at 30°C in Cam-10 (A) and LB400-1 (B). The error bars indicate standard deviations (n = 3). The treatments consisted of 1 mM pyruvate alone (+), 1 mM pyruvate plus 0.01 mM biphenyl (⧫), 1 mM pyruvate plus 0.33 mM biphenyl, (▴), and 1 mM pyruvate plus 1 mM biphenyl (■).
At each concentration of biphenyl tested, the beta-galactosidase specific activity of Cam-10 initially increased with time and then decreased (Fig. 3A). This result suggested that biphenyl was depleted by Cam-10, thereby diminishing the concentration of the inducer. The utilization of biphenyl by Cam-10 was not surprising since the lacZ-Gmr cassette did not contain a transcription termination sequence and was inserted between the bphA and bphE genes in Cam-1 without disrupting either gene. To verify that Cam-10 transformed biphenyl, 0.1 mM biphenyl was added to cell suspensions of pyruvate-grown Cam-10, and then the cell suspensions were incubated at 30°C. After 3 and 6 h of incubation, 30 and 100% of the biphenyl added to cell suspensions of Cam-10 was removed, respectively. Biphenyl was not removed by killed cells or from medium without cells. Cam-10 also grew on 1 mM biphenyl. Biphenyl degradation by Cam-10 requires the bph gene products, since insertion of the transcription termination sequence containing the xylE-Gmr cassette from pX1918GT between bphA and bphE resulted in cells unable to grow on biphenyl. Thus, there do not appear to be any additional enzyme systems in Cam-1 catalyzing biphenyl degradation.
The observed decrease in beta-galactosidase activity in Cam-10 upon biphenyl depletion is consistent with observations by other workers suggesting that repeated addition of biphenyl to soil microcosms is necessary for PCB biodegradation (5). The decrease in induction of bphA with biphenyl depletion may also explain why pure cultures of certain PCB-degrading bacteria remove more PCBs when cells are growing on biphenyl than when resting cells are used (33). Interestingly, although the solubility of biphenyl is approximately 0.044 mM (19), greater induction of beta-galactosidase activity was consistently observed in cell suspensions of Cam-10 containing 1 mM biphenyl than in cell suspensions containing 0.33 mM biphenyl (Fig. 3A). This result suggests that bacteria may use biphenyl via direct contact with the crystals instead of, or in addition to, via uptake of dissolved biphenyl.
In contrast to induction of beta-galactosidase activity in Cam-10, the level of beta-galactosidase activity in Burkholderia sp. strain LB400-1 did not depend on the presence of biphenyl (Fig. 3B). This suggests that regulation of the bphA gene in LB400 is constitutive. A gradual increase in beta-galactosidase specific activity over time was consistently observed. This result may reflect recovery from a decrease in beta-galactosidase activity during harvesting and preparation of cell suspensions of LB400-1. Like Cam-10, LB400-1 completely transformed 0.1 mM biphenyl after 6 h. Biphenyl degradation by LB400-1 is believed to require the bph gene products, since many attempts by other workers to find more than one biphenyl dioxygenase in LB400 have not been successful (26).
Generally, PCB-degrading bacteria are prepared for bioaugmentation of PCB-contaminated soil by growing the bacteria on biphenyl. The rates of growth and the final cell densities of bacteria are often lower when the organisms are grown on biphenyl than when they are grown on certain alternative substrates, such as pyruvate. Our results demonstrate that a PCB-degrading bacterium can be grown on pyruvate (or a cheaper substrate) quickly and to high optical densities and then induced within hours to remove biphenyl. Thus, it is possible that this method can be used to prepare bacterial inocula for bioremediation of PCB-contaminated soil, particularly in cases where the bacterial inoculum is defined and where catabolic genes are located on chromosomes rather than on plasmids which can be lost during growth on substrates other than biphenyl (20, 29). However, it may be important to determine the effect of biphenyl on other physiological parameters, such as membrane composition, and to determine how these parameters affect PCB biodegradation.
Inducers of beta-galactosidase activity in Cam-10.
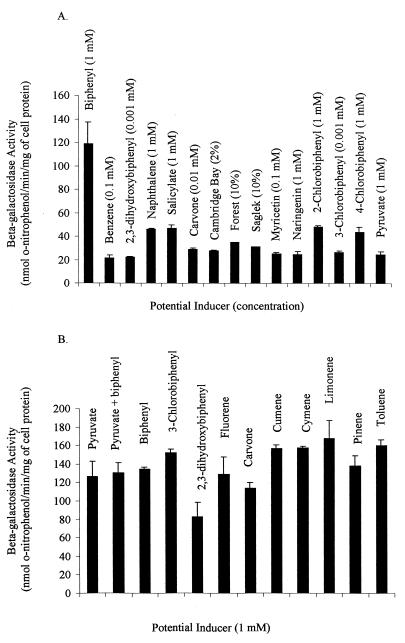
Biphenyl induced beta-galactosidase activity in Cam-10 to a level approximately six times greater than the basal level of expression in cells grown with pyruvate (Fig. 4A). At a concentration of 1 mM, 2-chlorobiphenyl, 4-chlorobiphenyl, salicylate, and naphthalene induced beta-galactosidase activity to levels greater than the basal levels. Thus, these compounds appear to be inducers of bphA in Cam-1. However, none of them appears to be as strong an inducer as biphenyl, as none of them induced beta-galactosidase to the same level of activity in Cam-10 as biphenyl did. At noninhibitory concentrations (Table 2), none of the other potential inducers tested induced beta-galactosidase activity to levels greater than the basal levels in Cam-10. The levels of beta-galactosidase activity after exposure to benzene, carvone, 3-chlorobiphenyl, and pyruvate (Fig. 4A) were typical of those observed after exposure to other noninducing aromatic compounds, terpenoids, chlorobiphenyls, sugars, alcohols, and organic acids (data not shown) (the compounds tested are listed in Table 2).
FIG. 4.
Induction of beta-galactosidase activity for 3 h at 30°C in Cam-10 (A) and LB400-1 (B). The error bars indicate standard deviations (n = 3). The concentrations of potential inducers used to test induction of beta-galactosidase activity in Cam-10 are indicated in parentheses. All preparations were supplemented with 1 mM pyruvate.
TABLE 2.
Percentages of inhibition of beta-galactosidase activity in Cam-10 by potential inducers at various concentrationsa
| Potential inducer | % Inhibition at the following concn:
|
|||
|---|---|---|---|---|
| 1 mM | 0.1 mM | 0.01 mM | 0.001 mM | |
| Aromatic compounds | ||||
| Benzene | 98 | 0 | —b | — |
| Toluene | 97 | 70 | 5 | — |
| Benzoate | 51 | 0 | — | — |
| Catechol | 100 | 64 | 15 | 0 |
| 2,3-Dihydroxybiphenyl | 100 | 70 | 52 | 2 |
| Acenaphthalene | 50 | 19 | 0 | — |
| Fluorene | 100 | 69 | 68 | 10 |
| Dioxin | 100 | 100 | 85 | 43 |
| 2-Methylnaphthalene | 100 | 58 | 0 | — |
| 1,4-Dimethylnaphthalene | 100 | 67 | 0 | — |
| Anthracene (crystals) | 41 | 0 | — | — |
| Phenanthrene | 100 | 37 | 0 | — |
| Terpenoids | ||||
| Camphor | 100 | 55 | 10 | — |
| (s)-(+)-Carvone | 99 | 76 | 0 | — |
| beta-Citronellol | 95 | 70 | 5 | — |
| Cumene | 99 | 0 | 0 | — |
| p-Cymene | 100 | 56 | 8 | — |
| Dehydroabietic acid | 45 | 0 | — | |
| (+)-Limonene | 97 | 33 | 24 | — |
| Linoleic acid | 81 | 0 | — | — |
| Pinene | 88 | 49 | 0 | — |
| Soil extract and flavenoids | ||||
| Cambridge Bay soil (10%, 2%) | 100 | 30 | — | — |
| Forest soil (10%) | 52 | 0 | — | — |
| Saglek soil (10%) | 55 | 0 | — | — |
| Myricetin | 90 | 90 | — | — |
| Chlorinated biphenyls | ||||
| 3-Chlorobiphenyl | 100 | 100 | 32 | 0 |
| 2,2′-Dichlorobiphenyl | — | 20 | — | — |
| 4,4′-Dichlorobiphenyl | — | 0 | — | — |
| Aroclor 1242 (100 ppm) | 60 | 23 | — | — |
Inhibition of beta-galactosidase activity in Cam-10 by potential inducers was determined by incubating cells with 1 mM pyruvate plus 1 mM biphenyl plus the potential inducer and then determining beta-galactosidase activity. Percentages of inhibition were determined by comparing the beta-galactosidase activities in cell suspensions containing pyruvate and biphenyl without a potential inducer to the activities in cell suspensions containing pyruvate and biphenyl plus the potential inducers.
—, Not measured.
Since Cam-10 grew with naphthalene and since salicylate is a metabolite of naphthalene degradation, it is possible that the observed induction by naphthalene of beta-galactosidase activity in Cam-10 was due to salicylate or its catabolites (41). Induction of beta-galactosidase activity by salicylate is consistent with the observation that certain Pseudomonas species readily oxidize biphenyl when they are grown on salicylate and readily oxidize salicylate when they are grown on biphenyl (21).
Other workers have proposed that naphthalene could be used as a growth substrate for PCB-degrading bacteria (40). Naphthalene is a natural component of soil and has been used in solvents and motor oils. Consequently, naphthalene frequently occurs as a co-contaminant at PCB-contaminated sites (31). Pellizari et al. (40) found that bacteria isolated on biphenyl remove more PCBs than bacteria isolated on naphthalene. In these experiments, PCB removal was assayed with resting cells of bacteria grown on the substrate used for isolation. Our results are consistent with the findings of Pellizari et al. (40) since naphthalene induced bphA in Cam-1, although at lower levels than biphenyl did. As has been proposed previously (40), the stimulatory effect of naphthalene on PCB degradation may be sufficient for PCB bioremediation in cases where extensive initial dechlorination has occurred.
Inducers of beta-galactosidase activity in LB400-1.
In contrast to the results obtained with Cam-10, the beta-galactosidase activities in cell suspensions of LB400-1 containing 1 mM biphenyl, carvone, cumene, cymene, pinene, limonene, fluorene, 3-chlorobiphenyl, or toluene were similar to the beta-galactosidase activity observed in cell suspensions containing only pyruvate (Fig. 4B). Interestingly, 1 mM 2,3-dihydroxybiphenyl had a slight inhibitory effect on induction of beta-galactosidase activity in LB400-1 (Fig. 4B). These results are consistent with S1 nuclease mapping studies of bph genes in LB400 done by other workers which identified three transcriptional initiation sites (16). Activation from the promoter region furthest upstream from the biphenyl dioxygenase translation start site (p3) is dependent on biphenyl. However, activation from the two proximal promoter regions (p2 and p1) is constitutive (16).
Despite constitutive expression of the bph genes in LB400 from p1 and p2, Mondello (36) showed that LB400 grown on biphenyl is able to degrade di-para-substituted PCBs and tetra- and pentachlorobiphenyls more effectively than LB400 grown on succinate or on biphenyl plus succinate. However, Brazil et al. (12) observed that expression of bphC, which is located downstream of p1, p2, and bphA and encodes 2,3-dihydroxybiphenyl 1,2-dioxygenase, was similar in LB400 grown on mineral medium supplemented with succinate and in LB400 grown in mineral medium supplemented with biphenyl. To examine the effect of pyruvate plus biphenyl on bphA gene induction in LB400, we compared the beta-galactosidase activities in cell suspensions of LB400-1 containing pyruvate alone, pyruvate plus biphenyl, and biphenyl alone. Similar levels of beta-galactosidase activity were detected for all treatments (Fig. 4B). These results suggest that bphA and bphC are coordinately and constitutively expressed. Constitutive expression of genes encoding the initial enzymes for biphenyl degradation by LB400 may explain why LB400 grown on glucose, glycerol (9), or terpenoid compounds (15) removes PCBs. It would be interesting to determine if regulation of the bph genes in other organisms that have been shown to be induced for PCB removal when they are grown on substrates other than biphenyl (9, 15, 27) is constitutive.
The activation of p3 by biphenyl in LB400 is correlated with increased efficiency of degradation of certain PCBs (36). Transcriptional activation from p3 results in transcription of orf0 (16). We verified the presence of orf0 in LB400 by PCR. The translation product of orf0 is 58% similar to BphS, a GntR-like negative regulator of bph genes in Ralstonia eutropha A5 (37). Recently, other investigators found that in the PCB degrader Pseudomonas pseudoalcaligenes KF707 the translation product of orf0 is autoregulated and is necessary for expression of genes encoding enzymes in the biphenyl degradation pathway downstream of bphC (48). Induction of genes downstream of bphC allows cells to grow on biphenyl and minimizes the accumulation of metabolites resulting from biphenyl and PCB catabolism. Decreased accumulation of metabolites from PCB transformation may explain why LB400 grown on biphenyl degrades di-para-substituted PCBs and tetra- and pentachlorobiphenyls more effectively than LB400 grown on other substrates (36). Also, other physiological effects of growth on biphenyl, such as changes in membrane composition, may be necessary for transformation of certain PCB congeners.
Inhibition effects of potential inducers.
At a concentration of 1 mM, most of the potential inducers tested actually inhibited induction by biphenyl of beta-galactosidase activity in Cam-10 (Table 2). The exceptions were compounds previously found to be inducers, naphthalene, salicylate, 2-chlorobiphenyl, and 4-chlorobiphenyl, as well as naringenin, fructose, glucose, and glycerol. With the exception of benzoate, acenaphthalene, fluorene, dioxin, anthracene, 3-chlorobiphenyl, 2-methylnaphthalene, and dimethylnaphthalene, compounds that inhibited induction also inhibited cell growth. At concentrations less than 0.1 mM, none of the potential inducers were inhibitory to cell growth, yet at such concentrations several of these compounds substantially inhibited induction of beta-galactosidase. Clearly, in complex environments, inhibitory effects such as those found here can be expected to modulate expression of genes essential for PCB biodegradation. The inhibitory effect of soil extracts (Table 2) is consistent with this expectation.
Metabolites of several potential inducers may have had a role in inhibition of beta-galactosidase induction in Cam-10. Transformation of 3-chlorobiphenyl by Cam-10 to 3-chlorocatechol was apparent from the formation of black catecholic polymers in cell suspensions (13). Since 3-chlorocatechol is a potent inhibitor of 2,3-dihydroxybiphenyl 1,2-dioxygenase (18, 45), inhibition of beta-galactosidase induction in Cam-10 by 3-chlorobiphenyl may result from negative regulation by accumulated metabolites. Cam-10 rapidly transformed 2,3-dihydroxybiphenyl to the meta-cleavage product, as indicated by production of a bright yellow metabolite. Thus, inhibition of beta-galactosidase induction in Cam-10 by 2,3-dihydroxybiphenyl may also result from negative regulation by accumulated metabolites. Fluorene, catechol, dioxin, and 2-methylnaphthalene were also transformed by Cam-10, as indicated by the production of colored metabolites. Interestingly, the compounds that were transformed by Cam-10 include the most potent inhibitors of beta-galactosidase induction (Table 2). Detailed biochemical studies will be necessary to determine if inhibition of bphA induction by particular compounds involves negative genetic regulation.
Temperature dependence of bphA induction in Cam-1 and LB400.
Cam-1 was isolated from PCB-contaminated arctic soil and was studied to determine the feasibility of bioremediating PCB-contaminated arctic soil with indigenous soil bacteria. We found that at 7°C Cam-1 removed PCBs at higher rates than LB400 removed PCBs (34). To investigate the role of bphA induction in the efficiency of PCB removal at low temperatures, we compared the beta-galactosidase activities in cell suspensions of Cam-10 and LB400-1 incubated at 7°C with pyruvate or biphenyl plus pyruvate. Cell suspensions of Cam-10 were prepared by using cells grown on pyruvate at 7°C. Since LB400 does not grow at 7°C (34), cell suspensions of LB400-1 were prepared by using cells grown on pyruvate at 15°C. Samples of the cell suspensions were obtained at several time points over 24 h and transferred to 28°C to measure beta-galactosidase activity.
After 24 h, the beta-galactosidase activity of Cam-10 cells incubated at 7°C with biphenyl was four times greater than that of cells incubated at 7°C with pyruvate (Table 3). Thus, bphA appears to be induced by biphenyl in Cam-1 at 7°C, which is consistent with Cam-1 being cold adapted. Interestingly, the initial beta-galactosidase activity was significantly less in LB400-1 cells grown at 15°C than in LB400-1 cells grown at 30°C (Table 3). As observed at 30°C, biphenyl did not induce beta-galactosidase activity in cell suspensions of LB400-1 at 7°C. These results further support the conclusion that bphA expression in LB400 is constitutive and indicate that the level of constitutive expression is temperature dependent.
TABLE 3.
Effect of temperature on induction of beta-galactosidase activities in Cam-10 grown at 7°C on pyruvate and in LB400-1 grown at 15°C on pyruvate
| Strain | Time | Beta-galactosidase activity (nmol of o-nitrophenol/min/mg of cell protein)a
|
|
|---|---|---|---|
| 7°C | 30°C | ||
| Cam-10 | Initialb | 18.0 ± 2.6 | 24.0 ± 0.6 |
| Final | 74.0 ± 8.3c | 118.6 ± 18.5d | |
| LB400-1 | Initialb | 15.5 ± 4.0 | 76.3 ± 4.5 |
| Final | 16.8 ± 1.0c | 83.9 ± 0.7d | |
Mean ± standard deviation (n = 3).
Activity at zero time.
Activity at 24 h.
Activity at 36 h.
Our research shows that regulation of the bphA gene is remarkably different in two PCB-degrading bacteria. The bphA gene in Cam-1 is inducible at 7 and 30°C, and induction is greatest with biphenyl. In contrast, expression of bphA in LB400 is constitutive and is lower at a lower temperature. These results indicate that available chemical inducers, as well as physical environmental conditions, can affect bphA expression in PCB-degrading bacteria. Consequently, knowledge of how physical and chemical environmental variables affect bphA induction in particular bacteria in a treatment system will be necessary to determine the optimal conditions for PCB bioremediation.
ACKNOWLEDGMENTS
We thank V. J. J. Martin and L. D. Eltis for helpful discussions and L. D. Eltis for providing Burkholderia strain LB400, pT7-7, and pT7-6a.
This research was supported by the Natural Science and Engineering Research Council of Canada, the Canadian Department of National Defense, and a Natural Science and Engineering Research Council graduate scholarship to E.R.M.
REFERENCES
- 1.Abramowicz D A. Aerobic and anaerobic biodegradation of PCBs: a review. Crit Rev Biotechnol. 1990;10:241–251. [Google Scholar]
- 2.Ahmad D, Masse R, Sylvestre M. Cloning and expression of genes involved in 4-chlorobiphenyl transformation by Pseudomonas testosteroni: homology to polychlorobiphenyl-degrading genes in other bacteria. Gene. 1990;86:53–61. doi: 10.1016/0378-1119(90)90113-6. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed M, Focht D D. Degradation of polychlorinated biphenyls by two species of Achromobacter. Can J Microbiol. 1972;19:47–52. doi: 10.1139/m73-007. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 2nd ed. New York, N.Y: Greene Publishing Associates and John Wiley & Sons; 1992. [Google Scholar]
- 5.Barriault D, Sylvestre M. Factors affecting PCB degradation by an implanted bacterial strain in soil microcosms. Can J Microbiol. 1993;39:594–602. doi: 10.1139/m93-086. [DOI] [PubMed] [Google Scholar]
- 6.Bedard D L, Unterman R, Bopp L H, Brennan M J, Haberl M L, Johnson C. Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl Environ Microbiol. 1986;51:761–768. doi: 10.1128/aem.51.4.761-768.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bedard D L, Haberl M L. Influence of chlorine substitution pattern on the degradation of polychlorinated biphenyls by eight bacterial strains. Microb Ecol. 1990;20:87–102. doi: 10.1007/BF02543870. [DOI] [PubMed] [Google Scholar]
- 8.Bedard D L, Wagner R E, Brennan M J, Haberl M L, Brown J F., Jr Extensive degradation of Aroclors and environmentally transformed polychlorinated biphenyls by Alcaligenes eutrophus H850. Appl Environ Microbiol. 1987;53:1094–1102. doi: 10.1128/aem.53.5.1094-1102.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Billingsley K A, Backus S M, Juneson C, Ward O P. Comparison of the degradation patterns of polychlorinated biphenyl congeners in Aroclors by Pseudomonas strain LB400 after growth on various carbon sources. Can J Microbiol. 1997;43:1172–1179. doi: 10.1139/m97-166. [DOI] [PubMed] [Google Scholar]
- 10.Bopp L H. Degradation of highly chlorinated PCBs by Pseudomonas strain LB400. J Ind Microbiol. 1986;1:23–29. [Google Scholar]
- 11.Boyle A W, Silvin C J, Hassett J P, Nakas J P, Tanenbaum S W. Bacterial PCB biodegradation. Biodegradation. 1992;3:285–298. [Google Scholar]
- 12.Brazil G M, Kenefick L, Callanan M, Haro A, de Lorenzo V, Dowling D N, O'Gara F. Construction of a rhizosphere pseudomonad with potential to degrade polychlorinated biphenyls and detection of bph gene expression in the rhizosphere. Appl Environ Microbiol. 1995;61:1946–1952. doi: 10.1128/aem.61.5.1946-1952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenner V, Arensdorf J J, Focht D D. Genetic construction of PCB degraders. Biodegradation. 1994;5:359–377. doi: 10.1007/BF00696470. [DOI] [PubMed] [Google Scholar]
- 14.Brunner W, Sutherland F H, Focht D D. Enhanced biodegradation of polychlorinated biphenyls in soil by analog enrichment and bacterial inoculation. J Environ Qual. 1985;14:324–328. [Google Scholar]
- 15.Donnelly P K, Hedge R S, Fletcher J S. Growth of PCB-degrading bacteria on compounds from photosynthetic plants. Chemosphere. 1994;28:981–988. [Google Scholar]
- 16.Erickson B D, Mondello F J. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J Bacteriol. 1992;174:2903–2912. doi: 10.1128/jb.174.9.2903-2912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Focht D D, Brunner W. Kinetics of biphenyl and polychlorinated biphenyl metabolism in soil. Appl Environ Microbiol. 1985;50:1058–1063. doi: 10.1128/aem.50.4.1058-1063.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Focht D D. Strategies for the improvement of aerobic metabolism of polychlorinated biphenyls. Curr Biol. 1995;6:341–346. [Google Scholar]
- 19.Foreman W T, Bidleman T F. Vapor pressure estimates of individual polychlorinated biphenyls and commercial fluids using gas chromatographic retention data. J Chromatogr. 1985;330:203–216. [Google Scholar]
- 20.Furukawa K, Chakrabarty A M. Involvement of plasmids in total degradation of chlorinated biphenyls. Appl Environ Microbiol. 1982;44:619–626. doi: 10.1128/aem.44.3.619-626.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furukawa K, Simon J R, Chakrabarty A M. Common induction and regulation of biphenyl, xylene/toluene, and salicylate catabolism in Pseudomonas paucimobilis. J Bacteriol. 1983;154:1356–1362. doi: 10.1128/jb.154.3.1356-1362.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furukawa K, Miyazaki T. Cloning of a gene cluster encoding biphenyl and chlorobiphenyl degradation in Pseudomonas pseudoalcaligenes. J Bacteriol. 1986;166:392–398. doi: 10.1128/jb.166.2.392-398.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furukawa K. Molecular genetics and evolutionary relationship of PCB-degrading bacteria. Biodegradation. 1994;5:289–300. doi: 10.1007/BF00696466. [DOI] [PubMed] [Google Scholar]
- 24.Gerhardt P, Murray R G E, Wood W A, Krieg N R. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. [Google Scholar]
- 25.Giacomini A, Corich V, Ollero F J, Squartini A, Nuti M P. Experimental conditions may affect reproducibility of the beta-galactosidase assay. FEMS Microbiol Lett. 1992;100:87–90. doi: 10.1111/j.1574-6968.1992.tb14024.x. [DOI] [PubMed] [Google Scholar]
- 26.Gibson D T, Cruden D L, Haddock J D, Zylstra G J, Brand J M. Oxidation of polychlorinated biphenyls by Pseudomonas sp. strain LB400 and Pseudomonas pseudoalcaligenes KF707. J Bacteriol. 1993;175:4561–4564. doi: 10.1128/jb.175.14.4561-4564.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert E S, Crowley D E. Plant compounds that induce polychlorinated biphenyl biodegradation by Arthrobacter sp. strain B1B. Appl Environ Microbiol. 1997;63:1933–1938. doi: 10.1128/aem.63.5.1933-1938.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernandez B B, Koh S-C, Chial M, Focht D D. Terpene-utilizing isolates and their relevance to enhance biotransformation of polychlorinated biphenyls in soil. Biodegradation. 1997;8:153–158. [Google Scholar]
- 29.Higson F K. Microbial degradation of biphenyls and its derivatives. Adv Appl Microbiol. 1992;37:135–164. doi: 10.1016/s0065-2164(08)70254-5. [DOI] [PubMed] [Google Scholar]
- 30.Hofer B, Eltis L D, Dowling D N, Timmis K N. Genetic analysis of a Pseudomonas locus encoding a pathway for biphenyl/polychlorinated biphenyl degradation. Gene. 1993;130:47–55. doi: 10.1016/0378-1119(93)90345-4. [DOI] [PubMed] [Google Scholar]
- 31.Jones K C, Stratford J A, Waterhouse K S, Vogt N B. Organic contaminants in Welsh soils: polynuclear aromatic hydrocarbons. Environ Sci Technol. 1989;5:540–550. [Google Scholar]
- 32.Kiyohara H, Nagao K, Yana K. Rapid screen for bacteria degrading water-insoluble, solid hydrocarbons on agar plates. Appl Environ Microbiol. 1982;43:454–457. doi: 10.1128/aem.43.2.454-457.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohler H-P E, Kohler-Staub D, Focht D D. Cometabolism of polychlorinated biphenyls: enhanced transformation of Aroclor 1254 by growing bacterial cells. Appl Environ Microbiol. 1988;54:1940–1945. doi: 10.1128/aem.54.8.1940-1945.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Master E R, Mohn W W. Psychrotolerant bacteria isolated from arctic soil that degrade polychlorinated biphenyls at low temperature. Appl Environ Microbiol. 1998;64:4823–4829. doi: 10.1128/aem.64.12.4823-4829.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 36.Mondello F J. Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polychlorinated biphenyl degradation. J Bacteriol. 1989;171:1725–1732. doi: 10.1128/jb.171.3.1725-1732.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mouz S, Merlin C, Spingael D, Toussaint A. A GntR-like negative regulator of the biphenyl degradation genes of the transposon Tn4371. Mol Gen Genet. 1999;262:790–799. doi: 10.1007/s004380051142. [DOI] [PubMed] [Google Scholar]
- 38.Park Y-I, So J-S, Koh S-C. Induction by carvone of the polychlorinated biphenyl (PCB)-degradative pathway in Alcaligenes eutrophus H850 and its molecular monitoring. J Microbiol Biotechnol. 1999;9:804–810. [Google Scholar]
- 39.Parsons J R, Sijm D T H M. Biodegradation kinetics of polychlorinated biphenyls in continuous cultures of Pseudomonas strain. Chemosphere. 1988;17:1755–1766. [Google Scholar]
- 40.Pellizari V H, Bezborodnikov S, Quensen III J F, Tiedje J M. Evaluation of strains isolated by growth on naphthalene and biphenyl for hybridization of genes to dioxygenase probes and polychlorinated biphenyl-degrading ability. Appl Environ Microbiol. 1996;62:2053–2058. doi: 10.1128/aem.62.6.2053-2058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schell M A. Transcriptional control of the nah and sal hydrocarbon-degradation operons by the nahR gene product. Gene. 1985;36:301–309. doi: 10.1016/0378-1119(85)90185-4. [DOI] [PubMed] [Google Scholar]
- 42.Schweizer H P. Small broad-host-range gentamycin resistance cassette for site specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–833. [PubMed] [Google Scholar]
- 43.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 44.Simons R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–790. [Google Scholar]
- 45.Sondossi M, Sylvestre M, Ahmad D. Effects of chlorobenzoate transformation on the Pseudomonas testosteroni biphenyl and chlorobiphenyl degradation pathway. Appl Environ Microbiol. 1992;58:485–495. doi: 10.1128/aem.58.2.485-495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 47.Viallard V, Poirier I, Cournoyer B, Haurat J, Wiebkin S, Ophel-Keller K, Balandreau J. Burkholderia graminis sp. nov., a rhizospheric Burkholderia species, and reassessment of Pseudomonas phenazinium, Pseudomonas pyrrocinia and Pseudomonas glathei as Burkholderia. Int J Syst Bacteriol. 1998;48:549–563. doi: 10.1099/00207713-48-2-549. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe T, Inoue R, Kimura N, Furukawa K. Versatile transcription of biphenyl catabolic bph operon in Pseudomonas pseudoalcaligenes KF707. J Biol Chem. 2000;275:31016–31023. doi: 10.1074/jbc.M003023200. [DOI] [PubMed] [Google Scholar]
- 49.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 50.Zon L I, Dorfman D M, Orkin S H. The polymerase chain reaction colony miniprep. BioTechniques. 1989;7:696–698. [PubMed] [Google Scholar]