Summary
Arbuscular mycorrhizal fungi (AMF) are keystone symbionts of agricultural soils but agricultural intensification has negatively impacted AMF communities. Increasing crop diversity could ameliorate some of these impacts by positively affecting AMF. However, the underlying relationship between plant diversity and AMF community composition has not been fully resolved.
We examined how greater crop diversity affected AMF across farms in an intensive agricultural landscape, defined by high nutrient input, low crop diversity and high tillage frequency. We assessed AMF communities across 31 field sites that were either monocultures or polycultures (growing > 20 different crop types) in three ways: richness, diversity and composition. We also determined root colonization across these sites.
We found that polycultures drive the available AMF community into richer and more diverse communities while soil properties structure AMF community composition. AMF root colonization did not vary by farm management (monocultures vs polycultures), but did vary by crop host.
We demonstrate that crop diversity enriches AMF communities, counteracting the negative effects of agricultural intensification on AMF, providing the potential to increase agroecosystem functioning and sustainability.
Keywords: agricultural diversification, agroecosystem multifunctionality, arbuscular mycorrhizal fungi (AMF), crop diversity, field studies
Introduction
Arbuscular mycorrhizal fungi (AMF) are a key component of the soil microbial community that contribute to the development of healthy soils and agricultural sustainability (Bender et al., 2016). Arbuscular mycorrhizal fungi establish associations with the majority of land plants, including most crops, and provide many ecosystem services to agriculture (Rillig et al., 2016; Bender et al., 2016; Thirkell et al., 2017). However, agricultural intensification, characterized by high nutrient input, low crop diversity and high tillage frequency, reduces the diversity of AMF taxa in agricultural soils (Helgason et al., 1998; Verbruggen & Toby Kiers, 2010; Rillig et al., 2016; Hontoria et al., 2019), compromising the potential functions and benefits of AMF in agricultural landscapes (Gottshall et al., 2017; Manoharan et al., 2017; Xu et al., 2017; de Graaff et al., 2019). In natural systems, a positive relationship between plant and AMF community composition has been well documented (Landis et al., 2004; Hiiesalu et al., 2014; Martínez‐García et al., 2015; Chen et al., 2017) but the underlying mechanisms are still unclear (Kokkoris et al., 2020). Plant communities could filter AMF (Šmilauer et al., 2020) or AMF could be driving plant community composition (Tedersoo et al., 2020). If plant communities can positively shape AMF communities in agricultural systems (Verbruggen & Toby Kiers, 2010), depauperate AMF communities could be bolstered by increasing crop diversity in intensive agricultural landscapes dominated by large areas of monocultures. Understanding whether increasing crop diversity can bolster AMF communities that could benefit sustainable agricultural systems requires a thorough investigation of the underlying mechanisms between crop and AMF diversity in agricultural landscapes.
Intensive agricultural production has often come at high environmental cost to soils, including increased soil erosion, greater nutrient leaching and lower water‐holding capacity (Foley et al., 2005). Ensuring agricultural sustainability requires strategies that prioritize multiple ecosystem services rather than just maximizing production (Kremen & Merenlender, 2018). Arbuscular mycorrhizal fungi are plant symbionts that are an important source of ecosystem services in agricultural landscapes (Gianinazzi et al., 2010; Thirkell et al., 2017; Rillig et al., 2019). Beyond nutrient acquisition (Smith & Read, 2008), AMF are helpful in pathogen protection (Veresoglou & Rillig, 2012; Jung et al., 2012), herbivore resistance (Middleton et al., 2015), drought tolerance (Leigh et al., 2009), nutrient cycling (van der Heijden, 2010) and soil formation and aggregation (Rillig & Mummey, 2006; Wilson et al., 2009). In this way, while AMF are not always tightly linked to increasing crop productivity (Ryan & Graham, 2018; but see Zhang et al., 2019), they are important to overall agroecosystem multifunctionality via ‘system performance and sustainability’ that can reduce negative external inputs (Rillig et al., 2019). Enhancing agroecosystem multifunctionality through AMF will depend in part on the composition of the AMF community (Verbruggen & Toby Kiers, 2010).
Multiple aspects of intensive agriculture adversely alter AMF communities. For example, intensively tilled agricultural soils tend to select for a less diverse, more ruderal AMF community, which includes taxa thought to have fewer mutualistic traits (Chagnon et al., 2013). Intensive tillage and bare fallows also decrease AMF colonization of crops by disrupting hyphal networks and leaving AMF without hosts, respectively (Bowles et al., 2017). Heavy fertilization can create less mutualistic and abundant AMF associations (Johnson et al., 1997; Johnson, 2010) and suppress colonization of roots by AMF. Specifically, high nutrient availability via fertilization decreases the dependency of plant hosts on AMF but selects for AMF that are more aggressive competitors for plant carbohydrates, leading to a net cost to plant hosts (Johnson, 2010; Thirkell et al., 2017). Farm management also influences AMF community composition indirectly via changes in other soil properties, such as soil pH and soil organic carbon (Vályi et al., 2016). For example, adding fertilizer acidifies soils (Geisseler & Scow, 2014) and tilling reduces soil organic carbon, both of which can drive changes in AMF communities (Fitzsimons et al., 2008; Bouffaud et al., 2016; Oehl et al., 2017). Apart from tillage, fertilization and other practices that change soil properties, agricultural soils may also have low numbers of AMF taxa because of the extremely low diversity of plant hosts when crops consist of monocultures in space and/or over time (Burrows & Pfleger, 2002; Oehl et al., 2003; Johnson et al., 2004; Strom et al., 2020).
By contrast, polycultures, which are more similar to biodiverse natural systems, have the potential to positively impact AMF communities by providing a more diverse set of plant hosts. While agriculture continues to shift towards monocultures, polycultures have traditionally been the dominant form of agriculture across many regions in the world (Altieri, 1999; Brooker et al., 2015) and have been promoted as a way to remedy the negative environmental impacts that intensive monoculture agriculture has had on soils (Power, 2010; Li et al., 2014; Iverson et al., 2014; Altieri et al., 2015), especially through AMF associations (Orrell & Bennett, 2013; Brooker et al., 2015). Yet surprisingly little is known about how AMF respond to polycultures (Verbruggen & Toby Kiers, 2010). More generally, the underlying mechanism driving the relationship between AMF and plant diversity in managed or natural ecosystems has not been fully resolved. Observational studies in natural ecosystems cannot differentiate whether AMF diversity supports greater plant diversity or AMF diversity is dependent on plant composition (Lekberg & Waller, 2016; Kokkoris et al., 2020). While AMF are generalists, there is a degree of selectivity in AMF associations with plant hosts (Bainard et al., 2014; Davison et al., 2016; Sepp et al., 2019; Davison et al., 2020), and thus plant hosts could filter AMF communities directly (Šmilauer et al., 2020). Alternatively, plant hosts could impart changes in the soil environment (e.g. pH, carbon, composition) that alter AMF communities indirectly (Vályi et al., 2016; Lekberg & Waller, 2016). Because the suite of plant hosts is carefully managed in agricultural systems, studies in agroecosystems might help to clarify the mechanism underlying the relationship between plant diversity and AMF communities.
Therefore, we sought to understand how AMF communities respond to greater crop diversity (i.e. polycultures) in an intensively managed agricultural landscape. Our study compared AMF communities (richness, diversity and composition) in soil, the legacy of previous management, as well as AMF colonization in roots in monoculture vs polyculture fields. Our experimental design allowed us to investigate the filtering effect of crop diversity (monoculture vs polyculture) on AMF communities, as well as how AMF communities could be influenced by soil properties. To reduce confounding effects of management practices on AMF, all field sites studied used similar tillage regimes and fertilizers, allowing us to focus on management differences in crop diversity (monoculture vs polyculture). Specifically, we asked: how does greater crop diversity via polyculture and its management legacy affect AMF richness and diversity relative to monoculture cropping; to what extent does AMF community composition differ between polyculture and monoculture field sites; how does AMF root colonization differ between crop plants grown in monoculture vs polyculture field sites; and, ultimately, do soil properties impact AMF community composition (richness, diversity, and composition) and colonization? We predicted that despite the legacy of intensive agricultural practices on soils in our study region, greater crop diversity would have a positive effect on richness and diversity. We also expected that the more diverse plant community in polycultures may foster more beneficial, host‐specific AMF taxa, which, in turn, lead to greater AMF root colonization for a given crop host when planted in polyculture rather than monoculture fields (Johnson et al., 2004). Based on previous literature, we also anticipated that soil properties would have an overriding effect on AMF community composition, but that crop diversity still plays an important role in shifting the AMF community. In this way, our study aims to increase our understanding of the relationship between plant diversity and AMF communities and to investigate whether greater crop diversity could support more sustainable agriculture systems via AMF communities.
Materials and Methods
Study system
Farm sites were located in Fresno County in California’s San Joaquin Valley, an agriculturally dominated region containing a wide range of annual and perennial crops, including row crops and orchards. While this region is dominated by large‐scale farms, our research focused on small‐scale farms (< 16.2 ha (40 acres)) embedded in this landscape. Some of these small‐scale farms grow a high diversity of specialty crops (including mycorrhizal and nonmycorrhizal crop plants) together, such as bok choy (Brassica rapachinensis), chard (Beta vulgarisvulgariscicla), Thai peppers (Capsicum annuum), jujube (Ziziphus jujuba) and bitter melon (Momordica charantia) (Molinar, 2012). Farmers frequently rotate these crops over space and time (i.e. ‘polycultures’) in previously intensively monocropped farmed land. Specifically, on polyculture farms, different crop types are planted in close proximity alternating as little as every couple of rows. Other small‐scale farmers grow only one crop per season of squash or other row crops (i.e. ‘monocultures’).
In order to compare monoculture and polyculture management and understand the effect of plant hosts on AMF communities, we selected farms if they grew at least two rows of the same ‘focal’ crop: a summer squash variety (zucchini, Cucurbita pepo L. var. cylindrica) or eggplant (Solanum melongena L. var. esculentum), which both associate with AMF (Smith & Read, 2008). In addition, all farms used conventional tillage and cultivation to prepare beds and synthetic inputs (e.g. chemical fertilizers and pesticides) to some extent but no soil fumigants. Farms were considered polycultures if they grew 20 or more different crop types and monoculture if they grew one crop type at the time of sample collection. The polyculture farms had a range of 7 to over 15 yr in this management whereas monoculture farms, except for one, had been in monoculture management for over 15 yr. Information on site history is provided in Supporting Information Table S1.
We sampled 25 farms (13 monoculture and 12 polyculture) during autumn 2017 (29 September to 17 October 2017) and early summer 2018 (28 May to 14 June 2018; Fig. 1; Table S1). Some eggplant farms were sampled in both years while others were sampled in just one year (2017 or 2018; Table S1), corresponding to peak productivity and the end of the eggplant growing season in this region. All squash farms, which have a shorter growing season, were only sampled in summer 2018. In total, we sampled 31 separate sampling units across both years (2017 and 2018): 11 eggplant polycultures, nine eggplant monocultures, five squash polycultures and five squash monocultures (Table S1).
Fig. 1.
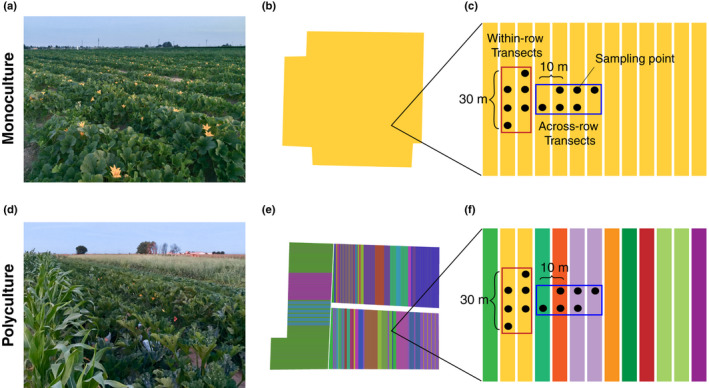
Representative images, row‐level crop maps and sampling scheme of monoculture fields (a–c) and polyculture fields (d–f). The row‐level crop maps (b, e) were hand‐digitized and each color represents a different crop type. The sampling scheme panels for monoculture (c) and polyculture (f) fields illustrate that samples collected from within‐row transects were always the same crop type (i.e. focal crop, eggplant or squash) and from across‐row transects were the same crop type on monocultures or distinct crop types on polycultures.
Sampling scheme
We used the same sampling scheme across all monoculture and polyculture field sites (Fig. 1). We set up two sets of 30 m transects arranged: within‐row and across‐row (Fig. 1). Within‐row transects ran along rows of the focal crop (squash or eggplant), with across‐row transects running perpendicular. For each transect, sampling occurred at three points 10 m apart for a total of three sampling points per transect and 12 sampling points per field site. In total, we collected 372 samples across 31 field sites.
Our sampling scheme was designed to compare AMF community composition on the same crop host (i.e. focal crop, eggplant or squash) whether it was grown in a polyculture or monoculture management through the within‐row transects. We localized our within‐row transects on eggplant or squash to limit plant species (on polyculture field sites) and varietal variation for plant host effects on AMF community composition. This design also allowed us to investigate how the interaction between farm management (polyculture or monoculture) and different crop hosts would impact the AMF community. On polyculture fields, across‐row transects would intersect crop hosts distinct from the focal crop, whereas on monoculture fields, these transects would still intersect the focal crop (eggplant or squash). In this way, we were able to investigate whether the legacy of farm management (monoculture or polyculture) on each field site was sustained across different crop hosts.
Soil and root sampling
Following the sampling scheme described earlier (Fig. 1), we collected root and soil samples to characterize the AMF community on the focal crops, squash and eggplant, and on other crops (polyculture farms). We collected a soil sample from the root zone using an auger (6 cm wide, 20 cm deep), avoiding areas where weeds were present. We separated roots from each soil sample to determine mycorrhizal colonization. However, we were unable to collect enough roots for molecular characterization; we were limited by our sampling because we sampled on working farms not experimental fields and thus were unable to attain farmer permission to harvest whole plants. The rest of the soil sample was divided into a subsample, with all visible roots and rocks removed, for molecular measurements of AMF communities and a subsample for measuring soil properties.
Measurements of soil properties
Soil samples for edaphic measurements were air‐dried and sieved in a 2 mm sieve. To determine total organic carbon (C), nitrogen content (N), and C : N each soil sample was ground and assessed using a combustion elemental analyzer, which is able to separate organic carbon from inorganic carbon (SoliTOC with a nitrogen detector; Elementar, Langenselbold, Germany). Soil texture for each sample was determined using the ‘micropipette’ method (Miller & Miller, 1987). The remaining soil chemical properties were measured at the University of Massachusetts Soil and Plant Testing Facility (Amherst, MA, USA). Soil chemistry data included pH, cation exchange capacity (CEC), and mg kg−1 of total extractable phosphorus (P), potassium (K), calcium (Ca), magnesium (Mg), zinc (Zn), boron (B), manganese (Mn), copper (Cu), iron (Fe), lead (Pb), aluminium (Al), sodium (Na) and sulfur (S).
A ‘soil properties index’ was created using the first principal component (PC1) scores of the principal component analysis (PCA) of all the edaphic measurement data. The PC1 axis explained 27.9% of the variation and had a negative loading for percentage sand and a positive loading for Ca (Fig. S1). The PC2 axis explained 15.5% variation and had a negative loading for Mg and a positive loading for N (Fig. S1).
AMF colonization of roots
We determined the percentage colonization by counting AMF composition in stained roots. Roots were cleared in 10% KOH, acidified in 1% HCl and stained with trypan blue (Koske & Gemma, 1989). Percentage colonization by AMF was determined using the intersections method at ×200 magnification (McGonigle et al., 1990). Arbuscular mycorrhizal fungi colonization in this study refers to percentage colonization by arbuscules, vesicles or hyphae over total intersections counted (c. 100 intersections per sample).
Molecular characterization of AMF communities
Soil samples for molecular measurements were immediately stored at −80°C upon return to the laboratory until DNA extractions could proceed. DNA was extracted from 0.25 g of soil using the DNeasy PowerSoil Kit (Qiagen). Detailed information about the molecular analysis, specifically primer selection, PCR conditions and amplicon library preparation, can be found in the Methods S1. Briefly, the ITS2 rRNA region (5.8Fun/ITS4Fun) was amplified to characterize the communities of fungi. In previous studies, ITS2 primers have also matched well with all lineages in Glomeromycotina (the subphylum AMF belong to Spatafora et al. (2016)) and, in the same study region, they have been successfully used to study fine‐scale patterns of AMF community succession (Gao et al., 2019). The (5.8S) forward and (ITS4) reverse primers contained a 29 (forward) or 25 (reverse) base linker, a 12 base barcode, a 29 (forward) or 34 (reverse) base pad, and a 0–8 base heterogeneity spacer (Fadrosh et al., 2014). Sequencing of amplicon libraries was performed on the Illumina MiSeq platform (Illumina, San Diego, CA, USA) with 300 bp paired‐end reads at the Vincent J. Coates Genomics Sequencing Laboratory (University of California, Berkeley, CA, USA).
The AMPtk pipeline (v.1.4.1) was used to process the fungal sequence data (Palmer et al., 2018). First, the forward and reverse sequences were demultiplexed and the primers were removed. Then, sequences were denoised into exact sequence variants using the Unoise3 algorithm, which removes artificial sequences including predicted sequence errors, contaminants such as putative PhiX carry‐over from Illumina sequencing, and putative chimeric sequences (Edgar, 2016). Next, the resulting exact sequence variants sequences were clustered in biologically relevant operational taxonomic units (OTUs) at 97% sequence similarity using the Uclust (Edgar, 2010) algorithm employed in Vsearch (Rognes et al., 2016). Then to account for index bleed (the percentage of possible reads that bled into other samples), the synthetic mock community was also used to calculate the observed rates of index bleed to remove spurious OTUs using the filter module in the AMPtk pipeline (Palmer et al., 2018). DNA extraction and PCR‐negative controls sequence reads present in samples were also subtracted from each sample (Nguyen et al., 2015). Finally, taxonomy was assigned using the AMPtk ‘last common ancestor’ approach with the combination of global sequence, Utax, and Sintax (Edgar, 2016) alignments against the Unite v.7.2.2 database (Kõljalg et al., 2013).
The resulting OTU table contained 243 AMF OTUs (1093 176 reads; OTUs annotated as Glomeromycota) and 3175 nonAMF fungal OTUs (12 081 866 reads). Raw sequence read files are available in NCBI SRA accession PRJNA650414. To account for sequence read depth variation per sample, the fungal OTU table was normalized by rarefying to equal fungal sequence reads (lowest sampling depth in a sample, 3258 reads), using the ‘rrarefy’ command in vegan in R (R Core Team, 2020) to account for uneven sequencing depth across samples.
Statistical analyses
We tested the effect of farm management (monoculture vs polyculture), transect type (within‐row vs across‐row), and their interaction plus focal crop (eggplant vs squash) and the soil properties index on AMF richness, diversity and the proportion of AMF colonization, using generalized linear mixed models with the lme4 and lmertest packages in R (Bates et al., 2015; Kuznetsova et al., 2017; R Core Team, 2020). An additional model was used to test only the effect of management legacy (the time in polyculture management: 0, < 10 and > 10 yr) on AMF richness and diversity removing one outlier monoculture site that had previously been under polyculture management for 10 yr. Arbuscular mycorrhizal fungi richness was estimated using the observed taxa (S obs) and using the Chao1 estimator (S chao) (Chao et al., 2006) with the vegan package (Oksanen et al., 2013) in R. The Chao1 estimator, which can account for unobserved taxa, is especially suited to microbial data because rarer taxa may not be adequately detected (Hughes et al., 2001; Willis, 2019). Arbuscular mycorrhizal fungi diversity was estimated using the Shannon diversity index (transformed as loge + 1). The soil properties index, derived from the first axis of the PCA of all edaphic variables was used to handle highly correlated soil properties. All models for AMF richness (negative binomial error), diversity (Gaussian error) and colonization (Poisson error) included a random effect of farm site, year and the farm site × year interaction to account for multiple sampling events across farm sites in different years.
To evaluate effects of farm management, transect type, their interactions and focal crop on AMF community composition, we used a permutational multivariate analysis of variance (PERMANOVA) and accounted for farm site × year differences using ‘strata’ in the ‘adonis’ function in the package vegan (Oksanen et al., 2013). PERMANOVA tests the compositional differences across group levels (e.g. farm management: monoculture vs polyculture) by examining whether the centroids of sample clusters by group level differ. PERMANOVA was carried out on AMF community matrices (Hellinger transformed) with Bray–Curtis dissimilarities created in the package vegan (Oksanen et al., 2013). To illustrate AMF community composition differences for farm management, the Bray–Curtis dissimilarity matrices were ordinated by a principle coordinates analysis (PCoA) using the ‘pcoa’ command in the ape package (Fig. 2a) (Paradis et al., 2004). In addition, we determined the relative abundance (based on read counts) of AMF taxa between monoculture and polyculture fields (Fig. 2b).
Fig. 2.
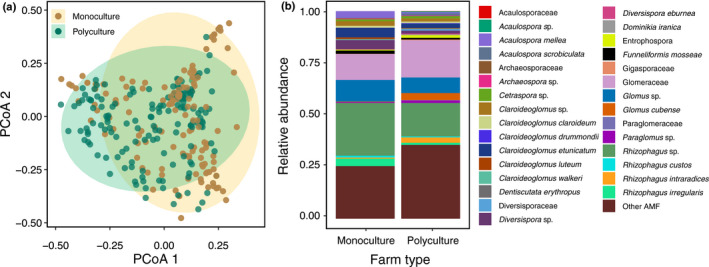
The principle coordinates analysis (PCoA) ordination (a) and relative abundance (based on read counts) (b) of arbuscular mycorrhizal fungi (AMF) taxa between types of farm management (monoculture vs polyculture). Relative abundances were partitioned by the highest taxonomic group available for each AMF taxa up to the family level.
To further understand differences in AMF community composition between monoculture and polyculture farm managements, we conducted indicator species analysis using the indicspecies package in R (R Core Team, 2020). The indicator species analysis uses a combination of taxa abundance and frequency to identify which AMF taxa may be most restricted (i.e. more specialized) to a certain farm management (Cáceres & Legendre, 2009).
To determine the edaphic variables that significantly influenced AMF community composition, we used a partial distance‐based redundancy analysis (dbRDA; based on the Bray–Curtis dissimilarity metric) using the ‘capscale’ function in the vegan package (Oksanen et al., 2013). The partial dbRDA tests how much variation within a community (i.e. AMF community composition) is explained by a group of explanatory variables (i.e. edaphic variables) (Legendre & Anderson, 1999). The number of variables in the partial dbRDA model were minimized via automatic stepwise model selection using the function ‘ordistep’ in R, and collinear variables were removed based on variance inflation factors calculated using the function ‘vif.cca’. Then a permutation‐based ANOVA, using 999 permutations, was performed on the partial dbRDA model to determine the significance of the coefficients. All analyses were carried out in the vegan package in R (Oksanen et al., 2013).
We then carried out a variance partitioning analysis on AMF community composition using the ‘varpart’ function (Legendre, 2008) of the vegan package in R to determine the contribution of soil properties vs farm management (monoculture vs polyculture) to the total variance of AMF community composition.
Lastly, we evaluated the differences in each edaphic variable (Gaussian error; pH, CEC, P, K, Ca, Mg, Zn, B, Mn, Cu, Fe, Pb, Al, Na, S, total organic C, N and C : N) between farm management, transect and their interaction using generalized linear mixed models with the lme4 and lmertest packages in R (Bates et al., 2015; Kuznetsova et al., 2017; R Core Team, 2020). All models included the interaction effect of farm management and transect type (farm management × transect) plus the random effect of farm site, year and the farm site × year interaction to account for the separate sampling events. Further, we also applied an analysis of multivariate homogeneity of group variances (‘betadisper’ function in the vegan package) to test if there were differences in the heterogeneity of edaphic properties between monocultures and polycultures.
Further, to determine differences in the heterogeneity of all the soil properties between monoculture and polyculture, a permutation‐based test for homogeneity of dispersion was performed (PERMDISP) on significant predictor variables in the vegan package (Oksanen et al., 2013). PERMDISP is a multivariate test that is analogous to the Levene’s test for homogeneity of variances.
Results
Across the 372 samples, the rarefied dataset contained a total of 244 AMF taxa belonging to the genera Acaulospora (eight OTUs), Archaeospora (one OTU), Cetraspora (one OTU), Claroideoglomus (21 OTUs), Dentiscutata (two OTUs), Diversipora (10 OTUs), Dominikia (one OTU), Funneliformis (two OTUs), Glomus (30 OTUs), Paraglomus (three OTUs) and Rhizophagus (33 OTUs). The remaining taxa (132 OTUs) did not have generic assignments (i.e. not assigned below AMF genus level; hereafter ‘unassigned’). Of the 244 AMF OTUs, 167 occurred in fewer than 10 samples (c. 70%; Fig. S2). In addition, the species accumulation curve reached its plateau, indicating that sampling was sufficient to reveal the AMF taxa present in this agricultural landscape (Fig. S3). On average eight AMF OTUs and a range of 0–53 AMF taxa were found per sample.
AMF richness and diversity
Farm management (monoculture vs polyculture) had a strong effect on AMF richness for both Chao1 richness (Fig. 3a) and observed richness (Fig. S4a; Table 1). On average, polyculture field sites contained a higher number of AMF taxa (S obs = 9.880 ± 0.651, mean ± SE) than monoculture sites (S obs = 5.122 ± 0.350). However, we found no effect of transect type (within‐row vs across‐row, S obs = 7.774 ± 0.571 vs 7.382 ± 0.548) on AMF richness (Table 1). Similarly, there was no interaction effect between farm management and transect type on AMF richness (Table 1). Results for Chao1 richness were similar (Table 1).
Fig. 3.
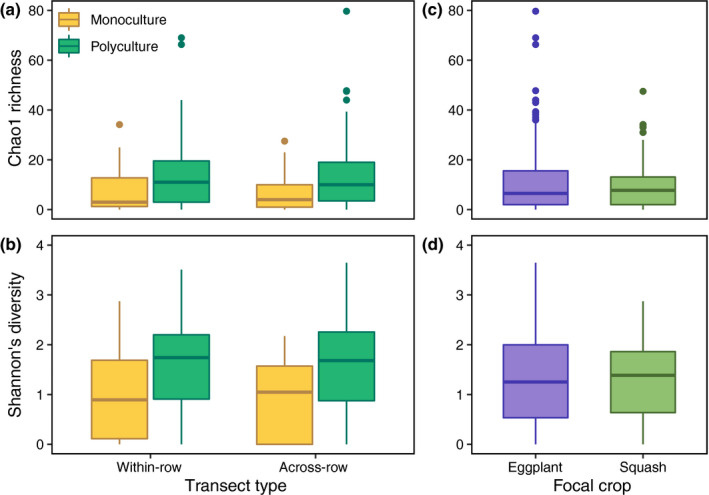
Boxplots for Chao1 richness and Shannon diversity of arbuscular mycorrhizal fungi (AMF) communities according to farm management (monoculture vs polyculture) and transect type (within‐row vs across‐row) (a, b) and focal crop (eggplant vs squash) (c, d). See Table 1 for details. The boxplot is bounded by the first and third quartile ranges, with the line in the box representing the median. The whiskers extend from the first and third quartiles to values that are not within 1.5 × interquartile range from both directions. Data beyond the whiskers are presented as individual circles.
Table 1.
Model outputs of the effect of farm management (monoculture vs polyculture), transect type (within‐row vs across‐row), and their interaction, plus focal crop (eggplant vs squash) and the soil properties index on Chao1 richness, observed richness, diversity and colonization of arbuscular mycorrhizal fungi (AMF).
| Explanatory variable | Chao1 richness | Observed richness | Shannon's diversity | Colonization | ||||
|---|---|---|---|---|---|---|---|---|
| χ2 | P | χ2 | P | χ2 | P | χ2 | P | |
| Farm management | 11.167 | < 0.001*** | 5.563 | 0.018* | 8.918 | 0.003** | 0.174 | 0.676 |
| Transect type | 0.545 | 0.460 | 0.208 | 0.649 | 0.004 | 0.952 | 80.138 | < 0.001*** |
| Soil properties index | 5.316 | 0.021* | 4.388 | 0.036* | 2.529 | 0.112 | 14.116 | < 0.001*** |
| Focal crop | 1.982 | 0.159 | 0.008 | 0.928 | 7.479 | 0.006** | 1.988 | 0.159 |
| Farm management × transect type | 0.743 | 0.389 | 1.266 | 0.261 | 0.034 | 0.853 | 109.486 | < 0.001*** |
*, P < 0.05; **, P < 0.01; ***, P < 0.001.
Similar to AMF richness, farm management (monoculture vs polyculture) had a significant effect on AMF diversity (Fig. 3b; Table 1). Arbuscular mycorrhizal fungi diversity in polyculture fields (1.518 ± 0.068, mean ± SE) was 50% greater than AMF diversity in monoculture fields (1.034 ± 0.057). Neither transect nor the farm management × transect interaction had an effect on AMF diversity (Table 1). The soil properties index also had a significant effect on AMF richness and diversity (Table 1).
Given that the sites varied by focal crop (eggplant vs squash), we also included the effect of focal crop on AMF richness and diversity in the same model. While field sites whose focal crop was eggplant had more AMF taxa on average than squash focal crop sites (S obs = 8.157 ± 0.526 and 6.383 ± 0.517, respectively), this trend was not significant (Fig. 3c; Table 1). Focal crop also did not have an effect on AMF diversity (eggplant, 1.284 ± 0.058; squash, 1.283 ± 0.076; Fig. 3d; Table 1).
Lastly, in a separate model, we tested the effect of management legacy (0, < 10 and > 10 yr in polyculture management) on AMF richness and diversity. While there was a trend of increasing AMF richness (S obs and S chao) and diversity with length under polyculture management, the trend was not significant (Fig. S5).
AMF community composition
Arbuscular mycorrhizal fungi community composition of polyculture field sites was significantly different from monoculture field sites (PERMANOVA: farm management, P = 0.019; Fig. 2) and its interaction with transect type (PERMANOVA: farm management × transect type, P = 0.022) but did not differ between transect type (within‐row vs across‐row) (PERMANOVA: transect type, P = 0.088). Specifically, we found 87 unique OTUs in polyculture field sites but only 19 unique OTUs in monoculture field sites. Focal crop (eggplant vs squash) also did not have an effect on AMF taxa assemblages (PERMANOVA: focal crop, P = 0.226).
There were 11 indicator taxa found for monoculture sites and 47 indicator taxa for polyculture sites (Table S2). Briefly, the top five indicator taxa found for monoculture field sites were OTUs in the genera Rhizophagus (indval = 0.429, P = 0.001), Acaulospora (N = 2, indval = 0.343, P = 0.001 and indval = 0.320, P = 0.001), Claroideoglomus (indval = 0.261, P = 0.048) and Diversipora (indval = 0.254, P = 0.003). By contrast, the top five indicator taxa found in polyculture sites were OTUs in the genera Glomus (indval = 0.552, P = 0.001), Rhizophagus (indval = 0.405, P = 0.001), and three unassigned taxa to genus (indval = 0.411, P = 0.001; indval = 0.405, P = 0.001; indval = 0.375, P = 0.001). Three of the 11 indicator species (c. 27%) for monoculture sites were unassigned (OTUs) whereas 27 of the 47 indicator species (c. 57%) for polyculture sites were unassigned OTUs.
The partial dbRDA revealed significant associations between the AMF community composition and edaphic variables (F 13 = 1.311, P = 0.001). The top edaphic predictor was K followed by clay, Na, pH and P (Table 2).
Table 2.
Results of partial distance‐based redundancy analysis (dbRDA) used to determine the edaphic variables that significantly influenced arbuscular mycorrhizal fungi (AMF) community composition.
| Edaphic variable | SS | F | P |
|---|---|---|---|
| pH | 0.527 | 1.453 | 0.022* |
| P | 0.489 | 1.349 | 0.046* |
| K | 0.630 | 1.736 | 0.001** |
| B | 0.353 | 0.974 | 0.504 |
| Mn | 0.351 | 0.967 | 0.519 |
| Cu | 0.462 | 1.275 | 0.126 |
| Pb | 0.524 | 1.445 | 0.076 |
| Na | 0.537 | 1.482 | 0.018* |
| S | 0.445 | 1.227 | 0.116 |
| % sand | 0.461 | 1.271 | 0.085 |
| % clay | 0.548 | 1.511 | 0.017* |
| TOC | 0.412 | 1.136 | 0.209 |
| N | 0.442 | 1.218 | 0.120 |
*, P < 0.05; **, P < 0.01.
P, phosphorus; K, potassium; B, boron; Mn, manganese; Cu, copper; Pb, lead; Na, sodium; S, sulfur; SS, sums of squares; TOC, total organic carbon; N, nitrogen.
The variance partitioning analysis revealed that the edaphic variables explained more of the variance in the AMF community composition (adjusted R 2 = 0.281) than farm management (adjusted R 2 = 0.037) with minimal shared variance (adjusted R 2 = 0.026).
AMF colonization
The average proportion of roots colonized by AMF did not differ between farm management (Table 1) yet there was an effect of transect (transect: χ 2 = 80.139, P < 0.001) as well as an interactive effect between farm management and transect (Table 1; Fig. 4a). On average, roots sampled from monoculture field sites had 16.1 ± 1.1% (mean ± SE) of roots colonized, and roots sampled from polyculture field sites had 14.8 ± 1.2% roots colonized, whereas average root colonization rates for within‐row and across‐row transects were 13.9 ± 1.0% and 17.0 ± 1.3%, respectively. Average root colonization rates for within‐row and across‐row transects in monoculture were similar (16.7 ± 1.6% and 15.5 ± 1.6%, respectively), whereas average root colonization in the across‐row transect (18.4 ± 1.9%) was higher than in the within‐row transect in polyculture sites (11.3 ± 1.2%). Soil properties also had a significant effect on AMF colonization (Table 1). There was no effect of focal crop (eggplant vs squash) on AMF colonization (Table 1; Fig. 4b).
Fig. 4.
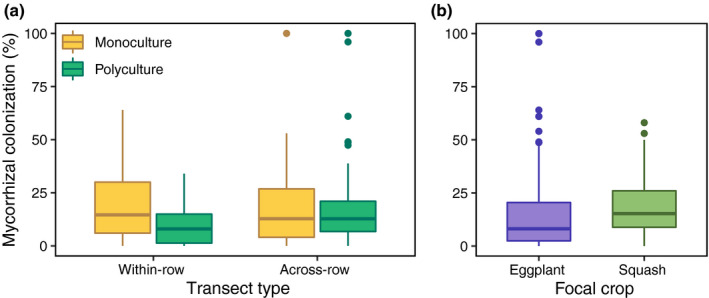
Percent mycorrhizal colonization according to farm management (monoculture vs polyculture) and transect type (within‐row vs across‐row) (a) and focal crop (eggplant) (b). See Table 1 for details.
Comparison of soil properties
There was no effect of farm management (monoculture vs polyculture) on pH (F 1,23 = 1.034, P = 0.319), total soil N (F 1,20 = 0.757, P = 0.394) or soil TOC (F 1,2 = 0.002, P = 0.968), but CEC (F 1,21 = 7.424, P = 0.013) and C : N (F 1,28 = 5.394, P = 0.028) were different. Total CEC for polyculture field sites was higher than that for monoculture field sites (7.425 ± 0.189 and 5.222 ± 0.148, respectively). We also found higher C : N on polyculture than on monoculture sites (14.250 ± 0.257 and 11.750 ± 0.255, respectively). For soil texture, only clay varied between farm management (F 1,28 = 19.119, P < 0.001) where polycultures had slightly higher clay content than monocultures (9.1 ± 0.2% and 5.4 ± 0.2%, respectively). For other macro‐ and micronutrients, there was a significant effect of farm management on K, Mg, Zn, Pb and S (Table S3) but not P, Ca, B, Mn, Al and Na. There was an effect of transect type (within‐row vs across‐row) on K plus an interactive effect of farm management and transect type on P, K, Mn and S (Table S3). For the effect of focal crop (eggplant vs squash), we only found an effect on Zn and Mn (Table S3). Lastly, we found that the soil properties were more heterogenous on polycultures than on monocultures (F = 40.339, P < 0.001; Fig. S6).
Discussion
Our study demonstrates that greater crop diversity in intensive agricultural systems drives a richer and more diverse AMF community. We observed nearly twice as many AMF taxa in polycultures than in monocultures, while accounting for variation in soil properties that also significantly affected AMF richness. The AMF community composition in polyculture sites was also distinct from that in monoculture field sites, but soil properties played a stronger role in structuring the AMF community. Contrary to our expectations, we also show that AMF colonization of roots is probably driven by plant host identity rather than farm management practices (monoculture vs polyculture). For both AMF diversity and colonization responses, soil properties were important factors that influenced the outcomes, but did not dominate relative to the important effect of higher crop diversity. Overall our findings indicate that managing for crop diversity in agricultural landscapes can strongly influence AMF community composition, including richness and diversity, across heterogeneous soils. Further, our results support the notion that plant diversity is key to belowground biodiversity, which in turn could support multifunctional agroecosystems (Bender et al., 2016; Isbell et al., 2017), including those that have been intensively managed in the past.
We show that polyculture fields harbor a richer and more diverse AMF community than do monoculture fields, suggesting that polyculture plantings may promote recovery of AMF richness following a long period of monoculture farming, which is known to be associated with decreased AMF diversity (Helgason et al., 1998; Daniell et al., 2001). Our polyculture sites were formerly farmed intensively as monocultures, as recently as 7 yr before sampling, and thus were likely to have had a depauperate AMF community (Druille et al., 2013; Manoharan et al., 2017; Williams et al., 2017). Soil properties also contributed to explaining some of the variance in AMF richness and diversity, but overall they played a minimal role. While AMF are ubiquitous across landscapes, AMF are obligate symbionts with a degree of host specificity; thus, AMF associations with plant hosts are typically not random (Martínez‐García et al., 2015; Werner & Kiers, 2015; Davison et al., 2016; Horn et al., 2017). Variation in plant traits, including phenology, root architecture and other factors, impacts the distribution and composition of AMF (Hart & Reader, 2002; Pringle & Bever, 2002; Oehl et al., 2004; Maherali & Klironomos, 2007), and thus functionally different plants can associate with distinct AMF communities (Davison et al., 2020). In our study, we found evidence that different AMF taxa occur in polycultures vs monocultures. A Rhizophagus taxon was the top indicator of monocultures, whereas the top indicator taxon in polycultures was in the genus Glomus. While research on AMF functional traits is still emerging (e.g. Chagnon et al., 2013), these taxonomic differences (Šmilauer et al., 2020), coupled with greater AMF diversity in polycultures, could indicate differences in AMF community functionality with implications for plant performance and ecosystem processes. Future research should focus on these possible functional differences among AMF taxa.
In polycultures, functionally distinct plant hosts are planted across space and time, creating a mosaic of diverse microhabitats, varying in microclimatic and microedaphic properties, as evidenced by our finding that polycultures have a more heterogenous soil environment compared with monocultures. Across polyculture fields in our study system, crop type can be distinct row by row in space, but single rows can also shift from crop to crop at different times throughout the year. For example, annuals and perennial crops are grown together at the same time, grasses and tubers can be grown adjacent to each other, and leafy greens and legumes could be grown sequentially in polyculture fields in this study system. In fact, the presence of perennials (Alguacil et al., 2012) and legumes (Drinkwater et al., 1998; Bünemann et al., 2004; Mathimaran et al., 2007) has been shown to increase AMF diversity. In the polyculture field sites, not only are legumes present, but functionally distinct leguminous species and cultivars are planted (e.g. long beans, faba, peanuts, peas, etc.). Therefore, the likely mechanism that fosters a richer and more diverse AMF community in polycultures is the heterogeneity in plant composition: over space, across time within a space, and as different species or varieties across and within functional types such as legumes. Arbuscular mycorrhizal fungi communities also depend, in part, on the composition and pattern of past plant communities (Bittebiere et al., 2020). This may explain why in polycultures we do not find a more diverse AMF community in the more plant‐rich transects when compared with the single species transects. Instead, the legacy of polyculture management, specifically the temporally, spatially and functionally heterogeneous plant community, leads to an overall richer and more diverse AMF community in polycultures than in monocultures.
Polycultures also harbored a distinct AMF community from monocultures. However, AMF communities were quite heterogeneous across both monoculture and polyculture field sites, reflecting a high turnover among sites. The heterogeneity and high turnover of the AMF community are evident in the fact that average site‐level AMF richness is much lower than total AMF richness recorded across all sites: the average AMF richness was c. 5 in monoculture and 10 in polyculture field sites, compared with a total AMF richness in the whole study of 244. Arbuscular mycorrhizal fungi communities tend to be heterogeneous even at fine scales (Pringle & Bever, 2002; Vályi et al., 2016; Mony et al., 2020). Our expectation that AMF communities on polyculture farms would be more heterogeneous at a fine scale than those on monocultures was not borne out; specifically, we found no interaction between farm management (monoculture vs polyculture) and transect (within‐row vs across‐row) for composition. This is further evidence that polycultures may impart a legacy effect on AMF communities.
Despite possible AMF compositional differences between monocultures and polycultures, our results show that soil properties played a larger role in explaining AMF community composition than farm management, with pH being a significant predictor, consistent with other studies (Fitzsimons et al., 2008; Bouffaud et al., 2016; Davison et al., 2016; Oehl et al., 2017; Van Geel et al., 2018), especially at finer spatial scales (Rasmussen et al., 2018). The greater heterogeneity in soil edaphic properties in polycultures than monocultures suggests that crop diversity may indirectly underlie these edaphic‐driven patterns of AMF community structure. Regardless, these findings suggest a role for soil properties in structuring AMF community composition across farms and a role for farm management in shifting the available AMF community into more or less diverse communities at the site level.
The role of plant diversity vs plant host identity on AMF associations was most evident in our measurements of AMF colonization in roots. Our study design allowed us to explore whether crop diversity (polyculture vs monoculture management) impacted AMF colonization in the same crop species, and also whether different crop plant hosts in polycultures play a role in determining AMF colonization. Contrary to our expectation, we found no difference in AMF colonization across the same crop host when planted in polyculture or monoculture fields. In part, this may be explained by fertilizer usage across all farms, which may mask or suppress changes in AMF root colonization as a result of increasing crop diversity because fertilization decreases the dependency of plants hosts on AMF (Johnson, 2010). But AMF colonization has been shown to increase in plant host roots within more diverse plant communities (Eriksson, 2001; Johnson et al., 2004; but see Burrows & Pfleger, 2002), especially when highly mycorrhizal plants are present (Chen et al., 2005). Instead, we found similar degrees of AMF colonization on the focal crop host between polyculture and monoculture farms, but greater degrees of AMF colonization on other crop hosts on polyculture fields. Recent research has found mixed results about the extent to which plant host identity determines the quantity of AMF colonization. Some studies show that plant identity rarely plays a role (Lekberg & Waller, 2016; Van Geel et al., 2018), while others, like this study, demonstrate that plant host identity does impact AMF colonization, especially at local scales (König et al., 2010; Davison et al., 2016; Šmilauer et al., 2020). Thus, our study strengthens the body of research showing that AMF colonization is dependent on specific AMF–plant host associations. While this finding could suggest that agricultural systems with higher plant diversity may not benefit from greater AMF colonization, AMF colonization may not actually be the most important indicator of AMF benefits and functions for crops in agricultural systems (Thirkell et al., 2017). Colonization does not indicate the extent of nutrient transfer or the degree of ecosystem services provided by AMF (Chagnon et al., 2013). Instead, there is a growing understanding that AMF composition is an important determinant of the benefits received by ecosystems from AMF communities (Chagnon et al., 2013).
A richer and more diverse AMF community could indicate differences in ecosystem functioning between monoculture and polyculture farming, with important implications for agricultural management. Previous research has shown that monocultures contribute to reducing AMF richness and can change community composition to favor less beneficial AMF taxa, in turn contributing to yield declines (Johnson et al., 1992). Although empirical evidence from field studies on AMF remains rare, a positive relationship between AMF diversity and ecosystem functioning is expected because AMF taxa differ in their functions (Powell & Rillig, 2018). For example, studies have shown differential plant productivity responses to different AMF taxa or communities (van der Heijden et al., 1998a, 1998b,1998a, 1998b, 2003; van der Heijden, 2002; Klironomos, 2003). Other studies have demonstrated that productivity, phosphorus uptake, soil aggregation and pathogen protection increase with AMF diversity (van der Heijden et al., 1998a, 1998b,1998a, 1998b; Sikes et al., 2009; Chen et al., 2017). In short, as AMF taxa are functionally heterogenous (Chagnon et al., 2013), a more diverse community could provide a wider array and/or stability of functions (Loreau et al., 2003; Isbell, 2015). Therefore, crops grown in polycultures may benefit from the enhanced and/or stabilized ecosystem functions and services of a richer and more diverse AMF community.
Conclusions
Through investigating the response of AMF communities to greater crop diversity, we have demonstrated that plant host diversity shifts the available AMF community into richer and more diverse communities while soil properties structure AMF community composition. Our on‐farm approach focused on the role of polycultures – the dominant form of agriculture across many regions in the world, especially among smallholder farmers (Altieri, 1999; Brooker et al., 2015) – allowing us to elucidate the important role that plant host diversity plays on AMF communities without the confounding reciprocal process (i.e. AMF communities influence plant communities), a common obstacle in observational studies of natural systems. Specifically, we show that polycultures doubled AMF richness in comparison to monocultures. We further find that AMF colonization is dependent on crop host identity. Together, the positive relationship between plant diversity and AMF community composition highlights the fact that vegetative diversity is essential to harnessing AMF functional diversity. Therefore, we conclude that plant diversity is key to enriching AMF communities, and that enhancing crop diversity locally on farms may allow multifunctionality to be re‐established via AMF communities in agricultural landscapes.
Author contributions
AG, MKF, TB, CK designed the study with support of RMDW. AG, MM, LH, GD and PY collected the data with assistance by AK. AG analyzed the data. AG wrote the first draft of the manuscript and all authors contributed to the final version of the manuscript.
Supporting information
Fig. S1 PCA of soil properties used to calculate the soil properties index.
Fig. S2 Frequency of AMF taxa found in as few as one to as many as 100 of all 372 communities sampled.
Fig. S3 AMF taxa accumulation curve reaching a plateau of 214.778 ± 9.546 out of 244 taxa after 167 samples.
Fig. S4 Boxplot for observed richness according to farm management type (monoculture vs polyculture) and transect type (within‐row vs across‐row) as well as focal crop (eggplant vs squash).
Fig. S5 Boxplot for AMF observed richness, Chao1 richness and diversity across the time in polyculture management (0, < 10 and > 10 yr).
Fig. S6 Boxplot for dispersion of the edaphic property dissimilarities from the centroid according to farm management type (monoculture vs polyculture).
Methods S1 Molecular analysis of AMF communities: primer selection, PCR conditions and amplicon library preparation.
Table S1 Site‐by‐site properties, including farm management (monoculture vs polyculture), focal crop (eggplant vs squash) and the first year of polyculture management, number of years in polyculture, monoculture or lying fallow.
Table S2 Results of the indicator species analysis for AMF taxa (listed by OTU plus their genus) of monoculture and polyculture fields.
Table S3 Model parameter estimates, with SE in parentheses, of all soil properties.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We are grateful to the farmers for collaborating on this project and so graciously sharing their expertise, time and resources. We are also thankful for the field and laboratory assistance provided by Martin Banuelos, Jennifer Becerra, Paulina Hernandez, Grace Pratt, Julian Samano, Lilliana Sandoval, Emily Wagner and Andrea Uribe, as well as the support of University of California Kearney Agricultural Research and Extension Center, and Dr Kent Daane. We are also grateful to Michael Mann and members of the Kremen, Berkeley Agroecology and Firestone laboratories for their support. We also want to thank the three anonymous reviewers for their invaluable feedback that strengthened the paper. Finally, we want to acknowledge and pay our respect to the indigenous people of the ancestral and unceded lands on which field and laboratory research took place: the Yokuts people and Chochenyo‐speaking Ohlone people. This research was funded by NSF Graduate Research Fellowship (to AG), Annie’s Sustainable Agriculture Scholarship (to AG), UCANR Graduate Student in Extension Fellowship (to AG) and grants from the Army Research Office (W911NF‐17‐1‐0231 to CK) and C.S. Fund (173‐010 to CK). The authors report there are no conflicts of interest.
Data availability
Sequence data generated in this study have been deposited at the NCBI SRA database under accession PRJNA650414.
References
- Alguacil MM, Torrecillas E, Roldán A, Díaz G, Torres MP. 2012. Perennial plant species from semiarid gypsum soils support higher AMF diversity in roots than the annual Bromus rubens . Soil Biology and Biochemistry 49: 132–138. [Google Scholar]
- Altieri MA. 1999. The ecological role of biodiversity in agroecosystems. Agriculture, Ecosystems & Environment 74: 19–31. [Google Scholar]
- Altieri MA, Nicholls CI, Henao A, Lana MA. 2015. Agroecology and the design of climate change‐resilient farming systems. Agronomy for Sustainable Development 35: 869–890. [Google Scholar]
- Bainard LD, Bainard JD, Hamel C, Gan Y. 2014. Spatial and temporal structuring of arbuscular mycorrhizal communities is differentially influenced by abiotic factors and host crop in a semi‐arid prairie agroecosystem. FEMS Microbiology Ecology 88: 333–344. [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting Linear Mixed‐Effects Models using lme4. Journal of Statistical Software 67: 1–48. [Google Scholar]
- Bender SF, Wagg C, van der Heijden MGA. 2016. An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends in Ecology & Evolution 31: 440–452. [DOI] [PubMed] [Google Scholar]
- Bittebiere A, Vandenkoornhuyse P, Maluenda E, Gareil A, Dheilly A, Coudouel S, Bahin M, Mony C. 2020. Past spatial structure of plant communities determines arbuscular mycorrhizal fungal community assembly. Journal of Ecology 108: 546–560. [Google Scholar]
- Bouffaud M‐L, Creamer RE, Stone D, Plassart P, van Tuinen D, Lemanceau P, Wipf D, Redecker D. 2016. Indicator species and co‐occurrence in communities of arbuscular mycorrhizal fungi at the European scale. Soil Biology and Biochemistry 103: 464–470. [Google Scholar]
- Bowles TM, Jackson LE, Loeher M, Cavagnaro TR. 2017. Ecological intensification and arbuscular mycorrhizas: a meta‐analysis of tillage and cover crop effects. Journal of Applied Ecology 54: 1785–1793. [Google Scholar]
- Brooker RW, Bennett AE, Cong W‐F, Daniell TJ, George TS, Hallett PD, Hawes C, Iannetta PPM, Jones HG, Karley AJ et al. 2015. Improving intercropping: a synthesis of research in agronomy, plant physiology and ecology. New Phytologist 206: 107–117. [DOI] [PubMed] [Google Scholar]
- Bünemann EK, Bossio DA, Smithson PC, Frossard E, Oberson A. 2004. Microbial community composition and substrate use in a highly weathered soil as affected by crop rotation and P fertilization. Soil Biology and Biochemistry 36: 889–901. [Google Scholar]
- Burrows RL, Pfleger FL. 2002. Arbuscular mycorrhizal fungi respond to increasing plant diversity. Canadian Journal of Botany 80: 120–130. [Google Scholar]
- Cáceres MD, Legendre P. 2009. Associations between species and groups of sites: indices and statistical inference. Ecology 90: 3566–3574. [DOI] [PubMed] [Google Scholar]
- Chagnon P‐L, Bradley RL, Maherali H, Klironomos JN. 2013. A trait‐based framework to understand life history of mycorrhizal fungi. Trends in Plant Science 18: 484–491. [DOI] [PubMed] [Google Scholar]
- Chao A, Shen TJ, Hwang WH. 2006. Application of Laplace’s boundary‐mode approximations to estimate species and shared species richness. Australian & New Zealand Journal of Statistics 48: 117–128. [Google Scholar]
- Chen X, Tang J, Zhi G, Hu S. 2005. Arbuscular mycorrhizal colonization and phosphorus acquisition of plants: effects of coexisting plant species. Applied Soil Ecology 28: 259–269. [Google Scholar]
- Chen Y‐L, Xu T‐L, Veresoglou SD, Hu H‐W, Hao Z‐P, Hu Y‐J, Liu L, Deng Y, Rillig MC, Chen B‐D. 2017. Plant diversity represents the prevalent determinant of soil fungal community structure across temperate grasslands in northern China. Soil Biology and Biochemistry 110: 12–21. [Google Scholar]
- Daniell TJ, Husband R, Fitter AH, Young JPW. 2001. Molecular diversity of arbuscular mycorrhizal fungi colonising arable crops. FEMS Microbiology Ecology 36: 203–209. [DOI] [PubMed] [Google Scholar]
- Davison J, García de León D, Zobel M, Moora M, Bueno CG, Barceló M, Gerz M, León D, Meng Y, Pillar VD et al. 2020. Plant functional groups associate with distinct arbuscular mycorrhizal fungal communities. New Phytologist 226: 1117–1128. [DOI] [PubMed] [Google Scholar]
- Davison J, Moora M, Jairus T, Vasar M, Öpik M, Zobel M. 2016. Hierarchical assembly rules in arbuscular mycorrhizal (AM) fungal communities. Soil Biology and Biochemistry 97: 63–70. [Google Scholar]
- Drinkwater LE, Wagoner P, Sarrantonio M. 1998. Legume‐based cropping systems have reduced carbon and nitrogen losses. Nature 396: 262–265. [Google Scholar]
- Druille M, Cabello MN, Omacini M, Golluscio RA. 2013. Glyphosate reduces spore viability and root colonization of arbuscular mycorrhizal fungi. Applied Soil Ecology 64: 99–103. [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2016. UNOISE2: improved error‐correction for Illumina 16S and ITS amplicon sequencing. BioRxiv. doi: 10.1101/081257. [DOI] [Google Scholar]
- Eriksson Å. 2001. Arbuscular mycorrhiza in relation to management history, soil nutrients and plant species diversity. Plant Ecology 155: 129–137. [Google Scholar]
- Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J. 2014. An improved dual‐indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimons MS, Miller RM, Jastrow JD. 2008. Scale‐dependent niche axes of arbuscular mycorrhizal fungi. Oecologia 158: 117–127. [DOI] [PubMed] [Google Scholar]
- Foley JA, Defries R, Asner GP, Barford C, Bonan G, Carpenter SR, Chapin FS, Coe MT, Daily GC, Gibbs HK et al. 2005. Global consequences of land use. Science 309: 570–574. [DOI] [PubMed] [Google Scholar]
- Gao C, Montoya L, Xu L, Madera M, Hollingsworth J, Purdom E, Hutmacher RB, Dahlberg JA, Coleman‐Derr D, Lemaux PG et al. 2019. Strong succession in arbuscular mycorrhizal fungal communities. The ISME Journal 13: 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisseler D, Scow KM. 2014. Long‐term effects of mineral fertilizers on soil microorganisms – a review. Soil Biology and Biochemistry 75: 54–63. [Google Scholar]
- Gianinazzi S, Gollotte A, Binet MN, van Tuinen D, Redecker D, Wipf D. 2010. Agroecology: the key role of arbuscular mycorrhizas in ecosystem services. Mycorrhiza 20: 519–530. [DOI] [PubMed] [Google Scholar]
- Gottshall CB, Cooper M, Emery SM. 2017. Activity, diversity and function of arbuscular mycorrhizae vary with changes in agricultural management intensity. Agriculture, Ecosystems and Environment 241: 142–149. [Google Scholar]
- de Graaff M‐A, Hornslein N, Throop HL, Kardol P, van Diepen LTA. 2019. Effects of agricultural intensification on soil biodiversity and implications for ecosystem functioning: A meta‐analysis. Advances in Agronomy 155: 1–44. [Google Scholar]
- Hart MM, Reader RJ. 2002. Taxonomic basis for variation in the colonization strategy of arbuscular mycorrhizal fungi. New Phytologist 153: 335–344. [Google Scholar]
- van der Heijden MGA. 2002. Arbuscular mycorrhizal fungi as a determinant of plant diversity: in search of underlying mechanisms and general principles. In: van der Heijden MGA, Sanders IR, eds. Mycorrhizal ecology. Ecological studies (analysis and synthesis), vol 157. Berlin, Germany: Springer. doi: 10.1007/978-3-540-38364-2_10. [DOI] [Google Scholar]
- van der Heijden MGA. 2010. Mycorrhizal fungi reduce nutrient loss from model grassland ecosystems. Ecology 91: 1171. [DOI] [PubMed] [Google Scholar]
- van der Heijden MGA, Boller T, Wiemken A, Sanders IR. 1998a. Different arbuscular mycorrhizal fungal species are potential determinants of plant community structure. Ecology 79: 2082–2091. [Google Scholar]
- van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf‐Engel R, Boller T, Wiemken A, Sanders IR. 1998b. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396: 69–72. [Google Scholar]
- van der Heijden MGA, Wiemken A, Sanders IR. 2003. Different arbuscular mycorrhizal fungi alter coexistence and resource distribution between co‐occurring plant. New Phytologist 157: 569–578. [DOI] [PubMed] [Google Scholar]
- Helgason T, Daniell TJ, Husband R, Fitter AH, Young JPW. 1998. Ploughing up the wood‐wide web. Nature 394: 431. [DOI] [PubMed] [Google Scholar]
- Hiiesalu I, Pärtel M, Davison J, Gerhold P, Metsis M, Moora M, Öpik M, Vasar M, Zobel M, Wilson SD. 2014. Species richness of arbuscular mycorrhizal fungi: associations with grassland plant richness and biomass. New Phytologist 203: 233–244. [DOI] [PubMed] [Google Scholar]
- Hontoria C, García‐González I, Quemada M, Roldán A, Alguacil MM. 2019. The cover crop determines the AMF community composition in soil and in roots of maize after a ten‐year continuous crop rotation. Science of the Total Environment 660: 913–922. [DOI] [PubMed] [Google Scholar]
- Horn S, Hempel S, Verbruggen E, Rillig MC, Caruso T. 2017. Linking the community structure of arbuscular mycorrhizal fungi and plants: a story of interdependence. The ISME Journal 11: 1400–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJ. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Applied and Environment Microbiology 67: 4399–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isbell F. 2015. Agroecology: agroecosystem diversification. Nature Plants 1: 15041. [DOI] [PubMed] [Google Scholar]
- Isbell F, Adler PR, Eisenhauer N, Fornara D, Kimmel K, Kremen C, Letourneau DK, Liebman M, Polley HW, Quijas S et al. 2017. Benefits of increasing plant diversity in sustainable agroecosystems. Journal of Ecology 105: 871–879. [Google Scholar]
- Iverson AL, Marín LE, Ennis KK, Gonthier DJ, Connor‐Barrie BT, Remfert JL, Cardinale BJ, Perfecto I. 2014. REVIEW: Do polycultures promote win‐wins or trade‐offs in agricultural ecosystem services? A meta‐analysis. Journal of Applied Ecology 51: 1593–1602. [Google Scholar]
- Johnson D, Vandenkoornhuyse PJ, Leake JR, Gilbert L, Booth RE, Grime JP, Young JPW, Read DJ. 2004. Plant communities affect arbuscular mycorrhizal fungal diversity and community composition in grassland microcosms. New Phytologist 161: 503–515. [DOI] [PubMed] [Google Scholar]
- Johnson NC. 2010. Resource stoichiometry elucidates the structure and function of arbuscular mycorrhizas across scales. New Phytologist 185: 631–647. [DOI] [PubMed] [Google Scholar]
- Johnson NC, Copeland PJ, Crookston RK, Pfleger FL. 1992. Mycorrhizae: possible explanation for yield decline with continuous corn and soybean. Agronomy Journal 84: 387–390. [Google Scholar]
- Johnson NC, Graham JH, Smith FA. 1997. Functioning of mycorrhizal associations along the mutualism–parasitism continuum. New Phytologist 135: 575–585. [Google Scholar]
- Jung SC, Martinez‐Medina A, Lopez‐Raez JA, Pozo MJ. 2012. Mycorrhiza‐induced resistance and priming of plant defenses. Journal of Chemical Ecology 38: 651–664. [DOI] [PubMed] [Google Scholar]
- Klironomos JN. 2003. Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology 84: 2292–2301. [Google Scholar]
- Kokkoris V, Lekberg Y, Antunes PM, Fahey C, Fordyce JA, Kivlin SN, Hart MM. 2020. Codependency between plant and arbuscular mycorrhizal fungal communities: what is the evidence. New Phytologist 228: 828–838. [DOI] [PubMed] [Google Scholar]
- Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson‐Palme J, Callaghan TM et al. 2013. Towards a unified paradigm for sequence‐based identification of fungi. Molecular Ecology 22: 5271–5277. [DOI] [PubMed] [Google Scholar]
- König S, Wubet T, Dormann CF, Hempel S, Renker C, Buscot F. 2010. TaqMan real‐time PCR assays to assess arbuscular mycorrhizal responses to field manipulation of grassland biodiversity: effects of soil characteristics, plant species richness, and functional traits. Applied and Environmental Microbiology 76: 3765–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koske RE, Gemma JN. 1989. A modified procedure for staining roots to detect VA mycorrhizas. Mycological Research 92: 486–505. [Google Scholar]
- Kremen C, Merenlender AM. 2018. Landscapes that work for biodiversity and people. Science 362: eaau6020. [DOI] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff PB, Christensen RHB. 2017. lmerTest Package: Tests in Linear Mixed Effects Models. Journal of Statistical Software 82: 1–26. [Google Scholar]
- Landis FC, Gargas A, Givnish TJ. 2004. Relationships among arbuscular mycorrhizal fungi, vascular plants and environmental conditions in oak savannas. New Phytologist 164: 493–504. [Google Scholar]
- Legendre P. 2008. Studying beta diversity: ecological variation partitioning by multiple regression and canonical analysis. Journal of Plant Ecology 1: 3–8. [Google Scholar]
- Legendre P, Anderson MJ. 1999. Distance‐based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecological Monographs 69: 1–24. [Google Scholar]
- Leigh J, Hodge A, Fitter AH. 2009. Arbuscular mycorrhizal fungi can transfer substantial amounts of nitrogen to their host plant from organic material. New Phytologist 181: 199–207. [DOI] [PubMed] [Google Scholar]
- Lekberg Y, Waller LP. 2016. What drives differences in arbuscular mycorrhizal fungal communities among plant species. Fungal Ecology 24: 135–138. [Google Scholar]
- Li L, Tilman D, Lambers H, Zhang FS. 2014. Plant diversity and overyielding: insights from belowground facilitation of intercropping in agriculture. New Phytologist 203: 63–69. [DOI] [PubMed] [Google Scholar]
- Loreau M, Mouquet N, Gonzalez A. 2003. Biodiversity as spatial insurance in heterogeneous landscapes. Proceedings of the National Academy of Sciences, USA 100: 12765–12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali H, Klironomos JN. 2007. Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316: 1746–1748. [DOI] [PubMed] [Google Scholar]
- Manoharan L, Rosenstock NP, Williams A, Hedlund K. 2017. Agricultural management practices influence AMF diversity and community composition with cascading effects on plant productivity. Applied Soil Ecology 115: 53–59. [Google Scholar]
- Martínez‐García LB, Richardson SJ, Tylianakis JM, Peltzer DA, Dickie IA. 2015. Host identity is a dominant driver of mycorrhizal fungal community composition during ecosystem development. New Phytologist 205: 1565–1576. [DOI] [PubMed] [Google Scholar]
- Mathimaran N, Ruh R, Jama B, Verchot L, Frossard E, Jansa J. 2007. Impact of agricultural management on arbuscular mycorrhizal fungal communities in Kenyan ferralsol. Agriculture, Ecosystems and Environment 119: 22–32. [Google Scholar]
- McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA. 1990. A new method which gives an objective measure of colonization of roots by vesicular—arbuscular mycorrhizal fungi. New Phytologist 115: 495–501. [DOI] [PubMed] [Google Scholar]
- Middleton EL, Richardson S, Koziol L, Palmer CE, Yermakov Z, Henning JA, Schultz PA, Bever JD. 2015. Locally adapted arbuscular mycorrhizal fungi improve vigor and resistance to herbivory of native prairie plant species. Ecosphere 6: art276. [Google Scholar]
- Miller WP, Miller DM. 1987. A micro‐pipette method for soil mechanical analysis. Communications in Soil Science and Plant Analysis 18: 1–15. [Google Scholar]
- Molinar RH. 2012. Indigenous Asian specialty vegetables in the central valley of California. HortScience 47: 835–838. [Google Scholar]
- Mony C, Brunellière P, Vannier N, Bittebiere A, Vandenkoornhuyse P. 2020. Effect of floristic composition and configuration on plant root mycobiota: a landscape transposition at a small scale. New Phytologist 225: 1777–1787. [DOI] [PubMed] [Google Scholar]
- Nguyen NH, Smith D, Peay K, Kennedy P. 2015. Parsing ecological signal from noise in next generation amplicon sequencing. New Phytologist 205: 1389–1393. [DOI] [PubMed] [Google Scholar]
- Oehl F, Laczko E, Oberholzer H‐R, Jansa J, Egli S. 2017. Diversity and biogeography of arbuscular mycorrhizal fungi in agricultural soils. Biology and Fertility of Soils 53: 777–797. [Google Scholar]
- Oehl F, Sieverding E, Ineichen K, Mader P, Boller T, Wiemken A. 2003. Impact of land use intensity on the species diversity of arbuscular mycorrhizal fungi in agroecosystems of Central Europe. Applied and Environmental Microbiology 69: 2816–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehl F, Sieverding E, Ineichen K, Ris E‐A, Boller T, Wiemken A. 2004. Community structure of arbuscular mycorrhizal fungi at different soil depths in extensively and intensively managed agroecosystems. New Phytologist 165: 273–283. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H et al. 2013. Package ‘vegan’. Community ecology package, version 2: 1–295. [Google Scholar]
- Orrell P, Bennett AE. 2013. How can we exploit above–belowground interactions to assist in addressing the challenges of food security. Frontiers in Plant Science 4: 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JM, Jusino MA, Banik MT, Lindner DL. 2018. Non‐biological synthetic spike‐in controls and the AMPtk software pipeline improve mycobiome data. PeerJ 6: e4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Powell JR, Rillig MC. 2018. Biodiversity of arbuscular mycorrhizal fungi and ecosystem function. New Phytologist 220: 1059–1075. [DOI] [PubMed] [Google Scholar]
- Power AG. 2010. Ecosystem services and agriculture: tradeoffs and synergies. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 365: 2959–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle A, Bever JD. 2002. Divergent phenologies may facilitate the coexistence of arbuscular mycorrhizal fungi in a North Carolina grassland. American Journal of Botany 89: 1439–1446. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2020. R: a language and environment for statistical computing, v. 4.0.0. Vienna, Austria: R Foundation for Statistical Computing. [WWW document] URL https://www.r‐project.org. [Google Scholar]
- Rasmussen PU, Hugerth LW, Blanchet FG, Andersson AF, Lindahl BD, Tack AJM. 2018. Multiscale patterns and drivers of arbuscular mycorrhizal fungal communities in the roots and root‐associated soil of a wild perennial herb. New Phytologist 220: 1248–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rillig MC, Aguilar‐Trigueros CA, Camenzind T, Cavagnaro TR, Degrune F, Hohmann P, Lammel DR, Roy J, van der Heijden MGA, Yang G. 2019. Why farmers should manage the arbuscular mycorrhizal symbiosis. New Phytologist 222: 1171–1175. [DOI] [PubMed] [Google Scholar]
- Rillig MC, Mummey DL. 2006. Mycorrhizas and soil structure. New Phytologist 171: 41–53. [DOI] [PubMed] [Google Scholar]
- Rillig MC, Sosa‐Hernández MA, Roy J, Aguilar‐Trigueros CA, Vályi K, Lehmann A. 2016. Towards an integrated mycorrhizal technology: harnessing mycorrhiza for sustainable intensification in agriculture. Frontiers in Plant Science 7: 1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4: e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MH, Graham JH. 2018. Little evidence that farmers should consider abundance or diversity of arbuscular mycorrhizal fungi when managing crops. New Phytologist 220: 1092–1107. [DOI] [PubMed] [Google Scholar]
- Sepp SK, Davison J, Jairus T, Vasar M, Moora M, Zobel M, Öpik M. 2019. Non‐random association patterns in a plant‐mycorrhizal fungal network reveal host‐symbiont specificity. Molecular Ecology 28: 365–378. [DOI] [PubMed] [Google Scholar]
- Sikes BA, Cottenie K, Klironomos JN. 2009. Plant and fungal identity determines pathogen protection of plant roots by arbuscular mycorrhizas. Journal of Ecology 97: 1274–1280. [Google Scholar]
- Šmilauer P, Košnar J, Kotilínek M, Šmilauerová M. 2020. Contrasting effects of host identity, plant community, and local species pool on the composition and colonization levels of arbuscular mycorrhizal fungal community in a temperate grassland. New Phytologist 225: 461–473. [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ. 2008. Mycorrhizal symbiosis, 3 rd edn. London, UK: Academic Press. [Google Scholar]
- Spatafora JW, Chang Y, Benny GL, Lazarus KL, Smith ME, Berbee ML, Bonito G, Corradi N, Grigoriev I, Gryganskyi A et al. 2016. A phylum‐level phylogenetic classification of zygomycete fungi based on genome‐scale data. Mycologia 108: 1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom N, Hu W, Haarith D, Chen S, Bushley K. 2020. Interactions between soil properties, fungal communities, the soybean cyst nematode, and crop yield under continuous corn and soybean monoculture. Applied Soil Ecology 147: 103388. [Google Scholar]
- Tedersoo L, Bahram M, Zobel M. 2020. How mycorrhizal associations drive plant population and community biology. Science 367: eaba1223. [DOI] [PubMed] [Google Scholar]
- Thirkell TJ, Charters MD, Elliott AJ, Sait SM, Field KJ. 2017. Are mycorrhizal fungi our sustainable saviours? Considerations for achieving food security. Journal of Ecology 105: 921–929. [Google Scholar]
- Vályi K, Mardhiah U, Rillig MC, Hempel S. 2016. Community assembly and coexistence in communities of arbuscular mycorrhizal fungi. The ISME Journal 10: 2341–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Geel M, Jacquemyn H, Plue J, Saar L, Kasari L, Peeters G, van Acker K, Honnay O, Ceulemans T. 2018. Abiotic rather than biotic filtering shapes the arbuscular mycorrhizal fungal communities of European seminatural grasslands. New Phytologist 220: 1262–1272. [DOI] [PubMed] [Google Scholar]
- Verbruggen E, Toby Kiers E. 2010. Evolutionary ecology of mycorrhizal functional diversity in agricultural systems. Evolutionary Applications 3: 547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veresoglou SD, Rillig MC. 2012. Suppression of fungal and nematode plant pathogens through arbuscular mycorrhizal fungi. Biology Letters 8: 214–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner GD, Kiers T. 2015. Partner selection in the mycorrhizal mutualism. New Phytologist 205: 1437–1442. [DOI] [PubMed] [Google Scholar]
- Williams A, Manoharan L, Rosenstock NP, Olsson PA, Hedlund K. 2017. Long‐term agricultural fertilization alters arbuscular mycorrhizal fungal community composition and barley (Hordeum vulgare) mycorrhizal carbon and phosphorus exchange. New Phytologist 213: 874–885. [DOI] [PubMed] [Google Scholar]
- Willis AD. 2019. Rarefaction, alpha diversity, and statistics. Frontiers in Microbiology 10: 2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GW, Rice CW, Rillig MC, Springer A, Hartnett DC. 2009. Soil aggregation and carbon sequestration are tightly correlated with the abundance of arbuscular mycorrhizal fungi: results from long‐term field experiments. Ecology Letters 12: 452–461. [DOI] [PubMed] [Google Scholar]
- Xu M, Li X, Cai X, Li X, Christie P, Zhang J. 2017. Land use alters arbuscular mycorrhizal fungal communities and their potential role in carbon sequestration on the Tibetan Plateau. Science Reports 7: 3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Lehmann A, Zheng W, You Z, Rillig MC. 2019. Arbuscular mycorrhizal fungi increase grain yields: a meta‐analysis. New Phytologist 222: 543–555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 PCA of soil properties used to calculate the soil properties index.
Fig. S2 Frequency of AMF taxa found in as few as one to as many as 100 of all 372 communities sampled.
Fig. S3 AMF taxa accumulation curve reaching a plateau of 214.778 ± 9.546 out of 244 taxa after 167 samples.
Fig. S4 Boxplot for observed richness according to farm management type (monoculture vs polyculture) and transect type (within‐row vs across‐row) as well as focal crop (eggplant vs squash).
Fig. S5 Boxplot for AMF observed richness, Chao1 richness and diversity across the time in polyculture management (0, < 10 and > 10 yr).
Fig. S6 Boxplot for dispersion of the edaphic property dissimilarities from the centroid according to farm management type (monoculture vs polyculture).
Methods S1 Molecular analysis of AMF communities: primer selection, PCR conditions and amplicon library preparation.
Table S1 Site‐by‐site properties, including farm management (monoculture vs polyculture), focal crop (eggplant vs squash) and the first year of polyculture management, number of years in polyculture, monoculture or lying fallow.
Table S2 Results of the indicator species analysis for AMF taxa (listed by OTU plus their genus) of monoculture and polyculture fields.
Table S3 Model parameter estimates, with SE in parentheses, of all soil properties.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
Sequence data generated in this study have been deposited at the NCBI SRA database under accession PRJNA650414.


