Abstract
Oxidative stress is known to inhibit osteogenesis and PKD1 is implicated in bone remodeling and skeletogenesis. In the present study, we explored the role of PKD1 in osteogenesis under oxidative stress. H2O2 was used to induce oxidative stress in rat bone marrow (BM)‐mesenchymal stem cells (MSCs) during osteoblast differentiation. Alkaline phosphatase (ALP) activity, calcium deposits, and the RUNX2 marker were assayed to determine osteogenic differentiation. The correlation of PKD1, Sirt1, c‐MYC, and TAZ was further confirmed by chromatin immunoprecipitation (ChIP) and dual‐luciferase reporter assay. We found that H2O2 induced the downregulation of PKD1 expression and the upregulation of c‐MYC, and Sirt1 was accompanied by decreasing cell viability in BM‐MSCs. During osteogenic differentiation, the expression of PKD1 was upregulated significantly whereas Sirt1 tended to be upregulated mildly under normal conditions. Both PKD1 and Sirt1 were upregulated upon oxidative stress. The positive correlation of PKD1 expression with osteogenic differentiation under normal conditions might be hindered by oxidative stress and PKD1 could interact with TAZ under oxidative stress to regulate osteogenic differentiation. Our results suggest that PKD1 may alleviate oxidative stress‐inhibited osteogenesis of rat BM‐MSCs through TAZ activation.
Keywords: bone marrow–derived mesenchymal stem cells, osteogenesis, PKD1, Sirt1, TAZ
1. INTRODUCTION
Mesenchymal stem cells (MSCs) can directly differentiate into mineralized matrix‐generating osteoblasts and condensate to form osteoblast‐mediated bone. 1 Polycystin‐1 (PC1) encodes PKD1, a chief mechanosensing molecule, which is implicated in bone remodeling and skeletogenesis. 2 As reported in previous studies, Runx2 may induce the differentiation of hypertrophic chondrocytes and osteoblasts from mesenchymal precursors. 3 Xiao et al. 1 reported that PKD1 may stimulate the osteoblast‐specific transcription factor RUNX2 to regulate skeletogenesis. In another study, mechanical load‐induced RUNX2 expression could regulate osteoblast differentiation and ultimately bone formation through the PKD1‐JAK2/STAT3 signaling pathway. 2 Through a cascade of calcineurin/nuclear factor of activated T cells signaling, PKD1 undergoes mechanical stimulation and modulates bone‐cell differentiation by regulation of osteoblast gene transcription. 4
Oxidative stress is believed to play an important role in osteogenesis. Peroxide anion free radicals may be important in the bone‐like formation of disease related to ossification, indicated by the results of fourier transform infrared spectra. 5 Oxidative stress is reported to inhibit osteogenesis by decreasing Wnt‐related gene expression. 6 In addition, oxidative stress is known to inhibit activated protein kinase activation as well as FOXO3a and RUNX2 expression in osteogenic differentiation of human MSCs. 7 The NAD+‐dependent deacetylase Sirt1 is reported to be associated with cell longevity following oxidative stress. Singh et al. 8 found that Sirt1 was correlated with α‐synuclein aggregate formation and cell survival, and Sirt1 activity was reduced under oxidative stress, which suggested that Sirt1 is a pro‐survival protein that is downregulated under cellular stress. In other studies, Sirt1 was found to have potential antioxidative stress activity in vascular endothelial cells, the mechanisms of which included Sirtuin1/nitrogen oxide, Sirtuin1/Superoxide Dismutase, Sirtuin1/Forkhead box O, Sirtuin1/endothelium nitric oxide synthase, and Sirtuin1/nuclear factor kappa‐B pathways. 9 In renal epithelial cells and tissues of embryonic and postnatal PKD1‐mutant mice, Sirt1 was upregulated through c‐MYC. 10 A previous report found that there is a feed‐forward regulatory PKD1‐c‐Myc loop mechanism between c‐Myc and PKD1. 11 Studies on whether PKD1 is associated with Sirt1 in the osteoblast differentiation of bone MSCs under oxidative stress remain relatively scarce. Perturbations of both PKD1 and TAZ expression, which may participate in a common signaling pathway, was considered to correlate with bone and kidney phenotypes. 12
In the present study, we explored the role of PKD1 in osteogenesis inhibited by oxidative stress. Under oxidative stress, the effects of PKD1, Sirt1, and osteogenic‐related genes, such as RUNX2, were investigated in cell viability and osteogenesis.
2. MATERIALS AND METHODS
2.1. Cell isolation and culture
Bone marrow (BM)‐MSCs from rats were primarily separated and cultured according to previous reports. 13 , 14 MSCs were isolated from the BM of 4–6‐week‐old Wistar rats. The animals used in our study were obtained from the Shanghai Experimental Animal Center of the Chinese Academy of Science. The Shanghai Jiao Tong University Medical Animal Ethics Committee approved the animal studies. In brief, the marrow sample was flushed and isolated from the bone using low glucose Dulbecco's modified eagle medium (DMEM) and then filtered through 63‐µm meshes. The cells were centrifuged at 1500 rpm for 20 min and then collected, resuspended, and cultured in plates. Cells of three to five passages were used for further study.
2.2. CCK8 assays cell viability of H2O2‐treated BM‐MSCs
A single cell suspension of three passage BM‐MSCs (P3) was seeded in 96‐well plates with 4 × 104 cells per well. After 6 h incubation at 37°C with 5% CO2, 0–400 µM H2O2 was added to cells followed by sustained incubation for 24, 48, and 72 h. A Cell Counting Kit‐8 (CCK8) (Dojindo) was then used to measure cell viability according to the manufacturer's instructions. A Multiskan GO microplate reader (Thermo Fisher Scientific) was used to measure the absorbance of cells at 460 nm.
2.3. Food and Drug Administration (FDA) labeling of H2O2‐treated BM‐MSCs
The BM‐MSCs on the plates were incubated with 10 µM FDA for 1 h at 37°C in the dark, then soaked and washed with phosphate‐buffered saline (PBS) three times for 3 min each time. The liquid on the slide was dried with absorbent paper, and the slide was sealed with sealing liquid containing an anti‐fluorescence quenching agent. Images were then observed and collected under a fluorescence microscope.
2.4. Osteoblast differentiation and H2O2 treatment
For osteogenic differentiation, rat BM‐MSCs (P3) were plated in six‐well plates at a density of 5 × 105 cells/well and incubated in low glucose DMEM (Gibco) with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin and streptomycin (Sigma‐Aldrich) until the cells reached 75% confluence. The medium was replaced by osteogenic differentiation medium (ODM; Cyagen) composed of 10% FBS, 1% penicillin and streptomycin, glutamine (4 mM), ascorbate (50 µg/ml), β‐Glycerophosphate (10 mM), and Dexamethasone (0.1 µM) in DMEM. Fresh ODM was replenished every 3 days. To explore the association of oxidative stress with osteogenic differentiation, BM‐MSCs were pre‐exposed to 100 μM H2O2 diluted in DMEM for 24 h followed by incubation in ODM. Cells were transfected with c‐Myc, PKD1, and TAZ‐siRNA, and a selective SIRT1 inhibitor (Selisistat, EX 527; Sigma‐Aldrich) was used to treat BM‐MSCs.
2.5. ALP activity assay
We performed an alkaline phosphatase (ALP) activity assay to assess the early osteogenic differentiation of rat BM‐MSCs. After osteogenic differentiation for 21 days, cells were stained using a 5‐Bromo‐4‐chloro‐3‐indolyl phosphate/Tetrazole nitro blue Chromogenic Kit (Sigma‐Aldrich) according to the manufacturer's instructions. Briefly, after being fixed, the cells were then washed three times for 5 min each time and then stained using a dye working solution for 20 min in the dark at room temperature. After washing two times with PBS, cells were stained with nuclear solid red staining solution, then dehydrated by anhydrous ethanol, made transparent by xylene, and sealed in a fume hood after neutral drying. Photos of stained cells were taken under a microscope.
2.6. Alizarin red S staining
Alizarin red S staining was performed in the presence of calcium deposits to assess the osteogenic differentiation of rat BM‐MSCs. In brief, the cells were dipped and washed three times with PBS for 1 min each time, fixed using 4% formaldehyde for 15 min, and then washed three times with PBS for 3 min each time, followed by staining with 0.2% (w/v) Alizarin red S for 30 min. Photos of stained cells were taken under a microscope. Cells that were positive for calcium deposition were stained orange‐red.
2.7. Real‐time quantitative PCR (RT‐PCR)
Trizol (Invitrogen) was used to isolate total RNA from BM‐MSCs according to the manufacturer's instructions. Equal amounts of RNA were added to the reverse transcriptase reaction mix with oligo‐dT primers. The templates obtained were subjected to the following PCR with compliant specific primers, as shown in Table 1.
Table 1.
Primers used in qPCR assay
| Genes | Primer | Primer sequence(5′‐3′) | Product |
|---|---|---|---|
| PKD1 | Forward | TGGGTGCTGATGTTCGTAGA | 134 bp |
| Reverse | CGTAATGCAAATGGGCGGAC | ||
| Sirt1 | Forward | GATCTCCCAGATCCTCAAGCC | 189 bp |
| Reverse | TTCCACTGCACAGGCACATA | ||
| c‐MYC | Forward | ACTCGGTGCAGCCCTATTTC | 187 bp |
| Reverse | GTAGCGACCGCAACATAGGA | ||
| RUNX2 | Forward | GCGGTGCAAACTTTCTCCAG | 196 bp |
| Reverse | GTCACTGCACTGAAGAGGCT | ||
| COL1A1 | Forward | TTTCCCCCAACCCTGGAAAC | 287 bp |
| Reverse | CAGTGGGCAGAAAGGGACTT | ||
| BGLAP | Forward | TGAGGACCCTCTCTCTGCTC | 150 bp |
| Reverse | GGTAGCGCCGGAGTCTATTC |
Abbreviation: BGLAP, bone gammacarboxyglutamate (gla) protein (osteocalcin); qPCR, quantitative polymerase chain reaction.
2.8. Western blot analysis
The total protein from BM‐MSCs was extracted using radio immunoprecipitation assay buffer (50 mM Tris‐HCl, pH 7.4,1% NP‐40, 150 mM NaCl, and 0.5% sodium deoxycholate). A Bicinchoninic Acid Protein Assay Kit (Thermo Fisher Scientific) was then used to determine the protein concentration. Total protein samples of cells were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS‐PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). The membranes were incubated with rabbit monoclonal anti‐PKD1 antibody (1:1000; Abcam; ab51246), and rabbit polyclonal anti‐c‐MYC antibody (1:1000; CST; #9402) and anti‐Sirt1 antibody (1:1000; CST; #2493) overnight at 4°C. After being washed, the membranes were incubated with the corresponding horseradish peroxidase‐conjugated secondary antibody for 1 h at 25°C. Protein blots were visualized using enhanced chemiluminescence reagents (ECL; Invitrogen) and quantified using ImageJ software. A rabbit monoclonal anti‐Actin antibody (1:1000; Abcam; ab179467) was used to correct for loading.
2.9. Chromatin immunoprecipitation (ChIP)
We performed a chromatin immunoprecipitation (ChIP) assay according to a previous study. 10 , 15 A ChIP Kit (Merck‐Millipore) was used following the manufacturer's instructions. Briefly, chromatin DNA was immunoprecipitated with rabbit polyclonal anti‐c‐MYC antibody (1:50; CST; #9402) or normal rabbit IgG. After being washed, the DNA and protein‐antibodies were cross‐linked. quantitative polymerase chain reaction (qPCR) was performed to analyze the recovered DNA for the presence of c‐MYC binding motifs at the Sirt1 promoter.
2.10. Dual‐luciferase reporter assay and transfection
According to previous studies, 12 , 16 after transfection using Lipofectamine 3000 (Invitrogen), rat BM‐MSCs were seeded to 24‐well plates to achieve 70% confluency. Cells were then cotransfected with TAZ‐Luc (Luciferase), pRL‐TK (Renilla), PKD1‐CTT or PKD1‐CTT mutant (PKD1‐CTTm), and Lipofectamine for 12 h. The transfected cells were harvested and luciferase activity indicating the relative TAZ activity was assayed using a dual‐luciferase assay kit (Promega) according to the manufacturer's instructions.
2.11. siRNA treatment
Rat BM‐MSCs were seeded into 24‐well plates, and cotransfected with target‐specific siRNA of PKD1 or TAZ, and control siRNA using Lipofectamine 3000. The siRNA sequences that target PKD1 (Gene Bank accession no, NM_001257352.1) or TAZ (Gene Bank accession no, NM_001025748.1) were designed using an internet‐based application (Invitrogen).
2.12. Statistical analysis
GraphPad Prism 8.0 software was used for data analysis. All results are presented as the mean ± SD. Differences between two groups were analyzed using the Student's t test. Differences were considered statistically significant when p <.05.
3. RESULTS
3.1. Expression of PKD1 and cell viability was reduced in BM‐MSCs treated with H2O2
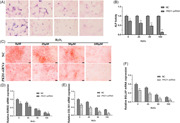
Hydrogen peroxide (H2O2) associated with the production of reactive oxygen species (ROS) is believed to affect MSC differentiation in osteocytes. We first determined the cell viability of BM‐MSCs treated with 0–400 µM H2O2. We found that viability tended to decrease in a dose‐dependent manner and the difference was more significant over time from 24 to 72 h (Figure 1A, B). Moreover, there was no significant difference in the viability of cells treated with either 200 or 400 µM H2O2 for 24 and 72 h. We next measured the expression of PKD1 in H2O2‐treated cells by qPCR and western blot analysis. The results indicated that both the mRNA and protein levels of PKD1 were significantly downregulated in a dose‐dependent manner within a dose range of 0–400 µM H2O2. This was similar to the cell viability results where the two doses of 200 and 400 µM H2O2 exhibited almost no significant difference in PKD1 expression (Figure 1C, D).
Figure 1.
Reduced expression of PKD1 and cell viability in BM‐MSCs treated with H2O2. Primary rat BM‐MSCs were treated with 0–400 µM H2O2. (A) Cell viability was measured using CCK8. (B) FDA was used to fluorescently label BM‐MSCs. (C) Western blot analysis was performed to measure the protein expression of PKD1. (D) qPCR was performed to measure the mRNA expression of PKD1. Data were expressed as the mean ± SD, n = 3. *p < .05, **p < .01 versus 0 µM H2O2‐treated cells. BM‐MSCs, bone marrow‐mesenchymal stem cells; FDA, Food and Drug Administration; mRNA, messenger RNA; qPCR, quantitative polymerase chain reaction
3.2. Upregulation of Sirt1 targets c‐MYC associated with PKD1 down regulation in BM‐MSCs to resist oxidative stress
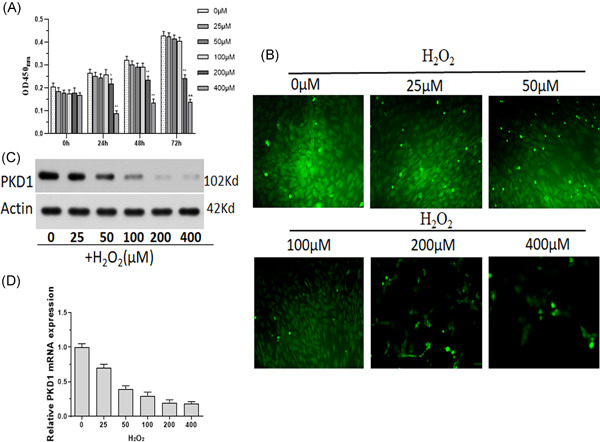
The expression of Sirt1 and c‐MYC in rat BM‐MSCs under oxidative stress was measured. In contrast to the expression of PKD1, both mRNA and protein levels of Sirt1 and c‐MYC were upregulated in BM‐MSCs treated with H2O2 in a dose‐dependent manner (Figure 2A, B). As shown in Figure 2C, D, the RNA interference fragment siRNA2 was the most effective in the knockdown of PKD1 mRNA and protein. Oxidative stress‐induced Sirt1 and c‐MYC upregulation in BM‐MSCs, which was further increased by siRNA PKD1 transfection (Figure 2E, F). Moreover, c‐MYC may bind to the promoter of Sirt1 to upregulate its expression, as confirmed by the results of the ChIP assay (Figure 2G). An RNA interference fragment of siRNA was chosen to knock down c‐MYC expression (Figure 2H, I). BM‐MSCs transfected with PKD1‐siRNA and undergoing oxidative stress showed significantly increased Sir1 expression, whereas c‐MYC‐siRNA transfection decreased Sir1 expression. It is worth noting that c‐MYC‐siRNA could upregulate, but not reverse Sir1 expression, which induced a decrease in PKD1‐siRNA (Figure 2J, K). The viability of BM‐MSCs transfected with c‐MYC‐siRNA was further decreased under oxidative stress (Figure 2L).
Figure 2.
Sirt1 is targeted by c‐MYC to inhibit PKD1 expression in BM‐MSCs after oxidative stress injury. Primary rat BM‐MSCs treated with 0–100 µM H2O2. (A) The protein expression of Sirt1 and c‐MYC was measured using western blot analysis. (B) The mRNA expression of Sirt1 and c‐MYC was measured by qPCR. n = 3. *p < .05, **p < .01 versus 0 µM H2O2‐treated BM‐MSCs. (C) Western blot analysis was performed to measure PKD1 protein expression in BM‐MSCs transfected with RNA interference fragments (siRNA) of PKD1. (D) PKD1 mRNA expression of BM‐MSCs transfected with RNA interference fragments of PKD1 was detected by qPCR. (E, F) mRNA expression of Sirt1 and c‐MYC in PKD1‐siRNA transfected BM‐MSCs simultaneously treated with 0–100 µM H2O2. (G) ChIP assay was used to explore whether c‐MYC may bind to the promoter of Sirt1 to upregulate its expression. (H, I) Western blot analysis and qPCR assays were used to measure the c‐MYC expression of BM‐MSCs transfected with RNA interference fragments (siRNA) of c‐MYC. (J, K) Western blot analysis and qPCR assay was performed to measure Sirt1 expression in BM‐MSCs transfected with PKD1‐siRNA and/or c‐MYC‐siRNA undergoing oxidative stress induced by 100 µM H2O2. (L) Cell viability of BM‐MSCs transfected with PKD1‐siRNA and/or c‐MYC‐siRNA undergoing oxidative stress was measured using CCK8. Data are expressed as the mean ± SD, n = 3. *p < .05, **p < .01 versus negative control RNA fragment (NC) transfected cells. BM‐MSCs, bone marrow‐mesenchymal stem cells; ChIP, chromatin immunoprecipitation; mRNA, messenger RNA; qPCR, quantitative polymerase chain reaction
3.3. PKD1 expression is associated with cell differentiation under oxidative stress
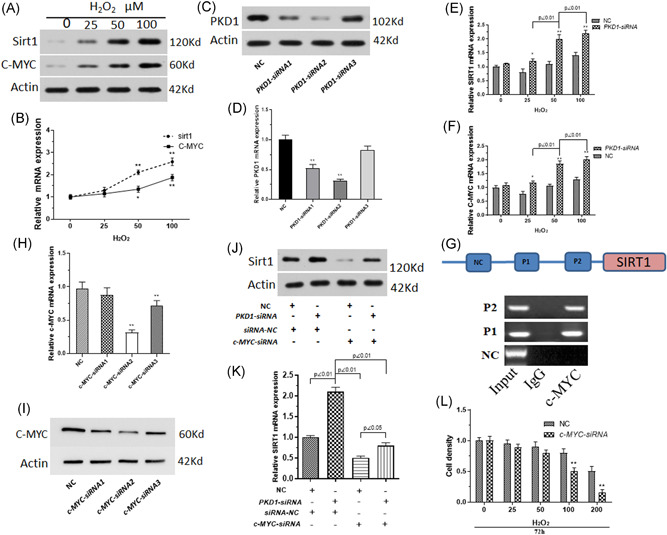
ALP activity is concerned with early osteogenic differentiation. ALP activity was assayed to explore the effect of PKD1 on early osteogenic differentiation of rat BM‐MSCs. As shown in Figure 3A,B, the downregulation of PKD1 was correlated with ALP activity in BM‐MSCs treated with H2O2 in a dose‐dependent manner. The downregulation of PKD1 expression with siRNA aggravated the inhibition of early osteogenic differentiation by H2O2 in BM‐MSCs. Alizarin red S staining indicated that H2O2‐induced calcium deposits decreased during the osteogenic differentiation of rat BM‐MSCs, whereas the siRNA inhibition of PKD1 expression confirmed the induction of oxidative stress by H2O2 (Figure 3C). This was similar to osteogenic differentiation. H2O2 treatment decreased the mRNA expression of RUX2, COL1A, and bone gammacarboxyglutamate (gla) protein (osteocalcin) (BGLAP) whereas the siRNA inhibition of PKD1 expression enhanced the downregulating effects of H2O2 significantly (Figure 3D–F).
Figure 3.
PKD1 is associated with cell differentiation of BM‐MSCs under oxidative stress. Primary rat BM‐MSCs transfected with PKD1‐siRNA and treated with 0–100 µM H2O2. (A, B) ALP activity assays were performed to assess the early osteogenic differentiation of BM‐MSCs. (C) Alizarin red S staining was used to detect the presence of calcium deposits. qPCR was used to measure the mRNA expression of RUNX2 (D), COL1A1, (E), and BGLAP (F). Data are expressed as the mean ± SD, n = 3. *p < .05, **p < .01 versus cells transfected with negative control RNA fragments (NC). BGLAP, bone gammacarboxyglutamate (gla) protein (osteocalcin); BM‐MSCs, bone marrow‐mesenchymal stem cells; mRNA, messenger RNA; qPCR, quantitative polymerase chain reaction
3.4. PKD1 interacted with TAZ under oxidative stress to regulate osteogenic differentiation
To evaluate how PKD1 was involved in osteogenic differentiation under oxidative stress, we further measured PKD1 and Sirt1 expression in BM‐MSCs during osteogenic differentiation under normal conditions or oxidative stress. Our results showed that the mRNA and protein expression of PKD1 was upregulated in osteogenic differentiated cells from 0 to 21 days under normal conditions or oxidative stress. Sirt1 was upregulated in osteogenic differentiated cells from 0 to 14 days, whereas it was downregulated on Day 14 under oxidative stress (Figure 4A, B). It was noted that the mRNA and protein expression of Sirt1 increased mildly during osteogenic differentiation under normal conditions. Calcium deposits and ALP activity of BM‐MSCs transfected with PKD1‐siRNA was significantly decreased compared with negative control cells (Figures 4C, 4D, and 4F). c‐MYC‐siRNA and EX527, an inhibitor of Sirt1, was used to inhibit c‐MYC and Sirt1 in our study. c‐MYC‐siRNA and EX 527 inhibited the ALP activity of BM‐MSCs. The effect of c‐MYC‐siRNA was similar to that of PKD1‐siRNA on calcium deposits and ALP activity (Figure 4C, D), and ALP activity was lower in cells transfected with PKD1‐siRNA and treated with EX 527 than in cells treated with PKD1‐siRNA or EX 527 (Figure 4F). The results of a RUNX2 mRNA expression assay indicated that c‐MYC‐siRNA, PKD1‐siRNA, or EX 527 induced a lower expression of RUNX2, both of which had a synergistic effect on the downregulation of RUNX2 expression (Figure 4E, G). The results of dual‐luciferase reporter assays suggested that PKD1 may associate with TAZ activity (Figure 4H). We further investigated the effect of PKD1 combined with TAZ on the osteogenic differentiation of BM‐MSCs under oxidative stress. Both PKD1‐siRNA and TAZ‐siRNA inhibited ALP activity, RUNX2 expression, and calcium deposits (Figure 4J, K). TAZ‐siRNA especially enhanced the inhibition by PKD1‐siRNA.
Figure 4.
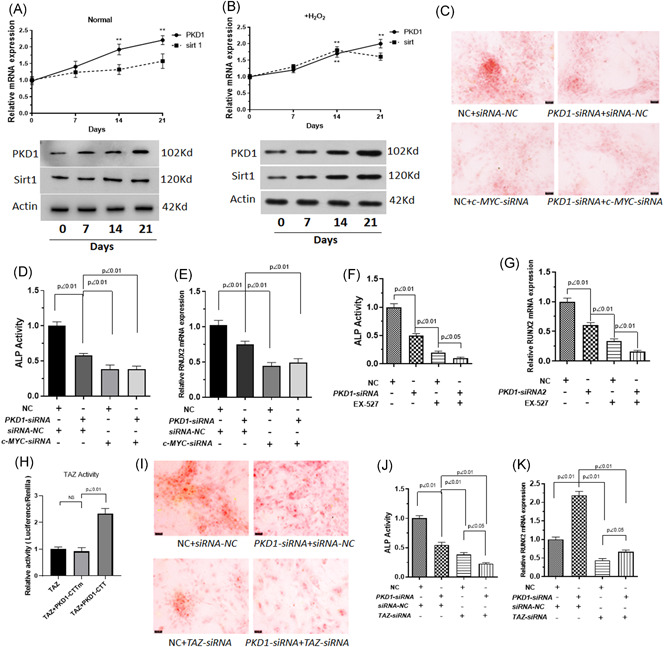
PKD1 regulates osteogenic differentiation by interacting with TAZ under oxidative stress. BM‐MSCs were pre‐exposed or not to 100 μM H2O2 diluted in DMEM for 24 h followed by incubation in differentiation medium (ODM) for 0–20 days and/or treated with the Sirt1 inhibitor, Selisistat (EX 527). Cells were transfected with PKD1‐siRNA, c‐MYC‐siRNA, or TAZ‐siRNA. (A, B) Protein and mRNA expression of PKD1 and Sirt1 was measured by western blot analysis and qPCR under normal conditions or oxidative stress. (C) Alizarin red S staining for the presence of calcium deposits was performed to investigate the effect of PKD1 combined with c‐MYC on the osteogenic differentiation of BM‐MSCs under oxidative stress. (D, F) ALP activity was measured. (E, G) qPCR assays were used to measure RUNX2 mRNA expression. (H) Dual‐luciferase reporter assays were used to evaluate the relation between PKD1 and TAZ activity. (I) Alizarin red S staining was used to detect the presence of calcium deposits, (J) ALP activity, and (K) RUNX2 mRNA expression were obtained from qPCR detection to investigate the effect of PKD1 combined with TAZ on the osteogenic differentiation of BM‐MSCs under oxidative stress. Data are expressed as the mean ± SD, n = 3. NS, not significant. BM‐MSCs, bone marrow‐mesenchymal stem cells; DMEM, Dulbecco's modified eagle medium; mRNA, messenger RNA; qPCR, quantitative polymerase chain reaction
4. DISCUSSION
Osteogenesis is necessary for bone formation. As a type of multipotent stem cell, MSCs can differentiate into osteocytes, myocytes, adipocytes, and adipocytes. Osteogenesis has been reported to be regulated by many conventional pathways. The MSC microenvironment provides dynamic physical and chemical cues essential to cell proliferation, survival, and function for bone regeneration and repair. 17 Stimulation of osteogenesis tends to be regulated by many processes, including ROS. Hydrogen peroxide (H2O2) is a ROS described in many living organisms. 18 During MSC differentiation, ROS appear to be generated and this may be related to some signaling cascades. Oxidants are reported to regulate signaling cascades associated with differentiation to affect the differentiation of MSCs into osteocytes. 19 Wnt is considered as a molecular switch for adipogenic/osteogenic differentiation, 18 and disruption of the Wnt/β‐catenin pathway impairs osteogenesis. 18 , 20 Increased oxidative stress production correlates with decreased bone formation, which was attenuated by the activation of the FOXO family transcription to suppress Wnt signaling. 21 This was similar to the findings of another study, where FOXOs impaired bone formation by antagonizing Wnt signaling in C57BL/6 mice in vivo. 22 Hh signaling was reported to be associated with osteoinduction. Oxidative stress inhibits Hh‐induced osteogenic differentiation in bone marrow‐derived MSCs. 18 , 23 In the present study, bone marrow‐derived MSCs were isolated from rats and induced by H2O2 to explore the factors affecting osteogenesis. Our results indicate that cell viability, ALP activity, which is an early osteoblastic marker, and other osteoblastic markers, including RUX2, COL1A, and BGLAP dose‐dependently decreased BM‐MSCs induced by H2O2, which is similar to the findings of a previous study where H2O2‐induced ALP was significantly decreased in BM stromal cells and H2O2‐induced oxidative stress impaired MSC proliferation. 18 ROS inhibited Hh signaling‐mediated osteogenesis. 18
Many researchers have found that PKD1 has a protective effect against oxidative damage. Activation of PKD1 mediates a compensatory response during the early stage of neuronal degeneration induced by oxidative stress. 24 PKD1 protects intestinal cells from H2O2 induction through NF‐κB and p38 MAPK pathways. 25 Phosphorylation of cytoskeletal proteins mediated by PKD1 could prevent mitochondrial translocation to promote cell survival. 26 Activation of PKD1 may protect neurons from excitotoxicity‐induced damage in neurodegenerative disorders. 27 In another study, researchers found that the inhibition of PKD1 decreased insulin secretion and susceptibility to oxidative stress in p38delta null mice. 28 In the present study, we investigated the role of PKD1 in the osteogenesis differentiation of BM‐MSCs under normal conditions or oxidative stress. Our results indicated that the expression of PKD1 was downregulated in BM‐MSCs induced by H2O2, which was negatively correlated with cell viability. Moreover, cell viability and PKD1 expression were decreased by H2O2 dose‐ and time‐dependently.
Sirt1 is an NAD+‐dependent deacetylase known to be related to cell longevity under oxidative stress. Oxidative damage targets complexes containing Sirt1. 29 Sirt1 deacetylase regulates FOXO transcription factors stress‐dependently. Sirt1 could either increase the ability of FOXO3 to induce resistance to oxidative stress and cell‐cycle arrest or inhibit the ability of FOXO3 to induce cell death. 30 We also found that the expression of Sirt1 and c‐MYC were upregulated in BM‐MSCs induced by H2O2. PKD1 is reported to be involved in bone remodeling and skeletogenesis 2 and regulated by stimulation osteoblast‐specific transcription factor RUNX2. 1 PKD1 was negatively correlated with Sirt1 in renal epithelial cells and embryonic tissues in mice. 10 We further investigated the relation between PKD1, Sirt1, and c‐MYC. Our results showed that PKD1 expression was downregulated and both Sirt1 and c‐MYC expression were upregulated in BM‐MSCs induced by oxidative stress. As in previous studies, c‐MYC is strongly enriched on the PKD1 promoter with RNA pol II to upregulate PKD1 expression and β‐catenin activation, lending credence for the reciprocal crosstalk between c‐MYC and PKD1. 11 Therefore, there is a feed‐forward regulatory PKD1–c‐MYC loop mechanism. c‐MYC may bind to the promoter of Sirt1 to upregulate its expression. PKD1, Sirt1, and c‐MYC can also affect the cell viability of BM‐MSCs under oxidative stress. The effects of PKD1 and Sirt1 on the osteogenic differentiation of BM‐MSCs under oxidative stress were also explored. It was found that the mRNA and protein expression of PKD1 and Sirt1 was upregulated in osteogenic differentiated cells from 0 to 14 days under oxidative stress. EX 527, an inhibitor of Sirt1, inhibited ALP activity of BM‐MSCs even more. PKD1‐siRNA or EX 527 reduced the expression of RUNX2 in BM‐MSCs during osteogenic differentiation, and both EX 527 and RUNX2 had a synergistic effect on the downregulation of RUNX2 expression. Results of a dual‐luciferase reporter assay suggested that PKD1 could bind to TAZ to promote TAZ activity. We further investigated the effect of PKD1 combined with TAZ on the osteogenic differentiation of BM‐MSCs under oxidative stress. The downregulation of both PKD1 and TAZ inhibited ALP activity, RUNX2 expression, and calcium deposits. TAZ‐siRNA especially enhanced the inhibition of ALP activity, RUNX2 expression, and calcium deposits when PKD1 was under‐expressed.
In conclusion, the expression of PKD1 and cell viability in BM‐MSCs was dose‐ and time‐dependently decreased by H2O2. Expression of Sirt1 and c‐MYC was upregulated in H2O2‐treated BM‐MSCs. PKD1 expression was downregulated, while Sirt1 and c‐MYC expression was upregulated in BM‐MSCs under oxidative stress. c‐MYC may bind to the promoter of Sirt1 to upregulate its expression. It was also found that PKD1 was positively and Sirt1 and c‐MYC were negatively correlated with the cell viability of BM‐MSCs under oxidative stress. The reduced expression of PKD1 inhibited ALP activity, RUX2, COL1A, and BGLAP expression in BM‐MSCs induced by H2O2 dose‐dependently. The expression of PKD1 and Sirt1 was upregulated in osteogenic differentiated cells from 0 to 14 days. The inhibition of Sirt1 accompanied with the reduced expression of PKD1 inhibited ALP activity and RUNX2 expression in BM‐MSCs synergistically. PKD1 combined with TAZ may synergistically affect osteogenic differentiation of BM‐MSCs under oxidative stress. The reduced expression of both PKD1 and TAZ inhibited ALP activity, RUNX2 expression, and calcium deposits. Furthermore, TAZ‐siRNA enhanced the inhibition of PKD1 expression on osteogenic differentiation of BM‐MSCs, especially under oxidative stress. Our results suggested that PKD1 may alleviate oxidative stress‐inhibited osteogenesis of rat BM‐MSCs through promoting TAZ activity.
CONFLICT OF INTERESTS
All authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Planned experiments: Chaoyin Jiang, Yong Lu. Performed experiments: Tongtong Chen, Hanqi Wang. Analyzed data: Chaoyin Jiang, Yong Lu. Contributed reagents: Chaoyin Jiang, Yong Lu. Wrote the paper: Tongtong Chen, Hanqi Wang. All authors read and approved the final manuscript.
ACKNOWLEDGMENT
This study was funded by the Science Foundation of the Shanghai Science and Technology Committee (21ZR1439800); Project of the Action Plan of Major Diseases Prevention and Treatment (2017ZX01001‐S12); Project of Integrating Chinese and Western Medicine in General Hospital (ZHYY‐ZXYJHZX‐201901); Correlation between MRI characteristics of articular cartilage under different exercise loads and differentiation of chondrocytes into HTCs‐B (HKM201901).
Chen T, Wang H, Jiang C, Lu Y. PKD1 alleviates oxidative stress‐inhibited osteogenesis of rat bone marrow‐derived mesenchymal stem cells through TAZ activation. J Cell Biochem. 2021;122:1715‐1725. 10.1002/jcb.30124
T. Chen and H. Wang contributed equally to this study and should be considered cofirst authors.
Contributor Information
Chaoyin Jiang, Email: doctor_jiangcy@126.com.
Yong Lu, Email: 18917762053@163.com.
DATA AVAILABILITY STATEMENT
All the data involved in this study are included within the article.
REFERENCES
- 1. Xiao Z, Zhang S, Magenheimer BS, Luo J, Quarles LD. Polycystin‐1 regulates skeletogenesis through stimulation of the osteoblast‐specific transcription factor RUNX2‐II. J Biol Chem. 2008;283:12624‐12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dalagiorgou G, Piperi C, Adamopoulos C, et al. Mechanosensor polycystin‐1 potentiates differentiation of human osteoblastic cells by upregulating Runx2 expression via induction of JAK2/STAT3 signaling axis. Cell Mol Life Sci. 2017;74:921‐936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Komori T. Runx2, an inducer of osteoblast and chondrocyte differentiation. Histochem Cell Biol. 2018;149:313‐323. [DOI] [PubMed] [Google Scholar]
- 4. Dalagiorgou G, Piperi C, Georgopoulou U, Adamopoulos C, Basdra EK, Papavassiliou AG. Mechanical stimulation of polycystin‐1 induces human osteoblastic gene expression via potentiation of the calcineurin/NFAT signaling axis. Cell Mol Life Sci. 2013;70:167‐180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anastassopoulou J, Kyriakidou M, Kyriazis S, et al. Oxidative stress in ageing and disease development studied by FT‐IR spectroscopy. Mech Ageing Dev. 2018;172:107‐114. [DOI] [PubMed] [Google Scholar]
- 6. Wang F, Yin P, Lu Y, et al. Cordycepin prevents oxidative stress‐induced inhibition of osteogenesis. Oncotarget. 2015;6:35496‐35508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee S, Le NH, Kang D. Melatonin alleviates oxidative stress‐inhibited osteogenesis of human bone marrow‐derived mesenchymal stem cells through AMPK activation. Int J Med Sci. 2018;15:1083‐1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Singh P, Hanson PS, Morris CM. SIRT1 ameliorates oxidative stress induced neural cell death and is down‐regulated in Parkinson's disease. BMC Neurosci. 2017;18:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang W, Huang Q, Zeng Z, Wu J, Zhang Y, Chen Z. Sirt1 inhibits oxidative stress in vascular endothelial cells. Oxid Med Cell Longev. 2017;2017:7543973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou X, Fan LX, Sweeney WE, Jr. , Denu JM, Avner ED, Li X. Sirtuin 1 inhibition delays cyst formation in autosomal‐dominant polycystic kidney disease. J Clin Invest. 2013;123:3084‐3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parrot C, Kurbegovic A, Yao G, Couillard M, Cote O, Trudel M. c‐Myc is a regulator of the PKD1 gene and PC1‐induced pathogenesis. Hum Mol Genet. 2019;28:751‐763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Merrick D, Mistry K, Wu J, et al. Polycystin‐1 regulates bone development through an interaction with the transcriptional coactivator TAZ. Hum Mol Genet. 2019;28:16‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miladpour B, Rasti M, Owji AA, et al. Quercetin potentiates transdifferentiation of bone marrow mesenchymal stem cells into the beta cells in vitro. J Endocrinol Invest. 2017;40:513‐521. [DOI] [PubMed] [Google Scholar]
- 14. Choi KS, Shin JS, Lee JJ, Kim YS, Kim SB, Kim CW. In vitro trans‐differentiation of rat mesenchymal cells into insulin‐producing cells by rat pancreatic extract. Biochem Biophys Res Commun. 2005;330:1299‐1305. [DOI] [PubMed] [Google Scholar]
- 15. Yuan J, Minter‐Dykhouse K, Lou Z. A c‐Myc‐SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol. 2009;185:203‐211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiao Z, Baudry J, Cao L, et al. Polycystin‐1 interacts with TAZ to stimulate osteoblastogenesis and inhibit adipogenesis. J Clin Invest. 2018;128:157‐174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee CC, Hirasawa N, Garcia KG, Ramanathan D, Kim KD. Stem and progenitor cell microenvironment for bone regeneration and repair. Regen Med. 2019;14:693‐702. [DOI] [PubMed] [Google Scholar]
- 18. Atashi F, Modarressi A, Pepper MS. The role of reactive oxygen species in mesenchymal stem cell adipogenic and osteogenic differentiation: a review. Stem Cells Dev. 2015;24:1150‐1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shao JS, Aly ZA, Lai CF, et al. Vascular Bmp Msx2 Wnt signaling and oxidative stress in arterial calcification. Ann N Y Acad Sci. 2007;1117:40‐50. [DOI] [PubMed] [Google Scholar]
- 20. James AW. Review of signaling pathways governing MSC osteogenic and adipogenic differentiation. Scientifica. 2013;2013:684736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iyer S, Ambrogini E, Bartell SM, et al. FOXOs attenuate bone formation by suppressing Wnt signaling. J Clin Invest. 2013;123:3409‐3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Almeida M, Han L, Martin‐Millan M, O'Brien CA, Manolagas SC. Oxidative stress antagonizes Wnt signaling in osteoblast precursors by diverting beta‐catenin from T cell factor‐ to forkhead box O‐mediated transcription. J Biol Chem. 2007;282:27298‐27305. [DOI] [PubMed] [Google Scholar]
- 23. Kim WK, Meliton V, Bourquard N, Hahn TJ, Parhami F. Hedgehog signaling and osteogenic differentiation in multipotent bone marrow stromal cells are inhibited by oxidative stress. J Cell Biochem. 2010;111:1199‐1209. [DOI] [PubMed] [Google Scholar]
- 24. Asaithambi A, Kanthasamy A, Saminathan H, Anantharam V, Kanthasamy AG. Protein kinase D1 (PKD1) activation mediates a compensatory protective response during early stages of oxidative stress‐induced neuronal degeneration. Mol Neurodegener. 2011;6:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Song J, Li J, Qiao J, Jain S, Mark Evers B, Chung DH. PKD prevents H2O2‐induced apoptosis via NF‐kappaB and p38 MAPK in RIE‐1 cells. Biochem Biophys Res Commun. 2009;378:610‐614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xiang SY, Ouyang K, Yung BS, et al. PLCepsilon, PKD1, and SSH1L transduce RhoA signaling to protect mitochondria from oxidative stress in the heart. Sci Signal. 2013;6:ra108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pose‐Utrilla J, Garcia‐Guerra L, Del Puerto A, et al. Excitotoxic inactivation of constitutive oxidative stress detoxification pathway in neurons can be rescued by PKD1. Nat Commun. 2017;8:2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sumara G, Formentini I, Collins S, et al. Regulation of PKD by the MAPK p38delta in insulin secretion and glucose homeostasis. Cell. 2009;136:235‐248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Hagan HM, Wang W, Sen S, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606‐619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brunet A, Sweeney LB, Sturgill JF, et al. Stress‐dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011‐2015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data involved in this study are included within the article.


