Abstract
Norovirus (NoV) is the leading cause of acute gastroenteritis (AGE) worldwide. Globally, the GII.4 Sydney 2012 strain has predominated since 2012, although GII.4 variant strains have caused AGE outbreaks in China. Recent patterns of NoV genotype distributions in 6011 children with AGE in Tianjin, China were investigated. NoV was detected using real‐time reverse‐transcriptase polymerase chain reaction and sequencing of partial sequences of the viral capsid gene. NoV genotypes were determined, and phylogenetic analysis was conducted. Epidemiological and clinical data were compared between children infected with different NoV genotypes. NoV was detected in 27.6% of the specimens tested. GII.4 strains comprised 49.4% infections, followed by GII.3 at 39.9%. Genotypes GII.2, GII.13, GII.17, GII.1, GII.6, and GII.14 were also detected. NoV was detected during most of the year, with a peak season of cases in the winter. Diarrhea, vomiting, fever, abdominal pain, and dehydration were present in patients with NoV infection. The main genotypes were GII.4 and GII.3, with a slight increase in GII.2, beginning in March 2017. Among the GII.4 strains, GII.4 Sydney 2012 was the only epidemic strain in Tianjin. Patients with GII.4 genotypes were more likely to present with diarrhea and vomiting than those with GII.3. Children with GII. Others were prone to suffered from dehydration and abdominal pain than those with GII.3. NoV GII has become the main cause of viral AGE in Tianjin, China. The predominant genotypes of NoV were GII.4 and GII.3. Identification of emerging genotypes is crucial for the prevention and control of NoV‐caused AGE.
Keywords: acute gastroenteritis, capsid, children, genetic diversity, genotype, norovirus
1. INTRODUCTION
Norovirus (NoV) is considered to be the leading cause of human virus acute gastroenteritis (AGE) worldwide. 1 NoV was reported to be the second most important cause of AGE in children less than 5 years of age worldwide, following rotavirus. 2 It has been estimated that NoV is associated with 18% of all AGE cases and causes over 200,000 deaths in developing countries annually. 1 Its high morbidity in developing countries makes NoV infections an important public health issue with a substantial socioeconomic burden. 3 Following the introduction of the rotavirus vaccine, NoV is now the leading cause of diarrheal illness requiring medical attention in children less than 5 years of age in the United States. 4
The NoV single‐stranded RNA genome is approximately 7.5 kb in length and is divided into three open reading frames (ORFs). ORF1 encodes the nonstructural viral proteins, including RNA‐dependent RNA polymerase (RdRp). ORF2 encodes the major capsid protein (VP1), and ORF3 codes for the minor structural proteins (VP2). 5 NoV can be classified into at least seven recognized NoV genogroups (GI–GVII), based on the complete amino acid sequence of the capsid protein, with each genogroup subdivided into several genotypes. 6 , 7 Most infections in humans are caused by viruses of genogroups GI, which can be further divided into at least 9 genotypes, and GII, which includes 27 genotypes. 8
Members of genogroup GII are the most predominant and strains of genotype GII.4 have caused the majority of NoV‐related sporadic AGE worldwide since the mid‐1990s. 9 Over the past two decades, new GII.4 variants have emerged every 2–3 years, which resulted in the six global epidemics including in United States 1995/96 in the late 1990s, Farmington Hills virus in 2002, Hunter virus in 2004, Den Haag 2006b virus in late 2007, New Orleans virus in 2009, and the current predominant GII.4 strain in circulation, Sydney 2012. 10 Some studies have found that non‐GII.4 strains can also contribute significantly to the NoV disease burden. Recently, different genotypes (GII.17 and GII.2) emerged in China, Japan, and South Korea and replaced GII.4 strain predominance. 11 , 12 , 13 , 14 Meanwhile, related studies showed that GII.12, GII.1, and GI.6 viruses have been reported to cocirculate with GII.4 strains. 15
To better estimate the burden of NoV infections in Tianjin, we examined the epidemiological and molecular characteristics of NoV in AGE patients in Tianjin, China. Our epidemiological surveillance was initiated in January 2017, with the aim of monitoring the frequency and diversity of variations of NoV genotypes in strains over time. In this study, data from the epidemiological surveillance of NoV infection among children with AGE in Tianjin between 2017 and 2019 were collected and analyzed. Findings from the study could be useful in the early detection of NoV infection, and in initiating rapid responses to potential NoV‐AGE outbreaks.
2. MATERIALS AND METHODS
2.1. Patients and clinical samples
A total of 6011 stool specimens were collected from children who had been hospitalized with AGE at Tianjin Children's Hospital from January 2017 to December 2019. AGE was defined as follows: diarrhea (loose/watery stool) or vomiting with a frequency of three or more times in 24 h. 16 The age of the participants ranged from 1 month to 148 months. All stool specimens were stored at −80°C. The study was conducted according to the Helsinki guidelines and was approved by the Ethics Committee of Tianjin Children's Hospital.
2.2. RNA extraction and viral detection
Stool samples were diluted to 10% suspension with phosphate‐buffered saline and then clarified by centrifugation at 3000g for 10 min. Viral RNA was extracted using QIAamp Viral RNA Mini Kits (Qiagen) according to the manufacturer's instructions, and stored at −80°C. NoV was detected using the Probe real‐time polymerase chain reaction (RT‐PCR) Kit (Langde) on a 7500 Real‐Time PCR platform (ABI). The final volume for each reaction was 50 µl. Each reaction contained 35.8 µl of the reaction mixture, 4.2 µl of enzyme mix, and 10 µl of RNA. The amplification conditions were set as follows: reverse transcription 42°C for 30 min, followed by 95°C for 3 min, then 40 cycles of 95°C for 10 s, 60°C for 1 min. Targets with cycle threshold values below 36 were considered to be positive for that particular sample.
2.3. Genotyping of NoV
To identify the NoV genotype, positive samples were analyzed using conventional reverse‐ transcription polymerase chain reaction (RT‐PCR), using TaKaRa Ex Taq Hot Start RT‐PCR Kits (TaKaRa Shuzou). Partial capsid genes of NoV‐positive samples were amplified using the primer sets G1SKF/G1SKFR, to amplify 331 bp for NoV GI, and G2SKF/G2SKR to amplify 345 bp for the GII genogroup. 17 Each 25 µl reaction mixture contained 2.5 µl of 10× Ex Taq buffer, 2 µl of dNTPs (2.5 mM), 1 µl of each primer, 0.25 µl of Taq DNA polymerase (5 U/µl), and 5 µl of complementary DNA (cDNA), with 13.25 µl of ddH2O added, to reach a volume of 25 µl. PCR was performed under the following conditions: 94°C for 5 min followed by 35 cycles, 94°C for 3 min, 55°C for 45 s, 72°C for 1 min, and final extension at 72°C for 7 min. All RT‐PCR products were analyzed using 1.5% agarose gel electrophoresis. The PCR products were purified and sent to the GENEWIZ Company for sequencing. The genotypes of all NoV strains successfully sequenced were determined by nucleotide sequence analysis using the online Norovirus Typing Tool Version 2.0 (https://www.rivm.nl/mpf/typingtool/norovirus/).
2.4. Phylogenetic analysis
The phylogenetic relationships of NoV were analyzed by aligning sequences using ClustalW software with Kimura's parameter corrections for DNA or proteins. The phylogenetic tree of the nucleotide sequences of the capsid gene was constructed using the neighbor‐joining method with MEGA Version 6.0 software and validated using 1000 bootstrap replicates. 18
2.5. Nucleotide sequence submission
The nucleotide sequence data used in this study had been submitted to the GenBank database and had been assigned accession numbers MW8220022 to MW8220024, MW866524 to MW866528, MW866532 to MW866533, MW866536 to MW866537, MW866543 to MW866545, MW866554 to MW866556, and MW866541.
2.6. Statistical analysis
Data analysis was carried out using SPSS software version 19.0. Categorical variables were compared using χ 2 tests, and continuous variables were compared using t tests. p values <0.05 were considered to be statistically significant.
3. RESULTS
3.1. Detection of NoV infection in AGE patients
In total, 6011 AGE patients were recruited to our study between January 2017 and December 2019. The detection rate of NoV was 27.6%. NoV‐AGE occurred most frequently in the periods from November to February of the following year and reached a peak in December every year (Figure S1). For all 3 years, the genogroup of the AGE cases caused by NoV was GII (Table 1).
Table 1.
Genogroups of NoV detected in AGE patients from 2017 to 2019 in Tianjin, China
| Date | AGE cases | AGE NoV infections | GII | |
|---|---|---|---|---|
| n | % | |||
| 17‐Jan | 30 | 20 | 13 | 65 |
| 17‐Feb | 51 | 19 | 12 | 63.2 |
| 17‐Mar | 73 | 16 | 12 | 75 |
| 17‐Apr | 66 | 14 | 14 | 100 |
| 17‐May | 71 | 15 | 15 | 100 |
| 17‐Jun | 72 | 14 | 10 | 71.4 |
| 17‐Jul | 45 | 4 | 4 | 100 |
| 17‐Aug | 31 | 4 | 3 | 75 |
| 17‐Sep | 14 | 2 | 2 | 100 |
| 17‐Oct | 32 | 5 | 3 | 60 |
| 17‐Nov | 90 | 40 | 11 | 27.5 |
| 17‐Dec | 183 | 88 | 64 | 72.7 |
| 18‐Jan | 198 | 66 | 42 | 63.6 |
| 18‐Feb | 179 | 41 | 25 | 61 |
| 18‐Mar | 224 | 71 | 43 | 60.6 |
| 18‐Apr | 194 | 47 | 19 | 40.4 |
| 18‐May | 153 | 31 | 20 | 64.5 |
| 18‐Jun | 160 | 24 | 12 | 50 |
| 18‐Jul | 127 | 5 | 0 | 0 |
| 18‐Aug | 102 | 4 | 2 | 50 |
| 18‐Sep | 73 | 8 | 1 | 12.5 |
| 18‐Oct | 106 | 22 | 3 | 13.6 |
| 18‐Nov | 276 | 123 | 70 | 56.9 |
| 18‐Dec | 345 | 166 | 72 | 43.4 |
| 19‐Jan | 421 | 186 | 83 | 44.6 |
| 19‐Feb | 362 | 114 | 55 | 48.2 |
| 19‐Mar | 345 | 114 | 49 | 43 |
| 19‐Apr | 276 | 74 | 22 | 30 |
| 19‐May | 272 | 47 | 16 | 34 |
| 19‐Jun | 260 | 35 | 6 | 8.6 |
| 19‐Jul | 206 | 12 | 0 | 0 |
| 19‐Aug | 206 | 4 | 0 | 0 |
| 19‐Sep | 178 | 16 | 4 | 25 |
| 19‐Oct | 155 | 36 | 16 | 44.4 |
| 19‐Nov | 223 | 83 | 41 | 49.4 |
| 19‐Dec | 212 | 88 | 21 | 23.9 |
| Total | 6011 | 1658 | 785 | 47.3 |
Abbreviations: AGE, acute gastroenteritis; NoV, norovirus.
3.2. Epidemiological and clinical characteristics of NoV infection in AGE patients
Among the 6011 children with AGE enrolled in the study, 3674 (61.1%) were male and 2337 (38.9%) were female. The median age of the children with AGE was 12 months. NoV was detected at a higher rate in males (27.7%, 1018/3674) than females (27.4%, 640/2337), but this difference was not significant (p = 0.785).
In the study, AGE cases were divided into five age groups: 0–6, 290 (22%); 7–12 months, 830 patients (32%); 13–36 months, 352 (30.1%); 37–84 months, 127 (20.2%); and over 85 months, 59 (19.3%). Among all age groups, the NoV detection rate was highest in the 7–12 months group, and the difference was statistically significant (χ 2 = 77.744, p < 0.001).
The annual detection rates of NoV were 31.8% (241/758) in 2017, 28.5% (608/2137) in 2018, and 26% (809/3116) in 2019. NoV was more frequently detected in 2017 than in 2018 and 2019 (χ2 = 11.631, p = 0.003). According to the characteristics of the climate in China, 1 year is divided into four seasons: spring (March‐May), summer (June–August), autumn (September–November), and winter (December‐February). The seasonal pattern of NoV was analyzed, and GII NoV infection was prevalent in all seasons from 2017 to 2019. The highest NoV detection rate was in winter (39.8%, 788/1981), followed by autumn (29.2%, 335/1147), spring (25.6%, 429/1674), and summer (8.8%, 106/1209), and there was also a significant difference in NoV detection rate across seasons (χ2 = 366.485, p = 0.000). Clinical and epidemiological data are given in Table 2.
Table 2.
Demographic characteristics of the NoV‐AGE patients in Tianjin, 2017–2019
| AGE patients (n) | NoV‐AGE patients (n) | Detection rate (%) | χ 2 | p value | |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 3674 | 1018 | 27.7 | 0.074 | 0.785 |
| Female | 2337 | 640 | 27.4 | ||
| Age‐group | |||||
| 0–6 months | 1317 | 290 | 22 | ||
| 7–12 months | 2590 | 830 | 32 | ||
| 13–36 months | 1169 | 352 | 30.1 | ||
| 37–84 months | 629 | 127 | 20.2 | 77.744 | 0.000 |
| ≥85 months | 306 | 59 | 19.3 | ||
| Year | |||||
| 2017 | 758 | 241 | 31.8 | ||
| 2018 | 2137 | 608 | 28.5 | 11.631 | 0.003 |
| 2019 | 3116 | 809 | 26 | ||
| Season | |||||
| Spring (Mar–May) | 1674 | 429 | 25.6 | ||
| Summer (Jun–Aug) | 1209 | 106 | 8.8 | ||
| Autumn (Sep–Nov) | 1147 | 335 | 29.2 | 366.485 | 0.000 |
| Winter (Dec–Feb) | 1981 | 788 | 39.8 | ||
| Total | 6011 | 1658 |
Note: The bold values indicates p < 0.05.
Abbreviations: AGE, acute gastroenteritis; NoV, norovirus
3.3. Composition of GII NoV genotypes in AGE patients
A total of 785 NoV strains among 1658 NoV‐positive patients were further genotyped by sequencing the partial VP1. Eight genotypes were detected over the 3 years of the study (Table S1). The genotype diversity was dominated by the GII.4 strain at 49.4%, followed by GII.3 at 39.9%, GII.2 at 7.0%, GII.13 at 1.3%, GII.17 at 1.0%, GII.1 at 0.8%, GII.6 at 0.5%, and GII.14 at 0.1%. Genotype GII.4 Sydney 2012 was the only variant among GII.4 positive samples detected in the study from 2017 to 2019.
These strains dominated the NoV‐AGE epidemics jointly or in turn. The annual distribution of genotypes did not show major fluctuations over time, except for an increase in GII.4 in 2017 and GII.3 in 2018. Although GII.4 was the dominant strain over all four seasons from 2017 to 2019 in Tianjin, an unusual pattern was observed in 2018, when the GII.3 genotype was the most prevalent. The GII.6 strain was first detected in a case reported in January 2018 and was found only in 2018 (Figure 1).
Figure 1.
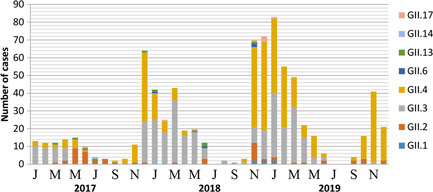
Monthly distribution of NoV genotypes in children with AGE in Tianjin from 2017 to 2019. NoV, Norovirus
3.4. Epidemiological and clinical characteristics of NoV‐AGE patients with different genotypes
Of 785 NoV‐positive samples successfully genotyped, the most common two genotypes, GII.3 and GII.4 were found from 2017 to 2019. The genotype distribution pattern differed according to the gender of the NoV‐AGE patients, but the difference was not statistically significant (χ2 = 2.440, p = 0.295). However, the genotype distribution was significantly different between the age groups (χ2 = 75.2, p = 0.000) (Table 3).
Table 3.
Demographic characteristics of GII NoV‐AGE patients with different genotypes
| GII.3 (n, %) | GII.4 (n, %) | GII. Others (n, %) | χ 2 | p value | |
|---|---|---|---|---|---|
| Age‐group | |||||
| 0–6 months | 76 (24.3) | 35 (9.0) | 8 (9.5) | ||
| 7–12 months | 146 (46.6) | 219 (56.4) | 35 (41.7) | 75.2 | 0.000 |
| 13–36 months | 70 (22.4) | 100 (25.8) | 18 (21.4) | ||
| 37–84 months | 14 (4.5) | 28 (7.2) | 12 (14.3) | ||
| ≥85 months | 7 (2.2) | 6 (1.6) | 11 (13.1) | ||
| Gender | |||||
| Male | 192 (61.3) | 237 (61.1) | 44 (52.4) | 2.440 | 0.295 |
| Female | 121 (38.7) | 151 (38.9) | 40 (47.6) | ||
| Year | |||||
| 2017 | 57 (18.2) | 72 (18.6) | 34 (40.5) | ||
| 2018 | 147 (47) | 128 (33.0) | 34 (40.5) | 44.540 | 0.000 |
| 2019 | 109 (34.8) | 188 (48.5) | 16 (19.0) | ||
| Season | |||||
| Spring (Mar–May) | 133 (42.5) | 58 (14.9) | 19 (22.6) | ||
| Summer (Jun–Aug) | 10 (3.2) | 3 (0.8) | 24 (28.6) | 245.263 | 0.000 |
| Autumn (Sep–Nov) | 12 (3.8) | 116 (29.9) | 23 (27.4) | ||
| Winter (Dec–Feb) | 158 (50.5) | 211(54.4) | 18 (21.4) | ||
| Total | 313 | 388 | 84 |
Note: The bold values indicates p < 0.05.
Abbreviations: AGE, acute gastroenteritis; NoV, norovirus
The distribution of genotypes was differed significantly over time across the 3 years (χ2 = 44.540, p = 0.000). GII.3 was mainly found in 2018, and GII.4 in 2019. Although NoV infections occurred throughout the year, the genotype distribution pattern showed significant seasonal variations across 3 years (χ2 = 245.263, p = 0.000). A major seasonal trend was observed in NoV GII.4 and GII.3, which peaked in the winter, while other GII genotypes were more prevalent in the summer (Table 3).
Further studies indicated that the genotype played a role in the severity of vomiting among the different groups (χ2 = 13.581, p = 0.009). Patients with genotype GII.4 were more likely to have diarrhea and vomiting than those with genotype GII.3 (χ2 = 13.366, p = 0.01; χ2 = 25.804, p = 0.000). However, children with other GII genotypes were more likely to suffer from dehydration and abdominal pain than those with GII.3 genotype (χ2 = 12.475, p = 0.002; χ2 = 9.395, p = 0.009) (Table 4).
Table 4.
Clinical manifestation of GII NoV‐AGE patients with different genotypes
| GII.3 (n, %) | GII.4 (n, %) | GII. Others (n, %) | χ 2 | p value | ||
|---|---|---|---|---|---|---|
| Diarrheal | ||||||
| Yes | 164 (52.4) | 254 (65.5) | 45 (53.6) | 13.366 | 0.01 a | |
| No | 149 (47.6) | 134 (34.5) | 39 (46.4) | |||
| Severity of diarrheal | ||||||
| 1–2 times per day | 95 (57.9) | 120 (47.2) | 18 (40) | |||
| 3–4 times per day | 28 (17.1) | 64 (25.2) | 14 (31.1) | 7.985 | 0.092 | |
| >5 times per day | 41 (25) | 70 (27.6) | 13 (28.9) | |||
| Vomiting | ||||||
| Yes | 122 (39.0) | 226 (58.2) | 43 (51.2) | 25.804 | 0.000 b | |
| No | 191 (61.0) | 162 (41.8) | 41 (48.8) | |||
| Severity of vomiting | ||||||
| 1–2 times per day | 80 (65.6) | 110 (48.7) | 26 (60.5) | |||
| 3–4 times per day | 23 (18.9) | 44 (19.5) | 5 (11.6) | 13.581 | 0.009 | |
| >5 times per day | 19 (15.6) | 72 (31.8) | 12 (27.9) | |||
| Fever (>38°C) | ||||||
| Yes | 191 (61.0) | 231 (59.5) | 40 (47.6) | 5.061 | 0.080 | |
| No | 122 (39.0) | 157 (40.5) | 44 (52.4) | |||
| Dehydration | ||||||
| Yes | 2 (0.6) | 20 (5.2) | 5 (6.0) | 12.475 | 0.002 c | |
| No | 311 (99.4) | 368 (94.8) | 79 (94.0) | |||
| Abdominal pain | ||||||
| Yes | 7 (2.2) | 22 (5.7) | 8 (9.5) | 9.395 | 0.009 d | |
| No | 306 (97.8) | 366 (94.3) | 76 (90.5) | |||
| Total | 313 | 388 | 84 | |||
Note: The bold values indicates p < 0.05.
χ 2 test, p < 0.05 by Bonferroni method multiple comparison test with group, GII.3 versus GII.4.
χ 2 test, p < 0.05 by Bonferroni method multiple comparison test with group, GII.3 versus GII.4.
χ 2 test, p < .0.05 by Bonferroni method multiple comparison test with group, GII.3 versus GII.4, and GII.3 versus GII.other.
χ 2 test, p < 0.05 by Bonferroni method multiple comparison test with group, GII.3 vs GII.Others.
3.5. Phylogenetic relationships among GII strains of NoV
A phylogenetic tree was constructed with 41 sequences from the strains collected from the study and 51 additional reference sequences collected worldwide. The partial VP1 sequences of the majority of strains identified in the present study were highly similar to the corresponding sequences of NoV strains that circulated worldwide. Figure 2 shows that 41 representative strains were highly homologous and clustered together, whereas genotype‐specific genetic clusters were formed in GII.4, GII.3, GII.2, GII.17, GII.13, GII.6, GII.1, and GII.14 branches. The GII.4 Sydney 2012 strain dominated during 2017–2019, and the nine GII.4 Sydney 2012 strains were closely related to the reference strain KP784696.1, with 92.9%–99.3% nucleotide identity. Replacing the GII.4 Sydney 2012 strain, the GII.3 strain emerged in 2018 and was clustered closely with the reference strain KY433576.1, sharing 92.1%–99.6% nucleotide identity.
Figure 2.

Phylogenetic analysis of partial nucleotide sequence of capsid gene of NoV strains detected in children with AGE in Tianjin, 2017–2019. The NoV strains detected in this study are indicated by black dots. The trees were constructed in MEGA 6.0 through the neighbor‐joining method using the Kimura 2‐parameter model. The bootstrap values (1000 replicates) are indicated in the phylogenetic tree. AGE, acute gastroenteritis; NoV, norovirus
4. DISCUSSION
Viral AGE remains an important cause of morbidity and mortality in developing countries. NoV is an important pathogen in humans, causing epidemics and sporadic AGE in both children and adults. 19 In this study, 3 years of data on NoV prevalence, clinical outcomes, and genetic diversity among hospitalized children with AGE from January 2017 to December 2019 in Tianjin were analyzed. The combination of epidemiological, etiological, and molecular data analysis could provide comprehensive evidence to support NoV infection prevention and control measures.
Analysis of the data found that the proportion of NoV in AGE children was 27.6% in Tianjin, similar to results previously reported elsewhere in China. 20 NoV‐AGE was prevalent throughout the year, and peaked in the winter, with a decline in summer, as previously reported. 21 , 22 A correlation between the seasonality of NoV infection and climatic conditions such as rainfall, humidity, and low temperatures has been proposed, due to the frequent transmission of NoV by waterborne sources, and high chances for human‐to‐human transmission due to people crowding together. 23 In Tianjin, the seasonal increase of NoV‐AGE started in October and usually lasted until March of the following year, consistent with the results of Xue et al. 24 In the study, NoV‐positive patients less than 3 years old accounted for 88.9% of all age groups, and children aged from 7 to 12 months seemed particularly vulnerable to NoV infection, a finding which was consistent with previous studies. 25
The circulating genotypes of NoV vary with time, place, and population in China, so these conditions might influence the size and severity of NoV‐AGE epidemics. Since 2002, GII.4 viruses have been the most common genotypes circulating in China. 26 A new GII.4 NoV strain was identified in Australia in March 2012 and named GII.4 Sydney 2012. As the predominant strain of the GII.4 genotype, GII.4 Sydney 2012 and its variants have been widely reported since 2012, 27 , 28 , 29 and has caused AGE outbreaks in multiple countries. 30 This phenomenon might be explained by the fact that the GII.4 strain has a 1.7‐fold higher rate of evolution on average within the capsid sequence, and a higher number of nonsynonymous changes than other NoVs, resulting in the emergence of a new strain of GII.4 every 2–3 years. 31 In this study, eight NoV GII genotypes—GII.4 (49.4%), GII.3 (39.9%), GII.2 (7.0%), and GII.13 (1.3%)—were the four most prevalent strains in Tianjin from 2017 to 2019, a result which is similar to those reported from other studies. 26 Genotypic and phylogenetic analysis showed that GII.4 Sydney 2012 strains were highly homologous. They were closely related to South Africa strain KP784696.1 (GII.P4‐GII.4), which indicated that GII.4 Sydney 2012 strains in our study might be recombinant strains.
Further studies were conducted to explore the epidemiological characteristics of the epidemic strains and provide a theoretical basis for the development of disease prevention strategies. Eight NoV genotypes were detected in this study. Each year, two to four genotypes cocirculated, with GII.4 strains detected most frequently. Children with GII.3 were concentrated into the 0–6 and 7–12 months groups, but children with GII.4 and GII. Others were more prevalent in the 7–12 and 13–36 months groups. The question of whether NoV genotype is linked to age‐group in children remains to be answered. There were no significant differences in gender among children infected with different NoV genotypes, indicating that gender was not a susceptibility factor in NoV infection. NoV was detected throughout the year, but GII.4 and GII.3 dominated during the cold winter, and GII. Others became the predominant genotypes during the summer. This trend is consistent with those identified in some of the published studies. 32 Patients infected with NoV in this study shared most of the common clinical symptoms of AGE, such as fever, vomiting, diarrhea, abdominal pain, and dehydration. However, the degree of severity varied between patients infected by different NoV genotypes. Patients with GII.4 genotypes were more likely to have diarrhea and vomiting than those with GII.3 genotypes. Children with GII. Others were more likely to suffer from dehydration and abdominal pain than those with GII.3 genotypes. Therefore, more clinical studies are necessary to conclusively identify the different clinical characteristics associated with infection with different NoV genotypes.
Our study had some limitations. First, NoV genotyping was conducted based only on the VP1 sequence. Other genes especially the RdRp gene should be analyzed to explain recombinant events and enhance the accuracy. Second, we only detected NoV in our study, other common gastroenteritis pathogens were not screened in our samples. Meanwhile, our study placed emphasis on molecular epidemiological analysis of NoV infection, which could only trace out the circulating patterns rather than capturing the genomic diversity. It would also become our research focus in the future.
In conclusion, this study reported an epidemiological and molecular investigation of NoV infection in children with AGE in Tianjin, China. NoV GII has been identified as the most frequent genogroup responsible for AGE in this area. Our study further found that genotype GII.4 was endemic, but genotypic diversity was high during the years of the study, suggesting that continuous surveillance is necessary, to monitor genotypes and the emergence of new strains, as a basis for the development of prevention and control strategies. Finally, further studies that include larger sample sizes from several regions of China are needed, to provide a better perspective into the strains circulating throughout the country.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
All authors contributed to the intellectual content of this manuscript and approved the final manuscript as submitted. Yulian Fang designed the study, performed the analysis, and drafted the manuscript. Yanzhi Zhang and Hong Wang conceived and designed the experiments, analyzed all the data. Ouyan Shi and Mengzhu Hou performed a phylogenetic analysis of the sequence and statistical analysis. Jinying Wu, Lu Wang, and Wei Wang searched the literature and collected the clinical samples. Yu Zhao revised the article critically for important intellectual content.
Supporting information
Supplementary information.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (Grant number 81771589), and the Program of Tianjin Science and Technology Plan (Grant number 18ZXDBSY00170).
Fang Y, Zhang Y, Wang H, et al. Molecular epidemiology of norovirus infections in children with acute gastroenteritis in 2017–2019 in Tianjin, China. J Med Virol. 2022;94:616‐624. 10.1002/jmv.27340
Yulian Fang, Yanzhi Zhang, and Hong Wang contributed equally to this study.
DATA AVAILABILITY STATEMENT
The datasets used in the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Lopman BA, Steele D, Kirkwood CD, Parashar UD. The vast and varied globalburden of norovirus: prospects for prevention and control. PLoS Med. 2016;13(4):e1001999. 10.1371/journal.pmed.1001999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Doll MK, Gagneur A, Tapiéro B, et al. Temporal changes in pediatric gastroenteritis after rotavirus vaccination in Quebec. Pediatr Infect Dis J. 2016;35(5):555‐560. 10.1097/INF.0000000000001077 [DOI] [PubMed] [Google Scholar]
- 3. Mugyia AE, Ndze VN, Akoachere J, et al. Molecular epidemiology of noroviruses in children under 5 years of age with acute gastroenteritis in Yaoundé, Cameroon. J Med Virol. 2019;91(5):738‐743. 10.1002/jmv.25380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Payne DC, Vinjé J, Szilagyi PG, et al. Norovirus and medically attended gastroenteritis in US children. N Engl J Med. 2013;368(12):1121‐1130. 10.1056/NEJMsa1206589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Green KY. Caliciviridae: the noroviruses. In: Knipe DM, Howley PM, eds. Fields Virology. Lippincott Williams & Wilkins; 2013:582‐604. [Google Scholar]
- 6. Mans J, Armah GE, Steele AD, Taylor MB. Norovirus epidemiology in Africa: a review. PLOS One. 2016;11(4):e0146280. 10.1371/journal.ppat.1002705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ouédraogo N, Kaplon J, Bonkoungou IJ, et al. Prevalence and genetic diversity of enteric viruses in children with diarrhea in Ouagadougou, Burkina Faso. PLOS One. 2016;11(4):e0153652. 10.1371/journal.pone.0153652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chhabra P, de Graaf M, Parra GI, et al. Updated classification of norovirus genogroups and genotypes. J Gen Virol. 2019;100(10):1393‐1406. 10.1099/jgv.0.001318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Siebenga JJ, Vennema H, Zheng DP, et al. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. J Infect Dis. 2009;200(5):802‐812. 10.1086/605127 [DOI] [PubMed] [Google Scholar]
- 10. Lim KL, Hewitt J, Sitabkhan A, et al. A multi‐site study of norovirus molecular epidemiology in Australia and New Zealand, 2013‐2014. PLOS One. 2016;11(4):e0145254. 10.1371/journal.pone.0145254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu J, Fang L, Zheng H, et al. The evolution and transmission of epidemic GII.17 noroviruses. J Infect Dis. 2016;214(4):556‐564. 10.1093/infdis/jiw208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Graaf M, van Beek J, Vennema H, et al. Emergence of a novel GII.17 norovirus‐end of the GII.4 era? Euro Surveill. 2015;20(26):21178. 10.2807/1560-7917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chan MCW, Lee N, Hung TN, et al. Rapid emergence and predominance of a broadly recognizing and fast‐evolving norovirus GII.17 variant in late 2014. Nat Commun. 2015;6:10061. 10.1038/ncomms10061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kwok K, Niendorf S, Lee N, et al. Increased detection of emergent recombinant norovirus GII.P16‐GII.2 strains in young adults, Hong Kong, China, 2016‐2017. Emerg Infect Dis. 2017;23(11):1852‐1855. 10.3201/eid2311.170561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vega E, Barclay L, Gregoricus N, Shirley SH, Lee D, Vinjé J. Genotypic and epidemiologic trends of norovirus outbreaks in the United States, 2009 to 2013. J Clin Microbiol. 2014;52(1):147‐155. 10.1128/JCM.02680-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chan MC, Leung TF, Chung TW, et al. Virus genotype distribution and virus burden in children and adults hospitalized for norovirus gastroenteritis, 2012‐2014, Hong Kong. Sci Rep. 2015;5:11507. 10.1038/srep11507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kojima S, Kageyama T, Fukushi S, et al. Genogroup‐specific PCR primers for detection of Norwalk‐like viruses. J Virol Methods. 2002;100(1‐2):107‐114. 10.1016/s0166-0934(01)00404-9 [DOI] [PubMed] [Google Scholar]
- 18. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA 6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725‐2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caddy S, Breiman A, le Pendu J, Goodfellow I. Genogroup IV and VI canine noroviruses interact with histo‐blood group antigens. J Virol. 2014;88(18):10377‐10391. 10.1128/JVI.01008-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fu JG, J, Zhang J, et al. Molecular epidemiology of genogroup II norovirus infection among hospitalized children with acute gastroenteritis in Suzhou (Jiangsu, China) from 2010 to 2013. J Med Virol. 2016;88(6):954‐960. 10.1002/jmv.24429 [DOI] [PubMed] [Google Scholar]
- 21. Zeng M, Xu X, Zhu C, et al. Chinese pediatric study group of norovirus. Clinical and molecular epidemiology of norovirus infection in childhood diarrhea in China. J Med Virol. 2012;84(1):145‐151. 10.1002/jmv.22248 [DOI] [PubMed] [Google Scholar]
- 22. Tan D, Deng L, Wang M, Li X, Ma Y, Liu W. High prevalence and genetic diversity of noroviruses among children with sporadic acute gastroenteritis in Nanning City, China, 2010‐2011. J Med Virol. 2015;87(3):498‐503. 10.1002/jmv.24103 [DOI] [PubMed] [Google Scholar]
- 23. Zhou N, Zhang H, Lin X, et al. A waterborne norovirus gastroenteritis outbreak in a school, Eastern China. Epidemiol Infect. 2016;144(6):1212‐1219. 10.1017/S0950268815002526 [DOI] [PubMed] [Google Scholar]
- 24. Xue Y, Pan H, Hu J, et al. Epidemiology of norovirus infections among diarrhea outpatients in a diarrhea surveillance system in Shanghai, China: a cross‐sectional study. BMC Infect Dis. 2015;15:183. 10.1186/s12879-015-0922-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oluwatoyin Japhet M, Adeyemi Adesina O, Famurewa O, et al. Molecular epidemiology of rotavirus and norovirus in Ile‐Ife, Nigeria: high prevalence of G12P[8] rotavirus strains and detection of a rare norovirus genotype. J Med Virol. 2012, 84(9):1489‐1496, 10.1002/jmv.23343 [DOI] [PubMed] [Google Scholar]
- 26. Zhou HL, Zhen SS, Wang JX, et al. Burden of acute gastroenteritis caused by norovirus in China: a systematic review. J Infect. 2017;75(3):216‐224. 10.1016/j.jinf.2017.06.004 [DOI] [PubMed] [Google Scholar]
- 27. Manso CF, Romalde JL. Molecular epidemiology of norovirus from patients with acute gastroenteritis in northwestern Spain. Epidemiol Infect. 2015;143(2):316‐324. 10.1017/S0950268814000740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jung S, Hwang BM, Jeong HJ, et al. Occurrence of norovirus GII. 4 Sydney variant‐related outbreaks in Korea. Osong Public Health Res Perspect. 2015;6(5):322‐326. 10.1016/j.phrp.2015.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Polkowska A, Ronnqvist M, Lepisto O, et al. Outbreak of gastroenteritis caused by norovirus GII. 4 Sydney variant after a wedding reception at a resort/activity centre, Finland, August 2012. Epidemiol Infect. 2014, 142, (9), 1877‐1883, 10.1017/S0950268813002847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Beek J, de Graaf M, Xia M, et al. Comparison of norovirus genogroup I, II and IV seroprevalence among children in the Netherlands, 1963, 1983 and 2006. J Gen Virol. 2016;97(9):2255‐2264. 10.1099/jgv.0.000533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bull RA, Eden JS, Rawlinson WD, White PA. Rapid evolution of pandemic noroviruses of the GII. 4 lineage. PLoS Pathog. 2010;6(3):e1000831. 10.1371/journal.ppat.1000831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xue C, Pan L, Zhu W, et al. Molecular epidemiology of genogroup II norovirus infections in acute gastroenteritis patients during 2014‐2016 in Pudong New Area, Shanghai, China. Gut Pathog. 2018;10:7. 10.1186/s13099-018-0233-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Data Availability Statement
The datasets used in the current study are available from the corresponding author on reasonable request.


