Abstract
Glucocorticoids enhance memory consolidation of emotionally arousing events via largely unknown molecular mechanisms. This glucocorticoid effect on the consolidation process also requires central noradrenergic neurotransmission. The intracellular pathways of these two stress mediators converge on two transcription factors: the glucocorticoid receptor (GR) and phosphorylated cAMP response element‐binding protein (pCREB). We therefore investigated, in male rats, whether glucocorticoid effects on memory are associated with genomic interactions between the GR and pCREB in the hippocampus. In a two‐by‐two design, object exploration training or no training was combined with post‐training administration of a memory‐enhancing dose of corticosterone or vehicle. Genomic effects were studied by chromatin immunoprecipitation followed by sequencing (ChIP‐seq) of GR and pCREB 45 min after training and transcriptome analysis after 3 hr. Corticosterone administration induced differential GR DNA‐binding and regulation of target genes within the hippocampus, largely independent of training. Training alone did not result in long‐term memory nor did it affect GR or pCREB DNA‐binding and gene expression. No strong evidence was found for an interaction between GR and pCREB. Combination of the GR DNA‐binding and transcriptome data identified a set of novel, likely direct, GR target genes that are candidate mediators of corticosterone effects on memory consolidation. Cell‐specific expression of the identified target genes using single‐cell expression data suggests that the effects of corticosterone reflect in part non‐neuronal cells. Together, our data identified new GR targets associated with memory consolidation that reflect effects in both neuronal and non‐neuronal cells.
Keywords: cyclic AMP response element binding, glucocorticoid receptor, learning, object location memory
Administration of corticosterone after object location training enhanced long‐term hippocampal memory. Glucocorticoid receptor and phosphorylated cAMP response element‐binding protein were co‐bound at the hippocampal genome and exclusively GR DNA‐binding levels were altered after corticosterone, yet binding of these two transcription factors was not affected by training nor was any interaction detected. Corticosterone altered the transcriptome in a context‐independent manner—partially via direct GR‐regulation—and reflects involvement of both neuronal and non‐neuronal cells in the male rat hippocampus.
Abbreviations
- ChIP‐seq
chromatin immunoprecipitation followed by sequencing
- CRE
cAMP response element
- Fc‐FC
fold change–fold change
- GR
glucocorticoid receptor
- GRE
glucocorticoid response element
- pCREB
phosphorylated cAMP response element‐binding protein
- rn6
Rattus Norvegicus genome version 6
- TSS
transcription start site
1. INTRODUCTION
Glucocorticoid hormones are well known to enhance memory consolidation of emotionally arousing experiences. Post‐training administration of glucocorticoids was previously shown to enhance memory formation in a variety of behavioural paradigms in rodents, including fear conditioning and inhibitory avoidance tasks (Atucha et al., 2015; Hui et al., 2004). Glucocorticoids also enhance the consolidation of object recognition memory. In conditions where training alone does not result in long‐term object recognition memory, post‐training corticosterone administration (mimicking a stressful event) acts as a switch, and enables consolidation of the otherwise neutral experience (Okuda et al., 2004). This corticosterone effect was found to be dependent on noradrenaline signalling as concomitant administration of the β‐adrenoceptor antagonist propranolol blocks the memory enhancement by corticosterone (Roozendaal et al., 2006). Prior habituation of the animal (preventing arousal‐induced increases in noradrenaline during training) also impedes the effect of corticosterone, confirming the involvement of endogenous noradrenaline (Okuda et al., 2004; Roozendaal et al., 2006). Taken together, there is a codependency of glucocorticoid and noradrenaline signalling in experimental settings where neither stress mediator on its own is able to induce long‐term memory.
This dependency may have its basis either at the cellular or the circuit level. One possibility is an interaction downstream in the intracellular pathways of these hormones during the memory consolidation process (Krugers et al., 2012). Corticosterone is known to exert its memory‐enhancing effects via the glucocorticoid receptor (GR) (Oitzl & de Kloet, 1992). The GR is a transcription factor that is widely expressed in the brain and its transcriptional effects are known to be modulated by the (cellular) context (Datson et al., 2013; John et al., 2011; Koning et al., 2019; Meijer et al., 2019; Provencal et al., 2020). Non‐genomic effects via membrane‐bound variants of the GR have been reported and corticosterone increases pCREB levels in multiple brain regions (Atsak et al., 2015; Joels et al., 2011; Roozendaal et al., 2010). These non‐genomic effects possibly interact with or prepare for genomic effects (Joels et al., 2013; Karst et al., 2010). Also, the transcriptional effects of GR are required for hippocampal memory, as mice lacking the ability to dimerize GR and therefore have impaired GR binding to DNA (GRdim/dim), exhibit impaired water‐maze spatial memory (Oitzl et al., 2001). The importance of GR's ability to bind the DNA for memory consolidation introduces the genome as another level of potential interaction with noradrenergic signalling. Upon release, noradrenaline binds to β‐adrenoceptors on the cell surface, activating various signalling cascades, one of which that leads to phosphorylation and thereby activation of cAMP response element‐binding protein (pCREB) (Hagena et al., 2016; Mayr & Montminy, 2001).
In addition, pCREB can also be activated by various non‐adrenergic signalling cascades that may be associated with the synaptic input during memory training (Gregory & Goudet, 2021; Lonze & Ginty, 2002). pCREB is a widely expressed, neuronal activity‐regulated transcription factor that is required for various forms of memory, including spatial memory (Silva et al., 1998; Tanis et al., 2008). Both GR and pCREB bind to specific motifs in the genome, the glucocorticoid response element (GRE) and cAMP response element (CRE) respectively, to exert their transcriptional effects by regulating the expression of specific target genes (Pooley et al., 2017; Tanis et al., 2008). A direct interplay between these two transcriptions factors that are both well known for their role in learning and memory is therefore plausible, yet not studied.
We studied the potential interaction of these two transcription factors in a two‐by‐two design, where object exploration training or no training was combined with post‐training administration of a memory‐enhancing dose of corticosterone or vehicle. Using this approach, we investigated the potential interaction of the two transcription factors at the hippocampal genome using chromatin immunoprecipitation followed by sequencing (ChIP‐seq) and transcriptome analysis on whole hippocampus. We hypothesized that GR and pCREB would independently be activated by corticosterone injection and training, but would show interactions under combined treatment. We confirmed glucocorticoid enhancement of hippocampal memory formation and observed substantial effects of corticosterone on GR binding and gene expression but observed minimal differences in pCREB binding in any of the groups.
2. MATERIALS AND METHODS
2.1. Animals
Adult male Sprague–Dawley rats (340–400 g at time of behavioural experiments) were obtained from Charles River Laboratories (Germany and Italy) and were single housed at 21 ± 1°C with a 12:12‐hr light‐dark cycle and ad libitum access to food and water. Training and testing was performed during the light phase (lights on at 07:00 h) between 10:00 and 15:00 h, when endogenous corticosterone levels are at the nadir of the circadian cycle. All procedures were in compliance with the European Communities Council Directive on the use of laboratory animals (2010/63/EU), the Dutch law on animal experiments, the D.L. 26/2014 of the Italian Ministry of Health and were approved by the Animal Ethics Committee of Radboud University, Nijmegen, the Netherlands.
For the behavioural experiments animals received object exploration training and retention was tested 24 hr later to determine the optimal memory‐enhancing dose of corticosterone. For the molecular work, rats were grouped according to a two‐by‐two study design with object exploration training versus no training and corticosterone (3.0 mg/kg) versus vehicle injection as factors resulting in four groups: (1) no training—vehicle, (2) no training—corticosterone, (3) training—vehicle and (4) training—corticosterone. The animals were sacrificed 45 min after injection for ChIP‐seq and 3 hr after injection for RNA‐seq.
2.2. Object location memory task
The experimental apparatus used for the object location task was a grey open‐field box (40 × 40 × 40 cm) with a sawdust‐covered floor placed in a dimly illuminated room. The objects used were white glass light bulbs (6 cm diameter by 11 cm length) and transparent glass vials (5.5 cm diameter by cm height). Starting 5 days before the training, the rats were handled for 1–2 min/day to habituate them to interactions with the experimenter. The rats were not habituated to the experimental apparatus to ensure novelty‐induced noradrenergic arousal during the training trial (Campolongo et al., 2013; Okuda et al., 2004). On the training trial, the rat was placed in the experimental apparatus and allowed to explore two identical, symmetrically placed objects for three minutes. Behaviour was recorded by a camera mounted above the box. The type of objects used and their corresponding locations (for retention testing) were counterbalanced to reduce potential bias of location and of object preference. Sawdust was stirred and the objects were cleaned with 70% ethanol in between animals to prevent the presence of olfactory trails that could influence object exploration. Immediately after object exploration training, rats received a subcutaneous injection (2.0 ml/kg) of corticosterone (0.3, 1.0, 3.0 mg/kg; Sigma‐Aldrich) dissolved in 5% ethanol in saline. Control animals received 5% ethanol in saline only. For behaviour, the animal was placed back in its home cage for 24 hr until the memory retention test. For molecular analysis, the animal was returned to its home cage until sacrifice 45 min or 3 hr after the injection. For the retention test, two copies of the familiar objects were placed in the experimental apparatus, one in the same location as during training and the other in a novel location. The rat was placed in the experimental apparatus for three minutes. Object exploration was analysed with the Observer XT software (Noldus Information Technology). The time spent exploring each object was measured on both the training and retention test trials. Exploration of an object was defined as pointing the nose to the object at a distance of <1 cm and/or touching it with the nose. Turning around, nibbling, climbing or sitting on an object was not considered exploratory behaviour. To analyse memory performance, a discrimination index was calculated as the difference in time exploring the object in the novel and familiar location, expressed as the ratio of the total time spent exploring both objects. Rats showing a total object exploration time less than 8 s on either training or testing (seven animals) or a discrimination index >2× standard deviation from group mean (two animals) were excluded from further analysis.
2.3. Plasma corticosterone
Trunk blood was collected in EDTA‐coated tubes to determine plasma corticosterone levels. Blood was centrifuged at 3.000 g for 15 min and plasma was transferred to new tubes and stored at −20°C. Corticosterone levels of the ChIP‐seq cohort were determined using a 125I radioimmunoassay kit, according to the manufacturer's instructions (MP Biomedicals) and for the RNA‐seq cohort an enzyme immune assay kit was used, according to the manufacturer's instructions (IDS).
2.4. ChIP‐sequencing
The hippocampus was freshly dissected, snap‐frozen in liquid nitrogen and stored at −80°C. Hippocampal DNA‐binding of transcription factors GR and pCREB during the post‐learning consolidation period was assessed using ChIP‐seq. ChIP was performed as described before (van Weert et al., 2017). All buffers used during tissue processing and the ChIP protocol were supplemented with protease and phosphatase inhibitors (Roche). Hippocampal tissue was fixated with 1% formaldehyde for 12–14 min and homogenized in Jiang buffer using a glass douncer (Kimble‐Chase). Chromatin of four hippocampi (i.e. bilateral hippocampi of two rats from the same experimental group) was pooled. Tissue of rats from different training days and times were pooled to prevent an effect of time and day. Hippocampi were resuspended in NP‐40 containing lysis buffer and fragmented by sonication for 32 cycles (30 s on/30 s off) using a Bioruptor (Diagenode). From each chromatin sample, an input aliquot was taken and pooled per group (n = 4 biological replicates per group), which resulted in a combined input sample per treatment group (50 µl total). The chromatin sample was split for a paired GR and pCREB ChIP (700 µl each), using 6 µg of anti‐GR antibody H‐300 (sc‐8992X, Santra Cruz) or 4 µg of anti‐phospho‐CREB Ser133 antibody (17–10131, Millipore), respectively. After several washing steps (van Weert et al., 2017), antibody‐bound DNA was collected with 250‐µl elution buffer (0.1 M NaHCO3, 1% SDS) while shaking at 37°C for 15 min. Input and eluted ChIP samples were decrosslinked (400 mM NaCl, overnight at 65°C), purified by phenolization and subsequently dissolved in 60‐µl TE buffer (10 mM Tris‐HCl, 1mM EDTA).
Before sequencing, adapters (Agilent) were ligated and samples were subjected to 15 rounds of polymerase chain reaction (PCR) for DNA library preparation (KAPA Biosystems). Single‐end sequencing was performed on a HiSeq 2500 (Illumina) at High Output. Due to overrepresentation of the input samples, the ChIP samples were sequenced over two runs to obtain the intended number of reads. In the first run, 51 bp were sequenced; as a result of developments at the sequencing facility (The Netherlands Cancer Institute), this was increased to 65 bp for the second run. Combined, the two runs gave a total of 11.0–22.5 million reads per GR ChIP sample and 13.5–24.8 million reads per pCREB ChIP sample. ChIP‐seq data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE160806.
2.5. ChIP‐sequencing data analysis
The Carp pipeline v0.8.0, published as part of Bio Pipeline Execution Toolkit (Biopet), was used for read quality control, read alignment and peak calling. Biopet contains the main sequencing analysis pipelines developed at Leiden University Medical Center with code being accessible at https://github.com/biopet/biopet. The rest of the analysis was done using custom scripts developed for this particular project.
Reads were aligned to Rattus norvegicus genome version 6 (rn6) with short read aligner bwa‐mem (version 0.7.10). Peaks were called based on unique reads using Model‐based Analysis of ChIP‐Seq (MACS2, version 2.1.1.20160309), invoking subcommand ‘callpeak’. Tool settings used were effective genome size = 2.00e + 09; q‐value cut‐off = 0.001; bdg true. For every sample, an input sample (one per treatment group) was provided. For both GR and pCREB, this step provided 16 (four replicates for each of the four treatment groups) BED files with peak (narrowPeak) locations in each sample. Separately for GR and pCREB, the corresponding 16 BED files were merged using mergeBed (version 2.26.0), resulting in a list with locations of all peaks found in any of the treatment groups. Overlapping peak regions were replaced by unions of the regions, leading to a single regions BED file for GR and one for pCREB. PCR duplicates were removed from the BAM files before the counting step. For the calculated regions and for each sample unique, non‐duplicated read counts were generated using HTSeq‐count (version 0.6.1). Tool settings used were as follows: ‐s no, ‐m intersection‐strict, ‐f bam (see Biopet website).
The goal of the analysis was to determine how glucocorticoid treatment, object exploration training and the combination affected DNA‐binding of each of the transcription factors. All GR samples were included in the analysis, and two of the samples for pCREB (1A and 4A) were identified as outliers based on sample‐to‐sample distance matrix and principal component analysis and excluded from further analysis. We used DEseq2 (version 1.29.4) (Love et al., 2014) for normalization of the read counts data (median of ratio's method) and identification of regions with differential counts (Love et al., 2014). For the differential binding analysis, we selected only regions which were present in a minimum of three replicates with more than ten reads for at least one of the groups to reduce false positive signals. For both GR and pCREB, four contrasts were analysed for differential binding in a pair‐wise comparison. A false discovery rate (FDR) adjusted p value of 0.05 was used as a cut‐off to classify a region as differentially bound in a contrast. Subsequently, all samples were pooled per treatment for each targeted transcription factor. Regions present in a minimum of three replicates were selected for the analyses.
The resulting peak files were annotated using HOMER to the rn6 genome using annotatePeaks.pl with default settings (Heinz et al., 2010). Promoter regions were defined as 5‐kb upstream to 100‐bp downstream transcription start sites (TSS). Cobinding of GR and pCREB was determined using the bedtools overlap (version 2.26.0). Coverage plots were created using Deeptools (version 3.5.0) (Ramirez et al., 2016) after bins per million mapped reads normalization and averaging of bw files with bamCoverage (version 3.5.0). De novo motif analysis was performed using MEME (version 5.0.5) (Bailey et al., 2009). Tool settings used were as follows: ‐dna ‐time 3,000, ‐maxsize 1,000,000 ‐mod zoops ‐nmotifs 20 ‐minw 6 ‐maxw 25 ‐revcomp.
2.6. Comparison to published ChIP‐seq data
Genomic coordinates of identified GR binding sites by Pooley et al. (2017) and pCREB binding sites by Lesiak et al. (2013) were obtained from the supplementary data. As described above, peaks were annotated using HOMER to the corresponding genome. Using UCSC liftover tool (Navarro Gonzalez et al., 2021) and corresponding chain files, genomic coordinates were transferred from the original annotation to Rattus Norvegicus 6 after 1.000‐bp extension for direct comparison at peak level with our data using bedtools overlap (version 2.26.0).
2.7. RNA‐sequencing
For RNA sequencing, the right hippocampal hemisphere was homogenized in TriPure (Roche) by shaking the tissue with 1.4‐mm diameter ceramic spheres (MP Biomedicals) at 4 m/s for 5 s in a FastPrep‐24 5G instrument (MP Biomedicals). Total RNA was isolated according to the manufacturer's protocol and RNA quality was assessed using the RNA 6000 Nano kit on Bioanalyzer (Agilent). All samples had a RNA Integrity Number (RIN) >8.5 and a 28/18s ratio >1.8 and were considered suitable for sequencing. Aliquots of total RNA samples were send for transcriptome sequencing at BGI Genomics, and the remaining RNA was stored at −80°C for (RT)‐PCR validation. Stranded mRNA libraries were constructed, and 100‐bp paired end sequencing was performed on the DNBseq platform resulting in >20 million reads per sample. RNA‐seq data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE160807.
2.8. RNA‐sequencing data analysis
The Gentrap pipeline, published as part of Bio Pipeline Execution Toolkit (Biopet), was used for read quality control, alignment and quantification. Quality control was performed using FastQC and MultiQC. Reads were aligned rn6 using STAR aligner (version 2.7.0b). Tool settings used were as follows: ‘‐‐runThreadN’ ‘8’ ‘‐‐outSAMunmapped’ ‘Within’ ‘‐‐twopassMode’ ‘Basic’ with an average of 95% alignment ratio. The gene‐read quantification was performed using HTSeq‐count (version 0.6.1). Tool settings used were as follows: ‘—format’ ‘bam’ ‘—order’ ‘name’ ‘‐‐stranded’ ‘reverse’ where an average of 60% of reads could be assigned uniquely to known genes based on Ensembl release 97 of rn6. HTSeq‐count output files were merged into a count matrix (24 samples, 32.883 genes) as input for differential gene expression analysis.
The goal of the analysis was to determine how glucocorticoid treatment, object exploration training and the combination affected the hippocampal transcriptome. DEseq2 (version 1.29.4) (Love et al., 2014) was used for normalization of the data (median of ratio's method) and identification of differentially expressed genes. For the differential expression analysis, we selected all genes which were expressed in a minimum of four out of six replicates with >20 normalized counts for at least one of the treatment groups, resulting in 13.514 genes in the analysis. Four contrasts were analysed for differential expression in a pair‐wise comparison. A FDR adjusted p value of 0.05 was used as a cut‐off to determine differentially expressed genes. Subsequently, all samples per treatment group were pooled and genes expressed in at least 8 out of 12 replicates in a group were selected, resulting in 13.297 genes in the pooled analysis. GOterm enrichment analysis was performed with the ViSEAGO package (version 1.4.0), using Fisher’s exact test with 0.01 as a significance cut‐off (Brionne et al., 2019).
2.9. Real‐time quantitative PCR
Total RNA was isolated as described above and 1,000 ng of RNA was used for cDNA synthesis using random hexamers and M‐MLV reverse transcriptase (Promega). Gene expression of the target genes was assessed with real‐time (RT)‐PCR using the GoTaq qPCR master mix (Promega) in a CFX96 RT‐PCR machine (Bio‐Rad) and normalized against expression of housekeeping gene Rplp0. Fold change expression was calculated according to the 2−ΔΔCT method. Primers sequences are available upon request.
2.10. Single‐cell expression data
Public mouse hippocampal single‐cell data was published by the Allen Institute for Brain Science (Yao et al., 2020) and downloaded from https://portal.brain‐map.org/atlases‐and‐data/rnaseq/mouse‐whole‐cortex‐and‐hippocampus‐10x. We extracted and analysed the hippocampal data, and our selection of target genes was displayed by dotpot using Seurat (version 3.1.5).
2.11. Statistical analysis
Discrimination index and total object exploration times were analysed using one‐way ANOVAs with Dunnett's post hoc tests compared to the vehicle group or a two sample t test to detect in‐between group differences. One‐sample t tests were performed to detect whether the discrimination index was different from chance level (zero). Corticosterone levels were analysed by two‐way ANOVA with Sidak's post hoc tests, using training and corticosterone treatment as factors. mRNA expression validation data were analysed by one‐way ANOVAs with Dunnett's post hoc tests, and genomic binding location data were analysed using chi‐square tests with Bonferroni correction. Statistical tests were performed using GraphPad Prism (version 8.4.2; GraphPad Software, San Diego, California, USA).
3. RESULTS
3.1. Corticosterone dose‐dependently enhanced object location memory
We determined the optimal dose of corticosterone to enhance object location memory in our set‐up. Rats were allowed to explore the experimental apparatus containing two identical objects for three minutes, after which they immediately received corticosterone (0.3, 1.0 and 3.0 mg/kg) or vehicle via subcutaneous injection. Retention was tested 24 hr later in the same experimental apparatus in which one of the objects was relocated. The discrimination index was used as a measure of memory and represents the preference for exploring the object in the novel location.
Total object exploration times of the two identical objects during the training trial were similar for all groups (F 3,36 = 0.99, p = 0.41), indicating no differences in exploration prior to corticosterone administration (Figure 1a). The discrimination index of the vehicle (t 7 = 0.05, p = 0.96) and 0.3 mg/kg corticosterone (t 6 = 0.38, p = 0.72) groups did not significantly differ from zero (comparable time spent exploring object in novel and familiar location), indicating no retention of location memory. Both the 1.0 mg/kg (t 8 = 3.14, p = 0.01) and 3.0 mg/kg (t 6 = 9.49, p < 0.01) corticosterone groups showed a preference for the object in the novel location (Figure 1b), revealing a dose‐dependent increase in memory retention upon corticosterone administration. A one‐way ANOVA for discrimination index showed a significant corticosterone effect (F 3,27 = 14.09, p < 0.01) and Dunnet's post hoc tests showed that the 1.0 mg/kg corticosterone group (p = 0.03) and the 3.0 mg/kg corticosterone group (p < 0.01) had a significantly higher discrimination index compared to the vehicle group. Subsequent molecular experiments were therefore performed with the 3.0 mg/kg dose of corticosterone.
FIGURE 1.
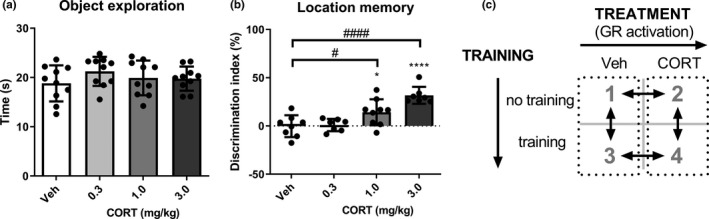
Corticosterone dose‐dependently increased object location memory retention. (a) Total object exploration time in seconds (s) of two identical objects during the object location memory training trial (n = 10 per group). (b) Dose‐dependent effect of corticosterone on the discrimination index at the 24 hr object location memory retention test (n = 7–9 per group). (c) The two‐by‐two design to study GR, pCREB and their potential interaction in corticosterone‐induced enhancement of hippocampus‐dependent memory. Data shown as mean ± SD. CORT, corticosterone; GR, glucocorticoid receptor; pCREB, phosphorylated cAMP response element‐binding protein; s, seconds; training, object exploration training; Veh, vehicle; *p < 0.05, ****p < 0.0001 compared to discrimination index of 0%; # p < 0.05, #### p < 0.0001 compared to vehicle group
3.2. Study design to investigate potential interaction between GR and pCREB in memory consolidation
In order to determine whether exposure to the training procedure may be of importance for transcriptional regulation when corticosterone induces memory enhancement, we used a two‐by‐two study design (Figure 1c). Post‐training administration of corticosterone was used to activate the GR and object exploration training was applied to induce novelty arousal in rats that were habituated to handling but not to the test apparatus. The study design allowed investigation of GR, pCREB and their potential interactions in corticosterone‐induced enhancement of hippocampus‐dependent memory.
3.3. Generation of ChIP‐sequencing cohort and data validation
To study GR, pCREB and their potential interaction at the hippocampal genome, a cohort of rats was subjected to the experimental paradigm and sacrificed 45 min after vehicle or corticosterone injection for ChIP‐seq. With this time point, we aimed to assess genomic binding of both GR and pCREB in the same animal. The animals were divided into four groups according to the study design (Figure 1c) with eight animals per group: (1) no training—vehicle, (2) no training—corticosterone, (3) training—vehicle and (4) training—corticosterone. Total object exploration times of the rats subjected to the training trial did not differ (t 14 = 1.66, p = 0.12) (Figure S1a). Corticosterone levels were significantly elevated 45 min after corticosterone injection (main effect of treatment: F 1,28 = 40.72, p < 0.01) (Figure S1b). ChIP‐seq identified a total of 4.343 GR DNA‐binding sites and 14.146 pCREB DNA‐binding sites. Principal component analysis revealed no clear clustering per experimental group (Figure S2).
In order to validate adequate technical performance of the ChIP‐seq experiment, direct binding of GR (Per1 and Camk2a) and pCREB (Fos and Cbwd1) at known target genes was confirmed by visual inspection of the sequencing data (Figure S3). In addition, we compared our data to published ChIP‐seq datasets of activated GR in adrenalectomized rats (Pooley et al., 2017) and of pCREB in primary mouse hippocampal neurons (Lesiak et al., 2013). After conversion of the public datasets to version 6 of the rat genome to enable comparison at peak level, 13% of the GR and 18% of the pCREB identified binding sites overlapped with the corresponding reference data (Figure S4a,b). This directly validated 630 GR and 2.635 pCREB DNA‐binding sites, despite the differences in experimental set‐up and analyses between datasets. Comparison at the more global level of annotated genes revealed a larger overlap of 52% for GR, and 70% for pCREB (Figure S4c,d).
3.4. Object exploration training minimally affects corticosterone‐induced differential GR DNA‐binding in the hippocampus
The analysis of differential hippocampal GR DNA‐binding revealed significant effects for corticosterone‐treated groups relative to vehicle: 52 loci in absence of object exploration training (50 up, 2 down) and 30 loci with object exploration training (30 up and 0 down) (Table S1). The fold change–fold change (Fc‐Fc) plot visualizes corticosterone‐induced GR DNA‐binding for three groups of loci (FDR adjusted p value < 0.05; Figure 2a): differential binding only in absence of object exploration training (red triangles), differential binding only with object exploration training (green circles) and differential binding irrespective of object exploration training (blue squares). At this FDR cut‐off, a total of 18 loci were differentially bound by GR (all increased binding) irrespective of the training condition. However, for the other loci, the absolute differences in GR DNA‐binding after corticosterone between the training and no training groups were mostly small (i.e., most data points in the Fc‐Fc plot are near the plotted diagonal in Figure 2a). No loci were differentially bound by GR after object exploration training, irrespective of treatment (Figure 2b). This finding is in line with the notion that corticosterone is required for GR activation and subsequent binding to the DNA, and corticosterone levels were not increased by training alone (Figure S1b). Visualization of the individual GR ChIP data confirmed that the call of ‘context‐specific GR DNA‐binding' was often based on increased variability in one of the groups (rather than clear quantitative differences), which was especially evident for loci that were called as training specific. Nevertheless, some loci showed more convincing specificity for a certain training condition (e.g., Nav3 and Mgst2; Figure S5).
FIGURE 2.
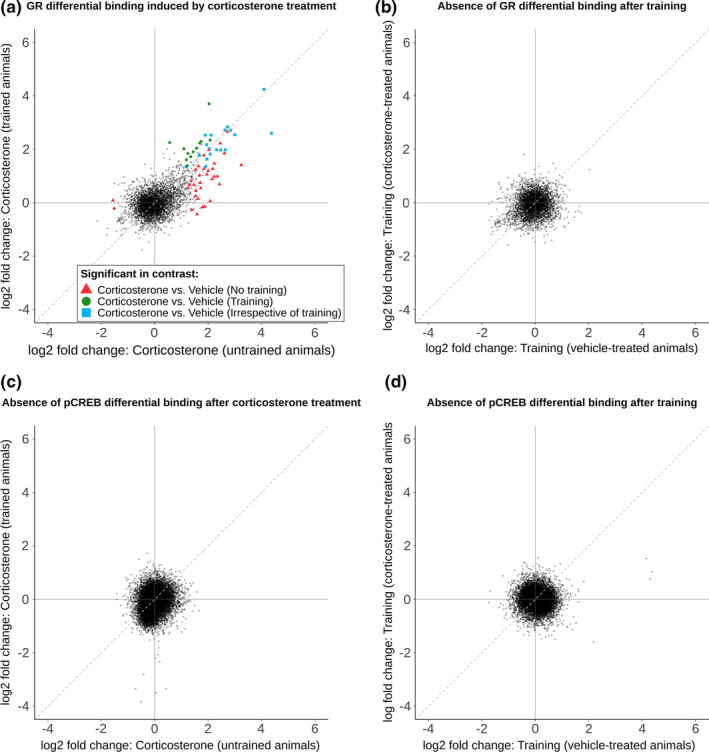
Object exploration training does not affect corticosterone‐induced GR and pCREB DNA‐binding. Fold change–fold change (Fc‐Fc) plots visualizing the effect of corticosterone treatment and training on DNA‐binding by GR or pCREB by plotting the fold changes of two contrasts against each other: (a) Differential GR DNA‐binding after corticosterone treatment in trained versus untrained animals, (b) no effect of training on GR DNA‐binding in corticosterone‐treated versus vehicle‐treated animals, (c) no effect of corticosterone treatment on pCREB DNA‐binding in trained versus untrained animals and (d) no effect of training on pCREB DNA‐binding in corticosterone‐treated versus vehicle‐treated animals. Black points represent all identified binding sites. Differentially GR bound binding sites (FDR adjusted p value < 0.05) are colour coded. Red triangles indicate differential GR binding in corticosterone versus vehicle without training only, green circles indicate differential GR binding in corticosterone versus vehicle with training only, and blue squares indicate GR differential binding in corticosterone versus vehicle irrespective of training. Dashed diagonal lines indicates equal fold change in both contrasts. GR, glucocorticoid receptor; pCREB, phosphorylated cAMP response element‐binding protein training, object exploration training
3.5. Absence of differential pCREB DNA‐binding in the hippocampus after training
ChIP‐seq data for pCREB revealed no loci with differential pCREB binding after corticosterone treatment versus vehicle (Figure 2c). To our initial surprise, but in line with other data (Carter et al., 2017; Everett et al., 2013; Tanis et al., 2008), assessment of differential pCREB binding after object exploration training did not result in differentially bound loci in training versus no training groups (Figure 2d) (Table S1).
3.6. Corticosterone treatment focused analysis identifies additional loci of interest
As the effects of object exploration training on DNA‐binding were minimal for GR and absent for pCREB, an additional corticosterone‐focused analysis was performed by pooling all animals per treatment group, irrespective of training background. Pooling increased the total number of loci included in the analysis, resulting in 4,807 GR and 14,465 pCREB DNA‐binding sites. We identified 133 loci (124 up, 9 down) differentially bound by GR (Figure 3a) and no loci that were differentially bound by pCREB (Figure 3b, Table S1). Ten loci were no longer differentially bound by GR after pooling (Figure S5, marked with red triangle), which may indicate that training did affect GR DNA‐binding at these sites.
FIGURE 3.
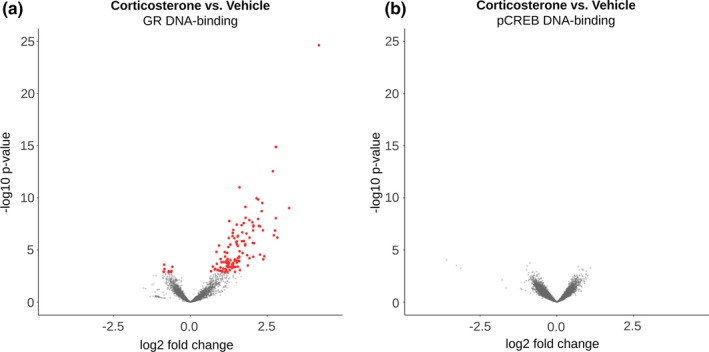
GR and pCREB DNA‐binding in the rat hippocampus after corticosterone administration. Volcano plots showing (a) GR and (b) pCREB DNA‐binding after corticosterone treatment versus vehicle. The graph displays the −log10 adjusted p value plotted against the log2‐fold change for all identified binding sites. Red dots indicate significantly increased or decreased DNA‐binding. GR, glucocorticoid receptor; pCREB, phosphorylated cAMP response element‐binding protein
To visualize DNA‐binding in more detail, average coverage plots of the normalized ChIP‐signal flanking the peak summits were constructed for GR, pCREB and input samples, subdivided into four groups: (I) loci differentially bound by GR after corticosterone, (II) loci not differentially bound by GR, (III) pCREB loci overlapping GR and (IV) pCREB loci not overlapping GR (Figure 4a,b visualizes loci with increased GR ChIP‐signal after corticosterone treatment). Peak widths were similarly distributed for GR and pCREB with median peak width of 409 and 466, respectively (Figure S6a). To determine if the identified loci were directly bound by GR and pCREB, we performed de novo motif analysis under the hypothesis of finding enrichment of GRE and CRE motifs, respectively. However, the presence of simple repeat regions in the sequences precluded motif discovery, with and without masking of repeat regions. De novo motif analysis performed specifically on loci differentially bound by GR after corticosterone identified a GRE in 126 of 133 loci, but no motifs of other transcription factors (Figure 4c,d).
FIGURE 4.
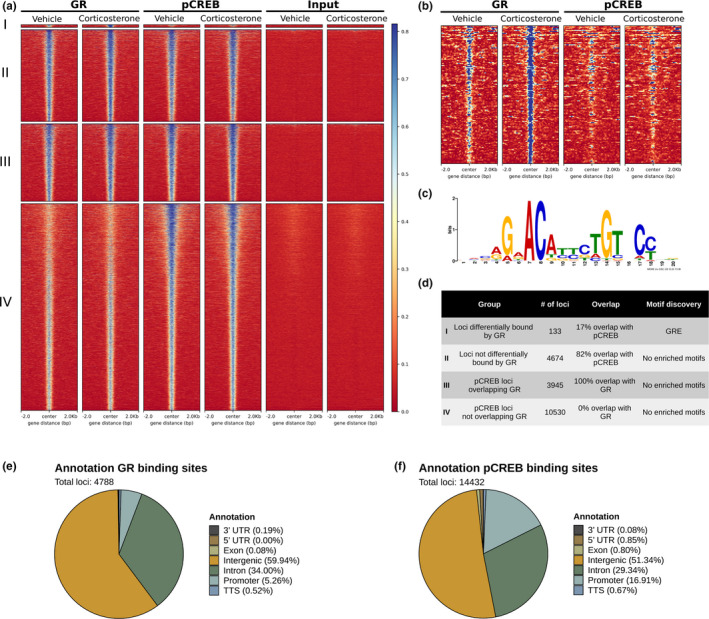
Visualization of GR and pCREB ChIP‐signal, co‐binding and genomic locations. (a) Heatmaps with average coverage plots of the normalized ChIP‐signal 2‐kB flanking the summits of the called GR and pCREB peaks and the corresponding input signal with vehicle and corticosterone treatment. Heatmaps are subdivided into four groups: (I) loci differentially bound by GR after corticosterone, (II) loci not differentially bound by GR, (III) pCREB loci overlapping GR and (IV) pCREB loci not overlapping GR. (b) Enlarged heatmap of loci differentially bound by GR after corticosterone. High ChIP‐signal is visualized in blue. (c) Motif discovered by de novo motif discovery performed on sequences of loci differentially bound by GR after corticosterone. (d) Overview table with the number of loci per group, the percentage overlap with called peaks of the other transcription factor (pCREB for GR and GR for pCREB) indicating co‐binding and the result of the de novo motif discovery per group. Annotation to genomic regions of the identified (e) GR and (f) pCREB binding sites. GR, glucocorticoid receptor; GRE, glucocorticoid response element; pCREB, phosphorylated cAMP response element‐binding protein; TTS, transcription termination site; UTR, untranslated region
Cobinding of GR and pCREB peaks was assessed to identify a potential genomic interaction of the two transcription factors underlying increased GR binding after corticosterone treatment. Direct overlap of the genomic coordinates of the called peaks was considered as cobinding, and this revealed that 82% of the non‐differentially bound GR loci overlapped with pCREB, while only 17% of the differentially bound GR loci cobound with pCREB (Figure 4d). Significant cobinding of GR at pCREB peaks was less frequent with 27%.
The identified GR and pCREB DNA‐binding sites were annotated relative to the nearest TSS (Figure S6b) and were associated to the corresponding genes. Genomic distribution of GR and pCREB DNA‐binding sites was comparable with binding predominantly occurring in intergenic and intronic regions. The fraction of promoter binding was approximately three times higher for pCREB compared to GR (Figure 4e,f). We separately assessed whether cobinding affected the genomic location of GR loci, in particular because the percentage of pCREB cobinding was lower at the loci that were differentially bound by GR after corticosterone. The overall distribution of binding sites did not differ extensively. There was a significant increase of GR binding in 3′ UTR regions in absence of pCREB cobinding (Bonferroni corrected p = 0.01) (Figure S6c,d). However, the biological relevance of this result is uncertain, as it concerns only nine loci.
3.7. Corticosterone altered the hippocampal transcriptome, whereas training did not
We next analysed the hippocampal transcriptome to determine if the loci differentially bound by GR were associated with expression of the nearby genes. Rats were trained conform the study design (Figure 1c) and hippocampal RNA was collected 3 hr after corticosterone or vehicle injection allowing to assess transcriptional effects of GR. In this cohort, total object exploration times of the rats subjected to object exploration training did not differ (t 14 = 0.58, p = 0.57) (Figure S7a), and corticosterone levels did not significantly differ anymore 3 hr after injection in the corticosterone groups relative to vehicle (main effect of treatment: F 1,29 = 0.63, p = 0.43) (Figure S7b).
Clustering of the transcriptome data revealed two distinct clusters not explained by the experimental groups (Figure S8a,b) and no clear clustering in subsequent principal components (Figure S8c). Evident clustering was explained by a degree of contamination with choroid plexus tissue based on high Ttr counts in these samples (Figure S8d,e). Differential gene expression analysis identified 86 differentially expressed genes (70 upregulated and 16 downregulated) after corticosterone treatment without object exploration training and 61 differentially expressed genes (46 upregulated and 15 downregulated) after corticosterone treatment with object exploration training (Table S2). The Fc‐Fc plot (Figure 5a) indicates that object exploration training only modestly affected the corticosterone induced transcriptome, as most points show very comparable fold changes for corticosterone versus vehicle with and without object exploration training. This is in line with the analysis of the effect of training on the transcriptome, as 3 min of object exploration training, which did not result in any long‐term memory, did not lead to differential gene expression, either without or with corticosterone treatment (Figure 5b). The absence of transcriptome changes after object exploration training with or without corticosterone and the minimal effect of training on the corticosterone‐induced transcriptome did not provide leads to further understand the hypothesized interactions between GR and pCREB. We therefore focused on the GR‐mediated effects discovered after corticosterone treatment.
FIGURE 5.
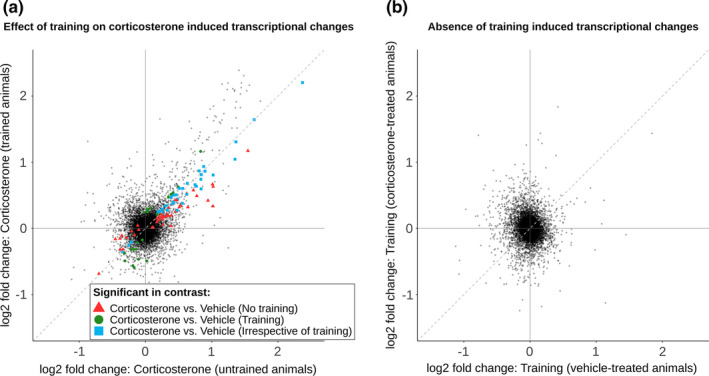
Object exploration training does not change the hippocampal transcriptome. Fold change–fold change (Fc‐Fc) plots visualizing the effect of corticosterone treatment and training on the hippocampal transcriptome by plotting the fold changes of two contrasts against each other: (a) Effect of corticosterone treatment in trained versus untrained animals and (b) effect of training in corticosterone‐treated versus vehicle‐treated animals. Black points represent all genes expressed in the hippocampus. Differentially expressed genes (FDR adjusted p value < 0.05) are colour coded. Red triangles indicate differential expression in corticosterone versus vehicle without training only, green circles indicate differential expression in corticosterone versus vehicle with training only, and blue squares indicate differential expression in corticosterone versus vehicle irrespective of training. Dashed diagonal lines indicate equal fold change in both contrasts. Training, object exploration training
3.8. Corticosterone treatment‐focused analysis identifies additional genes of interest
We also performed a pooled analysis on corticosterone induced changes in gene expression as we found no effect of object exploration training on the hippocampal transcriptome, in concordance with the absence of effects on DNA‐binding. Differential gene expression analysis identified 201 differentially expressed genes (137 upregulated and 64 downregulated) after corticosterone treatment (Figure 6a, Table S2). Pooling resulted in 15 genes no longer being differentially expressed, some of which again indicating training‐dependent effects (Figure S9). A large portion of these genes (170 out of 201) showed merely a small induction or reduction (<50%) upon corticosterone stimulation, potentially indicative of a dilution effect of differential expression in subsets of hippocampal cell types. GO‐term analysis performed on all differentially expressed genes resulted in a top 10 of enriched terms, without clear links to learning and memory (Figure 6b). Comparison with a previously published dataset 1 hr after corticosterone injection in intact animals suggested that 20 transcripts can be also induced at this much earlier time point (Table S3) (Gray et al., 2014). The data provide a set of direct and indirect glucocorticoid regulated genes in intact animals under conditions were glucocorticoids enhanced memory and may therefore contain the genes responsible for this effect.
FIGURE 6.
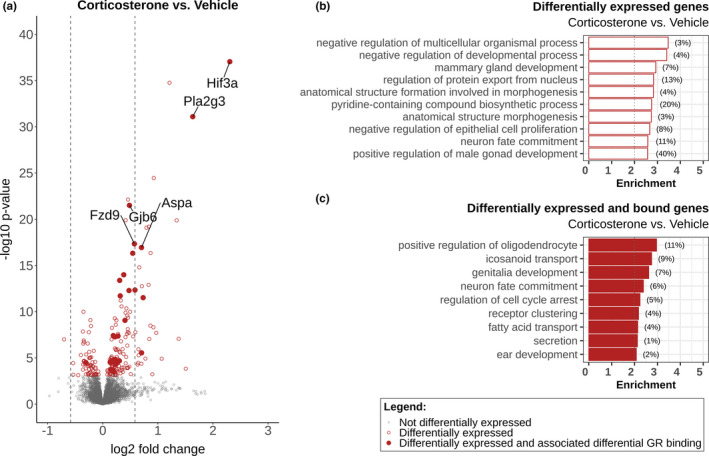
Corticosterone induces transcriptional changes in the rat hippocampus. (a) Volcano plot showing differential gene expression after corticosterone treatment versus vehicle. The graph displays the −log10‐adjusted p value plotted against the log2‐fold change for all genes expressed in the hippocampus. Red circles indicate significantly upregulated and downregulated genes (FDR‐adjusted p value < 0.05). Differentially expressed genes which had differentially GR bound DNA loci associated are marked with red dots; the five most significant genes are annotated. Dashed lines mark a log2‐fold change of (−)0.58 (50% increase or decrease in expression). Results of GOterm biological processes enrichment analysis on (b) all differentially expressed genes and (c) differentially expressed genes which had differentially GR bound DNA loci associated. Enrichment is the −log10 (p value), with a significance cut‐off of p = 0.01 (enrichment of two as indicated by grey dotted line). The percentage of genes differentially expressed of all genes in each enriched GO term is shown behind the corresponding bar
3.9. Identification and validation of direct GR target genes linked to memory enhancement by corticosterone
To identify primary GR target genes, we combined the GR ChIP‐seq and hippocampal transcriptome data. GR loci were linked to genes based on the nearest TSS and their vicinity near (intergenic)/ localization in (intronic and exonic) genes (providing a maximum of three associated genes per locus). This identified 27 genes that were differentially expressed by corticosterone treatment (highlighted by filled red circles in Figure 6a) linked to 26 linked differentially bound GR loci. GO‐term analysis performed on this set of primary GR target genes highlighted the role of oligodendrocytes and icosanoids (Figure 6c). Details of these targets are summarized in Table 1, ordered based on the log2‐fold change of differential expression analysis. GR binding sites associated to the differentially expressed genes were mainly located in intergenic and intronic regions, with four loci located in promoter regions. GREs were discovered in all associated loci and cobinding with pCREB was only detected for the binding site linked to Olig1 and Olig2.
TABLE 1.
Overview of transcriptionally responsive differential GR binding sites
| Gene | Associated GR binding site | Annotation | Distance from TSS (bp) | pCREB co‐binding | ChIP log2 FC | RNA log2 FC | qPCR validated |
|---|---|---|---|---|---|---|---|
| Hif3a | chr1:78999395‐78999862 | Promoter‐TSS | −1,760 | No | 2.05 | 2.31 | <0.0001 |
| Pla2g3 | chr14:83721049‐83721421 | Promoter‐TSS | −3,698 | No | 1.38 | 1.63 | — |
| Lmod1 | chr13:52169413‐52169878 | Intron | 22,078 | No | 1.08 | 0.73 | <0.0001 |
| Aspa | chr10:59884799‐59885334 | Intron | 3,131 | No | 4.18 | 0.70 | <0.0001 |
| Micalcl | chr1:177149010‐177149324 | Intron | −44,568 | No | 1.17 | 0.70 | <0.0001 |
| Eva1a | chr4:112734550‐112734850 | Intergenic | −27,430 | No | 1.48 | 0.58 | — |
| Fzd9 | chr12:24446543‐24446837 | Intergenic | −27,291 | No | 2.23 | 0.57 | <0.0001 |
| Mfsd2a | chr5:140655216‐140655593 | Intron | 2,340 | No | 1.16 | 0.54 | — |
| Fkbp5 |
chr20:8091823‐8092175 chr20:7998838‐7999373 |
Intergenic Intron |
−17,692 −55,531 |
No No |
2.83 2.32 |
0.48 | <0.0001 |
| Gjb6 | chr15:37409719‐37409969 | Promoter‐TSS | −176 | No | 3.21 | 0.48 | <0.0001 |
| Mertk | chr3:121282845‐121283172 | Intron | 47,779 | No | 0.98 | 0.40 | — |
| Svil | chr17:55203186‐55203543 | Intergenic | 142,914 | No | 1.78 | 0.38 | — |
| Mrpl41 | chr3:2251775‐2252102 | Intron | −10,314 | No | 2.34 | 0.32 | 0.0238 |
| Pnpla7 | chr3:2251775‐2252102 | Intron | −10,314 | No | 2.34 | 0.31 | 0.0117 |
| Nxn | chr10:64458325‐64458655 | Intron | 60,151 | No | 1.79 | 0.30 | — |
| Idh2 | chr1:141944200‐141944597 | Intergenic | −41,746 | No | 1.23 | 0.28 | — |
| Sema4b | chr1:141944200‐141944597 | Intergenic | −41,746 | No | 1.23 | 0.28 | — |
| Ralgds | chr3:7099006‐7099290 | Intergenic | −10,772 | No | 1.04 | 0.23 | — |
| Olig1 | chr11:31424285‐31424773 | Promoter‐TSS | −3,848 | Yes | 1.60 | 0.23 | 0.5848 |
| Htra1 | chr1:201515758‐201516104 | Intron | 16,864 | No | 1.74 | 0.22 | — |
| Tsc22d3 | chrX:111855808‐111856239 | Intergenic | 31,882 | No | 1.23 | 0.20 | — |
| Plcl1 | chr9:62313386‐62313692 | Intron | 297,427 | No | 2.78 | 0.20 | 0.0601 |
| Ptpn11 | chr12:40898549‐40898905 | Intron | 3,212 | No | 2.02 | 0.19 | — |
| Wdr19 | chr14:44774230‐44774484 | Intergenic | −7,237 | No | 1.87 | 0.15 | — |
| Glul |
chr13:71575065‐71575656 chr13:71606937‐71607641 |
Intergenic Intergenic |
244,309 276,237 |
No No |
1.27 1.51 |
0.15 | — |
| Slc25a33 | chr5:166744420‐166744747 | Intergenic | −17,790 | No | 2.04 | 0.13 | — |
| Olig2 | chr11:31424285‐31424773 | Promoter‐TSS | −3,848 | Yes | 1.60 | −0.31 | — |
Differential GR binding sites of which the associated gene(s) are differentially expressed based on the transcriptome data identifying putative direct GR‐target genes. Associated gene is based on annotation to the closest TSS using HOMER. A subset of the genes is validated by qPCR, for which the p value of a one‐way ANOVA is displayed.
Abbreviations: bp, base pairs; ChIP, chromatin immunoprecipitation; FC, fold change; GR, glucocorticoid receptor; training, object exploration training; TSS, transcription start site.
Transcriptional regulation of a subset of the GR‐binding associated genes was validated using qPCR, which included a treatment naïve group (n = 16–17 per treatment group and n = 8 for naïve group). Analysis of the qPCR data by one‐way ANOVA confirmed that nine out of the 11 genes analysed showed significant changes in expression and one at trend level (Table 1). Nine transcripts had significantly higher expression after corticosterone compared to the vehicle group according to Dunnet's post hoc tests (Figure S10).
Post‐hoc analysis identified three genes (Hif3a [p = 0.01], Lmod1 [p = 0.02], Micalcl [p = 0.01]) that were significantly upregulated in the vehicle group compared to the naïve group, indicating that the handling/vehicle injection affected the expression by itself.
3.10. Various cell types implicated in memory enhancement by corticosterone
The GO enrichment of a cell type specific biological process introduced the question which cell types were affected by corticosterone. Analysis of single‐cell transcriptome data of mouse hippocampus (Yao et al., 2020) indicated that the putative GR targets are (under basal conditions) expressed in multiple and for some targets distinct cell types (Figure 7). Transcription factors of interest (Nr3c1 and Creb1) are expressed to a varying level throughout the different cell types in the mouse hippocampus, with coexpression confirming a possibility for interaction. Our identified GR targets could be divided into three subclasses based on their expression patterns: widely expressed (13 genes), predominantly neuronal (three genes) or predominantly non‐neuronal (11 genes). These basal expression patterns indicate that a subset of the hippocampal targets of corticosterone is almost exclusively expressed in non‐neuronal cells, highlighting a potential role of these cells in the memory enhancement by corticosterone.
FIGURE 7.
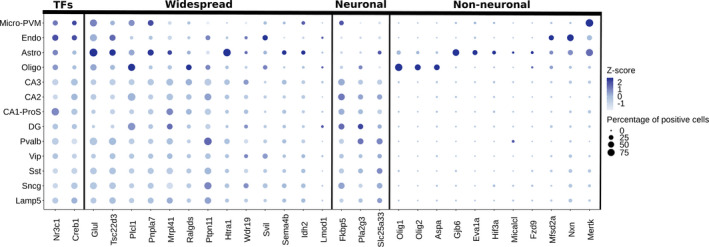
GR target genes are cell type specifically expressed under basal condition. Dotplot visualizing single‐cell expression data of the investigated transcription factors GR (Nr3c1) and pCREB (Creb1) and identified GR target genes in the mouse hippocampus. Target genes are grouped based on basal expression pattern: widespread expression, predominant neuronal expression (CA3, CA2, CA1‐ProS, DG, Pvalb, Vip, Sst, Sncg, Lamp5) and predominant non‐neuronal expression (Micro‐PVM, Endo, Astro, Oligo). Colour intensity indicates expression level per cell type based on Z scores and dot size shows the percentage of cells expressing the gene. Astro, astrocytes; CA1‐ProS, cornu ammonis 1/pro‐subiculum pyramidal cells; CA2, cornu ammonis 2 pyramidal cells; CA3, cornu ammonis 3 pyramidal cells; DG, dentate gyrus granule cells; Endo, endothelial cells; GR, glucocorticoid receptor; Lamp5, lysosomal associated membrane protein family 5 positive GABA neurons; Micro‐PVM, microglial cells and perivascular macrophages; Oligo, oligodendrocytes; pCREB, phosphorylated cAMP response element‐binding protein; Pvalb, parvalbumin positive GABA neurons; Sncg, synuclein gamma positive GABA neurons; Sst, somatostatin positive GABA neurons; TFs, transcription factors; Vip, vasoactive intestinal peptide positive GABA neurons
4. DISCUSSION
In this study, we set out to investigate the effects of glucocorticoids at a dose that induces memory consolidation of emotionally arousing training experiences, and a potential genomic interaction with pCREB. We hypothesized that an interaction could affect the extent of GR DNA‐binding and transcription, in association with the observed memory enhancement after glucocorticoid administration. Therefore, using a hippocampus‐dependent learning task (Barker & Warburton, 2011) and post‐training corticosterone administration, we studied the chromatin occupancy of GR and pCREB in whole hippocampus and the effect on the transcriptome. We demonstrate in adrenally intact rats that a memory‐enhancing dose of corticosterone induced GR DNA‐binding and concomitant gene transcription. These effects were largely independent of the training experience, indicating context‐independent effects of GR. We observed no changes in pCREB DNA‐binding or gene expression after this training, in line with the GR effects being independent of the training context. Despite GR effects being context independent, our data provide a short list of likely primary GR target genes that may underlie the enhancement of memory consolidation, under the assumption that the genomic effects of GR are essential for enhanced memory consolidation after corticosterone treatment.
We selected a hippocampus‐dependent learning task for two reasons. First, GR DNA‐binding was demonstrated to be necessary for long‐term memory on an hippocampus‐dependent water‐maze task (Oitzl et al., 2001). Second, and more practical, the hippocampus allowed sufficient and well‐defined tissue for the genomic analysis opposed to, for example, the amygdala (another brain region likely implicated in GR‐pCREB cross‐talk). The transcriptional outcome of glucocorticoid exposure can depend on cellular context (Provencal et al., 2020). The extracellular and intracellular signalling molecules and associated transcription factors that would mediate this contextual dependence of hippocampal GR signalling in the current setting are not known. However, pCREB is a reasonable candidate, because it is downstream of noradrenaline and glutamate signalling (Gregory & Goudet, 2021; Hagena et al., 2016). The interaction between corticosterone and noradrenaline in particular has been studied in multiple types of memory (McReynolds et al., 2010; Roozendaal et al., 2006), but not at the level of their transcription factors GR and pCREB.
Of note, all rats in our experiment received a subcutaneous injection, and this precludes us to consider untrained rats as being non‐aroused. In mice, basic experimental procedures as picking up the animal and administration of injections activate corticotropin‐releasing hormone neurons in the paraventricular nucleus, a hallmark of activation of the stress system (Kim et al., 2019). This can be translated to rats, in which post‐training injection or handling is sufficiently arousing/stressful to strengthen consolidation of fear conditioning (Hui et al., 2006). Therefore, effects of training on pCREB DNA‐binding and the transcriptome may have been masked by the subcutaneous injection procedure, despite handling of the animals to minimize stress. An intrinsic effect of vehicle injection on gene expression is evident based on the increased expression of Hif3a, Lmod1 and Micalcl in our qPCR validation and the full extent of this effect is not known. However, regardless of the potential absence of differential pCREB DNA‐binding due to the route of corticosterone administration, the pCREB cistrome still allowed the assessment of potential genomic interactions with GR.
As DNA‐binding and subsequent transcription of target genes is a dynamic process, selection of appropriate time points to assess the effect of training and corticosterone injection is difficult. Potentially interesting effects will inevitably be missed based on the time point selected, and the data represents a snapshot. For this study, in order to evaluate both GR and pCREB DNA‐binding in the same animal, we performed ChIP‐seq 45 min after corticosterone injection. A time point clearly suited for detecting GR DNA‐binding, yet in retrospect perhaps less optimal for pCREB. Hippocampal noradrenaline levels (as a proxy for pCREB activation) after object exploration training in earlier studies peaked after 15 min and amygdala levels of noradrenaline restore to baseline 30 min after a footshock (Mello‐Carpes et al., 2016; Quirarte et al., 1998). It is therefore possible that differential binding events occurred prior to the selected time point and that only constitutively bound pCREB loci were identified. Similarly, consolidation of memories is a complex and temporal process that requires multiple waves of transcription, starting with the response of well characterized immediate early genes such as cFos and Egr1 (Alberini, 2009; Igaz et al., 2002; O'Sullivan et al., 2007). Our transcriptome analysis 3 hr after injection aimed to investigate a second wave of transcription during which GR is able to exert its transcriptional effects (Morsink et al., 2006). Comparison of our data to Gray et al., (2014) showed that only 20 genes were also differentially expressed 1 hr after corticosterone, confirming this temporal aspect of transcription. Of note, the result of this comparison needs to be interpreted cautiously, as a very high dose of corticosterone (15 mg/kg) was used in the cited study and the data were not corrected for multiple testing.
Because we did not observe differential pCREB DNA‐binding in the current study, we critically assessed the quality of the data. Validity of our ChIP‐seq datasets was confirmed by comparison to previously published datasets of genome‐wide GR and pCREB DNA‐binding, showing a high percentage of overlap in genes associated to bound loci and DNA‐binding at proven GR and pCREB target genes (Lesiak et al., 2013; Pooley et al., 2017). Overlap at a peak level was lower than we would have intuitively expected. This can be partially attributed to differences in experimental design and data analyses of the reference sets, which limit the value of a direct comparison. Reference data of GR were based on adrenalectomized animals and for pCREB on a neuronal cell culture model, both of which likely imply different transcription factor‐binding locations and levels compared to whole hippocampus from adrenally intact rats. The comparison does confirm that our dataset likely contains numerous true positive binding loci. Therefore, we consider the absence of differential pCREB DNA‐binding as biological and not technical, for which a multitude of reasons can be postulated.
Apart from the possibility that a subcutaneous injection might have masked the effect of training, it is possible that differential pCREB binding to the DNA is not the mechanism by which pCREB regulates transcription of its target genes. In line with this, a published study detected no differential pCREB binding in the hippocampus at known target genes c‐Fos and Egr‐1 at 30 min after a forced swim stressor, while the hippocampal expression of these genes was altered (Carter et al., 2017). A similar observation was found in livers of fasted mice and in the rat hippocampus after electroconvulsive therapy, in which pCREB DNA‐binding was not changed (Everett et al., 2013; Tanis et al., 2008). While this would explain the absence of differential pCREB DNA‐binding, it does not explain the absence of transcriptome changes after training.
Alternatively, the absence of differential pCREB binding in our data could also be explained by a dilution effect due to performing ChIP‐seq on the whole hippocampus. The hippocampus consists of a large number of different cell types as evident from recent advances in single‐cell RNA‐seq (Saunders et al., 2018; Yao et al., 2020). Moreover, interaction may take place in specific cell populations that are activated during the learning process. In Arc reporter mice, a sparse subset of neurons in the lateral amygdala and hippocampus was activated after fear conditioning (Gouty‐Colomer et al., 2016; Rao‐Ruiz et al., 2019). Transcriptome analysis revealed gene expression differences related to pCREB regulation after contextual fear conditioning specifically in the activated hippocampal neurons. In all likelihood, object exploration training will also only activate a subset of the hippocampal cells. The pCREB effects that are responsible for consolidation may occur only in the activated neurons, and bulk ChIP and RNA analyses used in this study were too crude to detect these.
Another explanation is that training does lead to global differential pCREB DNA‐binding and gene expression but not in the hippocampus. While there are convincing data on the involvement of both pCREB and GR in hippocampus‐dependent learning, noradrenergic blockade in the basolateral amygdala prevented enhancement of inhibitory avoidance memory after intrahippocampal GR agonist administration (Roozendaal et al., 1999). Similarly, administration of the noradrenaline receptor antagonist propranolol into the basolateral amygdala blocked increased hippocampal Arc expression after corticosterone administration in inhibitory avoidance training (McReynolds et al., 2010). In this neural circuitry‐based model, pCREB‐dependent transcriptional effects that are responsible for strengthening of the memory take place in the basolateral amygdala, while the GR target genes crucial for memory enhancement are regulated in the hippocampus. These GR‐mediated effects would not have to be specific for the training situation but could rather be permissive for effects that depend on synaptic activation.
To focus the analysis on the effects of corticosterone, the treatment groups were pooled. The increased statistical power doubled the number of detected loci that were differentially bound by GR and the amount of differentially expressed genes after corticosterone treatment. This strengthened the observation that training only minimally affected GR signalling. A hand full of loci and genes lost significance upon pooling of data, and these targets might hint towards a true effect of training background.
Comparison of the genome‐wide binding of GR and pCREB revealed an interesting observation regarding cobinding. A high percentage of all GR peaks in our dataset overlapped with pCREB peaks (82%), but only 17% of the differentially bound GR‐binding sites after additional corticosterone overlapped with pCREB. Of these GR binding sites cobound by pCREB, only one was associated to differentially expressed genes (Olig1 and Olig2) of which the Olig1 expression change could not be validated with qPCR. This finding hints towards an interaction where cobinding of pCREB limits additional DNA‐binding by GR and limits the ability of GR to increase transcription of the linked target gene.
The total number of corticosterone‐responsive genes was relatively small, probably in part reflecting the use of adrenally intact rats. For the identification of genes responsible for the memory enhancing effects, this is actually an advantage, given that shortlists are more informative than longlists. We acknowledge that the expression of some genes likely peaked before the 3 hr time point, as is known for glucocorticoid responsive gene Sgk‐1 (van Gemert et al., 2006) and other immediate early genes. Focussing on a later wave of transcription, GOterm analysis of all differentially expressed genes did not reveal enriched biological process classically linked to learning and memory. This might imply that enhancement of memory by glucocorticoids depends on non‐canonical processes, but this notion warrants further investigation. Our analysis combining GR DNA‐binding and transcriptome data yielded a set of new direct GR target genes that qualify as potential mediators of memory consolidation—or in fact other adaptations after stressful events. Some of the identified GR target genes, for example, Gjb6, Fkbp5 and Fzd9, have already been implicated in synaptic plasticity or learning processes (Lutz et al., 2009; Qiu et al., 2019; Zhao et al., 2005). For many other targets, the connection to learning and memory is not clear and requires further study.
Perhaps the study of cell types involved in the effects of glucocorticoids on hippocampal function merits more attention than the study of individual gene function. The cellular composition of the hippocampus adds a level of complexity and raises the question which cell types are affected by corticosterone and to what goal. Even though we do not have a single‐cell resolution in our experiments, available single‐cell data of the mouse hippocampus enabled us to determine which cell types express the identified target genes under basal conditions. Interestingly, while about half of the genes showed widespread expression, only three genes were predominantly expressed in neurons and the others displayed a clear non‐neuronal pattern. This highlights a potential role for the—so far understudied—non‐neuronal cells in the effects of corticosterone on learning and memory, such as microglial cells (Sanguino‐Gomez et al., 2021). As for the current study, further work will have to establish either additional filters to reduce the current shortlist of target genes and/or individually evaluate the effects of reduced expression of these putative mediators on memory.
5. CONCLUSION
Altogether, we were unable to find strong evidence for a genomic interaction between GR and pCREB in the male rat hippocampus after object exploration training. We identified novel GR target genes that may be permissive for the enhancement of memory after emotionally arousing training experiences. Further studies are required to pinpoint in which cell types these expression changes occur and how it results in enhanced performance in memory retention.
CONFLICT OF INTEREST
The authors declare no conflict of interest regarding the publication of this paper.
AUTHOR CONTRIBUTIONS
Conceived and designed study: JCB, LW, BR and OM; carried out data collection: JCB, LW, MG, EV, LK, RS, SL and PC; technical supervision: JB; conducted data analysis: JCB, LW, IM, HM and SK; interpreted the findings: JCB, LW, BR and OM; wrote the manuscript: JCB, BR and OM.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/ejn.15226.
Supporting information
Fig S1‐S10
ACKNOWLEDGEMENTS
We would like to thank Trea Streefland for her assistance with the lab work and Dr. J Kroon for reading and providing feedback on initial draft version of the manuscript. This research was supported by NWO ALW grant 823.02.002.
Buurstede JC, van Weert LTCM, Colucci P, et al. Hippocampal glucocorticoid target genes associated with enhancement of memory consolidation. Eur J Neurosci. 2022;55(9):2666–2683. 10.1111/ejn.15226
Edited by: Carmen Sandi
Jacobus C. Buurstede and Lisa T. C. M. van Weert contributed equally.
DATA AVAILABILITY STATEMENT
Sequencing data are available via NCBI's Gene Expression Omnibus and are accessible through GEO reference series accession number GSE160808. Further data could be obtained upon reasonable request to the corresponding author.
REFERENCES
- Alberini, C. M. (2009). Transcription factors in long‐term memory and synaptic plasticity. Physiological Reviews, 89(1), 121–145. 10.1152/physrev.00017.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsak, P. , Hauer, D. , Campolongo, P. , Schelling, G. , Fornari, R. V. , & Roozendaal, B. (2015). Endocannabinoid signaling within the basolateral amygdala integrates multiple stress hormone effects on memory consolidation. Neuropsychopharmacology, 40(6), 1485–1494. 10.1038/npp.2014.334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atucha, E. , Zalachoras, I. , van den Heuvel, J. K. , van Weert, L. T. , Melchers, D. , Mol, I. M. , Belanoff, J. K. , Houtman, R. , Hunt, H. , Roozendaal, B. , & Meijer, O. C. (2015). A mixed glucocorticoid/mineralocorticoid selective modulator with dominant antagonism in the male rat brain. Endocrinology, 156(11), 4105–4114. 10.1210/en.2015-1390 [DOI] [PubMed] [Google Scholar]
- Bailey, T. L. , Boden, M. , Buske, F. A. , Frith, M. , Grant, C. E. , Clementi, L. , Ren, J. , Li, W. W. , & Noble, W. S. (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Research, 37(Web Server), W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, G. R. , & Warburton, E. C. (2011). When is the hippocampus involved in recognition memory? Journal of Neuroscience, 31(29), 10721–10731. 10.1523/JNEUROSCI.6413-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brionne, A. , Juanchich, A. , & Hennequet‐Antier, C. (2019). ViSEAGO: A bioconductor package for clustering biological functions using gene ontology and semantic similarity. BioData Mining, 12, 16. 10.1186/s13040-019-0204-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campolongo, P. , Morena, M. , Scaccianoce, S. , Trezza, V. , Chiarotti, F. , Schelling, G. , Cuomo, V. , & Roozendaal, B. (2013). Novelty‐induced emotional arousal modulates cannabinoid effects on recognition memory and adrenocortical activity. Neuropsychopharmacology, 38(7), 1276–1286. 10.1038/npp.2013.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, S. D. , Mifsud, K. R. , & Reul, J. (2017). Acute stress enhances epigenetic modifications but does not affect the constitutive binding of pCREB to immediate‐early gene promoters in the rat hippocampus. Frontiers in Molecular Neuroscience, 10, 416. 10.3389/fnmol.2017.00416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datson, N. A. , van den Oever, J. M. , Korobko, O. B. , Magarinos, A. M. , de Kloet, E. R. , & McEwen, B. S. (2013). Previous history of chronic stress changes the transcriptional response to glucocorticoid challenge in the dentate gyrus region of the male rat hippocampus. Endocrinology, 154(9), 3261–3272. 10.1210/en.2012-2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett, L. J. , Le Lay, J. , Lukovac, S. , Bernstein, D. , Steger, D. J. , Lazar, M. A. , & Kaestner, K. H. (2013). Integrative genomic analysis of CREB defines a critical role for transcription factor networks in mediating the fed/fasted switch in liver. BMC Genomics, 14, 337. 10.1186/1471-2164-14-337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouty‐Colomer, L. A. , Hosseini, B. , Marcelo, I. M. , Schreiber, J. , Slump, D. E. , Yamaguchi, S. , Houweling, A. R. , Jaarsma, D. , Elgersma, Y. , & Kushner, S. A. (2016). Arc expression identifies the lateral amygdala fear memory trace. Molecular Psychiatry, 21(8), 1153. 10.1038/mp.2016.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, J. D. , Rubin, T. G. , Hunter, R. G. , & McEwen, B. S. (2014). Hippocampal gene expression changes underlying stress sensitization and recovery. Molecular Psychiatry, 19(11), 1171–1178. 10.1038/mp.2013.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, K. J. , & Goudet, C. (2021). International Union of Basic and Clinical Pharmacology. CXI. Pharmacology, signaling, and physiology of metabotropic glutamate receptors. Pharmacological Reviews, 73(1), 521–569. 10.1124/pr.119.019133 [DOI] [PubMed] [Google Scholar]
- Hagena, H. , Hansen, N. , & Manahan‐Vaughan, D. (2016). Beta‐adrenergic control of hippocampal function: Subserving the choreography of synaptic information storage and memory. Cerebral Cortex, 26(4), 1349–1364. 10.1093/cercor/bhv330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz, S. , Benner, C. , Spann, N. , Bertolino, E. , Lin, Y. C. , Laslo, P. , Cheng, J. X. , Murre, C. , Singh, H. , & Glass, C. K. (2010). Simple combinations of lineage‐determining transcription factors prime cis‐regulatory elements required for macrophage and B cell identities. Molecular Cell, 38(4), 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, G. K. , Figueroa, I. R. , Poytress, B. S. , Roozendaal, B. , McGaugh, J. L. , & Weinberger, N. M. (2004). Memory enhancement of classical fear conditioning by post‐training injections of corticosterone in rats. Neurobiology of Learning and Memory, 81(1), 67–74. 10.1016/j.nlm.2003.09.002 [DOI] [PubMed] [Google Scholar]
- Hui, I. R. , Hui, G. K. , Roozendaal, B. , McGaugh, J. L. , & Weinberger, N. M. (2006). Posttraining handling facilitates memory for auditory‐cue fear conditioning in rats. Neurobiology of Learning and Memory, 86(2), 160–163. 10.1016/j.nlm.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Igaz, L. M. , Vianna, M. R. , Medina, J. H. , & Izquierdo, I. (2002). Two time periods of hippocampal mRNA synthesis are required for memory consolidation of fear‐motivated learning. Journal of Neuroscience, 22(15), 6781–6789. 10.1523/JNEUROSCI.22-15-06781.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joels, M. , Fernandez, G. , & Roozendaal, B. (2011). Stress and emotional memory: A matter of timing. Trends in Cognitive Sciences, 15(6), 280–288. 10.1016/j.tics.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Joels, M. , Pasricha, N. , & Karst, H. (2013). The interplay between rapid and slow corticosteroid actions in brain. European Journal of Pharmacology, 719(1–3), 44–52. 10.1016/j.ejphar.2013.07.015 [DOI] [PubMed] [Google Scholar]
- John, S. , Sabo, P. J. , Thurman, R. E. , Sung, M. H. , Biddie, S. C. , Johnson, T. A. , Hager, G. L. , & Stamatoyannopoulos, J. A. (2011). Chromatin accessibility pre‐determines glucocorticoid receptor binding patterns. Nature Genetics, 43(3), 264–268. 10.1038/ng.759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst, H. , Berger, S. , Erdmann, G. , Schutz, G. , & Joels, M. (2010). Metaplasticity of amygdalar responses to the stress hormone corticosterone. Proceedings of the National Academy of Sciences USA, 107(32), 14449–14454. 10.1073/pnas.0914381107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. , Lee, S. , Fang, Y. Y. , Shin, A. , Park, S. , Hashikawa, K. , Bhat, S. , Kim, D. , Sohn, J. W. , Lin, D. , & Suh, G. S. B. (2019). Rapid, biphasic CRF neuronal responses encode positive and negative valence. Nature Neuroscience, 22(4), 576–585. 10.1038/s41593-019-0342-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning, A. , Buurstede, J. C. , van Weert, L. , & Meijer, O. C. (2019). Glucocorticoid and mineralocorticoid receptors in the brain: A transcriptional perspective. Journal of the Endocrine Society, 3(10), 1917–1930. 10.1210/js.2019-00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krugers, H. J. , Karst, H. , & Joels, M. (2012). Interactions between noradrenaline and corticosteroids in the brain: From electrical activity to cognitive performance. Frontiers in Cellular Neuroscience, 6, 15. 10.3389/fncel.2012.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesiak, A. , Pelz, C. , Ando, H. , Zhu, M. , Davare, M. , Lambert, T. J. , Hansen, K. F. , Obrietan, K. , Appleyard, S. M. , Impey, S. , & Wayman, G. A. (2013). A genome‐wide screen of CREB occupancy identifies the RhoA inhibitors Par6C and Rnd3 as regulators of BDNF‐induced synaptogenesis. PLoS One, 8(6), e64658. 10.1371/journal.pone.0064658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonze, B. E. , & Ginty, D. D. (2002). Function and regulation of CREB family transcription factors in the nervous system. Neuron, 35(4), 605–623. 10.1016/s0896-6273(02)00828-0 [DOI] [PubMed] [Google Scholar]
- Love, M. I. , Huber, W. , & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15(12), 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz, S. E. , Zhao, Y. , Gulinello, M. , Lee, S. C. , Raine, C. S. , & Brosnan, C. F. (2009). Deletion of astrocyte connexins 43 and 30 leads to a dysmyelinating phenotype and hippocampal CA1 vacuolation. Journal of Neuroscience, 29(24), 7743–7752. 10.1523/JNEUROSCI.0341-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr, B. , & Montminy, M. (2001). Transcriptional regulation by the phosphorylation‐dependent factor CREB. Nature Reviews Molecular Cell Biology, 2(8), 599–609. 10.1038/35085068 [DOI] [PubMed] [Google Scholar]
- McReynolds, J. R. , Donowho, K. , Abdi, A. , McGaugh, J. L. , Roozendaal, B. , & McIntyre, C. K. (2010). Memory‐enhancing corticosterone treatment increases amygdala norepinephrine and Arc protein expression in hippocampal synaptic fractions. Neurobiology of Learning and Memory, 93(3), 312–321. 10.1016/j.nlm.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer, O. C. , Buurstede, J. C. , & Schaaf, M. J. M. (2019). Corticosteroid receptors in the brain: Transcriptional mechanisms for specificity and context‐dependent effects. Cellular and Molecular Neurobiology, 39(4), 539–549. 10.1007/s10571-018-0625-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello‐Carpes, P. B. , da Silva de Vargas, L. , Gayer, M. C. , Roehrs, R. , & Izquierdo, I. (2016). Hippocampal noradrenergic activation is necessary for object recognition memory consolidation and can promote BDNF increase and memory persistence. Neurobiology of Learning and Memory, 127, 84–92. 10.1016/j.nlm.2015.11.014 [DOI] [PubMed] [Google Scholar]
- Morsink, M. C. , Steenbergen, P. J. , Vos, J. B. , Karst, H. , Joels, M. , De Kloet, E. R. , & Datson, N. A. (2006). Acute activation of hippocampal glucocorticoid receptors results in different waves of gene expression throughout time. Journal of Neuroendocrinology, 18(4), 239–252. 10.1111/j.1365-2826.2006.01413.x [DOI] [PubMed] [Google Scholar]
- Navarro Gonzalez, J. , Zweig, A. S. , Speir, M. L. , Schmelter, D. , Rosenbloom, K. R. , Raney, B. J. , Powell, C. C. , Nassar, L. R. , Maulding, N. D. , Lee, C. M. , Lee, B. T. , Hinrichs, A. S. , Fyfe, A. C. , Fernandes, J. D. , Diekhans, M. , Clawson, H. , Casper, J. , Benet‐Pages, A. , Barber, G. P. , … Kent, W. J. (2021). The UCSC genome browser database: 2021 update. Nucleic Acids Research, 49(D1), D1046–D1057. 10.1093/nar/gkaa1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oitzl, M. S. , & de Kloet, E. R. (1992). Selective corticosteroid antagonists modulate specific aspects of spatial orientation learning. Behavioral Neuroscience, 106(1), 62–71. 10.1037//0735-7044.106.1.62 [DOI] [PubMed] [Google Scholar]
- Oitzl, M. S. , Reichardt, H. M. , Joels, M. , & de Kloet, E. R. (2001). Point mutation in the mouse glucocorticoid receptor preventing DNA binding impairs spatial memory. Proceedings of the National Academy of Sciences USA, 98(22), 12790–12795. 10.1073/pnas.231313998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda, S. , Roozendaal, B. , & McGaugh, J. L. (2004). Glucocorticoid effects on object recognition memory require training‐associated emotional arousal. Proceedings of the National Academy of Sciences USA, 101(3), 853–858. 10.1073/pnas.0307803100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan, N. C. , McGettigan, P. A. , Sheridan, G. K. , Pickering, M. , Conboy, L. , O'Connor, J. J. , Moynagh, P. N. , Higgins, D. G. , Regan, C. M. , & Murphy, K. J. (2007). Temporal change in gene expression in the rat dentate gyrus following passive avoidance learning. Journal of Neurochemistry, 101(4), 1085–1098. 10.1111/j.1471-4159.2006.04418.x [DOI] [PubMed] [Google Scholar]
- Pooley, J. R. , Flynn, B. P. , Grontved, L. , Baek, S. , Guertin, M. J. , Kershaw, Y. M. , Birnie, M. T. , Pellatt, A. , Rivers, C. A. , Schiltz, R. L. , Hager, G. L. , Lightman, S. L. , & Conway‐Campbell, B. L. (2017). Genome‐wide identification of basic helix‐loop‐helix and NF‐1 motifs underlying GR binding sites in male rat hippocampus. Endocrinology, 158(5), 1486–1501. 10.1210/en.2016-1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provençal, N. , Arloth, J. , Cattaneo, A. , Anacker, C. , Cattane, N. , Wiechmann, T. , Röh, S. , Ködel, M. , Klengel, T. , Czamara, D. , Müller, N. S. , Lahti, J. , Räikkönen, K. , Pariante, C. M. , & Binder, E. B. (2020). Glucocorticoid exposure during hippocampal neurogenesis primes future stress response by inducing changes in DNA methylation. Proceedings of the National Academy of Sciences USA, 117(38), 23280–23285. 10.1073/pnas.1820842116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, B. , Xu, Y. , Wang, J. , Liu, M. , Dou, L. , Deng, R. , Wang, C. , Williams, K. E. , Stewart, R. B. , Xie, Z. , Ren, W. , Zhao, Z. , Shou, W. , Liang, T. , & Yong, W. (2019). Loss of FKBP5 affects neuron synaptic plasticity: An electrophysiology insight. Neuroscience, 402, 23–36. 10.1016/j.neuroscience.2019.01.021 [DOI] [PubMed] [Google Scholar]
- Quirarte, G. L. , Galvez, R. , Roozendaal, B. , & McGaugh, J. L. (1998). Norepinephrine release in the amygdala in response to footshock and opioid peptidergic drugs. Brain Research, 808(2), 134–140. 10.1016/s0006-8993(98)00795-1 [DOI] [PubMed] [Google Scholar]
- Ramirez, F. , Ryan, D. P. , Gruning, B. , Bhardwaj, V. , Kilpert, F. , Richter, A. S. , Heyne, S. , Dundar, F. , & Manke, T. (2016). deepTools2: A next generation web server for deep‐sequencing data analysis. Nucleic Acids Research, 44(W1), W160–165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao‐Ruiz, P. , Couey, J. J. , Marcelo, I. M. , Bouwkamp, C. G. , Slump, D. E. , Matos, M. R. , van der Loo, R. J. , Martins, G. J. , van den Hout, M. , van IJcken, W. F. , Costa, R. M. , van den Oever, M. C. , & Kushner, S. A. (2019). Engram‐specific transcriptome profiling of contextual memory consolidation. Nature Communications, 10(1), 2232. 10.1038/s41467-019-09960-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal, B. , Hernandez, A. , Cabrera, S. M. , Hagewoud, R. , Malvaez, M. , Stefanko, D. P. , Haettig, J. , & Wood, M. A. (2010). Membrane‐associated glucocorticoid activity is necessary for modulation of long‐term memory via chromatin modification. Journal of Neuroscience, 30(14), 5037–5046. 10.1523/JNEUROSCI.5717-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal, B. , Nguyen, B. T. , Power, A. E. , & McGaugh, J. L. (1999). Basolateral amygdala noradrenergic influence enables enhancement of memory consolidation induced by hippocampal glucocorticoid receptor activation. Proceedings of the National Academy of Sciences USA, 96(20), 11642–11647. 10.1073/pnas.96.20.11642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal, B. , Okuda, S. , Van der Zee, E. A. , & McGaugh, J. L. (2006). Glucocorticoid enhancement of memory requires arousal‐induced noradrenergic activation in the basolateral amygdala. Proceedings of the National Academy of Sciences USA, 103(17), 6741–6746. 10.1073/pnas.0601874103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguino‐Gomez, J. , Buurstede, J. C. , Abiega, O. , Fitzsimons, C. P. , Lucassen, P. J. , Eggen, B. J. , Lesuis, S. L. , Meijer, O. C. , & Krugers, H. J. (2021). An emerging role for microglia in stress‐effects on memory. European Journal of Neuroscience, 10.1111/ejn.15188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, A. , Macosko, E. Z. , Wysoker, A. , Goldman, M. , Krienen, F. M. , de Rivera, H. , Bien, E. , Baum, M. , Bortolin, L. , Wang, S. , Goeva, A. , Nemesh, J. , Kamitaki, N. , Brumbaugh, S. , Kulp, D. , & McCarroll, S. A. (2018). Molecular diversity and specializations among the cells of the adult mouse brain. Cell, 174(4), 1015–1030 e1016. 10.1016/j.cell.2018.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, A. J. , Kogan, J. H. , Frankland, P. W. , & Kida, S. (1998). CREB and memory. Annual Review of Neuroscience, 21, 127–148. 10.1146/annurev.neuro.21.1.127 [DOI] [PubMed] [Google Scholar]
- Tanis, K. Q. , Duman, R. S. , & Newton, S. S. (2008). CREB binding and activity in brain: Regional specificity and induction by electroconvulsive seizure. Biological Psychiatry, 63(7), 710–720. 10.1016/j.biopsych.2007.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gemert, N. G. , Meijer, O. C. , Morsink, M. C. , & Joels, M. (2006). Effect of brief corticosterone administration on SGK1 and RGS4 mRNA expression in rat hippocampus. Stress, 9(3), 165–170. 10.1080/10253890600966169 [DOI] [PubMed] [Google Scholar]
- van Weert, L. , Buurstede, J. C. , Mahfouz, A. , Braakhuis, P. S. M. , Polman, J. A. E. , Sips, H. C. M. , Roozendaal, B. , Balog, J. , de Kloet, E. R. , Datson, N. A. , & Meijer, O. C. (2017). NeuroD factors discriminate mineralocorticoid from glucocorticoid receptor DNA binding in the male rat brain. Endocrinology, 158(5), 1511–1522. 10.1210/en.2016-1422 [DOI] [PubMed] [Google Scholar]
- Yao, Z. , Nguyen, T. N. , van Velthoven, C. T. J. , Goldy, J. , Sedeno‐Cortes, A. E. , Baftizadeh, F. , Bertagnolli, D. , Casper, T. , Crichton, K. , Ding, S.‐L. , Fong, O. , Garren, E. , Glandon, A. , Gray, J. , Graybuck, L. T. , Hirschstein, D. , Kroll, M. , Lathia, K. , Levi, B. , … Zeng, H. (2020). A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. bioRxiv. 10.1101/2020.03.30.015214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C. , Aviles, C. , Abel, R. A. , Almli, C. R. , McQuillen, P. , & Pleasure, S. J. (2005). Hippocampal and visuospatial learning defects in mice with a deletion of frizzled 9, a gene in the Williams syndrome deletion interval. Development, 132(12), 2917–2927. 10.1242/dev.01871 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S10
Data Availability Statement
Sequencing data are available via NCBI's Gene Expression Omnibus and are accessible through GEO reference series accession number GSE160808. Further data could be obtained upon reasonable request to the corresponding author.


