Abstract
Temperature is a critical driver of ectotherm life‐history strategies, whereby a warmer environment is associated with increased growth, reduced longevity and accelerated senescence. Increasing evidence indicates that thermal adaptation may underlie such life‐history shifts in wild populations. Single nucleotide polymorphisms (SNPs) and copy number variants (CNVs) can help uncover the molecular mechanisms of temperature‐driven variation in growth, longevity and senescence. However, our understanding of these mechanisms is still limited, which reduces our ability to predict the response of non‐model ectotherms to global temperature change.
In this study, we examined the potential role of thermal adaptation in clinal shifts of life‐history traits (i.e. life span, senescence rate and recruitment) in the Columbia spotted frog Rana luteiventris along a broad temperature gradient in the western United States.
We took advantage of extensive capture–recapture datasets of 20,033 marked individuals from eight populations surveyed annually for 14–18 years to examine how mean annual temperature and precipitation influenced demographic parameters (i.e. adult survival, life span, senescence rate, recruitment and population growth). After showing that temperature was the main climatic predictor influencing demography, we used RAD‐seq data (50,829 SNPs and 6,599 putative CNVs) generated for 352 individuals from 31 breeding sites to identify the genomic signatures of thermal adaptation.
Our results showed that temperature was negatively associated with annual adult survival and reproductive life span and positively associated with senescence rate. By contrast, recruitment increased with temperature, promoting the long‐term viability of most populations. These temperature‐dependent demographic changes were associated with strong genomic signatures of thermal adaptation. We identified 148 SNP candidates associated with temperature including three SNPs located within protein‐coding genes regulating resistance to cold and hypoxia, immunity and reproduction in ranids. We also identified 39 CNV candidates (including within 38 transposable elements) for which normalized read depth was associated with temperature.
Our study indicates that both SNPs and structural variants are associated with temperature and could eventually be found to play a functional role in clinal shifts in senescence rate and life‐history strategies in R. luteiventris. These results highlight the potential role of different sources of molecular variation in the response of ectotherms to environmental temperature variation in the context of global warming.
Keywords: adaptation, amphibian, copy number variants, life history, senescence, single nucleotide polymorphisms, temperature, transposable elements
This paper combines high‐quality demographic and genomic data to examine how climate drives clinal variation in life‐history strategies and local adaptation.

1. INTRODUCTION
Global temperatures are projected to increase up to 4℃ by the end of the current century (IPCC, 2014), with dramatic consequences for the individual life history and long‐term viability of animal populations (Selwood et al., 2015; Stenseth & Mysterud, 2002; Walsh et al., 2019). The demographic impact of global warming is expected to be particularly severe in ectothermic animals that have limited capacities to use metabolic heat to maintain their body temperature, making their life‐history traits highly sensitive to temperature variation (Burraco et al., 2020; Seebacher et al., 2015). Studies on free‐ranging and experimental populations have shown that warmer environmental temperatures decrease ectotherm longevity (Flouris & Piantoni, 2015; Keil et al., 2015; Munch & Salinas, 2009) and accelerate their actuarial senescence (i.e. increase in mortality with age; Jacobson et al., 2010; Valenzano et al., 2006; Yen & Mobbs, 2010). In parallel, ectotherm growth and reproductive rate increase with temperature (Angilletta et al., 2004; Burraco et al., 2020; Huey & Berrigan, 2001). Together, these temperature‐dependent processes regulate the position of populations and species along the fast–slow continuum of life history (sensu, Promislow & Harvey, 1990). In ectothermic vertebrates (e.g. fish: Blanck & Lamouroux, 2007, amphibians: Morrison & Hero, 2003), individuals from cold environments (at high altitude and latitude) have a slower life history (i.e. reduced growth rate, delayed sexual maturity, low fecundity and long adult life span) than those experiencing warmer conditions. The evolutionary mechanisms underlying these temperature‐driven shifts in life history, however, are still poorly understood, which limits our ability to accurately predict the fate of ectotherm populations in the face of global warming (Mims et al., 2018).
For a long time, temperature‐dependent changes in life history have been thought to be regulated by adaptive plasticity. Thermoregulatory behaviours indeed allow ectotherms to plastically adjust their body temperature and buffer the detrimental impact of thermal variation on survival (Bodensteiner et al., 2021; Gunderson & Stillman, 2015). Furthermore, warm environments induce thermal plasticity in most ectotherms, resulting in fast growth at the expense of adult life span, possibly via the rapid accumulation of oxidative damage (Burraco et al., 2020). However, an increasing number of molecular studies suggests that temperature‐driven changes in life history could be also partially regulated by thermal adaptation. Genotype‐environment associations (GEAs) have revealed molecular signals of thermal adaptation in a broad range of ectotherms by identifying single nucleotide polymorphisms (SNPs) whose allelic frequencies are correlated with temperature (e.g. lacustrine and marine fish: Perrier et al., 2017, Cayuela, Rougemont, et al., 2020; amphibians: Rödin‐Mörch et al., 2019; reptiles: Rodríguez et al., 2017; crustaceans: Benestan et al., 2016; molluscs: Lehnert et al., 2019; insects: Parker et al., 2018). In addition, genome‐wide association studies (GWAS) showed that SNPs were associated with changes in thermal preference (Campbell‐Staton et al., 2020), thermal tolerance (i.e. survival in response to thermal stress; Amish et al., 2019; Rajpurohit et al., 2018), cardiac performances (e.g. Chen & Narum, 2021), growth rate (Quigley et al., 2020) and mitochondrial metabolism (Mallard et al., 2018) among ectotherm populations experiencing contrasting temperature conditions.
Genomic structural variation (Mérot et al., 2020; Wellenreuther et al., 2019) could also contribute to the thermal adaptation underlying temperature‐dependent change in life history. Copy number variants (CNVs) are structural variants in which a segment of DNA can be absent or present in two or more paralogous copies (in diploid species) due to either gene duplication, deletion or transposable elements (TEs; Mérot et al., 2020). Studies have identified molecular signatures of adaptation involving CNVs (i.e. including duplicated coding and regulatory regions) whose copy number covaried with local temperature in marine fish and crustaceans (Cayuela et al., 2021; Dorant et al., 2020). Furthermore, interspecific comparisons between the long‐lived Antarctic toothfish and its short‐lived relatives from warm water identified dozens of genes in multiple copies that were overexpressed and likely improved survival and growth of toothfish in cold and oxygen‐rich waters of the Antarctic zone (Chen et al., 2008). In addition, thermal stress experienced during an individual's lifetime may lead to temperature‐dependent accumulation of TEs (Carducci et al., 2020), which are known to generate a broad variety of mutations that may lead to phenotypic and fitness variation through the modification of gene expression, the inactivation of genes and the alteration in gene sequence and reading frame (Casacuberta & González, 2013; Chuong et al., 2017).
To date, few studies have investigated whether life history and senescence acceleration driven by temperature are associated with molecular signatures of thermal adaptation among wild ectotherm populations. Here, we address this issue in the Columbia spotted frog Rana luteiventris, a pond‐breeding anuran from the western United States and Canada that is threatened by increasing environmental temperatures and decreasing precipitation during its active season (April to October; Pilliod et al., 2015), particularly in the southern end of its distribution where drought is increasingly prevalent (Pilliod et al., 2021). This species is an excellent model for our study because it is widely distributed (distribution range size: 1,322,432 km2, encompassing parts of eight US states and two Canadian provinces) and its populations are spread along a broad gradient of mean annual temperature (ranging from 1.71 to 8.35℃ in our study area). Taking advantage of long‐term (from 14 to 18 years, depending on site) capture–recapture data collected in eight populations of R. luteiventris in the United States (Figure 1), we tested the hypothesis that senescence rate increases with mean annual temperature while adult survival and longevity decrease. Furthermore, we predicted that recruitment increases with mean annual temperature, leading to an acceleration of life‐history strategies along the thermal gradient. Then, generating RAD‐seq data from DNA samples collected across 31 breeding sites (Figure 1), we examined SNP‐temperature and CNV‐temperature associations to detect the genomic signatures of thermal adaptation and to identify the candidate genes that might be involved in the life‐history shifts detected using demographic analyses. In the light of these analyses, we then discuss the potential role of adaptation in life‐history shifts and an organism's response to global warming.
FIGURE 1.
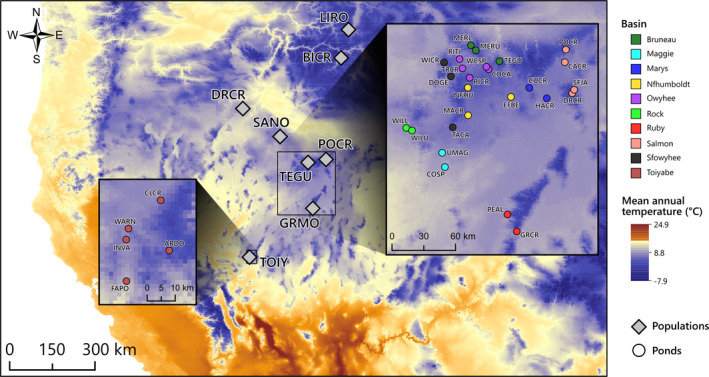
Map of the study area showing the eight in Rana luteiventris populations surveyed using the capture–recapture method (grey diamonds) and the 31 breeding ponds (located in 10 hydrographic basins) considered in the genomic analyses. State lines were omitted for clarity; study sites occurred in Montana (LIRO), Idaho (BICR and SANO), Oregon (DRCR) and Nevada (POCR, TEGU, GRMO and TOIY) [Colour figure can be viewed at wileyonlinelibrary.com]
2. MATERIALS AND METHODS
2.1. Demographic analyses
2.1.1. Capture–recapture data collection
We collected capture–recapture data at eight populations or metapopulations from four states within the range of R. luteiventris in the western United States (Figure 1). The survey period differed among populations, ranging from 14 to 18 years (see detailed information in Table S1). The surveyed populations were generally spread across multiple breeding waterbodies, thus behaving like ‘metapopulations’ (sensu lato). The studied populations were generally not selected randomly, but represent a convenient and opportunistic sample of R. luteiventris populations. Populations were typically large enough to conduct mark–recapture studies, conveniently located for researchers and workers, and relatively free from major anthropogenic threats.
This study focused on adult frogs (with a snout‐vent length longer than 45 mm) that were captured during annual visual encounter surveys. Surveys were conducted during a consistent 1‐ to 3‐week period each year (usually July or August), depending on the studied population. For each population, wetland surveys consisted of one to eight workers searching for animals along the shoreline of deeper water bodies or the entire surface area of wadable water bodies. Most populations were surveyed two to three times during this annual survey period to increase the capture rates. Frogs were surveyed during the day when air temperatures were above 10°C. Sampling was time constrained whereby surveys ended either when the entire area was thoroughly searched or about 30 min after the last animal was captured.
All captured animals were checked for an individual mark or tagged if they were unmarked. Unmarked individuals were injected with a passive integrated transponder (PIT) tag into the dorsal subcutaneous tissue or marked with a unique pattern of toe clips (Montana population only, prior to 2013). The PIT tag number of all newly marked animals as well as all recaptured individuals was recorded. After handling, frogs were released approximately at the location where they were captured or at least within the same water body, and always with minutes or an hour of capture. The number of individual frogs marked per population ranged from 370 in population POCR to 8,215 in population TOIY (Table S1). The number of captures ranged from 707 in population GRMO to 13,067 in population TOIY.
Climate data were compiled from 800‐m Parameter‐elevation Relationships on Independent Slopes Model (PRISM) Climate Group, Oregon State University (http://prism.oregonstate.edu, accessed 5 October 2020). For the 30‐year period, 1990–2019, we calculated the mean and standard deviation of annual temperature, and the mean and standard deviation of annual cumulative precipitation for each population. Because populations occupy complex waterbody networks spread across sometimes large areas (e.g. lakes within mountain basins or several kilometre of stream), we selected the coordinates of the area of highest animal density to use for our modelled climate data.
2.1.2. Estimating adult survival, seniority and population growth rate
We estimated adult survival (φ), seniority (γ) and population growth rate (λ) for the eight populations using the Bayesian formulation of the Pradel model (Pradel, 1996) proposed by Tenan et al. (2014). Seniority probability is the probability that, at each year y, an individual was already present in the population at y − 1; hence, 1 − γ corresponds to the probability that an individual was recruited in the population between y − 1 and y (Pradel, 1996). Population was assumed to be stable if λ = 1, to experience a decline if λ < 1 or to be growing if λ was >1. We included a random effect of year on those three parameters to quantify their temporal variance. As the number of study years differed among populations, the eight populations were analysed separately. Summaries of posterior parameter distributions were calculated from three independent Markov chains of 1,000,000 iterations initialized with random starting values, run 500,000 times after a 50,000 burn‐in and resampling every 20 draws. We evaluated chain convergence using the R package coda (Plummer et al., 2006). The models were implemented in program JAGS (Plummer, 2003) that we executed from R (R Core Team, 2019) with the package r2 jags (Su & Yajima, 2012).
2.1.3. Estimating senescence rate and life span
Senescence rate and adult life span were calculated using the modelling procedure proposed by Lemaître et al. (2020). Bayesian survival trajectory analyses implemented in the R package basta were used to estimate those mortality metrics (Colchero & Clark, 2012; Colchero et al., 2012). Simulations by Colchero and Clark (2012) showed that BaSTA models are highly efficient to investigate age‐dependent mortality when dates of birth and death are unknown and recapture probability is imperfect. BaSTA therefore allowed us to account for imperfect detection, left‐truncated [i.e. unknown birth date (age)] and right‐censored (i.e. unknown death date) capture–recapture data in our analysis. Again, the eight populations were analysed separately. In an effort to be conservative, we specified time‐dependent recapture probability for all species. Four MCMC chains were run with 50,000 iterations and a burn‐in of 5,000. Chains were thinned by a factor of 50. Model convergence was evaluated using the diagnostic analyses implemented in BaSTA, which calculate the potential scale reduction for each parameter to assess convergence.
We fitted a Siler model on age‐specific mortality data (Siler, 1979) for each species to obtain comparable metrics. The five‐parameter Siler model is given by:
where a 0, a 1, b 0, b 1 and c ≥ 0 are the parameters of the mortality function and x the age in years. The first exponential function with parameters a describes mortality in early adult stage, whereas c gives the lower limit of mortality during the adult stage. The second exponential function with b parameters corresponds to the mortality increase during the senescent stage. The parameter b 1 of the Siler model measures the exponential increase in mortality rate with age, and is therefore commonly used as a metric of rate of mortality ageing in vertebrates (Lemaître et al., 2020; Ronget & Gaillard, 2020).
Following the procedure presented in Lemaître et al. (2020), we estimated adult life span 80% (in years), corresponding to the age at which 80% of the individuals alive at the onset of adulthood (i.e. when first reproduction occurs) were dead (i.e. when cumulative survivorship reaches 0.8). Adult life span 80% was calculated from Siler model parameters by solving numerically the following equation:
2.1.4. Statistical analyses
We used linear models to examine the effect of mean temperature and precipitation (calculated for the period 1990–2019) on ageing rate, life span 80%, mean annual adult survival, mean annual seniority, seniority and mean annual population growth rate. The demographic parameters were included as response variables (log‐transformed), whereas the climate variables were included as explanatory variables. The effect of the temperature and precipitation was examined separately. We used likelihood ratio tests performed between the models with and without the climatic covariate to assess the significance of the relationships. R 2 were also calculated. Furthermore, we used a similar approach to investigate the effect of temporal variability of temperature and precipitation (standard deviation calculated for the period 1990–2019) on the temporal variance (standard deviation) of annual adult survival, seniority and population growth rate. Siler model parameters appropriately converged for all populations except POCR; hence, ageing rate and life span 80% of this population were not included in the analyses.
2.2. Molecular analyses
2.2.1. Sampling area and DNA collection
Toe clips (from metamorph and adult frogs) and tail clips (5–10 mm, from tadpoles) were collected between 2010 and 2019. Individuals were captured at 31 breeding ponds across the Nevada range (Figure 1). The ponds were located within 10 hydrographic catchments (Figure 1). All individuals were released at their capture location after processing. The average number of individuals sampled in the breeding ponds was 11 (range: 4–14; see Table S2 for sampling, geographical and temperature information).
2.2.2. DNA extraction, library preparation and RAD‐sequencing
We extracted DNA from tissue samples using Qiagen DNeasy Blood and Tissue Kits following the manufacturer's protocols, with the addition of 4 µl of RNase after tissue digestion. We quantified total DNA concentrations using Qubit dsDNA BR Assays and verified DNA quality in a subset of individuals using electrophoresis on agarose gels.
We tested five restriction enzyme digestions on a subset of individuals to find the enzyme combination that provided the appropriate distribution of fragment sizes (250–450 base pairs). We then digested ~1 μg of total DNA per individual using NspI and EcoRI. We followed Peterson et al. (2012), using a Pippin Prep size selection of 346–456 base pairs and Dynabeads cleanup. We used KAPA HiFi Hotstart Ready Mix for PCR amplification, and degenerate barcodes on the P2 adapter to facilitate the removal of PCR duplicates (Schweyen et al., 2014). We sequenced one library with the University of Oregon Genomics Core Facility (HiSeq‐4000, 100‐bp paired‐end reads), and the remaining nine libraries with Novogene Corp. (HiSeq‐4000 150‐bp paired‐end reads). The first two libraries contained 24 and 36 individuals respectively. All remaining libraries contained 40 individuals. Sites were split across libraries to avoid confounding site and library effects.
We used FastQC v. 0.11.8 (Andrews, 2019) to assess the data quality and identify the sequencing anomalies in each library. We removed PCR duplicates from the raw reads using clone_filter from Stacks v. 2.41 (Catchen et al., 2011, 2013; Rochette et al., 2019). Using Trim Galore! v. 0.6.4 (Krueger, 2019) and Cutadapt v. 2.5 (Martin, 2011), we removed the 10‐bp degenerative barcodes from the P2 reads and applied quality screening using a Phred score cut‐off of 20 and auto‐detect adapter screening with stringency set to 6. We then used Stacks process_radtags to demultiplex the raw data, trim reads to 90 bp, and run additional quality control (remove reads with an uncalled base; discard reads with low‐quality scores (default settings); rescue barcodes and cut sites with at most one mismatch).
We used within and cross‐library replicates to optimize three de novo Stacks parameters: minimum stack depth (‐m), distance between stacks (‐M) and the distance between catalogue loci (‐n). We used 19 individuals, each with a replicate sample, from 16 sites (total samples = 38), covering the geographic range of sampling effort. Thirteen combinations of ‐m (3–5), ‐M (2–6) and ‐n (1–6) were tested across the entire pipeline, setting the upper bound on the error rate for the SNP model (ustacks ‐‐bound.high) to 0.05. To determine the optimal combination of parameters, we evaluated mean coverage, the number of assembled loci, the number of polymorphic loci shared across 80% of individuals and the number of SNPs (Paris et al., 2017). We also assessed population/duplicate clustering in multivariate space using multidimensional scaling plots (Gugger et al., 2018) calculated in plink v. 1.90 (Chang et al., 2015).
After optimizing Stacks parameters, we ran the Stacks pipeline on all unique samples and filtered using the populations module: ‐p 1 (minimum number of populations a locus must be present in to process), ‐R 0.3 (minimum percentage of individuals across populations required to process a locus) and ‐‐min_mac 2 (minimum minor allele count required to process a SNP). We then ran the ‘populations’ module from Stacks to produce a vcf file defining the minimum number of populations a locus must be present in to process (p = 1), the minimum percentage of individuals across populations required to process a locus (R = 0.3) and the minimum minor allele count required to process a SNP (min_mac 2). The outputted vcf file was further filtered using python and bash scripts from the stacks workflow RAD‐seq pipeline analysis available online (https://github.com/enormandeau/stacks_workflow). The raw vcf was filtered keeping only SNPs that showed a minimum coverage depth of 10×, were called in at least 70% of the samples per sampling location (according a maximum of four locations that can fail this latter filter) and a minimum number of samples having the rare allele of four across all sampling locations. Ultimately, samples showing more than 60% of missing data and SNPs showing more than 40% of missing data were discarded.
2.2.3. SNP and CNV discovery
We discriminated SNPs and putative CNVs using the HDplot approach initially proposed by McKinney et al. (2017) and recently adapted to CNV studies by Dorant et al. (2020) and Cayuela et al. (2021). The duplicated loci detected by this method can be variant or invariant in copy number among the studied populations. A duplicated locus is considered as a CNV only if a correlation between the normalized read depth of a marker and a phenotypic trait or an environmental variable is detected. For this reason, we systematically use the term of ‘putative CNVs’ when we refer to the markers identified using the HDplot method.
The HDplot approach extended by Dorant et al. (2020) discriminates ‘singleton’ SNPs (i.e. non‐duplicated) from ‘duplicated’ SNPs (i.e. combining duplicated and diverged SNPs and SNPs with a high coverage) using the following four parameters: (a) proportion of heterozygotes (PropHet), (b) inbreeding coefficient (Fis), (c) median of allele ratio for heterozygotes (MedRatio) and (4) proportion of rare homozygotes. The parameters were calculated using a custom python script (available at https://github.com/enormandeau/stacks_workflow) parsing the filtered VCF file. The four parameters were plotted pairwise to visualize their distribution across all loci (see Figure S1). Based on the graphical demonstration by McKinney et al. (2017), we considered different combinations of parameters and graphically fixed the cut‐off of the five categories of SNPs (singleton SNPs, duplicated SNPs, diverged SNPs, high coverage SNPs and low confidence). We then kept all RAD loci associated with either duplicated, diverged and high‐coverage categories and extracted the loci read depth of to construct the dataset of putative CNVs using vcftools. For more simplicity, the term ‘putative CNV’ is used to define all loci classified as ‘duplicated’, ‘diverged’ and ‘high coverage’.
Following the procedure described in Dorant et al. (2020) and Cayuela et al. (2021), read counts of putative CNV loci were normalized to account for differences in sequencing effort across all samples. Normalization was performed using the trimmed mean of M‐values method originally described for RNA‐seq count normalization and implemented in the R package edger (Robinson & Oshlack, 2010). The correction accounts for the fact that for an individual with a higher copy number at a given locus, that locus will contribute proportionally more to the sequencing library than it will for an individual with a lower copy number at that locus.
Finally, two separated datasets were generated; the ‘SNP dataset’, composed of singleton SNPs only, and the ‘CNV dataset’, which is the matrix of normalized read depth described above. To construct the SNP dataset, we conserved the genotype calls from the VCF containing singleton SNPs, and post‐filtered these by keeping all unlinked SNPs within each locus using the python script 11_extract_unlinked_snps.py available in stacks workflow. Briefly, linkage was assessed for each pair of SNPs considering samples without missing data where at least one of the two genotypes contains the rare allele. If the two SNPs had identical genotypes in more than 30% among samples, the pair was considered linked and only the first SNPs was kept.
2.3. Putative CNV and DNA repeats in ranids
A previous study using similar RAD‐seq‐based CNV analyses showed that putative CNVs often correspond to DNA repeats (Cayuela et al., 2021). Therefore, we investigated the proportion of CNVs which fell into DNA repeats, relative to non‐CNV RAD tags. In addition, we examined if DNA repeats detected as putative CNVs diverged more recently than the other elements of their own repeat family located in the rest of the genome by analysing the relative age of their sequences.
We used RepeatMasker v.4.0.7 (Smit et al., 2015) to search RAD tag sequences against the custom repeat library which exists for the unpublished R. temporaria genome assembly (available at https://github.com/DanJeffries/Rana‐temporaria‐genome‐assembly/wiki). To reduce computational complexity, we randomly selected five subsets of 10,000 RAD tags from the full catalogue to act as a baseline expectation for a given set of RAD tags in the dataset. For each subset, we examined the proportion of tags that were assigned to each repeat family, and the relative age of the repeat sequences (Kimura distance). We then did the same for 6,599 sequences identified as putative CNVs and for 39 sequences identified as candidate CNVs associated with temperature (see Results section). All five subsets of randomly selected RAD tags were almost identical in their proportions and TE age landscapes and were thus combined and used for comparisons with the CNV set. The proportions of RAD tags falling into each major repeat family (DNA transposons, LTR retransposons and miniature inverted‐repeat transposable elements (MITEs); Bourque et al., 2018) were compared between the baseline and CNV subsets using Fisher's exact test. The distributions of TE age were compared for each minor TE family (DNA/DTA, DNA/DTC, DNA/DTH, DNA/DTM, DNA/DTT, DNA/Helitron, LTR/Copia, LTR/Gypsy, LTR/unknown, MITE/DTA, MITE/DTC, MITE/DTH, MITE/DTM, MITE/DTT) separately using Welch's t test for equal means and Kolmogorov–Smirnov test. All statistical tests were implemented in Scipy v.1.5.1 in Python 3.6.3.
2.3.1. SNP‐temperature associations
From our 50,829 SNP singletons (see Results section), we first examined population genetic structure using a principal component analysis (PCA) and level of genetic differentiation among sampling locations. We also calculated pairwise F ST using the R library StAMPP (Pembleton et al., 2013) with 95% CI estimated on 1,000 bootstraps.
Following the recommendations of Forester et al. (2018), SNP‐temperature associations were then evaluated using a combination of latent factor mixed models (LFMM; Frichot et al., 2013) and partial redundancy analysis (pRDA; Legendre & Legendre, 2012) to detect SNP candidates. To minimize the number of false positives, we corrected for population genetic structure in LFMMs (Forester et al., 2018). In the pRDA, we controlled for sequencing libraries to control for the potential bias caused by batch effect that we detected within the genomic data (see Figure S7). Next, we retained the outliers that were detected by both methods, and we represented the overlap with a Venn diagram. In the pRDA, an analysis of variance (ANOVA) with 1,000 permutations was performed to assess the significance of the model. SNP candidates were identified as those exhibiting a loading >2.25 standard deviations from the mean loading (p < 0.025). We tested for linear correlations between allele frequencies and mean annual temperature using latent factor mixed models implemented in the R package lfmm 2 (Caye et al., 2019). We first performed a PCA and assessed the global population structure in the genotypic data. After determining how many components (K) explained most of the genotypic variance, genomic inflation factors (λ) were processed with the function lfmm_ridge and z scores were calculated with the function lfmm_test. Resulted p‐values were adjusted based on λ and the χ 2 distribution (Caye et al., 2019). To control for false positives, we also applied a Benjamini–Hochberg correction algorithm with a false discovery rate (FDR) of 0.01 (Benjamini & Hochberg, 1995). To reduce the number of false positives and correct our GEA for both batch effect and population structure, we only retained the candidate SNPs that were detected by both LMMs (linear mixed models) and pRDA and represented the overlap with a Venn diagram. Then, to identify potential candidate genes involved in thermal adaptation, we performed BLAST analyses for the locus sequences bearing candidate SNPs against the NCBI NR database. To consider a SNP further, we required a blast length of at least 80 bp and an e‐value of 1e‐10.
2.3.2. CNV‐temperature associations
CNV‐temperature associations were evaluated by combining locus‐by‐locus regressions and multivariate analyses following the approach used in Dorant et al. (2020) and Cayuela et al. (2021). First, we used a partial redundancy analysis to detect CNV candidates that were associated with temperature after controlling for a sequencing batch effect detected with a PCA performed on the normalized read depth matrix (Figure S2). Global and marginal analyses of variance (ANOVA) with 1,000 permutations were performed to assess the significance of the models. Once CNVs were loaded against the pRDA axes, candidates for an association with temperature were determined as those exhibiting a loading >2.25 standard deviations from the mean loading (p < 0.025).
Next, we ran LMMs with restricted maximum likelihood optimization where the log‐transformed normalized read depth was incorporated as the response variable and temperature as the explanatory variable. The sequencing batch effect was included as a random effect (i.e. random intercepts) to account for the non‐independency of observations across batches. One model was performed for each putative CNV, and we used likelihood ratio tests and a FDR of 0.01 following the method of Benjamini and Hochberg (1995) to assess the significance of CNV‐temperature associations. We retained the outliers that were detected by both LMMs and pRDA to reduce the number of false positives and represented the overlap with a Venn diagram.
For the 20 strongest CNV loci putatively associated with temperature (those with highest R 2), we examined whether discrete copy number categories could be drawn from the distributions of normalized read depth using a model‐based unsupervised clustering approach implemented in the R library Mclust, V. 5.4.4 (Fraley et al., 2012). Mclust uses finite mixture estimation via iterative expectation maximization steps (EM) for a range of k components, and the best model is selected using the Bayesian information criterion (BIC). For each locus, individuals were classified in discrete clusters of normalized read depth (that we coined copy number groups) that likely reflect variation in copy number of any given locus.
3. RESULTS
3.1. Demographic analyses
3.1.1. Influence of temperature on life history and senescence
From 20,033 individual frogs captured 33,857 times at eight study areas (see Tables S1), we found that life‐history traits of R. luteiventris were strongly associated with mean annual temperature experienced by populations, whereas mean annual precipitation showed only a marginal correlation (Table 1). Ageing rate ranged from 0.11 to 1.51 and increased with increasing temperature (Table 1; Figure 2a; see Tables S3–S10). By contrast, life span 80% varied between 3.34 and 10.66 years and decreased with increasing temperature (Table 1; Figure 2b). Similarly, adult survival ranged from 0.26 and 0.78 and was negatively correlated with temperature (Table 1; Figure 2c). The increase in adult mortality was compensated by higher recruitment (1 − seniority) in populations experiencing warm conditions: seniority ranged from 0.26 and 0.94 and was negatively associated with increasing temperature (Table 1; Figure 2d). Although 95% credible intervals were broad, mean population growth was higher than 1 in seven of the eight populations, indicating that almost all populations (except POCR) were likely viable in the long term (Table S11). Mean population growth, which varied between 0.77 and 1.13, was not associated with mean annual temperature (Table 1). Furthermore, the annual variability of temperature and precipitation showed no significant association with the temporal variance of survival, seniority and population growth rate (Table 1). All demographic estimates from Pradel models are provided in Table S11.
TABLE 1.
Effect of temperature and precipitation on demographic parameters in eight populations of Rana luteiventris. Likelihood ratio tests were used to assess the influence of mean and temporal variance (SD) of temperature (calculated for the period 1990–2019) and precipitation on ageing rate, life span 80%, annual adult survival (mean μ and temporal variance σ), annual seniority (μ and σ) and annual population growth rate (μ and σ). Recruitment is approximated as 1 − seniority. R 2 values were calculated from the linear model. Relationships for which likelihood ratio test had a p‐value lower than 0.05 are shown in bold
| Parameter | df | χ 2 | p‐value | R 2 |
|---|---|---|---|---|
| Mean annual temperature | ||||
| Ageing rate | 1 | 10.13 | 0.001 | 0.73 |
| Life span 80% | 1 | 5.32 | 0.02 | 0.49 |
| Survival (μ) | 1 | 8.18 | 0.004 | 0.60 |
| Seniority (μ) | 1 | 7.69 | 0.005 | 0.58 |
| Population growth (μ) | 1 | 0.17 | 0.68 | 0.02 |
| Mean annual cumulative precipitation | ||||
| Ageing rate | 1 | 3.05 | 0.08 | 0.31 |
| Life span 80% | 1 | 0.88 | 0.35 | 0.10 |
| Survival (μ) | 1 | 2.15 | 0.14 | 0.21 |
| Seniority (μ) | 1 | 2.14 | 0.14 | 0.21 |
| Population growth (μ) | 1 | 0.15 | 0.69 | 0.02 |
| Standard deviation of annual temperature | ||||
| Survival (σ) | 1 | 0.008 | 0.93 | 0.05 |
| Seniority (σ) | 1 | 0.31 | 0.57 | 0.03 |
| Population growth (σ) | 1 | 0.26 | 0.61 | 0.03 |
| Standard deviation of n annual cumulative precipitation | ||||
| Survival (σ) | 1 | 0.47 | 0.49 | 0.05 |
| Seniority (σ) | 1 | 0.05 | 0.82 | 0.001 |
| Population growth (σ) | 1 | 1.97 | 0.16 | 0.19 |
FIGURE 2.
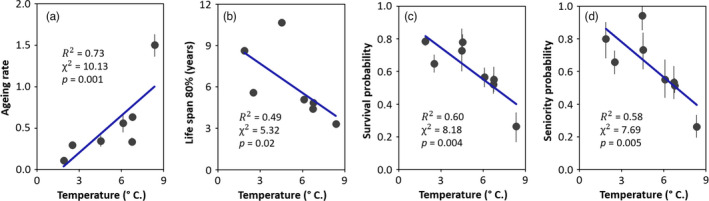
Influence of mean annual temperature (calculated for the period 1990–2019) on ageing rate, life span 80% (age at which 80% of the individuals alive at the onset of adulthood die), mean annual adult survival probability and mean annual seniority probability in Rana luteiventris. Estimates (black points) and 95% credible intervals (error bars) extracted from Siler models (a and b) and Pradel models (c and d) are given. Siler model parameters appropriately converged for all populations except POCR; hence, ageing rate and life span 80% of this population were not included in the analyses. We show the regression line (in blue) extracted from the linear models built to test the effect of temperature on demographic parameters. In addition, we provide R 2 and the statistics of the likelihood ratio tests (χ 2 and p) [Colour figure can be viewed at wileyonlinelibrary.com]
3.2. Molecular analyses
3.2.1. SNP and CNV discovery
RAD‐seq produced 7,095,898 reads per sample on average (range: 1,428,670–10,553,796) before any quality filtering. The mean coverage was 25.56 (range: 8–41). The SNP calling process identified 109,542 SNPs that were successfully genotyped in 352 out of 360 samples initially sequenced (eight samples >60% missing data were discarded), with a rate of missing data lower than 40% among SNPs. The HDplot approach identified 7,631, 5,787 and 1,523 SNP markers as duplicated, diverged and high coverage respectively (Figure S2). All of these markers were distributed over 6,599 putative CNV loci and form the so‐called ‘CNV dataset’. Moreover, 13,820 markers were classified as low confidence, and 80,658 markers were classified as singleton SNPs. A total of 29,829 SNPs were pruned during the linkage disequilibrium procedure. The final dataset used in subsequent analyses contained 50,829 SNP singletons, with a global 18.3% missing genotypes.
3.2.2. Putative CNV repeat content
The comparison of the repeat content of putative CNVs relative to the randomly selected sets of RAD tags showed that CNVs more commonly fall into LTR elements than expected (OR = 0.65, p < 0.001), and consequently, occur less frequently in DNA and MITE elements. Repeat elements containing CNVs were also generally younger (KS test: D = 0.02, p < 0.001; Figure S8) although this is seemingly driven by younger DNA elements (Figures S9 and 10). In contrast, LTRs containing CNVs tended to be older than expected (Figures S9 and 10).
3.2.3. Population genetic structure
Using the 50,829 SNP singletons, we detected very strong population genetic structure. Pairwise F ST calculated between breeding sites were all highly significant and ranged from 0.05 to 0.84, with an average of 0.50 (Figure 3a; Table S12). The PCA revealed a strong genetic differentiation among hydrographic basins (Figure 3a).
FIGURE 3.
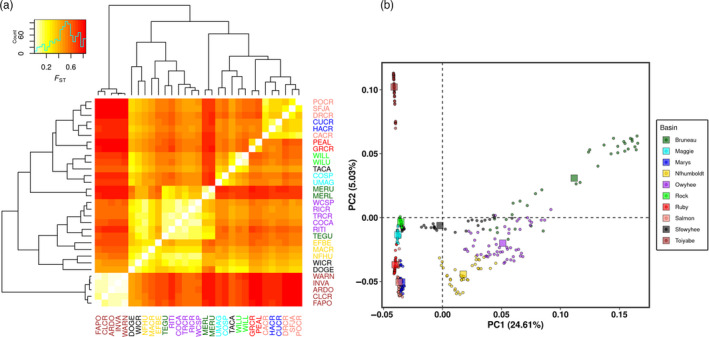
Population structure based on a F ST matrix (a) and a principal component analysis (b) in Rana luteiventris. (a) Pairwise F ST were calculated for each pair of breeding sites and were all significant (p‐values are provided in Table S12). Breeding sites (Table S2) have been ordered based on a UPGMA cluster algorithm. (b) Principal component analysis represents the genetic variation within our SNP dataset. We coloured the individuals according to their hydrographic basin of origin [Colour figure can be viewed at wileyonlinelibrary.com]
3.2.4. SNP‐temperature associations
SNP‐temperature associations revealed a strong signature of thermal adaptation in R. luteiventris. The pRDA (Figure 4a) designed to detect SNPs associated with temperature was significant (df = 1, F = 17.67, p < 0.001), although the coefficient of determination was low (adjusted R 2 = 0.045). A total of 1889 and 234 SNP candidates were associated with temperature based on pRDA and LFMM2 respectively (Figure 4a; p‐values before and after genomic inflation factor calibration are provided in Figure S6). A set of 148 candidate SNPs located on 147 RAD loci were identified by both methods.
FIGURE 4.
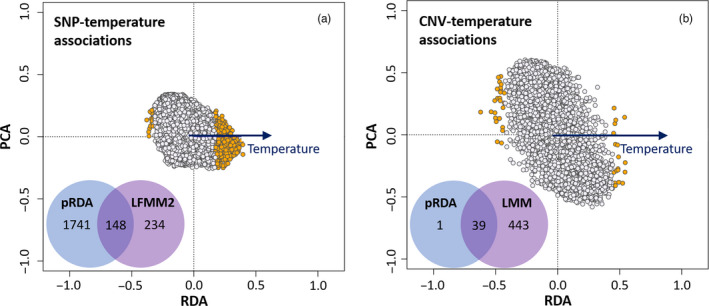
Single nucleotide polymorphism (SNP)‐temperature (a) and copy number variant (CNV)‐temperature (b) associations using partial redundancy analyses (pRDA) in Rana luteiventris. We also provide Venn diagrams showing the number of candidate SNPs and CNVs detected using either RDA, or latent factor mixed models (LFMM2) or both methods combined [Colour figure can be viewed at wileyonlinelibrary.com]
Among these 147 RAD loci, BLAST identified 10 loci for which nucleotide sequence had a high similarity with published sequences in other anurans or vertebrates (Table S14). BLAST indicated that seven candidate SNPs were located within loci having a high level of similarity with genes regulating resistance to cold and anoxia (locus 155392; Figure 5), gonadotropin‐releasing hormone receptor activity (locus 120066; Figure 5), bradykinin production (locus 2796132; Figure 5), organelle biogenesis associated with melanosomes (locus 27203), cell proliferation activity (locus 27781), RNA‐binding (locus 2279721) and transcription coactivator activity (locus 117543). Three other loci (2786826, 1770849 and 73386) corresponded to microsatellites described for R. temporaria.
FIGURE 5.
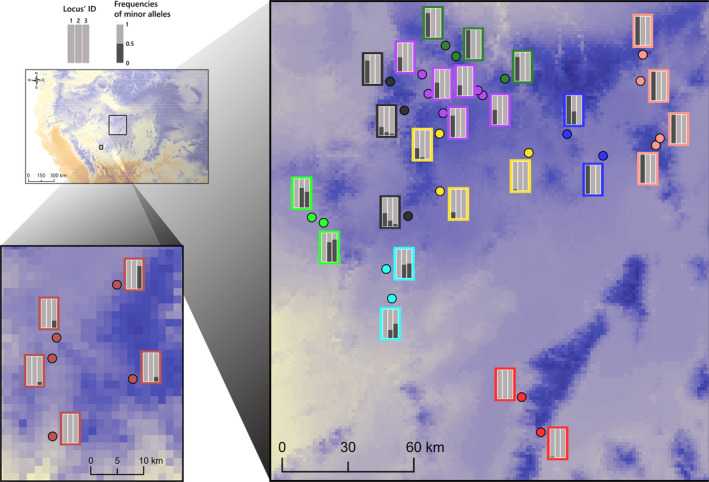
Minor allele frequency of three single nucleotide polymorphism candidates associated with temperature and located in protein‐coding genes that could be involved in temperature‐dependent life‐history variation observed in Rana luteiventris: (1) locus 155392 is located within a gene regulating resistance to cold and anoxia, (2) locus 120066 within the gene regulating gonadotropin‐releasing hormone receptor activity and (3) locus 2796132 within the gene regulating the bradykinin production [Colour figure can be viewed at wileyonlinelibrary.com]
3.2.5. CNV‐temperature associations
CNV‐temperature associations showed strong associations between the normalized read depth of putative CNVs and mean annual temperature. The pRDA (Figure 4b) built to detect CNVs associated with temperature was significant (df = 1, F = 9.38, p < 0.01), although the coefficient of determination was low (adjusted R 2 = 0.023). The pRDA detected 40 candidate CNVs associated with temperature, whereas LMMs identified 482 candidate CNVs (Figure 4b). Here, 39 out of 40 loci detected using the pRDA were also detected by LMMs (Figure 4b) and were thus considered as strong candidates. Among the 39 candidate CNVs, 15 CNVs had their normalized read depth positively correlated with temperature while 24 CNVs displayed negative correlations (see Figure 6; Table S15). The sequences of the 39 candidate CNVs had a high similarity with those of DNA repeats identified in the genome of R. temporaria (Table S16). Most candidates (38) were TEs, including 28 TIR transposons of four families (DTA, DTC, DTM and Helitron), one MITE transposon (DTH family) and five LTRs (two of the Gypsy family and three of unknown families).
FIGURE 6.
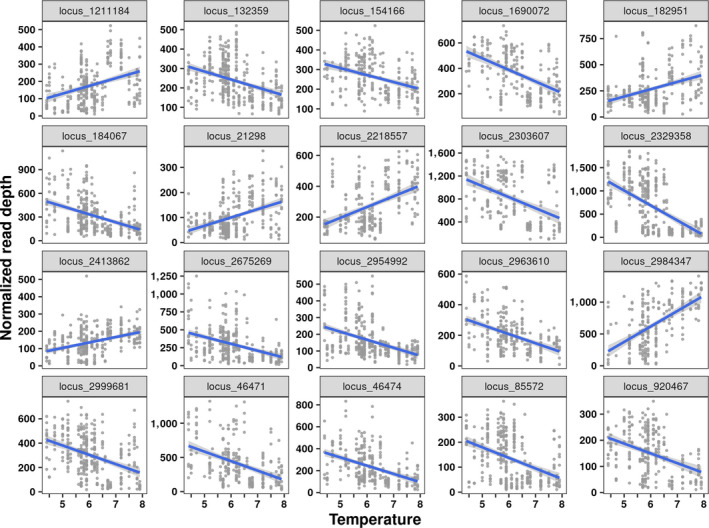
Copy number variant‐temperature associations in Rana luteiventris: relationships between the normalized read depth and mean annual temperature for the 20 strongest copy number variant candidates (i.e. those with the high R 2) [Colour figure can be viewed at wileyonlinelibrary.com]
The identification of discrete groups of copy number categories was examined using the model‐based clustering procedure for the 20 strongest CNV candidates (those with highest R 2 for each category) associated with temperature. This analysis showed that up to three discrete categories can be delineated for candidate CNVs, where each category may represent a specific number of copies for a given locus (Figures S3 and S4).
4. DISCUSSION
Our analyses showed that R. luteiventris life history accelerates with an increase in mean annual temperature; survival was lower, life span was shorter, senescence was faster and recruitment was higher in populations experiencing warmer mean annual temperatures than those exposed to colder conditions. We also found signature of thermal adaptation at the molecular level; we identified SNPs associated with mean annual temperature that were located within the body of protein‐coding genes regulating diverse biological processes including cold resistance. Furthermore, we found several CNV candidates (especially transposable elements) whose normalized read depth was associated with temperature, suggesting that thermal conditions may influence copy number variation within the R. luteiventris genome.
4.1. Temperature‐dependent life‐history variation
Our results indicated that the position of R. luteiventris populations along the fast–slow continuum of life history was strongly associated with temperature, whereas mean annual precipitation had a marginal effect on demographic parameters. In populations experiencing low mean annual temperature, which typically occurred at higher elevations and latitudes, individuals had higher adult survival, longer life span and lower senescence rate than those from warmer environments. Interestingly, increased adult mortality was compensated by high recruitment in populations experiencing warm conditions. This compensatory recruitment (a mechanism also observed in amphibian response to parasites, see Valenzuela‐Sánchez et al., 2021) resulted in positive population growth rate in seven of the eight R. luteiventris populations we studied.
The temperature‐dependent increase in mortality found using our long‐term capture–recapture data is congruent with the results of many skeletochronological studies that have highlighted an increase in median age and maximal longevity with altitude and latitude at the intraspecific level (Sinsch, 2015; Zhang & Lu, 2012; Zhong et al., 2018). The increase in recruitment with increasing temperature in our study system is also congruent with the reduction of breeding frequency and clutch size (relative to body size) reported in many anuran populations at high altitude and latitude (Morrison & Hero, 2003). Overall, our study and previous ones indicate that temperature produces similar effects on anuran life history at intraspecific and interspecific levels (Stark & Meiri, 2018; Zhang & Lu, 2012), promoting slow strategies under cold conditions and fast strategies in warmer environments.
Our results showed for the first time a positive effect of temperature on senescence rate among wild populations, thus confirming senescence acceleration with temperature observed under laboratory conditions in model organisms of ageing (Drosophila melanogaster, Jacobson et al., 2010; Nothobranchius furzeri, Valenzano et al., 2006; Caenorhabditis elegans, Yen & Mobbs, 2010). We found that senescence rate was 14 times higher in the population experiencing the warmest conditions (8.35℃) compared to the one with the coldest temperature (1.71℃). Senescence rate varied from 0.11 to 1.51, a broader range of values than that has been reported for female mammals (senescence rate calculated from Siler models: 0.01–1.17) at a broad phylogenetic scale (Lemaître et al., 2020). Overall, our analyses and a previous study on the yellow‐bellied toad Bombina variegata (Cayuela et al., 2020) suggest that amphibian senescence patterns can display high intraspecific variation (but see also Cayuela et al., 2019) and may be highly sensitive to environmental variation.
4.2. Genomic signatures of thermal adaptation
We identified 148 strong SNP candidates associated with temperature using a conservative approach combining two GEA methods. Despite the large number of SNPs (50,829) and non‐duplicated loci (37,499) discovered in our study, the number of SNP candidates potentially involved in thermal adaptation is likely underestimated since we used a reduced representation method (Lowry et al., 2017) in an amphibian with a large genome size (~9 Gbp in the Oregon spotted frog Rana pretiosa, a sister species of R. luteiventris, http://www.genomesize.com/). Among the 148 SNP candidates, BLAST identified 10 loci for which nucleotide sequence displayed a high similarity with published sequences in other anurans. This limited number of BLAST hits probably resulted from the absence of annotated reference genome for anurans phylogenetically related to R. luteiventris.
We identified three candidate SNPs located within the body of protein‐coding genes that could play a critical role in thermal adaptation and might be involved in temperature‐dependent life‐history variation. One SNP could especially contribute to the recruitment changes detected in our study. This marker was located within a locus having a high sequence similarity with the gene GnRH receptor‐3 that regulates gonadotropin‐releasing hormone receptor activity in another Nearctic anuran, the American bullfrog Lithobates catesbeianus. This gonadotropin‐releasing hormone is a peptide hormone responsible for the release of follicle‐stimulating hormone (FSH) and luteinizing hormone (LH) from the anterior pituitary in vertebrates (Norris & Carr, 2020). In amphibians, FSH and LH regulate hormone steroid secretion (progesterone, 17β‐estradiol, testosterone and 5a‐dihydrotestosterone), gametogenesis and mating behaviour (Aranzábal, 2011; Moore, 1987; Propper, 2011). Mutations on this gene could modulate breeding effort in both sexes along the temperature gradient and might be involved in the temperature‐related life‐history patterns highlighted in our demographic analyses.
Two candidate SNPs were also located within the body of genes that regulate resistance to cold and hypoxia in anurans and might contribute to the extended life span and the slow senescence observed in R. luteiventris populations experiencing low mean annual temperature. One of the two SNP candidates was positioned on a locus which had high sequence similarity to the gene Aat (Matsuba et al., 2007), which codes for ADP/ATP translocase (AAT), an enzyme involved in freeze and hypoxia resistance in the wood frog, Lithobates sylvaticus (Cai et al., 1997). The other SNP candidate was located within the body of the bradykinin precursor gene that regulates bradykinin production. This gene may be directly involved in adaptation to cold since bradykinin‐like peptides secreted from amphibian skin may act to increase microvascular permeability (Xi et al., 2015). This physiological response could facilitate skin respiration (Tattersall, 2007) and limit mortality due to hypoxia in cold‐adapted populations experiencing long periods of aquatic overwintering, such as R. luteiventris (Bull & Hayes, 2002; Tattersall & Ultsch, 2008).
The bradykinin precursor gene could also regulate immune response to pathogen‐climate interactions in R. luteiventris. Bradykinin is one of the most abundant (and well‐studied, see Xi et al., 2015) antimicrobial peptides in ranid skin secretion that inhibits the growth of the pathogenic chytrid fungus (Batrachochytrium dendrobatidis, Bd; Rollins‐Smith et al., 2006) causing chytridiomycosis. Bd is broadly spread across the western United States and can have high prevalence level in R. luteiventris populations (Adams et al., 2010; Pearl et al., 2007; Russell et al., 2010). Bd prevalence increases with elevation in ranid populations of this region (Pearl et al., 2009; Piovia‐Scott et al., 2011), leading to higher infection risk in cold environments where anurans are less likely to clear Bd infections by increasing their body temperature (Richards‐Zawacki, 2010; Russell et al., 2010; Woodhams et al., 2003). Research indicates that antimicrobial peptides inhibit Bd (in bioassays) in R. luteiventris, although inhibition is not necessarily correlated with Bd intensity in frogs (Loudon et al., 2020). We propose that mutations in the bradykinin precursor gene could play a role in temperature‐mediated evolutionary response to Bd in R. luteiventris. Further studies could investigate how this sequence polymorphism may influence bradykinin secretion and modulate the evolution of tolerance and resistance to Bd in cold‐adapted populations.
Although the candidate SNPs detected by the GEA are likely hitchhiking markers rather than causal variants, our results suggest that sequence polymorphisms on several candidate genes regulating reproductive activity, immunity and resistance to cold and hypoxia/anoxia could eventually be found to contribute to thermal adaptation in R. luteiventris. This SNP‐based signal is congruent with the findings of three previous studies that have highlighted molecular signatures of thermal adaptation in ranid and bufonid species from Europe (Common frog R. temporaria, Rödin‐Mörch et al., 2019; Common toad Bufo bufo Rödin‐Mörch et al., 2021) and Asia (Yang et al., 2016). In particular, a comparative transcriptome study (Yang et al., 2016) that focused on three anuran species (Asiatic toad Bufo gargarizans, Plateau brown frog Rana kukunoris and Asiatic grass frog Rana chensinensis) from the Tibetan region found that altitude determined the allele frequency of large set of genes notably regulating immune response, metabolism and longevity. Interestingly, skeletochronological data revealed that maximal longevity and/or median age increased with altitude or latitude in several of these species (R. temporaria, Hjernquist et al., 2012; B. gargarizans, Yu & Lu, 2013; R. chensinensis, Ma et al., 2009).
Our study adds to a growing body of work suggesting that temperature‐dependent life‐history acceleration commonly observed in anurans could be partially regulated by SNPs evolving under polygenic selection. However, the functional links between this life‐history variation and the candidate SNPs detected by GEA still remain to be deciphered using whole‐genome and transcriptome sequencing data. Novel GWAS approaches based on capture–recapture modelling, where genotype‐survival and genotype‐senescence associations can control for imperfect detection probability, still need to be developed to tackle this issue in wild animal populations.
4.3. Temperature‐dependent accumulation of transposable elements
Our study also highlighted CNV‐temperature associations: a total of 39 candidate CNVs were detected using a conservative approach combining regression and redundancy analyses. The normalized read depth of 24 and 15 duplicated regions were negatively and positively correlated with temperature respectively. Thirty‐eight candidates were TEs, and 63% (24) were transposons that have diverged more recently than the other elements of their own family. Overall, these results suggest that relatively recent temperature changes determined TE accumulation patterns within R. luteiventris genome.
Although these mechanisms are still poorly understood in amphibians, temperature‐dependent TE accumulations have been recently documented in fish (Carducci et al., 2020; Cayuela et al., 2021). A study on a marine fish using similar RAD‐seq CNV analyses found associations between the normalized read depth of transposons and retrotransposons and water temperature (Cayuela et al., 2021). Furthermore, phylogenetic analysis performed on 39 teleost species highlighted an unexpected clusterization of a non‐LTR retrotransposon isolated from species living in cold waters compared with those isolated from species living in warm waters (Carducci et al., 2019). In R. luteiventris, temperature‐dependent accumulations of recently diverged TEs likely result from thermal stresses endured during individual lifetime, as proposed in fish (Carducci et al., 2020). This TE accumulation, potentially caused by thermo‐induced epigenetic changes (Rey et al., 2016), could be involved in the temperature‐dependent changes in senescence and life span observed in R. luteiventris since studies showed that activation of TE expression leads to an acceleration of cell ageing through inflammatory pathways (Andrenacci et al., 2020) and is partially responsible of sex‐related differences in longevity and ageing (Brown et al., 2020).
5. CONCLUSIONS
Our study highlighted that, in ectotherms like R. luteiventris, variation in thermal conditions leads to important clinal changes in life history, notably including an acceleration in senescence with increasing temperature. We also detected molecular signatures of thermal adaptation involving candidate genes that might play a role in this life‐history shift. In addition, temperature‐dependent accumulation of recently diverged transposons and retrotransposons in R. luteiventris genome might also have an impact on life‐history traits including senescence. Further studies based on whole‐genome sequencing data and novel GWAS approaches controlling for recapture probability will help clarify functional links between SNPs, CNVs (especially TEs) and individual mortality metrics across populations experiencing contrasting thermal conditions. A better understanding of these mechanisms will be crucial to understanding and predicting the impact of climate change on ectotherm populations.
AUTHORS' CONTRIBUTIONS
H.C. completed the demographic analyses and wrote the paper; B.R.F., D.L.J. and Y.D. contributed to the bioinformatics and genomic analyses; B.R.F., D.S.P., H.C., R.M.M. and W.C.F. initiated and conceptualized the project; H.C. coordinated the work; D.S.P., L.A.E., B.R.H. and R.M.M. collected demographic and DNA samples; J.M.W.G. conceived the maps and extracted environmental data. All the authors read and edited the final manuscript version.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank all the fieldworkers and students who helped to collect the capture–recapture data and DNA samples. Besides authors D.S.P. and R.M.M., lead personnel in these multi‐decade efforts include Jacqueline Cupples, Janice Engle, Hallie Lingo, Kristin Lohr, Megan McGuire, Bryce Maxell, Chad Mellison, Marisa Meyer, James Munger, Teri Slatauski and Rachel Van Horne. They also thank Jeffrey Petersen and Chad Mellison for coordinating tissue sample collection for genomic analyses. Justin Welty provided climate data. Stephen Spear provided helpful comments on an earlier version of this manuscript. Hugo Cayuela was supported as a postdoctoral researcher by the Swiss National Science Foundation (SNF grant number 31003A_182265). Funding for field work was supported by Boise State University, Bureau of Land Management, Idaho Department of Fish and Game, Nevada Department of Wildlife, University of Montana, University of Nevada Reno, US Fish and Wildlife Service, US Forest Service and US Geological Survey. Funding for genomic data collection and analysis was provided by the US Fish and Wildlife Service and the Nevada Department of Wildlife. Any use of trade, firm or product names is for descriptive purposes only and does not imply endorsement by the US Government. Open Access Funding provided by Universite de Lausanne.
Cayuela, H. , Dorant, Y. , Forester, B. R. , Jeffries, D. L. , Mccaffery, R. M. , Eby, L. A. , Hossack, B. R. , Gippet, J. M. W. , Pilliod, D. S. , & Chris Funk, W. (2022). Genomic signatures of thermal adaptation are associated with clinal shifts of life history in a broadly distributed frog. Journal of Animal Ecology, 91, 1222–1238. 10.1111/1365-2656.13545
Handling Editor Matthew Barbour
DATA AVAILABILITY STATEMENT
Genomic data are available on Dryad Digital Repository https://doi.org/10.5061/dryad.x3ffbg7k3 (Cayuela et al., 2021).
REFERENCES
- Adams, M. J. , Chelgren, N. D. , Reinitz, D. , Cole, R. A. , Rachowicz, L. J. , Galvan, S. , McCreary, B. , Pearl, C. A. , Bailey, L. L. , Bettaso, J. , Bull, E. L. , & Leu, M. (2010). Using occupancy models to understand the distribution of an amphibian pathogen, Batrachochytrium dendrobatidis . Ecological Applications, 20, 289–302. 10.1890/08-2319.1 [DOI] [PubMed] [Google Scholar]
- Amish, S. J. , Ali, O. , Peacock, M. , Miller, M. , Robinson, M. , Smith, S. , Luikart, G. , & Neville, H. (2019). Assessing thermal adaptation using family‐based association and FST outlier tests in a threatened trout species. Molecular Ecology, 28, 2573–2593. [DOI] [PubMed] [Google Scholar]
- Andrenacci, D. , Cavaliere, V. , & Lattanzi, G. (2020). The role of transposable elements activity in aging and their possible involvement in laminopathic diseases. Ageing Research Reviews, 57, 100995. 10.1016/j.arr.2019.100995 [DOI] [PubMed] [Google Scholar]
- Andrews, S. (2019). FastQC: A quality control tool for high throughput sequence data (version 0.11.8). http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- Angilletta, M. J. , Steury, T. D. , & Sears, M. W. (2004). Temperature, growth rate, and body size in ectotherms: Fitting pieces of a life‐history puzzle. Integrative and Comparative Biology, 44, 498–509. 10.1093/icb/44.6.498 [DOI] [PubMed] [Google Scholar]
- Aranzábal, M. C. U. (2011). Hormones and the female reproductive system of amphibians. In Norris D. O., & Lopez K. H. (Eds.), Hormones and reproduction of vertebrates (pp. 55–81). Academic Press. [Google Scholar]
- Benestan, L. , Quinn, B. K. , Maaroufi, H. , Laporte, M. , Clark, F. K. , Greenwood, S. J. , Rochette, R. , & Bernatchez, L. (2016). Seascape genomics provides evidence for thermal adaptation and current‐mediated population structure in American lobster (Homarus americanus). Molecular Ecology, 25, 5073–5092. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y. , & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological), 57, 289–300. [Google Scholar]
- Blanck, A. , & Lamouroux, N. (2007). Large‐scale intraspecific variation in life‐history traits of European freshwater fish. Journal of Biogeography, 34, 862–875. 10.1111/j.1365-2699.2006.01654.x [DOI] [Google Scholar]
- Bodensteiner, B. L. , Agudelo‐Cantero, G. A. , Arietta, A. A. , Gunderson, A. R. , Muñoz, M. M. , Refsnider, J. M. , & Gangloff, E. J. (2021). Thermal adaptation revisited: How conserved are thermal traits of reptiles and amphibians? Journal of Experimental Zoology Part A: Ecological and Integrative Physiology, 335, 173–194. 10.1002/jez.2414 [DOI] [PubMed] [Google Scholar]
- Bourque, G. , Burns, K. H. , Gehring, M. , Gorbunova, V. , Seluanov, A. , Hammell, M. , Imbeault, M. , Izsvák, Z. , Levin, H. L. , Macfarlan, T. S. , Mager, D. L. , & Feschotte, C. (2018). Ten things you should know about transposable elements. Genome Biology, 19, 1–12. 10.1186/s13059-018-1577-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, E. J. , Nguyen, A. H. , & Bachtrog, D. (2020). The Y chromosome may contribute to sex‐specific ageing in Drosophila. Nature Ecology & Evolution, 4, 853–862. 10.1038/s41559-020-1179-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull, E. L. , & Hayes, M. P. (2002). Overwintering of Columbia spotted frogs in Northeastern Oregon. Northwest Science, 76, 141–147. [Google Scholar]
- Burraco, P. , Orizaola, G. , Monaghan, P. , & Metcalfe, N. B. (2020). Climate change and ageing in ectotherms. Global Change Biology, 26, 5371–5381. 10.1111/gcb.15305 [DOI] [PubMed] [Google Scholar]
- Cai, Q. , Greenway, S. C. , & Storey, K. B. (1997). Differential regulation of the mitochondrial ADP/ATP translocase gene in wood frogs under freezing stress. Biochimica Et Biophysica Acta (BBA) ‐ Gene Structure and Expression, 1353(1), 69–78. 10.1016/S0167-4781(97)00057-2 [DOI] [PubMed] [Google Scholar]
- Campbell‐Staton, S. C. , Winchell, K. M. , Rochette, N. C. , Fredette, J. , Maayan, I. , Schweizer, R. M. , & Catchen, J. (2020). Parallel selection on thermal physiology facilitates repeated adaptation of city lizards to urban heat islands. Nature Ecology & Evolution, 4, 652–658. 10.1038/s41559-020-1131-8 [DOI] [PubMed] [Google Scholar]
- Carducci, F. , Barucca, M. , Canapa, A. , Carotti, E. , & Biscotti, M. A. (2020). Mobile elements in ray‐finned fish genomes. Life, 10, 221. 10.3390/life10100221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carducci, F. , Biscotti, M. A. , Forconi, M. , Barucca, M. , & Canapa, A. (2019). An intriguing relationship between teleost Rex3 retroelement and environmental temperature. Biology Letters, 15, 20190279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta, E. , & González, J. (2013). The impact of transposable elements in environmental adaptation. Molecular Ecology, 22(6), 1503–1517. 10.1111/mec.12170 [DOI] [PubMed] [Google Scholar]
- Catchen, J. M. , Amores, A. , Hohenlohe, P. , Cresko, W. , & Postlethwait, J. (2011). Stacks: Building and genotyping loci de novo from short‐read sequences. G3: Genes Genomes Genetics, 1, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen, J. , Hohenlohe, P. A. , Bassham, S. , Amores, A. , & Cresko, W. A. (2013). Stacks: An analysis tool set for population genomics. Molecular Ecology, 22, 3124–3140. 10.1111/mec.12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caye, K. , Jumentier, B. , Lepeule, J. , & François, O. (2019). LFMM 2: Fast and accurate inference of gene‐environment associations in genome‐wide studies. Molecular Biology and Evolution, 36, 852–860. 10.1093/molbev/msz008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayuela, H. , Dorant, Y. , Mérot, C. , Laporte, M. , Normandeau, E. , Gagnon‐Harvey, S. , Clément, M. , Sirois, P. , & Bernatchez, L. (2021). Thermal adaptation rather than demographic history drives genetic structure inferred by copy number variants in a marine fish. Molecular Ecology, 30, 1624–1641. 10.1111/mec.15835 [DOI] [PubMed] [Google Scholar]
- Cayuela, H. , Forester, B. R. , & Funk, W. C. (2021). Genomic data of Columbia spotted frog (Rana luteiventris). Dryad, 10.5061/dryad.x3ffbg7k3 [DOI] [Google Scholar]
- Cayuela, H. , Lemaître, J. F. , Bonnaire, E. , Pichenot, J. , & Schmidt, B. R. (2020). Population position along the fast–slow life‐history continuum predicts intraspecific variation in actuarial senescence. Journal of Animal Ecology, 89, 1069–1079. 10.1111/1365-2656.13172 [DOI] [PubMed] [Google Scholar]
- Cayuela, H. , Olgun, K. , Angelini, C. , Üzüm, N. , Peyronel, O. , Miaud, C. , Avcı, A. , Lemaitre, J.‐F. , & Schmidt, B. R. (2019). Slow life‐history strategies are associated with negligible actuarial senescence in western Palaearctic salamanders. Proceedings of the Royal Society B: Biological Sciences, 286, 20191498. 10.1098/rspb.2019.1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayuela, H. , Rougemont, Q. , Laporte, M. , Mérot, C. , Normandeau, E. , Dorant, Y. , Tørresen, O. K. , Hoff, S. N. K. , Jentoft, S. , Sirois, P. , Castonguay, M. , Jansen, T. , Praebel, K. , Clément, M. , & Bernatchez, L. (2020). Shared ancestral polymorphisms and chromosomal rearrangements as potential drivers of local adaptation in a marine fish. Molecular Ecology, 29, 2379–2398. 10.1111/mec.15499 [DOI] [PubMed] [Google Scholar]
- Chang, C. C. , Chow, C. C. , Tellier, L. C. , Vattikuti, S. , Purcell, S. M. , & Lee, J. J. (2015). Second‐generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience, 4. 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. , Cheng, C. H. C. , Zhang, J. , Cao, L. , Chen, L. , Zhou, L. , Jin, Y. , Ye, H. , Deng, C. , Dai, Z. , Xu, Q. , Hu, P. , Sun, S. , Shen, Y. , & Chen, L. (2008). Transcriptomic and genomic evolution under constant cold in Antarctic notothenioid fish. Proceedings of the National Academy of Sciences of the United States of America, 105, 12944–12949. 10.1073/pnas.0802432105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. , & Narum, S. R. (2021). Whole genome resequencing reveals genomic regions associated with thermal adaptation in redband trout. Molecular Ecology, 30, 162–174. 10.1111/mec.15717 [DOI] [PubMed] [Google Scholar]
- Chuong, E. B. , Elde, N. C. , & Feschotte, C. (2017). Regulatory activities of transposable elements: From conflicts to benefits. Nature Reviews Genetics, 18, 71. 10.1038/nrg.2016.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colchero, F. , & Clark, J. S. (2012). Bayesian inference on age‐specific survival for censored and truncated data. Journal of Animal Ecology, 81, 139–149. 10.1111/j.1365-2656.2011.01898.x [DOI] [PubMed] [Google Scholar]
- Colchero, F. , Jones, O. R. , & Rebke, M. (2012). BaSTA: An R package for Bayesian estimation of age‐specific survival from incomplete mark–recapture/recovery data with covariates. Methods in Ecology and Evolution, 3, 466–470. 10.1111/j.2041-210X.2012.00186.x [DOI] [Google Scholar]
- Dorant, Y. , Cayuela, H. , Wellband, K. , Laporte, M. , Rougemont, Q. , Mérot, C. , Normandeau, E. , Rochette, R. , & Bernatchez, L. (2020). Copy number variants outperform SNPs to reveal genotype–temperature association in a marine species. Molecular Ecology, 29, 4765–4782. 10.1111/mec.15565 [DOI] [PubMed] [Google Scholar]
- Flouris, A. D. , & Piantoni, C. (2015). Links between thermoregulation and aging in endotherms and ectotherms. Temperature, 2, 73–85. 10.4161/23328940.2014.989793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forester, B. R. , Lasky, J. R. , Wagner, H. H. , & Urban, D. L. (2018). Comparing methods for detecting multilocus adaptation with multivariate genotype–environment associations. Molecular Ecology, 27, 2215–2233. 10.1111/mec.14584 [DOI] [PubMed] [Google Scholar]
- Fraley, C. , Raftery, A. E. , Murphy, T. B. , & Scrucca, L. (2012). mclust version 4 for R: normal mixture modeling for model‐based clustering, classification, and density estimation (Vol. 597, p. 1). Technical report. [Google Scholar]
- Frichot, E. , Schoville, S. D. , Bouchard, G. , & François, O. (2013). Testing for associations between loci and environmental gradients using latent factor mixed models. Molecular Biology and Evolution, 30, 1687–1699. 10.1093/molbev/mst063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugger, P. F. , Liang, C. T. , Sork, V. L. , Hodgskiss, P. , & Wright, J. W. (2018). Applying landscape genomic tools to forest management and restoration of Hawaiian koa (Acacia koa) in a changing environment. Evolutionary Applications, 11, 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson, A. R. , & Stillman, J. H. (2015). Plasticity in thermal tolerance has limited potential to buffer ectotherms from global warming. Proceedings of the Royal Society B: Biological Sciences, 282, 20150401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjernquist, M. B. , Söderman, F. , Jönsson, K. I. , Herczeg, G. , Laurila, A. , & Merilä, J. (2012). Seasonality determines patterns of growth and age structure over a geographic gradient in an ectothermic vertebrate. Oecologia, 170, 641–649. 10.1007/s00442-012-2338-4 [DOI] [PubMed] [Google Scholar]
- Huey, R. B. , & Berrigan, D. (2001). Temperature, demography, and ectotherm fitness. The American Naturalist, 158, 204–210. 10.1086/321314 [DOI] [PubMed] [Google Scholar]
- IPCC . (2014). IPCC climate change 2014: Synthesis Report. Contribution of working groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. IPCC. p. 151.
- Jacobson, J. , Lambert, A. J. , Portero‐Otín, M. , Pamplona, R. , Magwere, T. , Miwa, S. , Driege, Y. , Brand, M. D. , & Partridge, L. (2010). Biomarkers of aging in Drosophila. Aging Cell, 9, 466–477. 10.1111/j.1474-9726.2010.00573.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil, G. , Cummings, E. , & de Magalhaes, J. P. (2015). Being cool: How body temperature influences ageing and longevity. Biogerontology, 16, 383–397. 10.1007/s10522-015-9571-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger, F. (2019). Trim Galore! (version 0.6.4). http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ [Google Scholar]
- Legendre, P. , & Legendre, L. (2012). Numerical ecology. Elsevier. [Google Scholar]
- Lehnert, S. J. , DiBacco, C. , Van Wyngaarden, M. , Jeffery, N. W. , Ben Lowen, J. , Sylvester, E. V. A. , Wringe, B. F. , Stanley, R. R. E. , Hamilton, L. C. , & Bradbury, I. R. (2019). Fine‐scale temperature‐associated genetic structure between inshore and offshore populations of sea scallop (Placopecten magellanicus). Heredity, 122, 69–80. 10.1038/s41437-018-0087-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaître, J. F. , Ronget, V. , Tidière, M. , Allainé, D. , Berger, V. , Cohas, A. , Colchero, F. , Conde, D. A. , Garratt, M. , Liker, A. , Marais, G. A. B. , Scheuerlein, A. , Székely, T. , & Gaillard, J. M. (2020). Sex differences in adult lifespan and aging rates of mortality across wild mammals. Proceedings of the National Academy of Sciences of the United States of America, 117, 8546–8553. 10.1073/pnas.1911999117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudon, A. H. , Kurtz, A. , Esposito, E. , Umile, T. P. , Minbiole, K. P. C. , Parfrey, L. W. , & Sheafor, B. A. (2020). Columbia spotted frogs (Rana luteiventris) have characteristic skin microbiota that may be shaped by cutaneous skin peptides and the environment. FEMS Microbiology Ecology, 96, fiaa168. 10.1093/femsec/fiaa168 [DOI] [PubMed] [Google Scholar]
- Lowry, D. B. , Hoban, S. , Kelley, J. L. , Lotterhos, K. E. , Reed, L. K. , Antolin, M. F. , & Storfer, A. (2017). Breaking RAD: An evaluation of the utility of restriction site‐associated DNA sequencing for genome scans of adaptation. Molecular Ecology, 17, 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Tong, L. , & Lu, X. (2009). Variation of body size, age structure and growth of a temperate frog, Rana chensinensis, over an elevational gradient in northern China. Amphibia‐Reptilia, 30, 111–117. 10.1163/156853809787392685 [DOI] [Google Scholar]
- Mallard, F. , Nolte, V. , Tobler, R. , Kapun, M. , & Schlötterer, C. (2018). A simple genetic basis of adaptation to a novel thermal environment results in complex metabolic rewiring in Drosophila. Genome Biology, 19, 1–15. 10.1186/s13059-018-1503-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. (2011). Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet.Journal, 17, 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Matsuba, C. , Uolevi Palo, J. , Kuzmin, S. L. , & Merilä, J. (2007). Evidence for multiple retroposition events and gene evolution in the ADP/ATP translocase gene family in Ranid frogs. Journal of Heredity, 98, 300–310. 10.1093/jhered/esm038 [DOI] [PubMed] [Google Scholar]
- McKinney, G. J. , Waples, R. K. , Seeb, L. W. , & Seeb, J. E. (2017). Paralogs are revealed by proportion of heterozygotes and deviations in read ratios in genotyping‐by‐sequencing data from natural populations. Molecular Ecology Resources, 17(4), 656–669. [DOI] [PubMed] [Google Scholar]
- Mérot, C. , Oomen, R. A. , Tigano, A. , & Wellenreuther, M. (2020). A roadmap for understanding the evolutionary significance of structural genomic variation. Trends in Ecology & Evolution, 35, 561–572. 10.1016/j.tree.2020.03.002 [DOI] [PubMed] [Google Scholar]
- Mims, M. C. , Olson, D. H. , Pilliod, D. S. , & Dunham, J. B. (2018). Functional and geographic components of risk for climate sensitive vertebrates in the Pacific Northwest, USA. Biological Conservation, 228, 183–194. 10.1016/j.biocon.2018.10.012 [DOI] [Google Scholar]
- Moore, F. L. (1987). Reproductive endocrinology of amphibians. In Chester‐Jones I., Ingleton P. M., & Phillips J. G. (Eds.), Fundamentals of comparative vertebrate endocrinology (pp. 207–221). Springer. [Google Scholar]
- Morrison, C. , & Hero, J. M. (2003). Geographic variation in life‐history characteristics of amphibians: A review. Journal of Animal Ecology, 72, 270–279. 10.1046/j.1365-2656.2003.00696.x [DOI] [Google Scholar]
- Munch, S. B. , & Salinas, S. (2009). Latitudinal variation in lifespan within species is explained by the metabolic theory of ecology. Proceedings of the National Academy of Sciences of the United States of America, 106, 13860–13864. 10.1073/pnas.0900300106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris, D. O. , & Carr, J. A. (2020). Vertebrate endocrinology. Academic Press. [Google Scholar]
- Paris, J. R. , Stevens, J. R. , & Catchen, J. M. (2017). Lost in parameter space: A road map for stacks. Methods in Ecology and Evolution, 8, 1360–1373. [Google Scholar]
- Parker, D. J. , Wiberg, R. A. W. , Trivedi, U. , Tyukmaeva, V. I. , Gharbi, K. , Butlin, R. K. , Hoikkala, A. , Kankare, M. , & Ritchie, M. G. (2018). Inter and intraspecific genomic divergence in Drosophila montana shows evidence for cold adaptation. Genome Biology and Evolution, 10, 2086–2101. 10.1093/gbe/evy147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl, C. A. , Bowerman, J. , Adams, M. J. , & Chelgren, N. D. (2009). Widespread occurrence of the chytrid fungus Batrachochytrium dendrobatidis on Oregon spotted frogs (Rana pretiosa). EcoHealth, 6, 209–218. 10.1007/s10393-009-0237-x [DOI] [PubMed] [Google Scholar]
- Pearl, C. A. , Bull, E. L. , Green, D. E. , Bowerman, J. , Adams, M. J. , Hyatt, A. , & Wente, W. H. (2007). Occurrence of the amphibian pathogen Batrachochytrium dendrobatidis in the Pacific Northwest. Journal of Herpetology, 41, 145–149. [Google Scholar]
- Pembleton, L. W. , Cogan, N. O. I. , & Forster, J. W. (2013). StAMPP: An R package for calculation of genetic differentiation and structure of mixed‐ploidy level populations. Molecular Ecology Resources, 13, 946–952. [DOI] [PubMed] [Google Scholar]
- Perrier, C. , Ferchaud, A. L. , Sirois, P. , Thibault, I. , & Bernatchez, L. (2017). Do genetic drift and accumulation of deleterious mutations preclude adaptation? Empirical investigation using RAD seq in a northern lacustrine fish. Molecular Ecology, 26, 6317–6335. 10.1111/mec.14361 [DOI] [PubMed] [Google Scholar]
- Peterson, B. K. , Weber, J. N. , Kay, E. H. , Fisher, H. S. , & Hoekstra, H. E. (2012). Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non‐model species. PLoS ONE, 7, e37135. 10.1371/journal.pone.0037135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilliod, D. S. , Arkle, R. S. , Robertson, J. M. , Murphy, M. A. , & Funk, W. C. (2015). Effects of changing climate on aquatic habitat and connectivity for remnant populations of a wide‐ranging frog species in an arid landscape. Ecology and Evolution, 5, 3979–3994. 10.1002/ece3.1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilliod, D. S. , Hausner, M. B. , & Scherer, R. D. (2021). From satellites to frogs: Quantifying ecohydrological change, drought mitigation, and population demography in desert meadows. Science of The Total Environment, 758, 143632. 10.1016/j.scitotenv.2020.143632 [DOI] [PubMed] [Google Scholar]
- Piovia‐Scott, J. , Pope, K. L. , Lawler, S. P. , Cole, E. M. , & Foley, J. E. (2011). Factors related to the distribution and prevalence of the fungal pathogen Batrachochytrium dendrobatidis in Rana cascadae and other amphibians in the Klamath Mountains. Biological Conservation, 144(12), 2913–2921. 10.1016/j.biocon.2011.08.008 [DOI] [Google Scholar]
- Plummer, M. (2003). JAGS: A program for analysis of Bayesian graphical models using Gibbs sampling. In Proceedings of the 3rd international workshop on distributed statistical computing (Vol. 124, No. 125.10, pp. 1–10). [Google Scholar]
- Plummer, M. , Best, N. , Cowles, K. , Vines, K. , Sarkar, D. , & Almond, R. (2006). CODA: Output analysis and diagnostics for MCMC. 0.16. R package version 0.19‐3. https://CRAN.R‐project.org/package=coda [Google Scholar]
- Pradel, R. (1996). Utilization of capture‐mark‐recapture for the study of recruitment and population growth rate. Biometrics, 52, 703–709. [Google Scholar]
- Promislow, D. E. , & Harvey, P. H. (1990). Living fast and dying young: A comparative analysis of life‐history variation among mammals. Journal of Zoology, 220, 417–437. 10.1111/j.1469-7998.1990.tb04316.x [DOI] [Google Scholar]
- Propper, C. R. (2011). Testicular structure and control of sperm development in amphibians. In Norris D. O., & Lopez K. H. (Eds.), Hormones and reproduction of vertebrates (pp. 39–53). Academic Press. [Google Scholar]
- Robinson, M. D. , & Oshlack, A. (2010). A scaling normalization method for differential expression analysis of RNA‐seq data. Genome Biology, 11(3), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley, K. M. , Bay, L. K. , & van Oppen, M. J. (2020). Genome‐wide SNP analysis reveals an increase in adaptive genetic variation through selective breeding of coral. Molecular Ecology, 29, 2176–2188. 10.1111/mec.15482 [DOI] [PubMed] [Google Scholar]
- R Core Team . (2019). R: A language and environment for statistical computing. R Foundation for Statistical Computing. http://www.R‐project.org/ [Google Scholar]
- Rajpurohit, S. , Gefen, E. , Bergland, A. O. , Petrov, D. A. , Gibbs, A. G. , & Schmidt, P. S. (2018). Spatiotemporal dynamics and genome‐wide association analysis of desiccation tolerance in Drosophila melanogaster . Molecular Ecology, 27, 3525–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey, O. , Danchin, E. , Mirouze, M. , Loot, C. , & Blanchet, S. (2016). Adaptation to global change: A transposable element–epigenetics perspective. Trends in Ecology & Evolution, 31, 514–526. 10.1016/j.tree.2016.03.013 [DOI] [PubMed] [Google Scholar]
- Richards‐Zawacki, C. L. (2010). Thermoregulatory behaviour affects prevalence of chytrid fungal infection in a wild population of Panamanian golden frogs. Proceedings of the Royal Society B: Biological Sciences, 277, 519–528. 10.1098/rspb.2009.1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochette, N. C. , Rivera‐Colón, A. G. , & Catchen, J. M. (2019). Stacks 2: Analytical methods for paired‐end sequencing improve RADseq‐based population genomics. Molecular Ecology, 28, 4737–4754. 10.1111/mec.15253 [DOI] [PubMed] [Google Scholar]
- Rödin‐Mörch, P. , Luquet, E. , Meyer‐Lucht, Y. , Richter‐Boix, A. , Höglund, J. , & Laurila, A. (2019). Latitudinal divergence in a widespread amphibian: Contrasting patterns of neutral and adaptive genomic variation. Molecular Ecology, 28, 2996–3011. 10.1111/mec.15132 [DOI] [PubMed] [Google Scholar]
- Rödin‐Mörch, P. , Palejowski, H. , Cortazar‐Chinarro, M. , Kärvemo, S. , Richter‐Boix, A. , Höglund, J. , & Laurila, A. (2021). Small‐scale population divergence is driven by local larval environment in a temperate amphibian. Heredity, 126, 279–292. 10.1038/s41437-020-00371-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez, A. , Rusciano, T. , Hamilton, R. , Holmes, L. , Jordan, D. , & Wollenberg Valero, K. C. (2017). Genomic and phenotypic signatures of climate adaptation in an Anolis lizard. Ecology and Evolution, 7, 6390–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins‐Smith, L. A. , Woodhams, D. C. , Reinert, L. K. , Vredenburg, V. T. , Briggs, C. J. , Nielsen, P. F. , & Conlon, J. M. (2006). Antimicrobial peptide defenses of the mountain yellow‐legged frog (Rana muscosa). Developmental & Comparative Immunology, 30, 831–842. 10.1016/j.dci.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Ronget, V. , & Gaillard, J. M. (2020). Assessing ageing patterns for comparative analyses of mortality curves: Going beyond the use of maximum longevity. Functional Ecology, 34, 65–75. 10.1111/1365-2435.13474 [DOI] [Google Scholar]
- Russell, D. M. , Goldberg, C. S. , Waits, L. P. , & Rosenblum, E. B. (2010). Batrachochytrium dendrobatidis infection dynamics in the Columbia spotted frog Rana luteiventris in north Idaho, USA. Diseases of Aquatic Organisms, 92, 223–230. 10.3354/dao02286 [DOI] [PubMed] [Google Scholar]
- Schweyen, H. , Rozenberg, A. , & Leese, F. (2014). Detection and removal of PCR duplicates in population genomic ddRAD studies by addition of a degenerate base region (DBR) in sequencing adapters. The Biological Bulletin, 227, 146–160. 10.1086/BBLv227n2p146 [DOI] [PubMed] [Google Scholar]
- Seebacher, F. , White, C. R. , & Franklin, C. E. (2015). Physiological plasticity increases resilience of ectothermic animals to climate change. Nature Climate Change, 5, 61–66. 10.1038/nclimate2457 [DOI] [Google Scholar]
- Selwood, K. E. , McGeoch, M. A. , & Mac Nally, R. (2015). The effects of climate change and land‐use change on demographic rates and population viability. Biological Reviews, 90, 837–853. 10.1111/brv.12136 [DOI] [PubMed] [Google Scholar]
- Siler, W. (1979). A competing‐risk model for animal mortality. Ecology, 60, 750–757. 10.2307/1936612 [DOI] [Google Scholar]
- Sinsch, U. (2015). Skeletochronological assessment of demographic life‐history traits in amphibians. The Herpetological Journal, 25, 5–13. [Google Scholar]
- Smit, A. F. A. , Hubley, R. , & Green, P. (2015). RepeatMasker Open‐4.0. http://www.repeatmasker.org [Google Scholar]
- Stark, G. , & Meiri, S. (2018). Cold and dark captivity: Drivers of amphibian longevity. Global Ecology and Biogeography, 27, 1384–1397. 10.1111/geb.12804 [DOI] [Google Scholar]
- Stenseth, N. C. , & Mysterud, A. (2002). Climate, changing phenology, and other life history traits: Nonlinearity and match–mismatch to the environment. Proceedings of the National Academy of Sciences of the United States of America, 99, 13379–13381. 10.1073/pnas.212519399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y. S. , & Yajima, M. (2012). R2jags: A package for running jags from R. R package version 0.03‐08. http://CRAN.R‐project.org/package=R2jags [Google Scholar]
- Tattersall, G. (2007). Skin breathing in amphibians. Endothelial Biomedicine: A Comprehensive Reference, 85–91. [Google Scholar]
- Tattersall, G. J. , & Ultsch, G. R. (2008). Physiological ecology of aquatic overwintering in ranid frogs. Biological Reviews, 83, 119–140. 10.1111/j.1469-185X.2008.00035.x [DOI] [PubMed] [Google Scholar]
- Tenan, S. , Pradel, R. , Tavecchia, G. , Igual, J. M. , Sanz‐Aguilar, A. , Genovart, M. , & Oro, D. (2014). Hierarchical modelling of population growth rate from individual capture–recapture data. Methods in Ecology and Evolution, 5, 606–614. 10.1111/2041-210X.12194 [DOI] [Google Scholar]
- Valenzano, D. R. , Terzibasi, E. , Cattaneo, A. , Domenici, L. , & Cellerino, A. (2006). Temperature affects longevity and age‐related locomotor and cognitive decay in the short‐lived fish Nothobranchius furzeri . Aging Cell, 5, 275–278. 10.1111/j.1474-9726.2006.00212.x [DOI] [PubMed] [Google Scholar]
- Valenzuela‐Sánchez, A. , Wilber, M. Q. , Canessa, S. , Bacigalupe, L. D. , Muths, E. , Schmidt, B. R. , Cunningham, A. A. , Ozgul, A. , Johnson, P. T. J. , & Cayuela, H. (2021). Why disease ecology needs life‐history theory: A host perspective. Ecology Letters, 24, 876–890. 10.1111/ele.13681 [DOI] [PubMed] [Google Scholar]
- Walsh, B. S. , Parratt, S. R. , Hoffmann, A. A. , Atkinson, D. , Snook, R. R. , Bretman, A. , & Price, T. A. (2019). The impact of climate change on fertility. Trends in Ecology & Evolution, 34, 249–259. 10.1016/j.tree.2018.12.002 [DOI] [PubMed] [Google Scholar]
- Wellenreuther, M. , Mérot, C. , Berdan, E. , & Bernatchez, L. (2019). Going beyond SNPs: The role of structural genomic variants in adaptive evolution and species diversification. Molecular Ecology, 28, 1203–1209. 10.1111/mec.15066 [DOI] [PubMed] [Google Scholar]
- Woodhams, D. C. , Alford, R. A. , & Marantelli, G. (2003). Emerging disease of amphibians cured by elevated body temperature. Diseases of Aquatic Organisms, 55, 65–67. 10.3354/dao055065 [DOI] [PubMed] [Google Scholar]
- Xi, X. , Li, B. , Chen, T. , & Kwok, H. F. (2015). A review on bradykinin‐related peptides isolated from amphibian skin secretion. Toxins, 7, 951–970. 10.3390/toxins7030951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W. , Qi, Y. , & Fu, J. (2016). Genetic signals of high‐altitude adaptation in amphibians: A comparative transcriptome analysis. BMC Genetics, 17, 1–10. 10.1186/s12863-016-0440-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, K. , & Mobbs, C. V. (2010). Evidence for only two independent pathways for decreasing senescence in Caenorhabditis elegans . Age, 32, 39–49. 10.1007/s11357-009-9110-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, T. , & Lu, X. (2013). Body size variation of four latitudinally‐separated populations of a toad species: Age and growth rate as the proximate determinants. Integrative Zoology, 8, 315–323. 10.1111/j.1749-4877.2012.00294.x [DOI] [PubMed] [Google Scholar]
- Zhang, L. , & Lu, X. I. N. (2012). Amphibians live longer at higher altitudes but not at higher latitudes. Biological Journal of the Linnean Society, 106, 623–632. 10.1111/j.1095-8312.2012.01876.x [DOI] [Google Scholar]
- Zhong, M. , Yu, X. , & Liao, W. (2018). A review for life‐history traits variation in frogs especially for anurans in China. Asian Herpetological Research, 9, 165–174. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
Genomic data are available on Dryad Digital Repository https://doi.org/10.5061/dryad.x3ffbg7k3 (Cayuela et al., 2021).


