Abstract
Systematic reviews of the scientific literature can be an important source of information supporting the daily work of the regulators in their decision making, particularly in areas of innovative technologies where the regulatory experience is still limited. Significant research activities in the field of nanotechnology resulted in a huge number of publications in the last decades. However, even if the published data can provide relevant information, scientific articles are often of diverse quality, and it is nearly impossible to manually process and evaluate such amount of data in a systematic manner. In this feasibility study, we investigated to what extent open‐access automation tools can support a systematic review of toxic effects of nanomaterials for health applications reported in the scientific literature. In this study, we used a battery of available tools to perform the initial steps of a systematic review such as targeted searches, data curation and abstract screening. This work was complemented with an in‐house developed tool that allowed us to extract specific sections of the articles such as the materials and methods part or the results section where we could perform subsequent text analysis. We ranked the articles according to quality criteria based on the reported nanomaterial characterisation and extracted most frequently described toxic effects induced by different types of nanomaterials. Even if further demonstration of the reliability and applicability of automation tools is necessary, this study demonstrated the potential to leverage information from the scientific literature by using automation systems in a tiered strategy.
Keywords: automation tools, knowledge management, nanomedicines, quality evaluation, systematic review automation, toxicity of nanomaterials
Short abstract
The regulation of innovative products such as nanotechnology‐based health products can be challenging because robust datasets characterising the physical chemical properties and resulting health effects of emerging materials are often lacking. At the same time, the scientific literature strongly increases and makes a systematic review in a conventional way nearly impossible. In this study, we presented a stepwise approach to automate systematic reviews with the aim to identify toxicity pattern of nanotechnology‐based health products.
1. INTRODUCTION
The regulation of innovative products that are based on emerging technologies is often challenging, because knowledge about their critical properties and eventually resulting health effects is limited. It is in particular complex in the area of nanotechnology, given the variety and heterogeneity of existing nanotechnological platforms and multitude of specific parameters that can influence biological effects. Innovative nanotechnological applications are developed across different industrial sectors, including the medical sector (Caster et al., 2017; Germain et al., 2020; Noorlander et al., 2015), yet regulatory experience with such products is limited, due to particularity of each product and the relatively low amount of robust datasets received so far from submissions of similar health products (Marques et al., 2019; Venditto & Szoka, 2013). Before receiving market authorisation health products need to be fully characterised according to given regulatory requirements. However, currently, only some draft/initial guidance has been provided on regulatory information needs for certain categories of nanotechnology‐based health products (EMA/CHMP, 2013b, 2013a, 2015; FDA, 2017; SCENIHR, 2015), creating a challenge and potential uncertainty for product developers (Halamoda‐Kenzaoui et al., 2019, 2020).
The open questions are related to the critical physical, chemical and biological parameters that are predictive for clinical outcomes and as such should be rigorously evaluated before the regulatory approval. On the other hand, the number of scientific reports on the physicochemical properties and biological effects of nanomaterials and nanomedicines has exploded in recent years (Figure 1), providing an additional source of information, potentially useful for the regulatory decision making. However, reviewing such high number of publications in a conventional way requires a lot of invested time and resources making systematic reviews in the field extremely rare and limited in scope.
FIGURE 1.
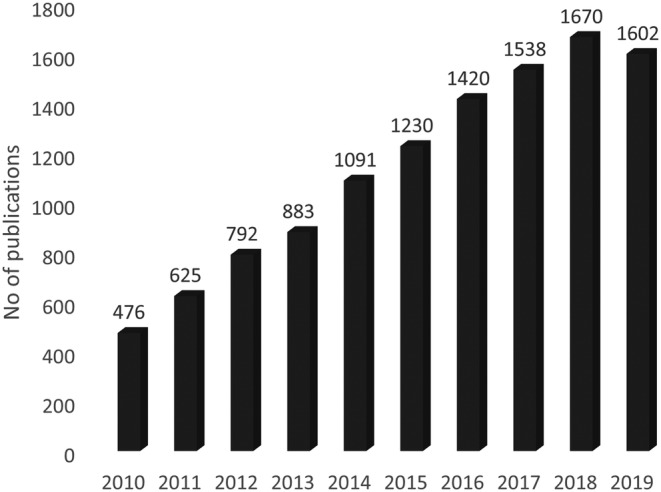
Number of publications with the term “nanomedicine” as author keyword, per year, in years 2010–2019 (source: Tools for Innovation Monitoring [TIM])
Despite an increased number of approaches and technologies for automation of a systematic review procedure (Marshall & Wallace, 2019), the uncertainty about their reliability as well as the lack of training and experience in text mining prevents broad applications of such tools in reviewing of the literature (Lau, 2019; O'Connor et al., 2019). Most widely used applications are related to initial steps of the systematic review procedure such as searches and identification of relevant publications. However, for more advanced steps of the systematic reviews such as extraction of information (textual data) from full texts and quality evaluations no ready‐to‐use systems are available, given the substantial level of specificity and complexity required at this stage.
In addition, considering the heterogeneity of published data, a quality evaluation step is necessary before high‐quality data can be identified and used to draw reliable conclusions. Several initiatives provide criteria and web‐based tools for the evaluation of reliability and relevance of published research studies in the area of chemicals assessment, in order to bridge the gap between academic research and chemicals regulation and policy (Science in Risk Assessment and Policy, n.d.; Genaidy & Shell, 2008; Schneider et al., 2009). Such criteria can guide researchers to design experiments and be used by reviewers to evaluate the quality of published studies. In the field of nanotechnology specific criteria for the reporting have also been proposed and gained a lot of attention from the scientific community (Faria et al., 2018). They are based on three types of reported characterisation: (1) nanomaterial characterisation, (2) biological characterisation and (3) details of experimental procedures. In particular, nanomaterial characterisation is frequently lacking in toxicological reports.
In this feasibility study, we aimed to verify whether suitable automation tools can be applied in a tiered approach to perform a systematic review of the literature on toxic effects of nanomaterials for medical applications. Following the steps of the systematic review procedure, 1 we used open‐access tools for the identification and curation of relevant dataset. In order to complement existing approaches, we developed a tool for article segmentation, which allowed us to extract predefined parameters (e.g., nanomaterial properties and toxic effects) from specific sections of the articles. By using this tool, we were able to rank the scientific articles based on quality criteria related to nanomaterial characterisation and to extract toxic effects of nanomaterials mostly reported in the scientific literature. Even if more validation steps are necessary, we could demonstrate that user‐friendly and open‐access tools used in a tiered approach can support the identification of potential health effects of products under evaluation and provide at the same time a collection of publications supporting the scientific evidence. Such approach can assist the regulatory community, in particular regarding products/technologies for which the scientific evidence is limited.
2. METHODOLOGY
2.1. Search strategy
Nine different searches related to specific toxic effects of nanomaterials relevant for health applications were performed using PubMed MeSH controlled terminology. The terminology of nanomaterials was based on following PubMed MeSH construct: “Nanostructures/adverse effects”[Majr] OR “Nanostructures/toxicity”[Majr]) OR (“Liposomes/adverse effects”[Majr] OR “Liposomes/toxicity”[Majr]) OR “Micelles”[Majr] OR “Nanoemulsion”[tiab], and was combined with terms related to different types of toxicity:
Cardiotoxicity (“Cardiotoxicity”[Majr])
Liver toxicity (“Chemical and Drug Induced Liver Injury”[Majr])
Neurotoxicity (“Neurotoxicity Syndromes”[Majr])
Biocompatibility/haemocompatibility (“Materials Testing”[Majr])
Immune effects (“Immune System Phenomena”[Majr] OR “Immune System Diseases”[Majr])
Lung toxicity (“Lung Injury”[Majr])
Genotoxicity (“Mutagenicity Tests”[Majr])
Nephrotoxicity (“renal toxicity”[tiab] OR “nephrotoxicity”[tiab])
Accumulation/biodistribution ((“biodistribution”[tiab] OR “ADME”[tiab] OR “toxicokinetics”[tiab])
No MeSH terms were found for nephrotoxicity and accumulation/biodistribution; therefore, the search of related keywords was used instead. Publications in languages other than English were not considered. The reviews were classified separately and used to retrieve additional records following a snowball approach. The latter was done manually. Review articles were also used to build up the ontology related to different types of toxicity.
2.2. Refining of dataset
The records originating from nine PubMed searches were processed through an open‐access tool Swift Review, 2 which allows screening for relevance based on titles and abstracts (Howard et al., 2016). The tool is based on a machine learning approach. Briefly, in each of the nine groups of records, a training set of 10 records was selected and qualified as relevant/irrelevant. The remaining records were screened automatically. The ranking of articles allowed to identify 90%–95% of most relevant records. The remaining ones, with the lowest score of relevance, were checked manually. The relevance was evaluated based on the defined criteria:
original article (not a review)
effect of a nanomaterial
effect on human health
The records evaluated as relevant were exported to the Mendeley 3 software and merged with the additional records retrieved in the snowball approach. The removal of duplicates was done automatically using Mendeley function.
2.3. Segmentation
The segmentation was performed on full texts (PDFs) obtained, from the journal websites and the EC library. The different sections of the articles were identified and extracted using an in‐house developed tool written in R (Leeper, 2018; Ooms, 2020; Straka, 2018; Wickham, 2019; Wickham et al., 2020; Wickham & Chang, 2016; Wickham & Hester, 2020; Wijffels, 2020) named segmenteR. In summary, first, the font and font size of the words that compose the PDF documents are identified using the cpp API of Poppler, the PDF rendering library. 4 This information was used to deduce the order and the name of the different sections, by matching the words from the PDF documents that have the most likely font and font size for a section title, to a list of pre‐defined section names (Table S1).
In a second step, the text of the articles was extracted from the PDF using tabuliser (Leeper, 2018) and annotated using UDPipe tool (Straka et al., 2016; Wijffels, 2020). This annotation converts the text to the CoNLL‐U format (Buchholz & Marsi, 2006), a format suitable for natural language processing (NLP). Using the properties of this format, the position of the different sections of the article (identified in the first step) inside the text is inferred, and the sections “Material and Methods” and “Results” are extracted and exported in CoNLL‐U format.
Inside an article, the section recognised as the section materials and methods is the first one of the sections containing one of the following words: “material,” “method,” “experimental,” “experiment” or “methodology.” The section recognised as the results section is the first one of the sections containing one the following words: “result” or “results.”The tool used for the segmentation of the articles, as well as the instructions on how to use and install it, are available online (https://github.com/ec-jrc/jrc_f2_refine/tree/master/segmenteR). In addition, a comparison of the manual and automatic segmentation has been performed on a sample of 33 articles (Table S2).
2.4. Scoring and ranking of articles based on the quality (materials and methods part)
After the extraction of the materials and methods section, the text of the section, in CoNLL‐U format, was parsed for specifics keywords reflecting different aspects of the characterisation of nanomaterial (Table S4), using syntactic parsing (Csardi & Nepusz, 2006; Dragulescu & Arendt, 2020; Pedersen, 2019; Wickham, 2007, 2016; Wickham & Henry, 2020).
If at least one of the keywords related to a nanomaterial property was found in the section, this characterisation was considered made by the author. The articles were ranked according to the number of nanomaterial properties characterised by the author, from a rank of 0 (none of the five characterisations were made) to 5 (at least one keyword for each of the properties were found).
The articles with a ranking equal or superior to two were kept for downstream analysis.
The same approach was used to estimate if an article describes in vitro or/and in vivo experiment. The materials and methods sections were screened for specifics keywords for in vivo and in vitro experiments (Table S5), using syntactic parsing. If an article contained one or more keywords for a type of experiment (in vitro or in vivo), the article was classified to a corresponding category. An article could belong to both categories.The original code used to produce these results and the charts, as well as the original charts, are available at (https://github.com/ec-jrc/jrc_f2_refine).
2.5. Extraction of toxic effects and nanomaterial type from the results section
The text of the extracted results sections, in CoNLL‐U format, of articles with a quality score equal or above to 2, were parsed for keywords related to specific, non‐specific toxicity and nanomaterial type using syntactic parsing (Tables S6 and S7).
In case of keywords composed of two or more words, the text was screened for sentences containing all the words included in the keyword term. If such sentence was found, the toxicity/nanomaterial was considered as reported/mentioned in the article.
Such screening allowed assigning the articles to different toxicity topics. An article could be assigned to more than one topic. In the topics with the highest number of articles, the additional analysis was performed using parsing with more specific terms related to particular toxicities (Table S8).The original code used to produce the result and the charts, as well as the original charts, are available online (https://github.com/ec-jrc/jrc_f2_refine ).
3. RESULTS
3.1. Identification of relevant articles related to toxicities of nanomaterials
The searches for specific toxicities of nanomaterials for health applications have been performed in the PubMed MeSH database, because its controlled vocabulary related to nanotoxicology improved the search. The specific searches were related to the following areas of toxicity: cardiotoxicity, liver toxicity, neurotoxicity, biocompatibility/haemocompatibility, immune effects, lung toxicity, genotoxicity, nephrotoxicity and accumulation (Figure 2). For the two last topics, no MeSH terms were found, and the search was performed using related keywords. Following the searches in PubMed MeSH, the review articles were kept separately for further use. The records of the original articles were exported directly to Swift Review, which is an open‐source interactive tool facilitating the screening of abstracts (Howard et al., 2016). It is based on a machine learning approach and helps to rapidly screen abstracts and automatically remove irrelevant entries. Around 35% of the search records from PubMed MeSH were removed in this step. In the next step, additional citations from the reviews articles were followed and added to the dataset, if relevant (Snowball approach). This particular step was performed manually. The citations from both sources were merged in the Mendeley reference manager, and duplicates were removed using the deduplication function. In the following step, the total number of references in all topic groups was equal to 821 (Figure 2); however, not all the publications were available as full texts, decreasing the number to 800 full texts obtained in PDF formats.
FIGURE 2.
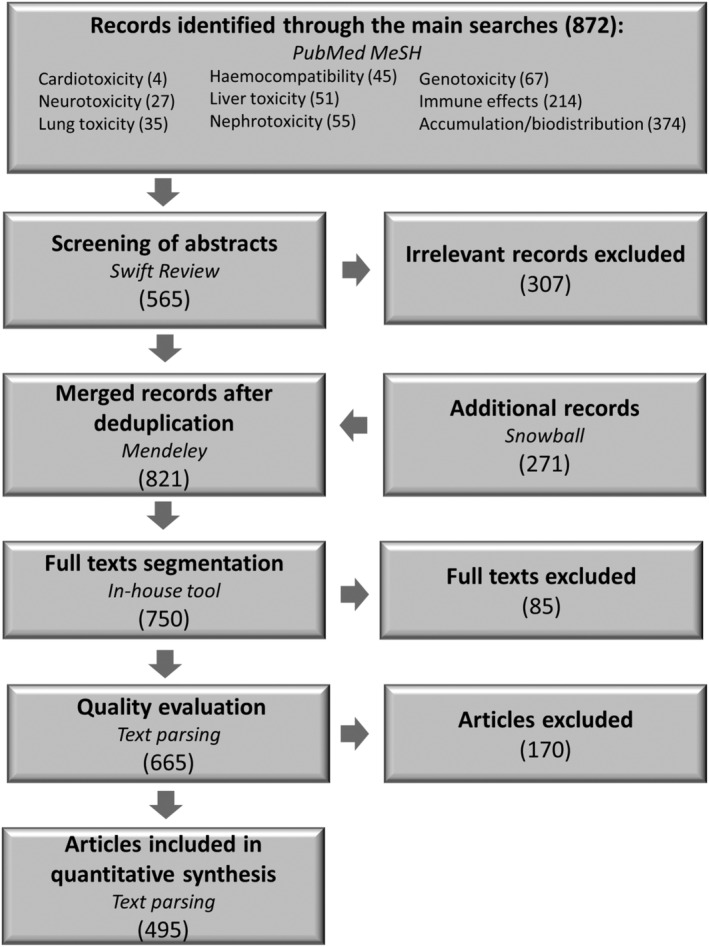
Main steps of the systematic review and corresponding numbers of publications
3.2. Article segmentation
In order to be able to automatically screen the text, it was necessary to extract specific sections of the article such as the materials and methods section (for evaluation of the quality of the articles) and the results section (for extraction of toxic effects). Limiting the text mining analysis to a specific section of an article ensured that the relevant experiments were performed in the given study and not coming from other sources, for example, in form of references.
According to our knowledge, no tools are currently available able to extract a specific section of an article. In order to close this gap we have developed a new tool, written in R, to perform this task. This tool can be applied to full texts (in PDF) and, based on the fonts, font sizes of the words inside the provided documents and the text annotation, gives as an output selected sections of the articles (materials and methods and results), in a format suitable for downstream analysis.
A sample of 50 has been randomly selected from 800 full‐text articles and left apart for validation purposes. Among them, 33 documents were used to perform a comparison between automated and manual extraction of the materials and methods section. This has been calculated using cosine similarity, a common metrics to measure the similarity between two texts (Jurafsky & Martin, 2020). The extraction failed on two articles, gave dissimilar results on two others and gave a similarity close to 1 on 29 articles (Table S2).
The segmentation tool has been applied to 750 articles and proofed to work successfully in 88% cases, resulting in the extraction of 665 sets of materials and methods section. Many of 85 documents on which segmentation tool did not work, appeared to belong to a different category of document than original research article, such as a review, a letter, a supporting information or an erratum; some of them were books or a scan of a paper (without any information on the font) (Table S3). Finally, some errors were due to articles with multiple font sizes inside the section names.
3.3. Evaluation of quality of scientific articles
A critical quality control of the collected articles is an important step in every systematic review procedure, as it ensures that the data used for the final analysis will lead to reliable and relevant conclusions. It is in particular important, if such evidence is used in the regulatory context. The evaluation of the quality of an article requires prior development of suitable criteria, which can be related to reporting quality, methodological quality or scientific relevance (Science in Risk Assessment and Policy, n.d.). Faria et al. (2018) developed a set of minimum information requirements for nanotoxicological studies. The authors identified three main types of characterisation that should be reported in the nanotoxicology papers: (1) material characterisation, (2) biological characterisation and (3) details of experimental procedures.
Following these recommendations, we focused on nanomaterial characterisation which is a part frequently lacking in toxicological studies even if detailed characterisation of nanomaterial properties is critical, to properly understand the impact of physical and chemical parameters on the observed biological effect. In our study, we focused on five basic nanomaterial properties, namely, size, surface charge, surface area, chemical composition and stability/aggregation, and investigated how many of these properties were characterised and reported in every article. Because the physicochemical characterisation of the nanomaterial is usually reported in materials and methods section of the articles, prior article segmentation step has been necessary to specifically mine this part of the text for the predefined quality criteria.
The text of the materials and methods section were parsed for developed terminology corresponding to each of the selected nanomaterial properties (Table S4) before the articles were scored and ranked according to number of characterised nanomaterial properties (Figure 3A). A threshold of minimal characterisation required before further analysis was set on at least two reported nanomaterial properties. The majority of articles reported on two out of the five selected nanomaterial properties whereas only a very small part of the articles (5%) analysed all five basic nanomaterial properties. Size was, by far, the most reported nanomaterial property, followed by stability/aggregation and chemical composition (Figure 3B).
FIGURE 3.
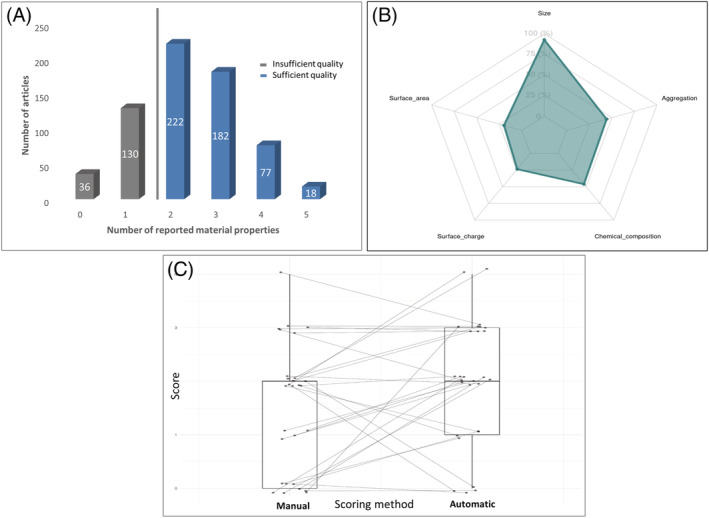
Evaluation of the quality of articles performed via the automatic scoring of reported physicochemical characteristics of nanomaterials, (A) ranking of the articles according to the number of reported nanomaterial properties with a threshold of quality set on at least two nanomaterial properties, (B) most frequently reported nanomaterial properties in all scored documents, (C) comparison of automatic versus manual scoring of articles performed on a sample of documents [Colour figure can be viewed at wileyonlinelibrary.com]
In addition, a comparison of automatic versus manual scoring was performed on a random sample of documents (Figure 3C and Figure S1). The automatic method resulted to be slightly over‐scoring in comparison with the manual method. Such over‐scoring could be, in major part, due to the presence of negative statements on the performed measurements that a parsing approach cannot discriminate from the other type of statements. On the other hand, some examples of under‐scoring were also found, probably resulting from the insufficient collection of terms for certain properties. Such limitation can be easily overcome by expanding the terms in the ontology. Addressing the over‐scoring issue is more complex, and additional text mining techniques should be probably explored here, such as sentiment analysis or context analysis.
3.4. Extraction of toxic effects
In total, 495 original articles met the quality criteria and were further used to mine toxicity studies of nanomaterials for health applications. The screening of the extracted materials and methods sections with a set of identified keywords specific to in vitro or in vivo experiments of different toxicities allowed the classification of studies describing the various toxic effects (Figure 4). Similar numbers of in vitro and in vivo studies were found for most of the toxicological areas, apart from the immune effects and genotoxicity, where more experiments have been performed in vitro.
FIGURE 4.
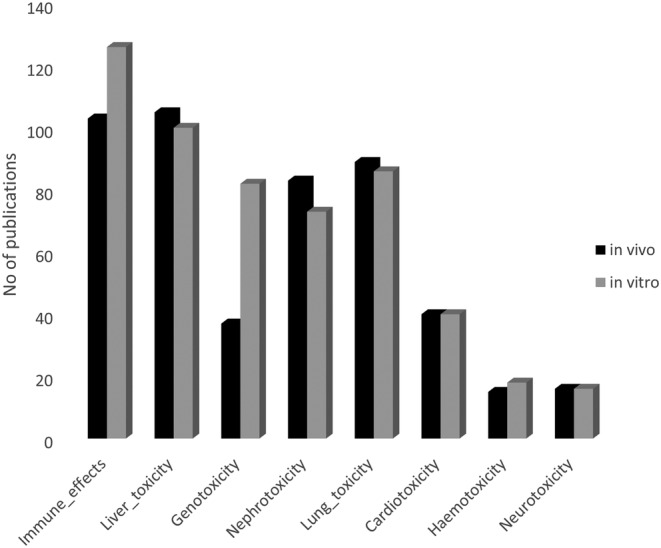
Number of toxicity studies in vitro and in vivo in various toxicological areas
In order to identify the main toxicity patterns reported for different types of nanomaterials a developed ontology (Tables S6 and S7) was used to apply text parsing of the results sections. All terms including more specific toxicity reactions as well as more general terms were counted and assigned the article to a given category (area of toxicity). Such screening allowed the identification of main toxicity patterns, such as genotoxicity, immune effects, liver toxicity, lung toxicity and nephrotoxicity, which have been reported for different types of nanomaterials (Figure 5). Most toxic effects in all groups were reported in association with metal‐based nanoparticles covering different types of metallic nanomaterials, such as copper, zinc oxide, titanium dioxide, iron oxide, gold, silver, aluminium oxide, cerium and manganese ‐based nanoparticles. In the category of articles related to lung toxicity, the most reported type of nanomaterials was the carbon‐based nanoparticles (Figure 5).
FIGURE 5.
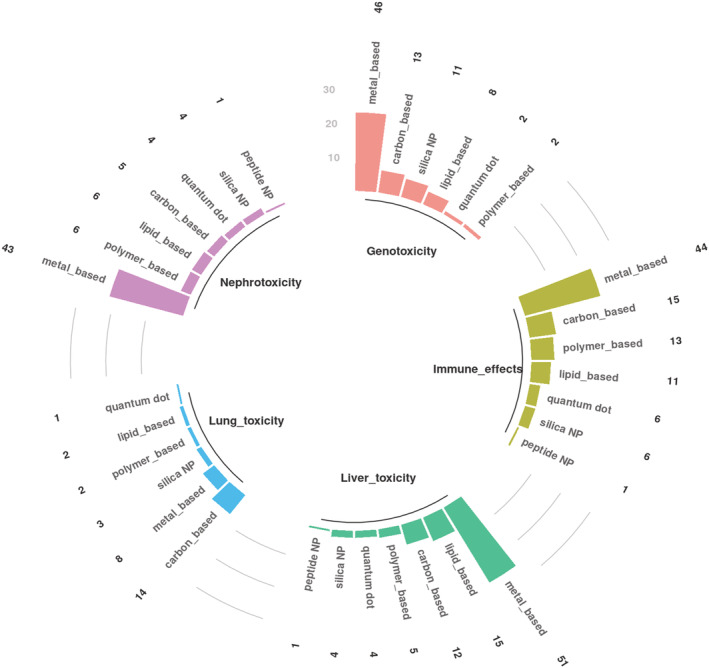
Main toxicity patterns associated with types of nanomaterials extracted from the results sections. The numbers above the bars represent numbers of corresponding articles
Additional analysis identified more specific biological reactions associated with different types of nanomaterials (Table S8). Analysis of the most reported immune effects highlighted the accumulation and toxicity of metal‐based and carbon‐based nanoparticles to the spleen and revealed the activation of macrophages as well as a modulation in cytokines release (Figure 6).
FIGURE 6.
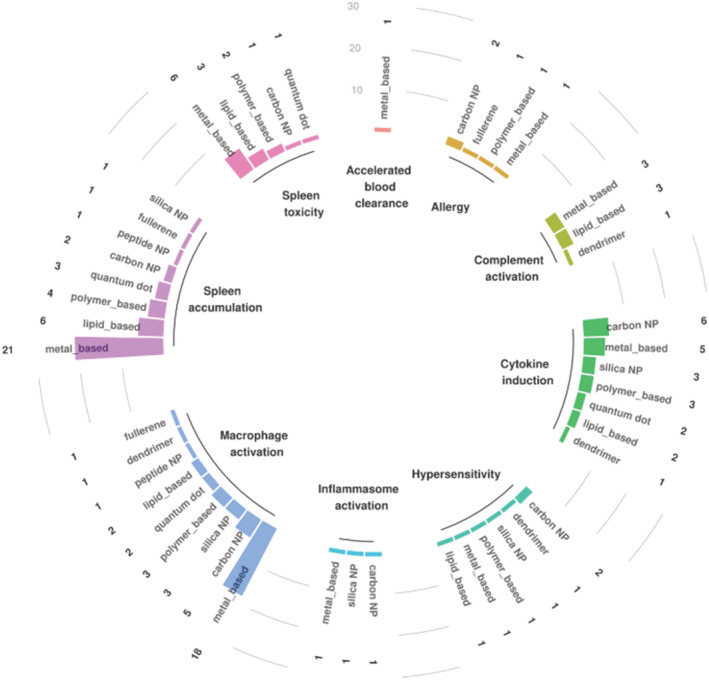
Main types of nanomaterials associated with most reported immune effects. The numbers above the bars represent numbers of corresponding articles
3.5. Available automation tools
Automation technologies have a huge potential to improve the process of a systematic reviews of the scientific literature and contribute as such to evidence‐based regulatory decisions (Barbui et al., 2017). The use of the scientific literature as an information source is particularly relevant in the field of innovative products/technologies, where the fast progress of the field is reflected in a continuously increasing number of scientific publications. However, the integration of automation in the systematic review procedures can only be achieved if the available tools demonstrate their reliability and ability to be used by scientific domain experts, which are not necessarily trained in computational technologies.
For the moment, suitable open‐access tools cover only initial steps of the systematic review, such as searches with controlled vocabulary or the curation of datasets. However, the use of controlled vocabulary searches requires pre‐existence of adequate terminology, related to the needs of a specific field. Development and expansion of such ontologies can not only enable controlled searches in innovative sectors but also improve the search recall and precision, resulting in the collection of relevant records. However, identification of relevant and well‐defined terminology in highly innovative fields such as nanotechnology can be a challenge on its own, due to the multitude and heterogeneity of terms used in regulatory and academic context (Arora et al., 2013; Quiros‐Pesudo et al., 2018).
Different automation systems exist already for the refinement of datasets (Jaspers et al., 2018; Marshall & Wallace, 2019; Tsafnat et al., 2014). They are mainly based on machine learning techniques, requiring some manual annotation from the reviewer to categorise abstracts from so‐called “training set” into relevant or irrelevant categories. Based on this information and subsequent mathematical algorithms, the remaining list of the documents is ranked according to the probability of their relevance. Even if a small number of relevant articles might be lost through automatic screening, the use of such tools can save considerable amount of time and effort compared to manually performed screening of records.
For the extraction of information from the full texts, no satisfying solutions exist for the moment. Currently available tools for data extraction, including textual data, are designed for randomised controlled trials reports (Jonnalagadda et al., 2015; Tsafnat et al., 2014), rather than original research articles. In our study, we extracted specific section of the articles prior to applying text mining techniques in order to improve the outcome of the analysis. This is particularly promising in the evaluation of the quality of articles, where a limited terminology describing defined parameters can facilitate the automation. Such approach can be adapted and tailored to any quality criteria suitable for a specific domain provided that corresponding ontology will be developed by a reviewer. Moreover, the delimitation to the specific section of the document can improve the reliability of the analysis. Linking such system with web‐based platforms for the quality evaluation would support regular and efficient quality evaluation in an automated way not requiring manual screening of the whole dataset with the quality criteria.
The extraction of toxic effects from the results section poses more challenges because the terminology varies and it can be difficult to predefine most relevant keywords. This, together with presence of negative statements that cannot be distinguished yet from the positive ones is the major sources of bias in the automatic text parsing. Therefore, additional more adapted approaches should be examined to extract the relevant information from the results section. Such approaches would ideally be designed to be used by specific domain experts.
Automatically extracted results do not contain a high level of granularity, which is achieved by manually performed reviews; however, they are sufficient to demonstrate the trends of toxicity patterns for specific types of nanomaterials or chemicals. Depending on the reviewer's need, the emphasis can be positioned on more specific toxic reactions or different domains of application. Linking the toxicity patterns with different types of nanotechnological platforms or their physicochemical properties can help to identify additional needs for the physical, chemical or toxicological characterisation that should be available for regulatory assessments.
A crucial aspect to be further elucidated is the level of trust and reliability of the reviews performed by using automation tools (O'Connor et al., 2019). Our study highlighted immune reactions, liver toxicity, genotoxicity and nephrotoxicity effects induced by nanotechnological platforms, which is in line with expert opinions in the nanotoxicology field (Dobrovolskaia, 2015; Magdolenova et al., 2014; Wolfram et al., 2015). However, more detailed results of the review pointing out to specific adversities induced by a given nanomaterial would require more extensive analysis and validation using conventional methods before being used for regulatory decision‐making. Here again, a wider use of automation tools in the routine literature reviews ideally performed, in parallel with conventional methods would provide more evidence on the performance, limitations and reliability of automation systems. Only with a high level of confidence in their reliability such tools can be integrated into the systematic review processes. Therefore, a more interdisciplinary effort involving domain experts and automation specialist is needed to advance the development and improvement of existing tools in order to make them more robust and fit for purpose, and to promote their application for evidence‐based regulatory applications and research needs (Beller et al., 2018).
4. CONCLUSIONS
Our analysis demonstrated the potential of open‐access automation tools to support a systematic review of the scientific literature. Whereas for the initial steps of the process relevant tools are available, for more advanced stages such as quality evaluation and information extraction, existing approaches require adaptation to the specific needs and additional validation of the results. Our in‐house developed tool for article segmentation fills an important gap in the strategy and allows extraction of specific sections of a scientific article prior applying text mining tools, which should improve the relevance of such analysis. Further refinements of automation systems should be pursued in an interdisciplinary collaboration leading to the development of fit for purpose and user‐friendly tools and their smooth integration in the routine literature reviews.
CONFLICT OF INTEREST
Any opinions expressed in this publication are those of the authors only, and this paper does not represent an official position of the European Commission. The authors report no conflicts of interest in this work.
Supporting information
Figure S1: Difference between the manual and automatic evaluation of the article quality, left: absolute difference in scoring, right: the notes of the manual evaluation were subtracted from the automatic evaluation.
Table S1: List of the predefined section names for segmentation.
Table S2: Comparison of manual end automatic extraction of Materials and Method section.
Table S3: Sources of errors in article segmentation
Table S4: List of keywords for the nanomaterial characterization regarding five basic properties of nanomaterials.
Table S5: List of keywords for the classification of in vivo vs in vitro experiments.
Table S6: List of keywords for the classification of the documents into different types of toxicity.
Table S7: List of keywords for the classification of nanomaterials.
Table S8: List of keywords for the extraction of specific toxicity effects.
Halamoda‐Kenzaoui, B. , Rolland, E. , Piovesan, J. , Puertas Gallardo, A. , Bremer‐Hoffmann, S. (2022). Toxic effects of nanomaterials for health applications: How automation can support a systematic review of the literature? Journal of Applied Toxicology, 42(1), 41–51. 10.1002/jat.4204
ENDNOTES
Preferred Reporting Items for Systematic Reviews and Meta‐Analyses (PRISMA) (http://www.prisma-statement.org/)
SWIFT‐Review: Sciome Workbench for Interactive computer‐Facilitated Text‐mining; available online (https://www.sciome.com/swift-review/).
Mendeley Reference Manager; available online (www.mendeley.com).
Poppler‐PDF rendering library; available online (https://poppler.freedesktop.org/).
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in the repository at https://github.com/ec-jrc/jrc_f2_refine.
REFERENCES
- Arora, S. K. , Porter, A. L. , Youtie, J. , & Shapira, P. (2013). Capturing new developments in an emerging technology: An updated search strategy for identifying nanotechnology research outputs. Scientometrics, 95(1), 351–370. 10.1007/s11192-012-0903-6 [DOI] [Google Scholar]
- Barbui, C. , Addis, A. , Amato, L. , Traversa, G. , & Garattini, S. (2017). Can systematic reviews contribute to regulatory decisions? European Journal of Clinical Pharmacology, 73(4), 507–509. 10.1007/s00228-016-2194-y [DOI] [PubMed] [Google Scholar]
- Beller, E. , Clark, J. , Tsafnat, G. , Adams, C. , Diehl, H. , Lund, H. , Ouzzani, M. , Thayer, K. , Thomas, J. , Turner, T. , Xia, J. , Robinson, K. , Glasziou, P. , Ahtirschi, O. , Christensen, R. , Elliott, J. , Graziosi, S. , Kuiper, J. , Moustgaard, R. , … Wedel‐Heinen, I. (2018). Making progress with the automation of systematic reviews: Principles of the international collaboration for the automation of systematic reviews (ICASR). Systematic Reviews, 7(1), 1–7. 10.1186/s13643-018-0740-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz, S. , & Marsi, E. (2006). CoNLL‐X shared task on multilingual dependency parsing. Proceedings of the Tenth Conference on Computational Natural Language Learning, CoNLL‐X, June, 149–164. 10.3115/1596276.1596305 [DOI]
- Caster, J. M. , Patel, A. N. , Zhang, T. , & Wang, A. (2017). Investigational nanomedicines in 2016: A review of nanotherapeutics currently undergoing clinical trials. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology, 9(1), e1416. 10.1002/wnan.1416 [DOI] [PubMed] [Google Scholar]
- Csardi, G. , & Nepusz, T. (2006). The igraph software package for complex network research. InterJournal, Complex Systems, 1695. http://igraph.org [Google Scholar]
- Dobrovolskaia, M. A. (2015). Pre‐clinical immunotoxicity studies of nanotechnology‐formulated drugs: Challenges, considerations and strategy. Journal of Controlled Release, 220(Pt B), 571–583. 10.1016/j.jconrel.2015.08.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragulescu, A. , & Arendt, C. (2020). xlsx: Read, write, format Excel 2007 and Excel 97/2000/XP/2003 files. https://cran.r-project.org/package=xlsx
- EMA/CHMP . (2013a). Joint MHLW/EMA reflection paper on the development of block copolymer micelle medicinal products . EMA/CHMP/13099/2013.
- EMA/CHMP . (2013b). Reflection paper on the data requirements for intravenous liposomal products developed with reference to an innovator liposomal product. EMA/CHMP/806058/2009/Rev.02.
- EMA/CHMP . (2015). Reflection paper on the data requirements for intravenous iron‐based nano‐colloidal products developed with reference to an innovator medicinal product. EMA/CHMP/SWP/620008/2012. https://www.ema.europa.eu/documents/scientific‐guideline/reflection‐paper‐data‐requirements‐intravenous‐iron‐based‐nano‐colloidal‐products‐developed_en.pdf
- Faria, M. , Björnmalm, M. , Thurecht, K. J. , Kent, S. J. , Parton, R. G. , Kavallaris, M. , Johnston, A. P. R. , Gooding, J. J. , Corrie, S. R. , Boyd, B. J. , Thordarson, P. , Whittaker, A. K. , Stevens, M. M. , Prestidge, C. A. , Porter, C. J. H. , Parak, W. J. , Davis, T. P. , Crampin, E. J. , & Caruso, F. (2018). Minimum information reporting in bio–nano experimental literature. Nature Nanotechnology, 13(9), 777–785. 10.1038/s41565-018-0246-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA . (2017). Drug products, including biological products, that contain nanomaterials . Guidance for Industry. https://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/default.htm [DOI] [PMC free article] [PubMed]
- Genaidy, A. M. , & Shell, R. L. (2008). The experimental appraisal instrument: A brief overview. Human Factors and Ergonomics in Manufacturing, 18(3), 270–292. 10.1002/hfm.20112 [DOI] [Google Scholar]
- Germain, M. , Caputo, F. , Metcalfe, S. , Tosi, G. , Spring, K. , Åslund, A. K. O. , Pottier, A. , Schiffelers, R. , Ceccaldi, A. , & Schmid, R. (2020). Delivering the power of nanomedicine to patients today. Journal of Controlled Release, 326, 164–171. 10.1016/j.jconrel.2020.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halamoda‐Kenzaoui, B. , Box, H. , van Elk, M. , Gaitan, S. , Geertsma, R. E. , Gainza Lafuente, E. , Owen, A. , del Pozo, A. , Roesslein, M. , & Bremer‐Hoffmann, S. (2019). Anticipation of regulatory needs for nanotechnology‐enabled health products—The REFINE white paper. Luxembourg: EUR 29919: Publications Office of the European Union. https://ec.europa.eu/jrc/en/publication/thematic-reports/anticipation-regulatory-needs-nanotechnology-enabled-health-products [Google Scholar]
- Halamoda‐Kenzaoui, B. , Box, H. , van Elk, M. , Gaitan, S. , Geertsma, R. E. , Gainza Lafuente, E. , Owen, A. , del Pozo, A. , Roesslein, M. , & Bremer‐Hoffmann, S. (2020). Launching stakeholder discussions on identified regulatory needs for nanotechnology‐enabled health products. Precision Nanomedicine, 3(3), 608–621. 10.33218/001c.13521 [DOI] [Google Scholar]
- Howard, B. E. , Phillips, J. , Miller, K. , Tandon, A. , Mav, D. , Shah, M. R. , Holmgren, S. , Pelch, K. E. , Walker, V. , Rooney, A. A. , Macleod, M. , Shah, R. R. , & Thayer, K. (2016). SWIFT‐review: A text‐mining workbench for systematic review. Systematic Reviews, 5(1), 87. 10.1186/s13643-016-0263-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers, S. , De Troyer, E. , & Aerts, M. (2018). Machine learning techniques for the automation of literature reviews and systematic reviews in EFSA. EFSA Supporting Publications, 15(6). 10.2903/sp.efsa.2018.en-1427 [DOI] [Google Scholar]
- Jonnalagadda, S. R. , Goyal, P. , & Huffman, M. D. (2015). Automating data extraction in systematic reviews: A systematic review. Systematic Reviews, 4(1), 78. 10.1186/s13643-015-0066-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurafsky, D. , & Martin, J. H. (2020). Speech and language processing (3rd draft). Chapter 6: Vector Semantics and Embeddings.
- Lau, J. (2019). Editorial: Systematic review automation thematic series. Systematic Reviews, 8(1), 4–5. 10.1186/s13643-019-0974-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeper, T. J. (2018). tabulizer: Bindings for Tabula PDF table extractor library. R package version 0.2.2. (0.2.2.). https://github.com/ropensci/tabulizer
- Magdolenova, Z. , Collins, A. , Kumar, A. , Dhawan, A. , Stone, V. , & Dusinska, M. (2014). Mechanisms of genotoxicity. A review of in vitro and in vivo studies with engineered nanoparticles. Nanotoxicology, 8(3), 233–278. 10.3109/17435390.2013.773464 [DOI] [PubMed] [Google Scholar]
- Marques, M. R. C. , Choo, Q. , Ashtikar, M. , Rocha, T. C. , Bremer‐Hoffmann, S. , & Wacker, M. G. (2019). Nanomedicines—Tiny particles and big challenges. Advanced Drug Delivery Reviews, 151‐152, 23–43. 10.1016/j.addr.2019.06.003 [DOI] [PubMed] [Google Scholar]
- Marshall, I. J. , & Wallace, B. C. (2019). Toward systematic review automation: A practical guide to using machine learning tools in research synthesis. Systematic Reviews, 8(1), 1–10. 10.1186/s13643-019-1074-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noorlander, C. W. , Kooi, M. W. , Oomen, A. G. , Park, M. V. , Vandebriel, R. J. , Geertsma, R. E. , & Agnes, G. (2015). Horizon scan of nanomedicinal products. Nanomedicine (London, England), 10(1), 1599–1608. 10.2217/nnm.15.21 [DOI] [PubMed] [Google Scholar]
- O'Connor, A. M. , Tsafnat, G. , Thomas, J. , Glasziou, P. , Gilbert, S. B. , & Hutton, B. (2019). A question of trust: Can we build an evidence base to gain trust in systematic review automation technologies? Systematic Reviews, 8(1), 1–8. 10.1186/s13643-019-1062-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms, J. (2020). Pdftools: Text extraction, rendering and converting of PDF documents. https://cran.r-project.org/web/packages/pdftools/index.html
- Pedersen, T. L. (2019). An Implementation of Grammar of Graphics for Graphs and Networks. 2018 (R package ggraph). Computer Science.
- Quiros‐Pesudo, L. , Balahur‐Dobrescu, A. , Gottardo, S. , Rasmussen, K. , Wagner, G. , Joanny, G. , & Bremer, S. (2018). Mapping nanomedicine terminology in the regulatory landscape. EUR 29291 EN; Publications Office of the European Union, Luxembourg, ISBN 978‐92‐79‐89872‐3, JRC112224. 10.2760/753829 [DOI]
- SCENIHR . (2015). Opinion on the guidance on the determination of potential health effects of nanomaterials used in medical devices. Final Opinion, 1–77. 10.2772/41391 [DOI]
- Schneider, K. , Schwarz, M. , Burkholder, I. , Kopp‐Schneider, A. , Edler, L. , Kinsner‐Ovaskainen, A. , Hartung, T. , & Hoffmann, S. (2009). “ToxRTool”, a new tool to assess the reliability of toxicological data. Toxicology Letters, 189(2), 138–144. 10.1016/j.toxlet.2009.05.013 [DOI] [PubMed] [Google Scholar]
- Science in Risk Assessment and Policy . (n.d.). Science in Risk Assessment and Policy http://scirap.org/#
- Straka, M . (2018). UDPipe 2.0 prototype at CONLL 2018 UD shared task. Proceedings of the CONLL 2018 Shared Task: Multilingual Parsing from Raw Text to Universal Dependencies, 197–207. 10.18653/v1/K18-2020 [DOI]
- Straka, M. , Hajič, J. , & Straková, J. (2016). UDPipe: Trainable pipeline for processing CoNLL‐U files performing tokenization, morphological analysis, POS tagging and parsing. Proceedings of the 10th International Conference on Language Resources and Evaluation, LREC 2016, 4290–4297.
- Tsafnat, G. , Glasziou, P. , Choong, M. K. , Dunn, A. , Galgani, F. , & Coiera, E. (2014). Systematic review automation technologies. Systematic Reviews, 3(1), 1–15. 10.1186/2046-4053-3-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venditto, V. J. , & Szoka, F. C. (2013). Cancer nanomedicines: So many papers and so few drugs! Advanced Drug Delivery Reviews, 65(1), 80–88. 10.1016/j.addr.2012.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. (2007). Reshaping data with the reshape package. Journal of Statistical Software, 21(12). http://www.jstatsoft.org/v21/i12/paper, 10.18637/jss.v021.i12 [DOI] [Google Scholar]
- Wickham, H. (2016). ggplot2: Elegant graphics for data analysis. New York: Springer‐Verlag. https://ggplot2.tidyverse.org, 10.1007/978-3-319-24277-4 [DOI] [Google Scholar]
- Wickham, H. (2019). Stringr: Simple, consistent wrappers for common string operations. https://cran.r-project.org/package=stringr
- Wickham, H. , & Chang, W. (2016). Devtools: Tools to make developing r packages easier. R Package Version, 1(0), 9000. [Google Scholar]
- Wickham, H. , François, R. , Henry, L. , & Müller, K. (2020). dplyr: A grammar of data manipulation. https://cran.r-project.org/package=dplyr
- Wickham, H. , & Henry, L. (2020). tidyr: Tidy messy data. https://cran.r-project.org/package=tidyr
- Wickham, H. , & Hester, J. (2020). readr: Read rectangular text data. https://cran.r-project.org/package=readr
- Wijffels, J. (2020). udpipe: Tokenization, parts of speech tagging, lemmatization and dependency parsing with the “UDPipe” “NLP” toolkit. https://cran.r-project.org/package=udpipe
- Wolfram, J. , Zhu, M. , Yang, Y. , Shen, J. , Gentile, E. , Paolino, D. , Fresta, M. , Nie, G. , Chen, C. , Shen, H. , Ferrari, M. , & Zhao, Y. (2015). Safety of nanoparticles in medicine. Current Drug Targets, 16(14), 1671–1681. 10.2174/1389450115666140804124808 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Difference between the manual and automatic evaluation of the article quality, left: absolute difference in scoring, right: the notes of the manual evaluation were subtracted from the automatic evaluation.
Table S1: List of the predefined section names for segmentation.
Table S2: Comparison of manual end automatic extraction of Materials and Method section.
Table S3: Sources of errors in article segmentation
Table S4: List of keywords for the nanomaterial characterization regarding five basic properties of nanomaterials.
Table S5: List of keywords for the classification of in vivo vs in vitro experiments.
Table S6: List of keywords for the classification of the documents into different types of toxicity.
Table S7: List of keywords for the classification of nanomaterials.
Table S8: List of keywords for the extraction of specific toxicity effects.
Data Availability Statement
The data that support the findings of this study are openly available in the repository at https://github.com/ec-jrc/jrc_f2_refine.


