Abstract
The success of Listeria monocytogenes as a food-borne pathogen owes much to its ability to survive a variety of stresses, both in the external environment prior to ingestion and subsequently within the animal host. Growth at high salt concentrations and low temperatures is attributed mainly to the accumulation of organic solutes such as glycine betaine and carnitine. We utilized a novel system for generating chromosomal mutations (based on a lactococcal pWVO1-derived Ori+ RepA− vector, pORI19) to identify a listerial OpuC homologue. Mutating the operon in two strains of L. monocytogenes revealed significant strain variation in the observed activity of OpuC. Radiolabeled osmolyte uptake studies, together with growth experiments in defined media, linked OpuC to carnitine and glycine betaine uptake in Listeria. We also investigated the role of OpuC in contributing to the growth and survival of Listeria in an animal (murine) model of infection. Altering OpuC resulted in a significant reduction in the ability of Listeria to colonize the upper small intestine and cause subsequent systemic infection following peroral inoculation.
Survival of the food-borne pathogen Listeria monocytogenes, both at high salt concentrations (29) and in low-temperature environments (45), is attributed mainly to the accumulation of the organic compounds glycine betaine (N,N,N-trimethylglycine [21, 32]) and carnitine (β-hydroxy-γ-N-trimethyl aminobutyrate [5]).
The preferred compatible solute for the majority of bacteria (9, 10) and the most important osmolyte in L. monocytogenes is the trimethylammonium compound glycine betaine (4). Present at relatively high concentrations in foods of plant origin (14), it has been shown to stimulate the growth of L. monocytogenes between 0.3 to 0.7 M NaCl (2) and at temperatures as low as 4°C (21). Recent studies identified genes encoding two glycine betaine transport systems in Listeria. The first of these, betL (37, 39), encodes a single-component membrane-bound protein belonging to a family of secondary transporters of which OpuD of Bacillus subtilis (18) and BetP of Corynebacterium glutamicum (34) are members. Transporters in this family couple ion motive force to solute transport across the cell membrane (36). The second system, encoded by the gbuABC operon (22), is a multicomponent, binding protein-dependent transport system, forming part of a superfamily of prokaryotic and eukaryotic ATP-binding cassette transporters (15). Members of this family, including OpuA (20) and OpuC (ProU) (25) of B. subtilis, couple ATP hydrolysis to substrate translocation across biological membranes.
After glycine betaine, l-carnitine is regarded as the most effective osmolyte in L. monocytogenes (21, 43). Playing a role in fatty acid transport across the inner mitochondrial membrane (17), carnitine can be accumulated to concentrations of up to 50 mM in some animal tissues (6), approximately 5,000-fold more than the previously calculated Km value (10 μM) for Listeria (42). However, carnitine is not as effective as glycine betaine in contributing to either the salt or chill stress response of L. monocytogenes (21). Nonetheless, the relative abundance of carnitine in mammalian tissues (6) makes it the most readily available and thus possibly the most important osmolyte contributing to the survival of L. monocytogenes both in foods of animal origin (40) and during subsequent intracellular growth following infection (42).
In this report we describe the isolation of mutants of L. monocytogenes unable to utilize carnitine as an osmoprotectant, using a modification of the system outlined by Law et al. (24) for generating chromosomal mutations. The isolated mutants were shown to carry a copy of pORI19 inserted into a region of the chromosome with extensive homology to the recently identified opuC operon of L. monocytogenes (13) and were used to determine the importance of OpuC-encoded osmolyte uptake in contributing to the growth and survival of L. monocytogenes in an animal (murine) model of infection.
MATERIALS AND METHODS
Media, chemicals, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli EC101 was grown at 37°C in Luria-Bertani (LB) medium (27). L. monocytogenes strains were grown either in brain heart infusion (BHI) broth (Oxoid, Unipath Ltd., Basingstoke, United Kingdom) or on Listeria-selective agar (Oxoid). Blood agar plates consisted of blood agar (Lab M) to which 5% sheep blood was added following autoclaving. When a defined medium (DM) was required, the medium described by Premaratne et al. (35) was used. Where indicated, carnitine and glycine betaine (Sigma Chemical Co., St. Lous, Mo.) were added to DM as filter-sterilized solutions to a final concentration of 1 mM. Radiolabeled l-[N-methyl-14C]carnitine (50 to 62 mCi/mmol) and N,N,N-[1-14C]trimethylglycine were purchased from NEN Life Sciences Products (Hoofddorp, The Netherlands) and Campo Scientific (Veenendaal, The Netherlands), respectively. Erythromycin (ERY) and chloramphenicol (CHL) were made up as described by Maniatis et al. (27) as concentrated stocks and were added to media at the required levels. Where necessary the medium osmolarity was adjusted by the addition of NaCl.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| L. monocytogenes | ||
| LO28 | Serotype 1/2c | P. Cossart, Institut Pasteur |
| LO28G | LO28 containing pVE6007 | 37 |
| LO28C | LO28 opuC::pCPL5, OpuC− | This study |
| ScottA | Wild type | T. Abee |
| ScottAG | ScottA containing pVE6007 | This study |
| ScottAC | ScottA opuC::pCPL5, OpuC− | This study |
| E. coli EC101 | E. coli JM101 with repA from pWVO1 integrated in the chromosome | 24 |
| Plasmids | ||
| pORI19 | Emr Ori+ RepA− derivative of pORI28 | 24 |
| pVE6007 | Cmr temperature-sensitive derivative of pWVO1 | 26 |
| pCPL5 | pORI19 containing 1.1 kb of L. monocytogenes genomic DNA | This study |
DNA manipulations and sequence analysis.
Routine DNA manipulations were performed as described by Maniatis et al. (27). Genomic DNA was isolated from L. monocytogenes by the method of Hoffman and Winston (16). Plasmid DNA was isolated using the Qiagen QIAprep Spin Miniprep Kit (Qiagen, Hilden, Federal Republic of Germany). E. coli was transformed by standard methods (27), while electrotransformation of L. monocytogenes was achieved by the protocol outlined by Park and Stewart (31). PCR reagents (Taq polymerase and deoxynucleoside triphosphates) were purchased from Boehringer and used according to the manufacturer's instructions with a Hybaid (Middlesex, United Kingdom) PCR express system. Where mentioned, colony PCR was carried out following cell lysis with Igepal CA-630 (Sigma). Oligonucleotide primers for PCR and sequence purposes were synthesized on a Beckman oligonucleotide 1000M DNA synthesizer (Beckman Instruments Inc., Fullerton, Calif.). Nucleotide sequence determination was performed on a Beckman CEQ 2000 DNA analysis system. Homology searches were performed against the GenBank database using the BLAST program (1).
Creation of a pORI19 integration bank in L. monocytogenes LO28.
A bank of L. monocytogenes LO28::pORI19 insertion mutants was generated essentially as described by Law et al. (24) with some minor modifications. A genomic DNA preparation from L. monocytogenes LO28 was partially digested with EcoRI and ligated to the Ori+ RepA− plasmid pORI19, which had been digested with EcoRI and dephosphorylated with shrimp alkaline phosphatase. The resulting recombinant plasmids were transformed into E. coli EC101 (RepA+), and colonies were selected on LB plates containing ERY (250 μg/ml), IPTG (isopropyl-1-β-d-thiogalactopyranoside) (1 mM), and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-gulactopyranoside) (40 μg/ml). Transformants were pooled and grown with shaking for 2 h in LB broth containing ERY (250 μg/ml). Plasmid DNA was then extracted and used to transform L. monocytogenes LO28G (LO28 harboring the temperature-sensitive RepA+ helper plasmid, pVE6007 [37]). Immediately following transformation, cells were incubated in BHI broth containing ERY (50 ng/ml) at 30°C for 180 min (to induce expression of Emr-encoding genes). To induce loss of pVE6007 and forced chromosomal integration of pORI19 at the points of homology with the cloned insert, 100 μl of the transformation mix was used to inoculate 10 ml of BHI broth prewarmed to 42°C (the nonpermissive temperature for pVE6007 replication in Listeria). Following overnight incubation at 42°C, transformants were plated onto prewarmed BHI-ERY plates and incubated at 42°C for 48 h. Loss of pVE6007 was confirmed by lack of growth of the transformants on BHI-CHL plates coupled with an inability to isolate replicating plasmids from the cytosol.
Isolation of osmolyte uptake mutants of L. monocytogenes LO28.
Putative osmolyte-deficient transport mutants were isolated by screening the pORI19 insertion mutant bank (by replica plating) on DM, DM plus 3% NaCl (DMS), DMS plus 1 mM carnitine (DMSC), and DMS plus 1 mM glycine betaine (DMSB). Mutants were confirmed by being restreaked onto DM agar plates to which either salt (3% [wt/vol]) or salt plus carnitine or glycine betaine (1 mM) was added.
Identification of disrupted genes.
The isolated osmolyte uptake mutants were electroporated with the RepA+ helper plasmid, pVE6007, recovered at 30°C on BHI-ERY-CHL plates, and passaged subsequently in BHI-ERY-CHL broth at 30°C. Inserts on the rescued plasmids, amplified by PCR with the Pharmacia (Uppsala, Sweden) universal and reverse primers, were subjected to restriction analysis before a representative plasmid (designated pCPL5) was chosen for sequence determination and homology studies.
Generation of L. monocytogenes LO28 and ScottA::pCPL5 insertion mutants.
L. monocytogenes strains LO28G and ScottAG (containing pVE6007) were transformed with pCPL5, and transformants were selected on BHI-ERY-CHL plates at 30°C. As before, temperature upshift from 30 to 42°C while selecting for ERY resistance resulted in the loss of pVE6007 and targeted chromosomal integration of pCPL5. Loss of pVE6007 was established by sensitivity to CHL, while chromosomal integration of pCPL5 was confirmed by PCR.
Uptake studies.
Radiolabeled osmolyte uptake studies were carried out essentially as described by Verheul et al. (43).
Virulence assays.
Bacterial virulence was determined by intraperitoneal and peroral inoculation of 8- to 12-week-old BALB/c mice. Intraperitoneal inoculations were carried out as described previously (39) using overnight cultures (6.5 × 105 cells) of mutant and wild-type Listeria suspended in 0.2 ml of phosphate-buffered saline. For peroral inoculations, mutant and wild-type strains suspended in buffered saline with gelatin (0.85% NaCl, 0.01% gelatin, 2.2 mM K2HPO4, and 4.2 mM Na2HPO4) were mixed at a ratio of 1:1 for LO28:LO28C and ScottA:ScottAC. Mice were infected with approximately 1010 cells (total) using a micropipette tip placed immediately behind the incisors. Three days postinfection mice were euthanatized and the listerial numbers in the small intestine wall and contents, Peyer's patches, liver, and spleen were determined by spread plating homogenized samples onto BHI and blood agar, with and without added ERY (5 μg/ml).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this study have been submitted to GenBank and assigned accession no. AF211851.
RESULTS
Generation and screening of an L. monocytogenes LO28::pORI19 insertion bank.
A genomic bank of L. monocytogenes LO28 was initially created in E. coli EC101 using the vector plasmid pORI19 as described in Materials and Methods. Analysis of the bank of 25,000 clones indicated that over 90% contained inserts with an average insert size of 1.5 kb (range, 500 bp to 2.5 kb). This number of clones is estimated to give more than 10× coverage of the entire LO28 genome (using a value of 3 Mb for the genome [44]). A plasmid bank was isolated from the EC101 clone set and electrotransformed into strain LO28G (a derivative of LO28 containing the helper plasmid pVE6007). A temperature upshift from 30°C to the nonpermissive 42°C, 180 min postelectroporation, resulted in transformation efficiencies of approximately 103 CFU/μg of plasmid DNA. Random transformants were screened and proved to be ERY resistant and CHL sensitive.
The parental strain LO28 can grow on DM but not on DMS. However, the addition of osmolytes to create DMSC or DMSB permits the growth of LO28. Screening approximately 2,000 colonies by replica plating led to the isolation of two isolates which grew on DM and DMSB but were incapable of growth on DMS or DMSC.
Restriction analysis of the pORI19 clones from both isolates following plasmid rescue from the chromosome revealed that both contained the same 1.1-kb insert; one such plasmid was chosen and designated pCPL5. Reintegration of pCPL5 into an LO28 wild-type background generated the same mutant phenotype, thus confirming the role of the inserted fragment in the observed phenotype. A representative mutant, designated LO28C, was chosen for further characterization. In addition, pCPL5 was used to create the corresponding mutant in L. monocytogenes ScottA, designated ScottAC. The stability of plasmid insertion in both mutants was confirmed by PCR analysis of cultures grown in the absence of Em at 30°C. No plasmid excision was observed, even after repeated subculture in the absence of antibiotic selection.
Genotypic analysis of the cloned insert on pCPL5.
Homology searches revealed significant similarity both at the nucleotide (99% identity) and protein level to the recently identified OpuC multicomponent osmolyte uptake system (comprised of opuCA-opuCB-opuCC-opuCD) in L. monocytogenes EGD reported by Fraser et al. (13). Further analysis of the 1.1-kb insert and surrounding chromosomal DNA confirmed that pORI19 had inserted into the opuCB gene in LO28 and that the gene organization reported for EGD is conserved in both LO28 and ScottA.
Physiological analysis of the listerial OpuC− mutants.
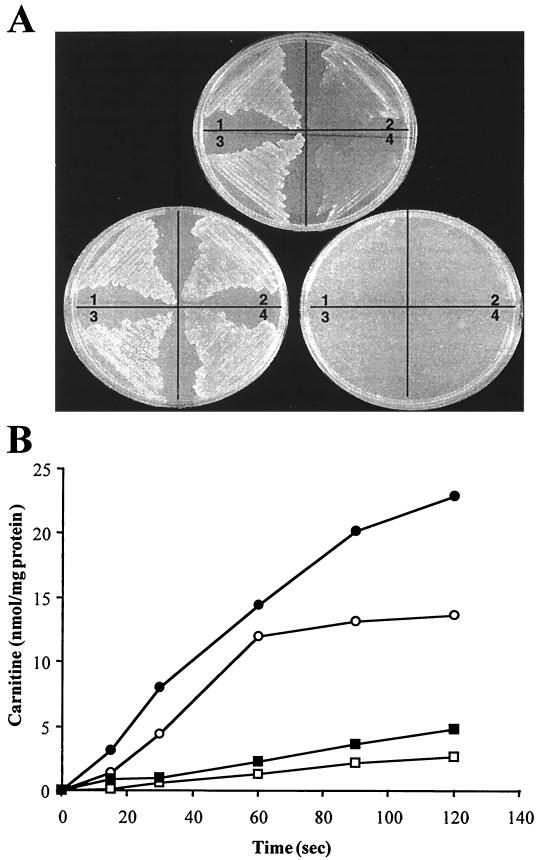
Inactivation of the Listeria opuC operon following pCPL5 insertion dramatically reduced the osmoprotective effects of carnitine, but not glycine betaine, on the growth of Listeria (both LO28 and ScottA) in DM of elevated osmolarity (Fig. 1A). Radiolabeled uptake studies revealed a dramatic reduction in the observed rates of carnitine uptake for ScottAC as expected, both in the presence and absence of salt stress, relative to the wild-type parent strain (Fig. 1B). However, we were only able to detect very low levels of carnitine uptake for the LO28 parent strain (∼10-fold lower than that for ScottA) under identical conditions.
FIG. 1.
(A) Growth of the OpuC− mutants, LO28C and ScottAC, relative to the parental wild-type strains on DM of elevated osmolarity. Plate A consists of DM containing 3% added NaCl plus 1 mM carnitine (DMSC), plate B consists of DMS plus 1 mM glycine betaine (DMSB), and plate C consists of DMS with no added osmolytes. For each plate, 1 is ScottA wild type, 2 is ScottAC, 3 is LO28 wild type, and 4 is LO28C. Clones were grown overnight in DM before being streaked onto the appropriate test plate; the photograph represents growth after 24 h. (B) l-Carnitine transport. ScottA (○, ●) and ScottAC (□, ■) were assayed for l-[N-methyl-14C] carnitine uptake, both in the presence (closed symbols) and absence (open symbols) of 3% NaCl.
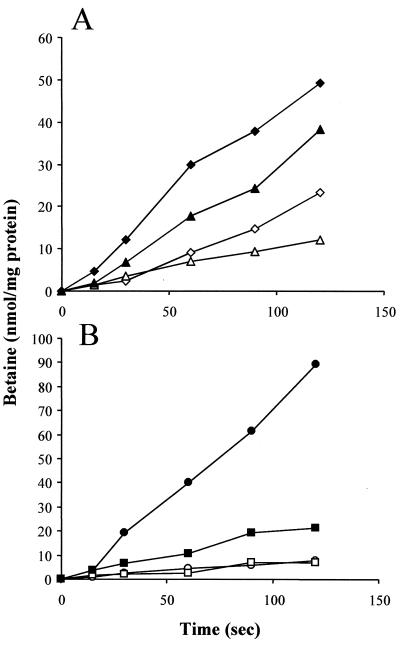
Since the OpuC homologue in B. subtilis is known to function in glycine betaine uptake (18, 25), both LO28C and ScottAC were analyzed for their ability to transport glycine betaine. In this instance, glycine betaine uptake was observed in both strains. While LO28C exhibited reduced glycine betaine uptake relative to the parent strain, at both reduced and elevated osmolarities (Fig. 2A) the ScottAC mutant appeared affected only in its ability to transport glycine betaine at high salt concentrations (Fig. 2B). These findings not only stress the importance of OpuC in contributing to osmolyte uptake but also serve to highlight significant strain variation in relation to osmolyte utilization in Listeria, a phenomenon previously observed by Dykes and Moorehead (12).
FIG. 2.
Glycine betaine transport. LO28C (▵, ▴) (A) and ScottAC (□, ■) (B) were assayed for N,N,N-[1-14C]trimethylglycine uptake either in the presence (closed symbols) or absence (open symbols) of 3% NaCl. The parental wild-type strains LO28 (◊, ⧫) and ScottA (○, ●) in the presence (closed symbols) or abscence (open symbols) of 3% NaCl are also represented.
Virulence studies.
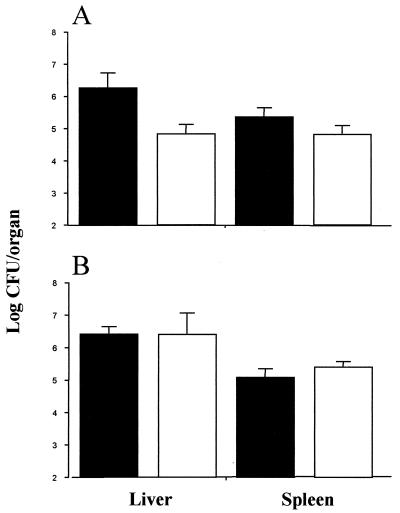
Our original premise was that carnitine may prove to be an important osmolyte for Listeria during infection. Therefore, in order to determine the effect of altering OpuC on the virulence of L. monocytogenes, strains were subjected to mouse virulence assays. The LO28C mutant strain reached significantly (P < 0.05) lower levels than the wild type in the livers and spleens of intraperitoneally infected animals after 3 days. Numbers of the mutant in infected spleens were more than threefold lower than those of the wild type, while numbers in the liver were over 20-fold lower than those of the parent strain (Fig. 3). However, in contrast, altering OpuC in L. monocytogenes ScottA had no significant effect on virulence following intraperitoneal infection.
FIG. 3.
Effect of altering OpuC on the survival of LO28C (A) and ScottAC (B) relative to that of the parent wild-type strains following intraperitoneal inoculation. Levels of Listeria in the livers and spleens of infected mice 3 days postinfection are shown (n = 4). Solid bars represent wild-type strains and open bars represent mutant strains.
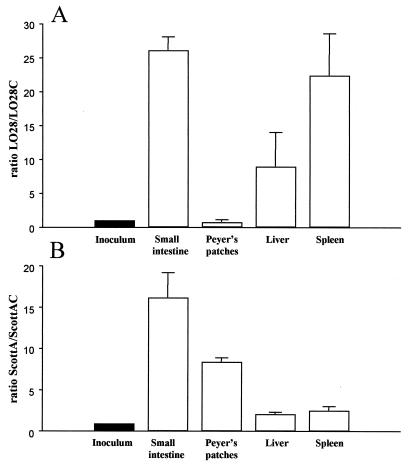
Since the lumen of the gastrointestinal tract (previously suggested to function as the human resevoir of the organism [28]) has an osmolarity approximately equal to 0.3 M NaCl (8), we examined the ability of the osmolyte uptake mutants LO28C and ScottAC to colonize the upper small intestine and cause systemic infection. Mice were coinoculated perorally with a 1:1 ratio of wild-type and mutant strains, and the numbers of mutant (Emr) and total bacteria were determined 3 days postinfection in the upper small intestine, Peyer's patches, liver, and spleen. The use of bacterial coinfection allowed direct comparison between mutant and parent strains in individual mice. Altering OpuC in the LO28 background significantly impaired the ability of this strain to colonize the small intestine (Fig. 4A). The ability of LO28C to infect mouse livers and spleens was also greatly reduced relative to that of the wild type via the oral route. Similarly, inactivation of this locus in ScottA impaired the capacity of the organism to colonize the small intestine and to subsequently replicate in Peyer's patches. Resultant infection and growth in organs was also reduced relative to that of the parent strain (Fig. 4B). However, in comparison to LO28C, ScottAC was only marginally affected in its ability to grow in the host liver and spleen following peroral infection, a result which reflects the data obtained following intraperitoneal infection (Fig. 3). The observed differences in the infectivity of the mutants (particularly ScottAC) when administered via either the intraperitoneal or peroral route mirror results obtained for salt-sensitive mutants of Salmonella enterica serovar Typhimurium (7). Chatfield et al. (7) proposed that the observed difference in virulence following intragastric as opposed to intraperitoneal inoculation reflects the osmotic stress imposed on the bacterium when entering the host by the oral route. The osmolarity of the intestinal lumen is equivalent to 0.3 M NaCl, while in the bloodstream bacteria encounter an osmolarity equivalent to only 0.15 M NaCl (8). Collectively, the results presented here suggest that OpuC is essential for efficient colonization of the small intestine and resulting systemic infection by L. monocytogenes.
FIG. 4.
Survival of LO28 relative to LO28C (A) and ScottA relative to ScottAC (B) following peroral coinoculation of BALB/c mice. The ratio of the strains was determined for both the inoculum (■) and the relevant tissues and organs (□) 3 days postinfection (n = 4).
DISCUSSION
Molecular characterization of the salt tolerance of L. monocytogenes has been the focus of much attention in recent times (13, 22, 37, 38, 41). Combined with previous physiological investigations, genetic analysis has provided new insights into the mechanisms of listerial osmotolerance. Glycine betaine, for example, previously assumed to be accumulated only by a single uptake system (33), is now known to be transported by at least three independent systems (22, 37, 39).
Originally identified as a chimeric proU operon conferring enhanced osmoprotection as a consequence of glycine betaine transport in B. subtilis LH45 (25), the opuC operon also encodes the only osmotically significant carnitine transporter in B. subtilis (19). Sequence analysis downstream of a recently constructed Tn1545 adhesion mutant (30) identified the opuC operon in L. monocytogenes EGD, the complete sequence of which has since been reported by Fraser et al. (13). In the present study, functional inactivation of this homologue in two distinct strains of Listeria, namely LO28 and ScottA, resulted in mutants exhibiting reduced glycine betaine uptake and an inability to use carnitine as an effective osmoprotectant. Uptake studies using radiolabeled substrate revealed significant variation in the observed rates of glycine betaine and carnitine transport not only between the mutants but also between the parental wild-type strains. Not restricted to Listeria (12), this phenomenon of strain variation in relation to osmolyte transport systems has previously been described in Bacillus. Disrupting the opuC operon in B. subtilis LH45 significantly reduces osmoprotection by glycine betaine (25), whereas a similar mutation in B. subtilis JH642 has only a minor effect on glycine betaine uptake (18).
The low levels of carnitine uptake observed for wild-type LO28 may reflect the absence of a dedicated carnitine transport system in this strain. The isolated opuC operon thus may encode a leaky system, which although primarily dedicated to the uptake of glycine betaine, transports the structurally related trimethyl amino acid carnitine at a level which, while too low to be detected under the conditions used in our assays, is nonetheless physiologically significant in terms of salt tolerance. Alternatively, the effect of altering OpuC on glycine betaine uptake may be indirect, and the low levels of carnitine uptake for LO28 may merely reflect strain-specific differences in gene expression. While uptake studies revealed a possible role for OpuC in the transport of glycine betaine for both strains tested, disrupting the operon had no significant effect on glycine betaine-mediated osmoprotection. Given that a number of nucleotide changes (one of which resulted in an amino acid substitution) were observed between the 1.1-kb insert of pCPL5 and the opuC sequence of EGD, it is tempting to speculate that the observed strain variation in the activity of OpuC is the consequence of strain-specific point mutations within the operon.
For many food-borne pathogens the ability to sense and respond to the high osmolarity of the gastrointestinal lumen is a key component of virulence. The shift in osmolarity between the external aqueous environment and the small intestine functions to trigger the synthesis of virulence factors essential for subsequent pathogenesis (8). In addition, in order to survive and grow in the lumen of the gastrointestinal tract bacteria must adapt to an environment with an osmolarity equivalent to 0.3 M NaCl (8), the concentration at which maximum carnitine uptake occurs in Listeria (40). We have determined that L. monocytogenes mutants in OpuC survive poorly in the upper small intestine, thus suggesting that carnitine may represent a key osmoprotectant facilitating growth in this otherwise limiting environment. The constant breakdown of the gastrointestinal epithelial layer (desquamation) may provide the source of carnitine for uptake by bacteria in this milieu of elevated osmolarity.
For OpuC− mutants in both LO28 and ScottA backgrounds, the reduced ability to colonize the small intestine is mirrored by lower bacterial levels in internal organs. This is especially evident for the OpuC− mutant in LO28 which demonstrates ∼20-fold lower levels in infected spleens relative to the parent strain. Interestingly, LO28C, but not ScottAC, exhibits reduced virulence when administered by the intraperitoneal route. This suggests that the ScottA strain either possesses a carnitine transporter other than OpuC (evidenced by the NaCl-inducible carnitine uptake observed against the OpuC− background of ScottAC [Fig. 1B]) or relies on mechanisms other than carnitine uptake to maintain turgor pressure during infection of internal organs. In contrast, the role of OpuC in LO28 is of key importance for efficient survival and growth in vivo. Barbour et al. (3) have previously shown significant variation in virulence of L. monocytogenes strains. Our data suggest that L. monocytogenes strains may differ in their reliance on specific systems for maintaining homeostasis in vivo.
ACKNOWLEDGMENT
We gratefully acknowledge the financial support of the National Food Biotechnology Centre, BioResearch Ireland.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amezaga M R, Davidson I, McLaggan D, Verheul A, Abee T, Booth I R. The role of peptide metabolism in the growth of Listeria monocytogenes ATCC 23074 at high osmolarity. Microbiology. 1995;141:41–49. doi: 10.1099/00221287-141-1-41. [DOI] [PubMed] [Google Scholar]
- 3.Barbour A H, Rampling A, Hormaeche C E. Comparison of the infectivity of isolates of L. monocytogenes following intragastric and intravenous inoculation in mice. Microb Pathog. 1996;20:247–253. doi: 10.1006/mpat.1996.0023. [DOI] [PubMed] [Google Scholar]
- 4.Bayles D O, Wilkinson B J. Osmoprotectants and cryoprotectants for Listeria monocytogenes. Lett Appl Microbiol. 2000;30:23–27. doi: 10.1046/j.1472-765x.2000.00646.x. [DOI] [PubMed] [Google Scholar]
- 5.Beumer R R, Te Giffel M C, Cox L J, Rombouts F M, Abee T. Effect of exogenous proline, betaine, and carnitine on growth of Listeria monocytogenes in a minimal medium. Appl Environ Microbiol. 1994;60:1359–1363. doi: 10.1128/aem.60.4.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bieber L L. Carnitine. Annu Rev Biochem. 1988;57:261–283. doi: 10.1146/annurev.bi.57.070188.001401. [DOI] [PubMed] [Google Scholar]
- 7.Chatfield S T, Dorman C J, Hayward C, Dougan G. Role of ompR-dependent genes in Salmonella typhimurium virulence: mutants deficient in both OmpC and OmpF are attenuated in vivo. Infect Immun. 1991;59:449–452. doi: 10.1128/iai.59.1.449-452.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury R, Sahu G K, Das J. Stress response in pathogenic bacteria. J Biosci. 1996;21:149–160. [Google Scholar]
- 9.Csonka L N. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev. 1989;55:476–511. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Csonka L N, Hanson A D. Prokaryotic osmoregulation: genetics and physiology. Annu Rev Microbiol. 1991;45:569–606. doi: 10.1146/annurev.mi.45.100191.003033. [DOI] [PubMed] [Google Scholar]
- 11.Davis B D, Mingioli E S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950;60:17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dykes G A, Moorhead S M. Survival of osmotic and acid stress by Listeria monocytogenes strains of clinical or meat origin. Int J Food Microbiol. 2000;56:161–166. doi: 10.1016/s0168-1605(99)00205-6. [DOI] [PubMed] [Google Scholar]
- 13.Fraser K R, Harvie D, Coote P J, O'Byrne C P. Identification and characterization of an ATP binding cassette l-carnitine transporter in Listeria monocytogenes. Appl Environ Microbiol. 2000;66:4696–4704. doi: 10.1128/aem.66.11.4696-4704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen A D, Rathinasabapathi B, Rivoal J, Burnet M, Dillon M O, Gage D A. Osmoprotective compounds in the plumbaginacease: a natural experiment in metabolic engineering of stress tolerance. Proc Natl Acad Sci USA. 1994;91:306–310. doi: 10.1073/pnas.91.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins C F. ABC transporters: from micro-organisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman C S, Winston F. Rapid DNA extraction procedure. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 17.Idell-Wenger J A. Carnitine: acylcarnitine translocase of rat heart mitochondria. J Biol Chem. 1981;256:5597–5603. [PubMed] [Google Scholar]
- 18.Kappes R M, Kempf B, Bremer E. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J Bacteriol. 1996;178:5071–5079. doi: 10.1128/jb.178.17.5071-5079.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kappes R M, Bremer E. Response of Bacillus subtilis to high osmolarity: uptake of carnitine, crotonobetaine and γ-butyrobetaine via the ABC transport system OpuC. Microbiology. 1998;144:83–90. doi: 10.1099/00221287-144-1-83. [DOI] [PubMed] [Google Scholar]
- 20.Kempf B, Bremer E. OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. J Bacteriol. 1995;270:16701–16713. doi: 10.1074/jbc.270.28.16701. [DOI] [PubMed] [Google Scholar]
- 21.Ko R, Smith L T, Smith G M. Glycine betaine confers enhanced osmotolerance and cryotolerance on Listeria monocytogenes. J Bacteriol. 1994;176:426–431. doi: 10.1128/jb.176.2.426-431.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ko R, Smith L T. Identification of an ATP-driven, osmoregulated glycine betaine transport system in Listeria monocytogenes. Appl Environ Microbiol. 1999;65:4040–4048. doi: 10.1128/aem.65.9.4040-4048.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;177:6874–6880. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 24.Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol. 1995;177:7011–7018. doi: 10.1128/jb.177.24.7011-7018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Y, Hansen J N. Characterization of a chimeric proU operon in a subtilin-producing mutant of Bacillus subtilis 168. J Bacteriol. 1995;177:6874–6880. doi: 10.1128/jb.177.23.6874-6880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. New thermosensitive plasmid for gram positive bacteria. J Bacteriol. 1992;174:5633–5638. doi: 10.1128/jb.174.17.5633-5638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 28.Marco A J, Altimira J, Prats N, López S, Dominguez L, Domingo M, Briones V. Penetration of Listeria monocytogenes in mice infected by the oral route. Microb Pathog. 1997;23:255–263. doi: 10.1006/mpat.1997.0144. [DOI] [PubMed] [Google Scholar]
- 29.McClure P J, Roberts T A, Oguru P O. Comparison of the effects of sodium chloride, pH and temperature on the growth of Listeria monocytogenes on gradient plates and liquid medium. Lett Appl Microbiol. 1989;9:95–99. [Google Scholar]
- 30.Milohanic E, Pron B, Berche P, Gaillard J-L the European Listeria Genome Consortium. Identification of new loci involved in adhesion of Listeria monocytogenes to eukaryotic cells. Microbiology. 2000;146:731–739. doi: 10.1099/00221287-146-3-731. [DOI] [PubMed] [Google Scholar]
- 31.Park F P, Stewart G S A B. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin treated cells. Gene. 1990;94:129–132. doi: 10.1016/0378-1119(90)90479-b. [DOI] [PubMed] [Google Scholar]
- 32.Patchett R A, Kelly A F, Kroll R G. Effect of sodium chloride on the intracellular solute pools of Listeria monocytogenes. Appl Environ Microbiol. 1992;58:3959–3963. doi: 10.1128/aem.58.12.3959-3963.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patchett R A, Kelly A F, Kroll R G. Transport of glycine betaine by Listeria monocytogenes. Arch Microbiol. 1994;162:205–210. doi: 10.1007/BF00314476. [DOI] [PubMed] [Google Scholar]
- 34.Peter H, Burkovski A, Krämer R. Isolation, characterization, and expression of the Corynebacterium glutamicum betP gene, encoding the transport system for the compatible solute glycine betaine. J Bacteriol. 1996;178:5229–5234. doi: 10.1128/jb.178.17.5229-5234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Premaratne R J, Lin W-J, Johnson E A. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl Environ Microbiol. 1991;57:3046–3048. doi: 10.1128/aem.57.10.3046-3048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reizner J, Reizner A, Saier M H. A functional superfamily of sodium/solute symporters. Biochim Biophys Acta. 1994;1197:133–166. doi: 10.1016/0304-4157(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 37.Sleator R D, Gahan C G M, Abee T, Hill C. Identification and disruption of BetL, a secondary glycine betaine transport system linked to the salt tolerance of Listeria monocytogenes LO28. Appl Environ Microbiol. 1999;65:2078–2083. doi: 10.1128/aem.65.5.2078-2083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sleator R D, Gahan C G M, Hill C. Molecular characterisation of the salt tolerance of Listeria monocytogenes LO28. In: Tuijtelaars A C J, Samson R A, Rombouts F M, Notermans S, editors. Food microbiology and food safety into the next millennium. Foundation Food Micro'99. Zeist, The Netherlands: TNO Nutrition and Food Research Institute; 1999. [Google Scholar]
- 39.Sleator R D, Gahan C G M, O'Driscoll B, Hill C. Analysis of the role of betL in contributing to the growth and survival of Listeria monocytogenes LO28. Int J Food Microbiol. 2000;60:261–268. doi: 10.1016/s0168-1605(00)00316-0. [DOI] [PubMed] [Google Scholar]
- 40.Smith L T. Role of osmolytes in adaptation of osmotically stressed and chill-stressed Listeria monocytogenes grown in liquid media and on processed meat surfaces. Appl Environ Microbiol. 1996;62:3088–3093. doi: 10.1128/aem.62.9.3088-3093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith G M, Smith L T, Gerhardt P N M, Ko R. Solute transport enzymes related to stress tolerance in Listeria monocytogenes: a review. J Food Biochem. 1998;22:269–285. [Google Scholar]
- 42.Verheul A, Rombouts F M, Beumer R R, Abee T. An ATP-dependent l-carnitine transporter in Listeria monocytogenes ScottA is involved in osmoprotection. J Bacteriol. 1995;177:3205–3212. doi: 10.1128/jb.177.11.3205-3212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verheul A, Glaasker E, Poolman B, Abee T. Betaine and l-carnitine transport by Listeria monocytogenes ScottA in response to osmotic signals. J Bacteriol. 1997;179:16979–16985. doi: 10.1128/jb.179.22.6979-6985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Von Both U, Otten S, Darbouche A, Domann E, Chakraborty T. Physical and genetic map of the Listeria mononcytogenes EGD serotype 1/2a chromosome. FEMS Microbiol Lett. 1999;175:281–289. doi: 10.1111/j.1574-6968.1999.tb13632.x. [DOI] [PubMed] [Google Scholar]
- 45.Walker S J, Archer P, Banks J G. Growth of Listeria monocytogenes at refrigeration temperatures. J Appl Bacteriol. 1990;68:157–162. doi: 10.1111/j.1365-2672.1990.tb02561.x. [DOI] [PubMed] [Google Scholar]