SUMMARY
Assessment of donor kidney quality is based on clinical scores or requires biopsies for histological assessment. Noninvasive strategies to identify and predict graft outcome at an early stage are, therefore, needed. We evaluated the perfusate of donation after brain death (DBD) kidneys during nonoxygenated hypothermic machine perfusion (HMP). In particular, we compared perfusate protein profiles of good outcome (GO) and suboptimal outcome (SO) 1‐year post‐transplantation. Samples taken 15 min after the start HMP (T1) and before the termination of HMP (T2) were analysed using quantitative liquid chromatography–tandem mass spectrometry (LC‐MS/MS). Hierarchical clustering of the 100 most abundant proteins showed discrimination between grafts with a GO and SO at T1. Elevated levels of proteins involved in classical complement cascades at both T1 and T2 and a reduced abundance of lipid metabolism at T1 and of cytoskeletal proteins at T2 in GO versus SO was observed. ATP‐citrate synthase and fatty acid‐binding protein 5 (T1) and immunoglobulin heavy variable 2‐26 and desmoplakin (T2) showed 91% and 86% predictive values, respectively, for transplant outcome. Taken together, DBD kidney HMP perfusate profiles can distinguish between outcome 1‐year post‐transplantation. Furthermore, it provides insights into mechanisms that could play a role in post‐transplant outcomes.
Keywords: biomarker discovery, complement activation, donation after brain death, hypothermic machine perfusion, kidney preservation, proteomics
Experimental design, workflow and results. Elevated complement, cytoskeleton and lipid metabolism proteins were identified.

Introduction
Kidney transplantation is the only curative treatment for patients suffering from end‐stage renal disease (ESRD) as it can permanently replace a patient’s kidney function. In 2019, almost 100 000 kidney transplantations were performed globally, and it is the most transplanted organ worldwide. [1]
Currently, donation after brain death (DBD) donors are the main source for donor kidneys [2]. Brain death causes various pathophysiological effects in the donor such as haemodynamic instability due to a severe reduction in blood flow, immune system activation including a cytokine storm and complement activation followed by leucocyte infiltration [3, 4. These effects have shown to have a negative impact on graft outcome [5].
Standard criteria donors (SCD) are preferred as they show favourable outcomes compared with other donor types [6, 7. The continuing shortage of donor organs forces the use of organs from donors that do not meet the standard criteria such as extended criteria donors (ECD). ECD are DBD with an age above 60, or an age above 50 with two of the following characteristics: history of hypertension, terminal serum creatinine levels of ≥ 1.5 mg/dl or cerebrovascular cause of death [8, 9. Unfortunately, ECD kidney grafts have worse outcomes compared with SCD [9, 10, 11, 12.
With the increased use of organs from ECD, the decision‐making process for suitable donor organs is becoming even more critical. Current strategies to assess donor organ quality are based on clinical characteristics like the kidney donor risk score or kidney donor profile index (KDPI), [13] 14] or require a biopsy for histological assessment. However, taking biopsies may cause unnecessary injury to the kidney, and the obtained information is intrinsically representative of a very focal site. Hence, quality assessment using biopsies is under debate [15]. Due to such limitations, the quality of the graft cannot always be correctly assessed, causing many organs to be discarded, possibly unnecessarily. Therefore, noninvasive strategies to identify and predict graft outcome are highly needed.
As of 2016, all donor kidneys derived from deceased donors in the Netherlands are preserved by means of nonoxygenated hypothermic machine perfusion (HMP) instead of static cold storage (SCS) [16]. HMP is a preservation method that allows continuous perfusion of the graft with perfusion solution in the time frame between donation and transplantation. The use of HMP itself is associated with improved graft outcome [17, 18, 19, 20, 21, 22. Furthermore, it creates a new horizon for measuring secreted predictive kidney graft biomarkers in a noninvasive manner, providing the opportunity to evaluate graft quality before transplantation. Several studies have demonstrated that organ preservation solution is a potential source for biomarkers associated with post‐transplantation organ function, although these studies did not look at HMP perfusate or only quantified selected proteins [23, 24, 25.
Our aim was to perform a proteomic perfusate analysis of DBD kidneys preserved using HMP by identifying the differences between proteomic profiles of kidneys with a good and suboptimal outcome 1‐year post‐transplantation. Furthermore, our aim was to identify potential biomarkers for organ quality and unravel underlying pathways that influence transplant outcome using an unbiased proteomics approach.
Materials and methods
Characteristics of donor cohort
Twenty‐two DBD kidneys with a good or suboptimal outcome 1‐year post‐transplantation were selected (n = 11 per group) that were transplanted between 11 January 2016 and 31 December 2017. Good outcome (GO) was defined as kidney function with an estimated glomerular filtration rate (eGFR) ≥ 45 ml/min/1.73 m2 and a serum creatinine level of ≤ 120 µmol/l, whereas suboptimal outcome (SO) was defined as kidneys with an eGFR ≤ 45 ml/min/1.73 m2 and a serum creatinine level of ≥ 120 µmol/l 1‐year post‐transplantation (Fig. 1). Selected kidneys were matched on various donor and recipient characteristics (Table 1). Characteristics were analysed using SPSS statistics 28. Noncategorical variables were tested for normal distribution using the Shapiro–Wilk normality test. Two‐sample t test or Mann–Whitney U test was used to test for two independent groups of continuous variables. Chi‐square test or Fisher’s exact test were used for discrete variables. Exclusion criteria were kidneys allocated to recipients receiving a multi‐organ transplant and donors with acute kidney injury.
Figure 1.
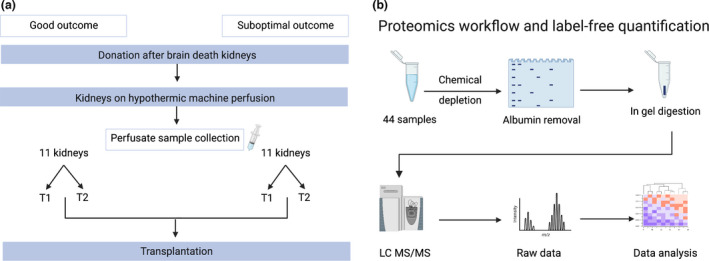
Experimental design and workflow. (a) Overview of kidney perfusate sample collection. Perfusate samples were selected on the basis of 1‐year transplant outcomes. Good outcome (GO) was defined as kidney function with an estimated glomerular filtration rate (eGFR) ≥ 45 ml/min/1.73 m2 and a serum creatinine level of ≤ 120 µmol/l, whereas suboptimal outcome (SO) was defined as kidneys with an eGFR ≤ 45 ml/min/1.73 m2 and a serum creatinine level of ≥ 120 µmol/l. Samples from each kidney were taken at 2 time points: 15 min after the start of HMP (T1) and at the end of HMP (T2). (b) Workflow for proteomics analysis. A total of 44 samples were analysed (11 kidneys per group, 22 samples per outcome). Perfusate samples were chemically depleted and loaded onto a gel for SDS‐PAGE. Albumin bands were removed, and samples were prepared for in‐gel digestion, desalted and analysed using LC‐MS/MS. Raw peaks and spectra were searched against Homo sapiens database and data were analysed. eGFR, estimated glomerular filtration rate; LC‐MS/MS, liquid chromatography–tandem mass spectrometry. (Image created using biorender.com).
Table 1.
Characteristics of donor and recipients.
| Donor | Good Outcome | Suboptimal Outcome | P ‐value |
|---|---|---|---|
| Age y | 53.6 (6.2) | 57.2 (6.2) | 0.195 |
| Male [n (%)] | 7 (63.6%) | 3 (27.3%) | 0.198 |
| BMI [kg/m−2] | 25.9 (4.9) | 26.8 (4.8) | 0.689 |
| History of hypertension [n (%)] | 5 (45.5%) | 4 (36.4%) | 1.000 |
| Terminal serum creatinine | 60.2 (16.6) | 68.6 (24.1) | 0.349 |
| Recipient | Good Outcome | Suboptimal Outcome | P ‐value |
| Age y | 51.6 (9.1) | 57.6(10.8) | 0.174 |
| Male [n (%)] | 7 (63.6%) | 8 (72.7%) | 1.000 |
| BMI [kg/m−2] | 24.0 (3.0) | 25.5 (3.1) | 0.373 |
| ≥3 HLA mismatches [n (%)] | 9 (81.8%) | 6 (54.5%) | 0.361 |
| Ischaemia times | |||
| Total CIT (minutes) | 969.9 (386.6) | 689.18 (323.7) | 0.086 |
| WIT 2 (minutes) | 32.4 | 37.8 (10.3) | 0.264 |
| Renal Outcome | Good Outcome | Suboptimal Outcome | P‐value |
| DGF | 0 (0%) | 5 (45.5%) | 0.035 |
| 3‐month serum creatinine | 95 (88–116) | 189 (176–189) | 0.000 |
| 3‐month eGFR | 68.6 (17.0) | 28 (12.0) | 0.000 |
| 12‐month serum creatinine | 103 (95–117) | 196 (182–230) | 0.000 |
| 12‐month eGFR | 67.4 (15.5) | 27.6 (9.7) | 0.000 |
Data are given as mean (SD), median (IQR) or n (%). n, number in group; BMI, body mass index; HLA, human leukocytes antigens; CIT, cold ischaemia time; WIT2, warm ischaemia time 2; DGF, delayed graft function.
Sample collection and storage
Nonoxygenated HMP was performed using either the LifePort Kidney Transporter machine (organ recovery systems) with a mean arterial pressure of 30 mmHg or the kidney assist transporter (organ assist) with a mean arterial pressure of 25 mmHg and a temperature between 1 and 8°C. Five hundred millilitres of University of Wisconsin machine perfusion solution (UW‐MPS; Bridge to Life) was used as the preservation solution. A total of 44 perfusate samples were collected; after 15 min of HMP (T1) and just before ending HMP (T2). The perfusate was centrifuged at 1000 g. for 12 min at 4°C. The supernatant was aliquoted and stored at −80°C until proteomic analysis (Fig. 1).
Ethical approval
The audit committee of the Dutch Transplant Foundation has approved the collection of HMP perfusate material and analysis of data as described within this manuscript. The research register number is 201600608 for Biobank perfusate donated kidneys.
Preparation of kidney perfusate samples
Perfusate samples were depleted of albumin by means of chemical depletion. In short, a 500 μl aliquot of each sample was precipitated with 42% ethanol 4°C for 1 h. Samples were centrifuged at 16 000 × g. for 45 min at 4°C, supernatant was removed and pellets were dried. Pellets were then resuspended in 50 μl 6 M urea buffer and 450 μL phosphate‐buffered solution, and the chemical depletion process was repeated as described above. Final pellets were then resuspended in 6 M urea and 100mM Tris/HCl (pH 8, 2). Protein concentration was determined using the bicinchoninic acid (BCA) Protein Assay Kit (Thermo Scientific) according to manufacturer’s protocol.
Next, 10 μG of protein per sample was applied on an SDS‐PAGE gel and stained with Coomassie Blue, after which the albumin‐containing bands were removed. The remaining bands were prepared for in‐gel digestion as previously described by Shevchenko et al. [26]. Protein digestion was performed using 20 ng/μl trypsin in ice‐cold 50 mM ammonium bicarbonate at 37°C overnight. Tryptic peptides were extracted using 50% acetonitrile, 5% formic acid in MilliQ‐H2O, followed by 85% acetonitrile, 5% formic acid in MilliQ‐H2O. Peptides were dried using a MiVac DUO concentrator (SP Scientific) and resuspended in 20 μl 2% acetonitrile, 0.1% TFA in MilliQ‐H2O. LC‐MS/MS analysis was performed using a Dionex Ultimate 3000 nano‐ultra high‐pressure reverse phase chromatography coupled online to a Q Exactive high field (HF) mass spectrometer (Thermo Scientific) as previously described by Fye et al. [27]. In short, samples were separated on an EASY‐Spray PepMap RSLC C18 column (500 mm × 75 mm, 2 mm particle size, Thermo Scientific) over a 60‐min gradient of 2–35% acetonitrile in 5% DMSO, 0.1% formic acid at 250 nl/min. MS1 scans were acquired at a resolution of 60 000 at 200 m/z and the top 12 most abundant precursor ions were selected for HCD fragmentation.
MS data analysis and label‐free quantitation
Raw MS data were searched against the Uniprot human sequence database (Homo sapiens) retrieved on 02.05.2017, and label‐free quantitation was performed using MaxQuant software v 1.6.0.16 [28]. The following parameters were used: Trypsin/P for enzyme specificity, allowing up to 2 missed cleavages and a minimum peptide length of 7, carbamidomethyl (C) was set as fixed a modification, and oxidation (M) and deamidation (NQ) were chosen as variable modifications included in protein quantification, precursor mass tolerance at 7 ppm and fragment mass tolerance at 0.01 Da and quantification with a false discovery rate (FDR) of 1%. Match between runs was used. Label‐free quantitation (LFQ) values were extracted from MS spectra in MaxQuant using the MaxLFQ algorithm and analysed using Perseus v1.6.12.0 [29]. In short, rows were log2 transformed, filtered based on 70% valid values and missing values were replaced using imputation of the normal distribution. Hierarchical clustering and heat maps were generated using Z‐score and Pearson’s correlation. For analysing the differences between the two groups, two‐sample t tests were performed. Volcano plots were generated using obtained p‐values and log2 intensity differences using GraphPad Prism v8 (GraphPad Software, USA). Analyses were considered statistically significant when P‐values <0.05. STRING v11 [30] and Cytoscape v3.8.2 [31] were used to visualize and interpret the molecular function of proteins and pathways represented by the significant different proteins. Prediction models were made in SPSS statistics 28 using multivariable logistic regression, with forward and backward stepwise inclusion of model factors. Different models were compared with a chi‐square test,the model discriminative potential was assessed by receiver operating characteristic (ROC) curves as previously described by van Balkom et al. [23], and P‐values < 0.05 were considered statistically significant.
Results
Clinical characteristics
There were no significant differences observed in donor and recipient characteristics (Table 1). DGF, 3‐month serum creatinine, 3‐month eGFR, 12‐month serum creatinine and 12‐month eGFR were significantly different between experimental groups (Table 1).
Kidney perfusate profiles
All 44 kidney perfusate samples were analysed using label‐free quantitative mass spectrometry analysis (LFQ MS). In total, 1255 proteins were identified and quantified across all different groups. These proteins were filtered based on valid values set at a minimum of 70%, leading to a total of 498 quantified proteins (Table S1).
Perfusate protein profiles after 15 min of HMP discriminate between good and suboptimal outcome and reveal enriched complement in good outcome group
Perfusate samples collected at T1 were first analysed (Fig. 2). Unsupervised hierarchical clustering for Pearson correlation distance using 100 most abundant proteins detected at T1 was performed to examine data uniformity. Distinctive clustering was observed between the GO and SO group with the exception of 2 samples per group (Fig. 2A). STRING pathway analysis and functional enrichment of these 100 most abundant proteins showed that 66 of these proteins are located in extracellular regions (FDR: 1.39E‐40) and that 23 of these proteins belong to complement and coagulation cascades (FDR: 2.31E‐32) (Fig. 2B).
Figure 2.
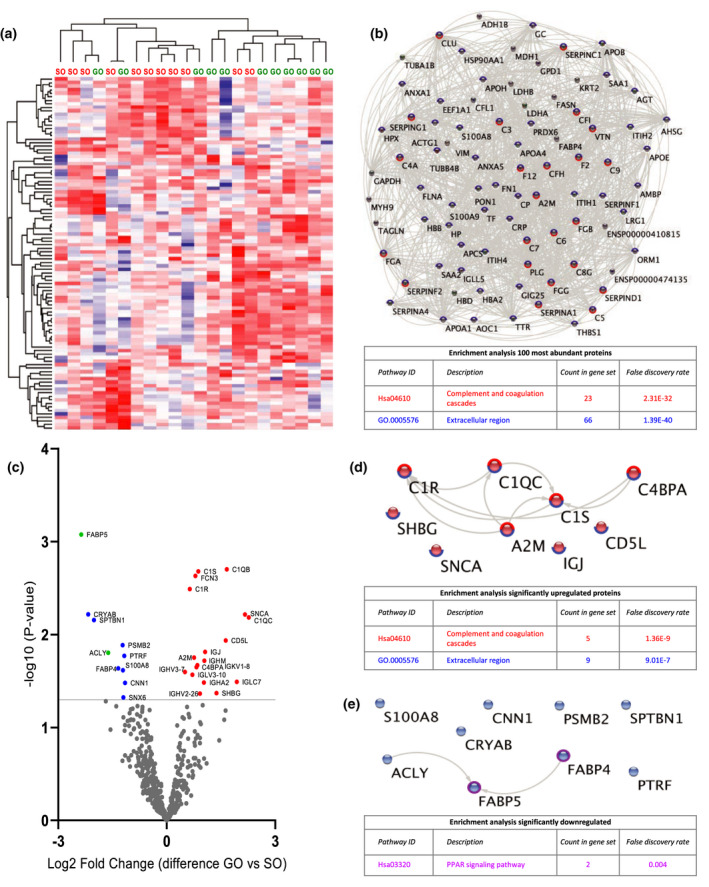
Quantitative proteomic analysis of perfusate samples after 15 min of HMP (T1). (a) Heat map and hierarchical clustering of 100 most abundant proteins at T1. (b) STRING analysis of 100 most abundant proteins at T1. Nodes circled with blue represent proteins located in extracellular regions (FDR: 1.39E‐40) and red represent functional enrichment of the KEGG complement and coagulation pathway (FDR: 2.31E‐32). (c) Volcano plot showing differential protein expression of 498 identified proteins at T1 between good outcome (GO) and suboptimal outcome (SO) at 1‐year post‐transplantation. X‐axis demonstrates protein level difference indicated by log2 fold change, and Y‐axis demonstrates statistical significance indicated by ‐log10 (P‐value). A ‐log10 (P‐value) of > 1.3, and a fold change of > 0 was considered significant. Blue dots represent significant downregulated proteins. Red dots represent significantly upregulated proteins. Green dots represent proteins identified using prediction models. (d) STRING pathway analysis of significantly upregulated proteins at T1. Nodes circled with blue represent proteins located in extracellular regions (FDR: 9.01E‐7) and red represent functional enrichment of the KEGG complement and coagulation pathway (FDR: 1.36E‐9). (e) STRING analysis of significantly downregulated proteins at T1. Nodes circled with purple represent proteins involved in PPAR signalling (FDR: 0.004). FDR, false discovery rate.
Figure 2C shows significant differentially expressed proteins between GO and SO at T1 (Table S1). Eighteen proteins were significantly upregulated in GO vs. SO. STRING pathway analysis and functional enrichment revealed that all significant proteins are located in extracellular regions (FDR: 9.01E‐7) (Fig. 2D). Furthermore, 5 of these proteins belong to complement and coagulation cascades including complement C1q subcomponent subunit B (C1QB), complement C1s subcomponent (C1S), complement C1r subcomponent (C1R) and C4b‐binding protein alpha chain (C4BPA) (FDR: 1.36E‐9 (Fig. 2D). Ten proteins were significantly downregulated in GO vs. SO. STRING pathway analysis and functional enrichment revealed that 2 of these proteins, fatty acid‐binding protein (FABP) 4 and 5, are involved in PPAR signalling (FDR: 0.004) (Fig. 2E).
Perfusate profiles at the end of HMP reveal enriched complement in good outcome group and enriched cytoskeletal proteins in suboptimal group
Perfusate samples at T2 were analysed (Fig. 3; Table S1). Unsupervised hierarchical clustering of 100 most abundant proteins at T2 showed no distinction between groups (Fig. 3A). STRING pathway analysis and functional enrichment of these 100 most abundant proteins showed that 67 of these proteins were located in extracellular regions (FDR: 2.28E‐41) and that 24 of these proteins belong to the complement and coagulation cascades (FDR: 4.2E‐34). (Fig. 3B).
Figure 3.
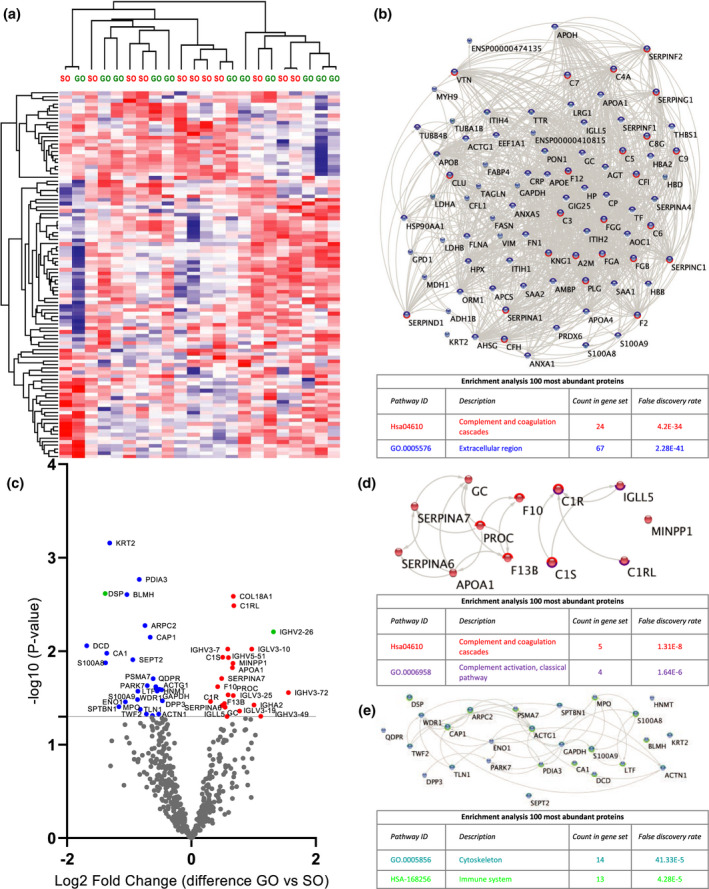
Quantitative proteomic analysis of perfusate samples taken before the termination of HMP (T2). (a) Heat map and hierarchical clustering of 100 most abundant proteins at T2. (b) STRING analysis of 100 most abundant proteins at T2. Nodes circled with blue represent proteins located in extracellular regions (FDR: 2.28E‐41) and red represent functional enrichment of the KEGG complement and coagulation pathway (FDR: 4.2E‐34). (c) Volcano plot showing differential protein expression of 498 identified proteins at T2 between good outcome (GO) and suboptimal outcome (SO) at 1‐year post‐transplantation. X‐axis demonstrates protein level difference indicated by log2 fold change; Y‐axis demonstrates statistical significance indicated by ‐log10 (P‐value). A ‐log10 (P‐value) of > 1.3, and a fold change of > 0 was considered significant. Blue dots represent significant downregulated proteins. Red dots represent significantly upregulated proteins. Green dots represent proteins identified using prediction models. (d) STRING pathway analysis of significantly upregulated proteins at T2. Nodes circled with red represent functional enrichment of the KEGG complement and coagulation pathway (FDR 1.21e‐8) and purple represents proteins involved in complement activation of the classical pathway (FDR: 1.6E‐6). (e) STRING analysis of significantly downregulated proteins at T2. Nodes circled with turquoise represent functional enrichment GO component cytoskeleton (FDR 1.33e‐5) and green represents proteins affiliated with the immune system (FDR: 4.28E‐5). FDR, false discovery rate.
Figure 3C shows significant differentially expressed proteins between GO and SO at T2 (Table S1). Twenty‐two proteins were significantly upregulated in GO vs. SO. STRING pathway analysis and functional enrichment revealed that 5 of these proteins belong to complement and coagulation cascades (FDR 1.21e‐8) (Fig. 3D), of which 4 are involved in complement activation of the classical pathway including complement C1r subcomponent‐like protein (C1RL), C1S and C1R (FDR: 1.6E‐6) (Fig. 3D). Twenty‐six proteins were significantly downregulated in GO vs. SO. STRING pathway analysis and functional enrichment revealed that 14 of these proteins are cytoskeletal proteins including desmoplakin (DSP), talin‐1 (TLN1) and alpha actinin‐1 (ACTN1) (FDR 1.33e‐5) (Fig. 3E). Thirteen of the downregulated proteins are affiliated with the immune system (FDR: 4.28E‐5) (Fig. 3D).
Outcome prediction based on differentially abundant proteins
To determine whether differentially abundant proteins are independent predictors of transplantation outcome, we performed a stepwise logistic regression analysis. At T1, ATP‐citrate synthase (ALCY) together with FABP5 provided a predictive value of 91%, corresponding to a ROC with an area under the curve (AUC) of 0.97 (Fig. 4A). Furthermore, immunoglobulin heavy variable 2‐26 (IGHV2‐26) together with DSP provided a predictive value of 86%, a corresponding AUC of 0.95 in the ROC analysis at T2 (Fig. 4B). When adding donor sex as an extra factor to logistic regression analysis, the predictive value at T2 becomes 100% with a corresponding AUC of 1 (data not shown).
Figure 4.
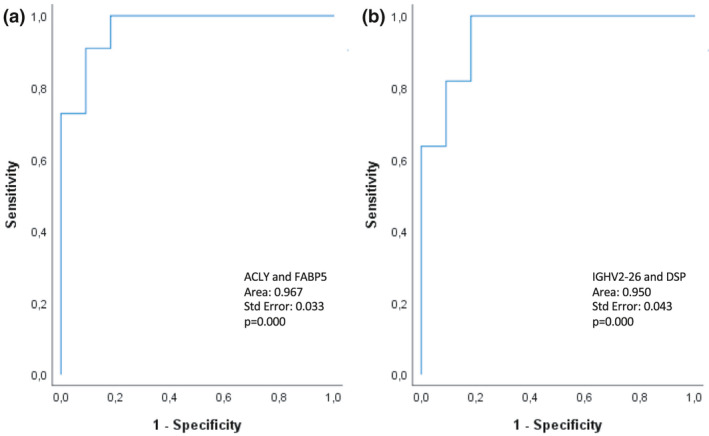
Receiver‐operated (ROC) curves for the prediction models based on T1 and T2. (a) Perfusate analysis at T1 shows a 91% predictive value for ATP‐citrate synthase (ALCY) and fatty acid‐binding protein 5 (FABP5). (b) Perfusate analysis at T2 shows an 86% predictive value for immunoglobulin heavy variable 2‐26 (IGHV2‐26) and desmoplakin (DSP).
Discussion
The use of ECD organs for transplantation is increasing due to the continuing donor organ shortage, urging novel noninvasive strategies to identify and predict graft outcome. The aim of this study was to evaluate the flushed‐out tissue‐resident proteins that end up in the perfusate of DBD kidneys preserved using HMP via an unbiased proteomics approach. We show that protein secretion profiles after 15 min of HMP results in distinctive clustering between DBD kidneys with a good and suboptimal outcome 1‐year post‐transplantation. To our knowledge, this is the first study that distinguishes between a good and suboptimal transplant outcome based on protein profiles identified in HMP perfusates of DBD kidneys.
Increase in complement proteins in HMP perfusate is associated with good kidney function at 1‐year post‐transplantation
It has been shown that brain death is associated with complement activation [32], and that the complement system is involved in transplant‐related pathologies [33, 34. Although the main function of the complement system is to aid the immune system to attack and clear pathogens, it also plays a role in apoptotic cell removal. Excessive complement activation can lead to cell lysis and necrosis and may, therefore, be linked to the allograft induced immune attack and apoptotic clearance. [35, 36 However, the exact underlying mechanisms of complement activation pathways during transplantation have been the topic of many studies and is still not fully resolved [32, 37.
Therefore, it was somewhat surprising to find significantly elevated levels of C1r, C1s, C1q and C4BPA in perfusate samples of donor kidneys with a good outcome 1‐year post‐transplantation. C1r, C1s and C1q are the three subcomponents of the C1 complex that represent upstream proteins of the classical complement system. The classical pathway is initiated when the C1 complex binds to an activator. One of the main roles of C1q is the removal of apoptotic cells and debris [44]. The deficiency of these three proteins, especially C1q, has been highly associated with renal failure caused by autoimmune diseases such as systemic lupus erythematosus [44]. Furthermore, deposition of C1q in the kidney is associated with C1q nephropathy [45]. Interestingly, studies have shown that treatment with C1 inhibitor has potential protective effects during renal transplantation [37, 38. However, C1 inhibitor also has anti‐inflammatory functions and anti‐apoptotic effects and inhibits coagulation, contact and kinin systems [46]. Therefore, these protective effects could also be the consequence of reduced inflammation, attenuated oedema and decreased formation of microthrombi during transplantation. C4BPA is a complement regulator that inhibits the complement system via the classical and the lectin pathway. Furthermore, it protects apoptotic cells from complement CD40‐dependent stimulation of B cells [47]. During ischaemia, the endothelium becomes activated by cytokines, resulting in the breakdown of the glycocalyx and subsequent loss of regulators of the complement system such as C4b‐binding protein [33]. The loss of these regulators means that the endothelial cell surface is no longer protected from attack by the complement, coagulation and contact systems. Our results suggest that a wash out of tissue‐resident complement proteins detected in perfusates is associated with a good outcome, perhaps due to detachment of remnant unproductive complement attack complexes.
The role of complement activation during brain death appears to be controversial. For instance, Brown et al. (2006) showed that differential expression of C3 alleles by donor renal cells appears to have an negative effect on late graft outcome [41]. However, these findings could not be reproduced in larger cohort studies, as Varaguman et al. (2009) did not show a difference in transplant outcomes between donors with different C3 allotypes [42]. Furthermore, Damman et al. (2012) showed that the whole lectin complement pathway gene profile of the donor does not influence outcome [43]. Damman et al. (2011) showed that elevated complement C5b‐9 levels in deceased donor plasma samples is associated with acute rejection in the recipient after renal transplantation. However, they did not find any significant association between donor complement activation levels and primary nonfunction (PNF), delayed graft function (DGF) or graft survival in the first year after transplantation [32]. Importantly, all of these reports were mainly focused on distinct components of the complement system in tissue and plasma, whereas our study was conducted in an unbiased manner using perfusate samples. Complement activation will inevitably occur in every donor kidney, but the amount of activation and/or complement composition may be a key determinator of the level of injury and repair. Understanding these aspects in the context of donor organ function warrants further investigation.
Disruption of cytoskeletal proteins is associated with a suboptimal transplant outcome
Proteomic perfusate analysis at the end of HMP revealed significant upregulation of cytoskeletal proteins in the suboptimal outcome group. This indicates that enriched cytoskeletal proteins in the perfusate are associated with a suboptimal outcome 1‐year post‐transplantation. These results are in line with a proteomics analysis performed by Coskun et al. (2016), who found secretion of cytoskeletal proteins of DBD donor kidneys during SCS. However, they did not include transplant outcome into their analysis [24].
Ischaemia has been associated with disruption of the cytoskeleton, leading to loss of tissue integrity [48, 49. However, the effects of brain death specifically on renal tissue integrity are still unknown. We observed enrichment of actinin‐1 and talin‐1 in the perfusate of the suboptimal outcome group. Actinin‐1 is one of four actin‐bundling proteins that seems to play a crucial role in podocyte architecture. Studies have shown that both the podocyte actin cytoskeleton, as well as podocyte‐associated talin‐1, play a crucial role in preserving an intact glomerular filtration barrier [50, 51. Enhanced excretion of cytoskeletal proteins could be the cause of lower eGFR rates in our suboptimal group due to loss of tissue integrity and disruption of the glomerular filtration barrier.
Desmoplakin together with IGHV2‐26 showed a predictive value in which an increased expression of desmoplakin in the perfusate was correlated with a suboptimal transplant outcome. Desmoplakin is the most abundant part of the desmosome or intercellular junctions of epithelia and cardiac muscle, providing mechanical strength to tissues. Desmoplakin, therefore, plays a key role in tissue morphogenesis and integrity [52], but studies on the role of desmoplakin in renal function and tissue integrity are scarce. It has been shown that structural alterations in desmosomal junctions could potentially influence renal function [53]. However, more research into the role of desmoplakin should be conducted.
Lipid metabolism and renal function
ACLY, FABP4 and FABP5 were significantly upregulated in the perfusate collected after 15 min of HMP of kidneys with a suboptimal outcome. Furthermore, FABP5 together with ALCY showed a predictive outcome of 91%. ACLY is a key enzyme of de novo fatty acid synthesis [54]. FABP4 and FABP5 are key proteins in lipid metabolism and play an important role in inflammation via the PPAR‐y signalling pathway [55]. The effect of FABP5 and ACLY on renal function is unclear,however, overexpression of both proteins has been associated with renal carcinoma [56, 57. Excreted FABP4 in the serum has been associated with renal dysfunction [58]. Furthermore, Huang et al. (2018) revealed elevated levels of FABP4 in rat kidneys exposed to ischaemia/reperfusion injury (IRI) [59]. Inhibition of FABP4 in an acute renal injury mouse model has shown to attenuate renal structural damage and improve renal function [60]. Several studies have shown that that renal transplant‐related IRI leads to disturbed metabolism due to energy deficit [59, 61. Our results suggest that this disturbed lipid metabolism plays a role in post‐transplant outcome, and oxygenated HMP may be the key to limit this metabolic disturbance and improve outcome.
Impact of donor characteristics, sample time point and total CIT
Perfusate samples obtained at T1 resulted in distinctive clustering between a good and suboptimal outcome. However, we did not observe distinctive clustering at T2. This could be caused by the variation in time point of sample collection at T2 between groups as shown by the total CIT (Table 1). Interestingly, kidneys with a good outcome had an average longer total CIT although these differences were not significant. Furthermore, samples collected at T1 better represent kidney quality as these samples can be considered a second flush‐out. Samples collected at T2 possibly also represent the effect of preservation.
Donor sex seems to influence the predictive values of proteins identified at T2, although there was no significant difference in male/female ratio between experimental groups (Table 1).
Limitations
This pilot study consisted of a small cohort, and the results are based on proteomics analysis only. Validation in a larger patient cohort will be needed to determine the impact of our discovered biomarkers and their predictive value for determining the level of injury of donor kidneys prior to transplantation.
Conclusion
In conclusion, this is the first study that shows that the proteomic perfusate analysis of DBD kidneys during HMP preservation can discriminate between a good and suboptimal post‐transplant outcome. Our results provide novel insights into pathophysiological mechanisms underlying brain death and organ preservation. The presence of markers in the perfusate reflecting complement activation and loss of epithelial and tissue integrity may be indicators for transplant outcome. However, additional research needs to be conducted to confirm these results. Ultimately, these insights provide a noninvasive way to assess donor organ quality and provide potential inroads for repair. Novel preservation techniques such as HMP are making their way into the clinics, and with this study, we show that HMP can provide a potential platform for noninvasive graft assessment and outcome prediction. Furthermore, HMP could be used as a platform for pharmaceutical interventions and repair of donor organs with a suboptimal quality.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest.
Supporting information
Table S1
Acknowledgements
We thank Letizia Lo Faro and members of the Ploeg and Kessler group for insightful discussions. We thank the transplant technicians for collecting the perfusate samples. We thank members of the Discovery Proteomics Facility (DPF), led by Roman Fischer, for expert help with mass spectrometry analysis. Honglei Huang and the experimental costs were supported by a grant from the Biomedical Research Centre (NIHR) awarded to Rutger Ploeg. Aukje Brat was supported by the Tekke Huizinga Foundation in the Netherlands.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [62] with the dataset identifier px‐submission PXD027127.
REFERENCES
- 1.No Title. 2019. http://www.transplant‐observatory.org/data‐charts‐and‐tables/chart/
- 2. Eurotransplant . Eurotransplant annual report 2019. Leiden: 2019. [Google Scholar]
- 3. Bos EM, Leuvenink HGD, van Goor H, Ploeg RJ. Kidney grafts from brain dead donors: Inferior quality or opportunity for improvement? Kidney Int 2007; 72(7): 797. [DOI] [PubMed] [Google Scholar]
- 4.[4]Chen EP, Bittner HB, Kendall SW, Van Trigt P . Hormonal and hemodynamic changes in a validated animal model of brain death. Crit Care Med 1996; 24: 1352. [DOI] [PubMed] [Google Scholar]
- 5.[5]Gavriilidis P, Inston NG. Recipient and allograft survival following donation after circulatory death versus donation after brain death for renal transplantation: a systematic review and meta‐analysis. Transpl Rev 2020; 34: 100563. [DOI] [PubMed] [Google Scholar]
- 6. Ma MKM, Lim WH, Craig JC, Russ GR, Chapman JR, Wong G. Mortality among younger and older recipients of kidney transplants from expanded criteria donors compared with standard criteria donors. Clin J Am Soc Nephrol 2016; 11: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Papachristou E, Provatopoulou S, Savvidaki E, et al. Outcome of transplantation in renal allograft recipients from cadaveric donors with standard and expanded criteria: a single‐center experience. Transplant Proc 2014; 46: 3172. [DOI] [PubMed] [Google Scholar]
- 8. Port FK, Bragg‐Gresham JL, Metzger RA, et al. Donor characteristics associated with reduced graft survival: an approach to expanding the pool of kidney donors. Transplantation 2002; 74: 1281. [DOI] [PubMed] [Google Scholar]
- 9. Merion RM. Expanded criteria donors for kidney transplantation. Transplant Proc 2005; 37: 3655. [DOI] [PubMed] [Google Scholar]
- 10. Harada KM, Mandia‐Sampaio EL, de Sandes‐Freitas TV, et al. Risk factors associated with graft loss and patient survival after kidney transplantation. Transplant Proc 2009; 41: 3667. [DOI] [PubMed] [Google Scholar]
- 11. Bonsignore P, Pagano D, Piazza S, et al. Crucial role of extended criteria donors in deceased donor single kidney transplantation to face chronic shortage in the heart of the mediterranean basin: a single‐center experience. Transplant Proc 2019; 51: 2868. [DOI] [PubMed] [Google Scholar]
- 12. Aubert O, Kamar N, Vernerey D, et al. Long term outcomes of transplantation using kidneys from expanded criteria donors: prospective, population based cohort study. BMJ 2015; 351: h3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Irish WD, Ilsley JN, Schnitzler MA, Feng S, Brennan DC. A risk prediction model for delayed graft function in the current era of deceased donor renal transplantation. Am J Transplant 2010; 10: 2279. [DOI] [PubMed] [Google Scholar]
- 14. Merion RM, Wolfe RA, Port FK, et al. A comprehensive risk quantification score for deceased donor kidneys: the kidney donor risk index. Transplantation 2009; 88: 231. [DOI] [PubMed] [Google Scholar]
- 15. Wang CJ, Wetmore JB, Crary GS, Kasiske BL. The donor kidney biopsy and its implications in predicting graft outcomes: a systematic review. Am J Transplant 2015; 15: 1903. [DOI] [PubMed] [Google Scholar]
- 16. Brat A, de Vries KM, van Heurn EWE, et al. Hypothermic Machine perfusion as a National standard preservation method for deceased donor kidneys. Transplantation 2021; 10.1097/TP.0000000000003845. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moers C, Smits JM, Maathuis M‐HJ, et al. Machine perfusion or cold storage in deceased‐donor kidney transplantation. N Engl J Med 2009; 360(1): 7. [DOI] [PubMed] [Google Scholar]
- 18. Jochmans I, Moers C, Smits JM, et al. Machine perfusion versus cold storage for the preservation of kidneys donated after cardiac death. Ann Surg 2010; 252: 756. [DOI] [PubMed] [Google Scholar]
- 19. Peng P, Ding Z, He Y, Zhang J, Wang X, Yang Z. Hypothermic machine perfusion versus static cold storage in deceased donor kidney transplantation: a systematic review and meta‐analysis of randomized controlled trials. Artif Organs 2019; 43: 478. [DOI] [PubMed] [Google Scholar]
- 20. Groen H, Moers C, Smits JM, et al. Cost‐effectiveness of hypothermic machine preservation versus static cold storage in renal transplantation. Am J Transplant 2012; 12: 1824. [DOI] [PubMed] [Google Scholar]
- 21. Jochmans I, Brat A, Davies L, et al. Oxygenated versus standard cold perfusion preservation in kidney transplantation (COMPARE): a randomised, double‐blind, paired, phase 3 trial. Lancet 2020; 396: 1653. [DOI] [PubMed] [Google Scholar]
- 22. Moers C, Pirenne J, Paul A, Ploeg RJ. Machine perfusion or cold storage in deceased‐donor kidney transplantation. N Engl J Med 2012; 366: 770. [DOI] [PubMed] [Google Scholar]
- 23. van Balkom BWM, Gremmels H, Ooms LSS, et al. Proteins in preservation fluid as predictors of delayed graft function in kidneys from donors after circulatory death. Clin J Am Soc Nephrol 2017; 12: 817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coskun A, Baykal AT, Kazan D, et al. Proteomic analysis of kidney preservation solutions prior to renal transplantation. PLoS One 2016; 11: e0168755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Parikh CR, Hall IE, Bhangoo RS, et al. Associations of perfusate biomarkers and pump parameters with delayed graft function and deceased donor kidney allograft function. Am J Transplant 2016; 16: 1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shevchenko A, Tomas H, Havliš J, Olsen JV, Mann M. In‐gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 2007; 1: 2856. [DOI] [PubMed] [Google Scholar]
- 27. Fye HKS, Mrosso P, Bruce L, et al. A robust mass spectrometry method for rapid profiling of erythrocyte ghost membrane proteomes. Clin Proteomics 2018; 15: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry‐based shotgun proteomics. Nat Protoc 2016; 11: 2301. [DOI] [PubMed] [Google Scholar]
- 29. Tyanova S, Temu T, Sinitcyn P, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 2016; 13: 731. [DOI] [PubMed] [Google Scholar]
- 30. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality‐controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res 2017; 45: D362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13(11): 2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Damman J, Seelen MA, Moers C, et al. Systemic complement activation in deceased donors is associated with acute rejection after renal transplantation in the recipient. Transplantation 2011; 92: 163. [DOI] [PubMed] [Google Scholar]
- 33. Biglarnia AR, Huber‐Lang M, Mohlin C, Ekdahl KN, Nilsson B. The multifaceted role of complement in kidney transplantation. Nat Rev Nephrol 2018; 14: 767. [DOI] [PubMed] [Google Scholar]
- 34. Nieuwenhuijs‐Moeke GJ, Pischke SE, Berger SP, et al. Ischemia and reperfusion injury in kidney transplantation: relevant mechanisms in injury and repair. J Clin Med 2020; 9(1): 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ogden CA, DeCathelineau A, Hoffmann PR, et al. C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J Exp Med 2001; 194: 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trouw LA, Bengtsson AA, Gelderman KA, Dahlbäck B, Sturfelt G, Blom AM. C4b‐binding protein and factor H compensate for the loss of membrane‐bound complement inhibitors to protect apoptotic cells against excessive complement attack. J Biol Chem 2007; 282: 28540. [DOI] [PubMed] [Google Scholar]
- 37. Poppelaars F, Jager NM, Kotimaa J, et al. C1‐inhibitor treatment decreases renal injury in an established brain‐dead rat model. Transplantation 2018; 102: 79. [DOI] [PubMed] [Google Scholar]
- 38. Berger M, Lefaucheur C, Jordan SC. Update on C1 esterase inhibitor in human solid organ transplantation. Transplantation 2019; 103: 1763. [DOI] [PubMed] [Google Scholar]
- 39. Jager NM, Poppelaars F, Daha MR, Seelen MA. Complement in renal transplantation: the road to translation. Mol Immunol 2017; 89: 22. [DOI] [PubMed] [Google Scholar]
- 40. Poppelaars F, Seelen MA. Complement‐mediated inflammation and injury in brain dead organ donors. Mol Immunol 2017; 1: 77. [DOI] [PubMed] [Google Scholar]
- 41. Brown KM, Kondeatis E, Vaughan RW, et al. Influence of donor C3 allotype on late renal‐transplantation outcome. N Engl J Med 2006; 354: 2014. [DOI] [PubMed] [Google Scholar]
- 42. Varagunam M, Yaqoob MM, Döhler B, Opelz G. C3 polymorphisms and allograft outcome in renal transplantation. N Engl J Med 2009; 360: 874. [DOI] [PubMed] [Google Scholar]
- 43. Damman J, Kok JL, Snieder H, et al. Lectin complement pathway gene profile of the donor and recipient does not influence graft outcome after kidney transplantation. Mol Immunol 2012; 50: 1. [DOI] [PubMed] [Google Scholar]
- 44. Macedo ACL, Isaac L. Systemic lupus erythematosus and deficiencies of early components of the complement classical pathway. Front Immunol 2016; 7: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vizjak A, Ferluga D, Rožič M, et al. Pathology, clinical presentations, and outcomes of C1q nephropathy. J Am Soc Nephrol 2008; 19(11): 2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Davis AE, Lu F, Mejia P. C1 inhibitor, a multi‐functional serine protease inhibitor. Thromb Haemost 2010; 104(11): 886. [DOI] [PubMed] [Google Scholar]
- 47. Blom A, Villoutreix B, Dahlbäck B. Functions of human complement inhibitor C4b‐binding protein in relation to its structure ‐ PubMed. Arch Immunol Ther Exp 2004; 52(2): 83. [PubMed] [Google Scholar]
- 48. Genescà M, Sola A, Hotter G. Actin cytoskeleton derangement induces apoptosis in renal ischemia/reperfusion. Apoptosis 2006; 11: 563. [DOI] [PubMed] [Google Scholar]
- 49. Han SJ, Kim J‐H, Kim JI, Park KM. Inhibition of microtubule dynamics impedes repair of kidney ischemia/reperfusion injury and increases fibrosis. Sci Rep 2016; 6: 27775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tian X, Ishibe S. Targeting the podocyte cytoskeleton: from pathogenesis to therapy in proteinuric kidney disease. Nephrol Dial Transplant 2016; 31: 1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tian X, Kim JJ, Monkley SM, et al. Podocyte‐associated talin1 is critical for glomerular filtration barrier maintenance. J Clin Invest 2014; 124: 1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Garrod D, Chidgey M. Desmosome structure, composition and function. Biochim Biophys Acta 2008; 1778: 572. [DOI] [PubMed] [Google Scholar]
- 53. Silberberg M, Charron AJ, Bacallao R, Wandinger‐Ness A. Mispolarization of desmosomal proteins and altered intercellular adhesion in autosomal dominant polycystic kidney disease. Am J Physiol ‐ Ren Physiol 2005; 288: 1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Elshourbagy NA, Near JC, Kmetz PJ, et al. Cloning and expression of a human ATP‐citrate lyase cDNA. Eur J Biochem 1992; 204(2): 491. [DOI] [PubMed] [Google Scholar]
- 55. Qiao Y, Liu L, Yin L, et al. FABP4 contributes to renal interstitial fibrosis via mediating inflammation and lipid metabolism. Cell Death Dis 2019; 10: 382. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56. Teng L, Chen Y, Cao Y, et al. Overexpression of ATP citrate lyase in renal cell carcinoma tissues and its effect on the human renal carcinoma cells in vitro. Oncol Lett 2018; 15: 6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lv Q, Wang G, Zhang Y, et al. FABP5 regulates the proliferation of clear cell renal cell carcinoma cells via the PI3K/AKT signaling pathway. Int J Oncol 2019; 54: 1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Furuhashi M, Ishimura S, Ota H, et al. Serum fatty acid‐binding protein 4 is a predictor of cardiovascular events in end‐stage renal disease. PLoS One 2011; 6:e27356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huang H, van Dullemen LFA, Akhtar MZ, et al. Proteo‐metabolomics reveals compensation between ischemic and non‐injured contralateral kidneys after reperfusion. Sci Rep 2018; 8:8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shinoda Y, Wang Y, Yamamoto T, Miyachi H, Fukunaga K. Analysis of binding affinity and docking of novel fatty acid‐binding protein (FABP) ligands. J Pharmacol Sci 2020; 143: 264. [DOI] [PubMed] [Google Scholar]
- 61. Lindeman JH, Wijermars LG, Kostidis S, et al. Results of an explorative clinical evaluation suggest immediate and persistent post‐reperfusion metabolic paralysis drives kidney ischemia reperfusion injury. Kidney Int 2020; 98: 1476. [DOI] [PubMed] [Google Scholar]
- 62. Perez‐Riverol Y, Csordas A, Bai J, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 2019; 47: D442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [62] with the dataset identifier px‐submission PXD027127.


