Abstract
The biosynthesis of pyrroindomycins A and B features a complexity-building [4 + 2] cycloaddition cascade, which generates the spirotetramate core under the catalytic effects of monofunctional Diels–Alderases PyrE3 and PyrI4. We recently showed that the main functions of PyrI4 include acid catalysis and induced-fit/conformational selection. We now present quantum mechanical and molecular dynamics studies implicating a different mode of action by PyrE3, which prearranges an anionic polyene substrate into a high-energy reactive conformation at which an inverse-electron-demand Diels–Alder reaction can occur with a low barrier. Stereoselection is realized by strong binding interactions at the endo stereochemical relationship and a local steric constraint on the endo-1,3-diene unit. These findings, illustrating distinct mechanisms for PyrE3 and PyrI4, highlight how nature has evolved multiple ways to catalyze Diels–Alder reactions.
Graphical Abstract

1. INTRODUCTION
The Diels–Alder (DA) reaction is a powerful transformation capable of constructing complex cyclic structures within a single synthetic step with remarkable regio- and stereocontrol.1 In the past decades, great attention has been paid to the discovery of natural Diels–Alderases (DAases),2–4 which was paralleled by the development of artificial enzymes5 and biomimetic catalysts6 for DA reactions. Mechanistic insights have been proven essential for understanding biocatalytic DA reactions2,3,7,8 and motivating the design of artificial DAases.5 However, the studies of DAases are often difficult because of enzyme multifunctionality,9 the multiplicity of mechanistic scenarios,10 and the occurrence of formal DA reactions that are stepwise in nature11,12 or display ambimodal reactivity.13–16 Because of these challenges, monofunctional DAases have received much interest. SpnF,14,15 the first monofunctional DAase discovered in nature,16 is also the most studied DAase. In 2015, two new monofunctional DAases, known as PyrE3 and PyrI4, were identified in the biosynthetic gene cluster of pyrroindomycins A and B.17 The products belong to the bacterial spirotetronate/spirotatramate family and display potent antimicrobial activities for MRSA and vancomycin-resistant bacteria .18 PyrE3 was found to catalyze the first step of a [4 + 2] cycloaddition cascade, converting a linear polyene precursor SM into a multi-substituted trans-decalin system P-1 in a highly endo-selective fashion (Scheme 1a). It is therefore one of many enzymes that catalyze decalin formation by intramolecular DA reactions.2,3d,19 Another DAase, PyrI4, then catalyzes the second cycloaddition to construct the spirotetramate core.17
Scheme 1.
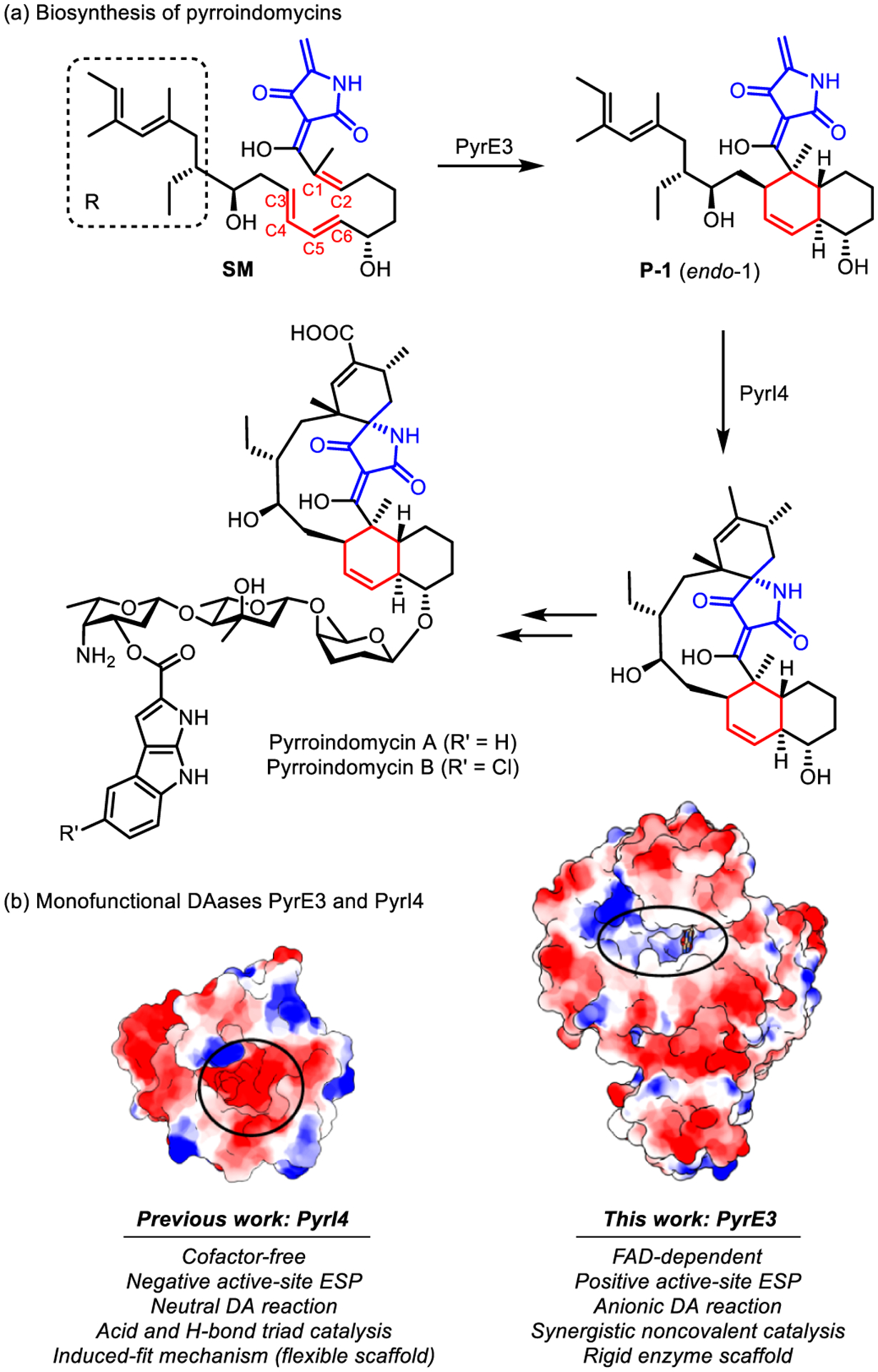
DAases, PyrE3 and PyrI4, in Pyrroindomycin Biosynthesis
The mechanism of catalysis by PyrI4 was recently studied by our group.20 However, PyrE3 is a structurally distinct FAD-dependent DAase, and a different mechanism is possible because the two enzymes exhibit contrasting active-site electrostatic environments21,28 and catalyze different types of DA reactions (Scheme 1b). This apparent contradiction presents a puzzle for the biosynthetic mechanism of pyrroindomycin. Although the structure of PyrE3 was disclosed by X-ray crystal analysis,21 it remains unclear how PyrE3 performs its function with a structure significantly different from PyrI4. Here, we present quantum mechanical and molecular dynamics (MD) explorations for the mechanism of the stereoselective catalysis of PyrE3. Unlike the case of PyrI4, in which acid catalysis and induced-fit/conformational selection comprise the main catalytic effects,20 our computations show that PyrE3, with a rigid enzyme scaffold, utilizes electrostatic effects, H-bonds, and shape complementarity to stereoselectively preorganize the deprotonated linear substrate into the endo reactive conformation (RC). These factors allow an inverse-electron-demand DA reaction to be facile at room temperature.
2. RESULTS AND DISCUSSION
2.1. DFT Studies for the Uncatalyzed Reaction.
To reveal the intrinsic DA reactivity of the polyene substrate (SM) and provide a basis for understanding PyrE3 catalysis, we first studied the uncatalyzed DA reaction using density functional theory (DFT).22 We employed the M06–2X method,23 and solvent effects of water were accounted for with the SMD implicit model24 (see Figure 1 for the full level of theory and Supporting Information for more details). As shown in Scheme 2, the DA reaction of SM can give four stereoisomers (P-1 through P-4) depending on the relative positions and orientations of the diene with the dienophile. To study the mechanism of catalysis, we concentrate on the endo-1 stereochemical pathway that leads to P-1. The stereoselectivity will be discussed later in the paper.
Figure 1.

Free-energy profile and transition state geometries for the uncatalyzed intramolecular DA reaction at the endo-1 stereochemical relationship.
Scheme 2.
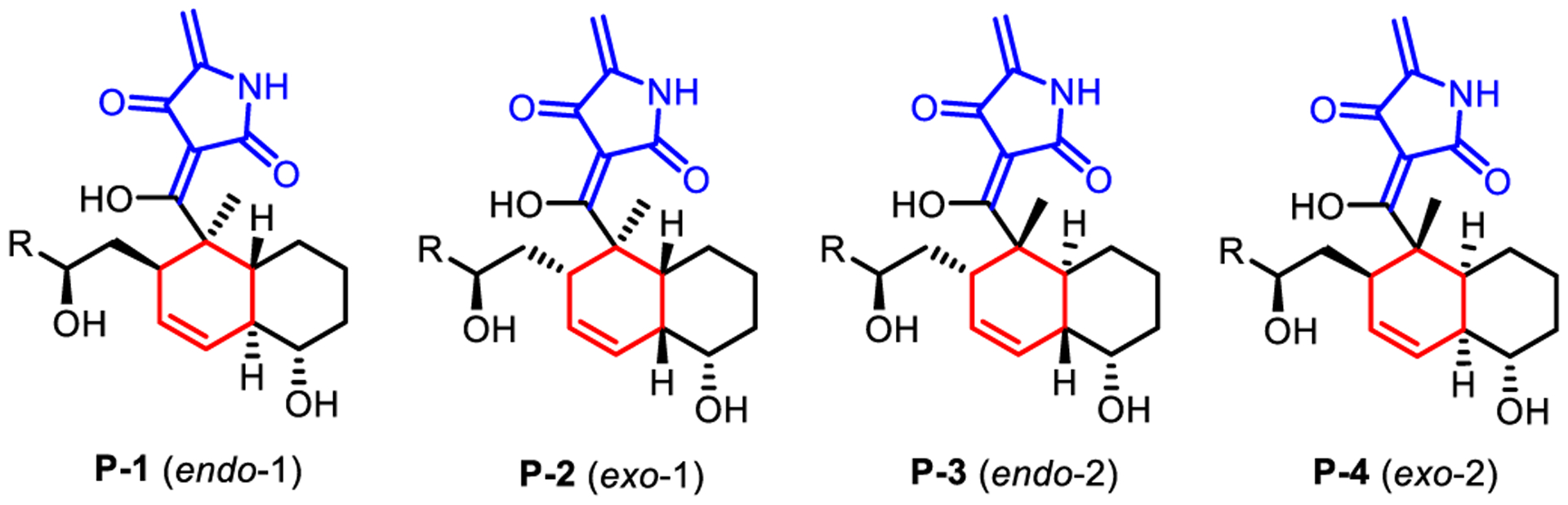
Possible DA Adducts
The substrate features a 3-acyltetramic acid structure as a part of the dienophile fragment. DFT calculations predict a pKa of 1.1 for SMn (the neutral form of SM, see Supporting Information for computations), which agrees with reported acidities of 3-acyltetramic acids (2.3–3.5).25 Therefore, the substrate is deprotonated and exists in the form of a tetramate anion SMa at a physiological pH. As shown in Figure 1, the formation of Pa-1 from SMa is exergonic by −23.4 kcal/mol. The barrier is 27.7 kcal/mol in the anionic manifold via TSa-1. TSa-1 is an asynchronous transition state; the shorter C2–C6 bond (2.0 Å) is much shorter than the longer partial bond C1–C3 (2.6 Å, see Scheme 1a for C atom numbering). In view of the high barrier of the anionic DA reaction, we also explored the effect of protonation. The cycloaddition barrier for the neutral pathway is only 21.1 kcal/mol (bidirectional blue arrow in Figure 1), 7 kcal/mol below that of the anion. However, the cost of protonation at pH 7.0 raises the overall barrier to 29.1 kcal/mol for TSn-1 in aqueous solution. The high barriers of both mechanisms explain the inertness of SM toward cycloaddition.17
2.2. Mechanism of Catalysis.
We next studied the catalytic mechanism of PyrE3. Although experimental efforts failed to co-crystallize PyrE3 with the substrate/product, the active site was identified by site-specific mutations to be a cavity between the middle domain and the FAD-binding domain (Figure 2a).21 How the substrate binds to the catalytic site is yet unknown. To study substrate binding, we docked RCa-1, the RC before TSa-1 (and thus it resembles TSa-1), into the crystal structure of apo PyrE3.21 The docked structure of RCa-1 was relaxed by 500 ns MD simulations to allow for induced-fit motions and to later evaluate the stability of the complex. Protein–ligand docking and MD simulations employed the AutoDock Vina and AMBER 16 programs,26 respectively. The protonation states were assigned by PROPKA,27 while the side-chain proton on H77 was manually moved from δ-N (i.e., HID) to ε-N (i.e., HIE) to facilitate hydrogen bonding. The predicted binding mode is presented in Figure 2b,c. At this pose, RCa-1 can bind PyrE3 with abundant hydrophobic contacts and hydrogen bonds (see Figure S4 for distance vs time plots). These binding forces, together with the shape complementarity between RCa-1 and PyrE3,21 cause a low average rmsd of 0.833 Å for the entire substrate and 0.185 Å for C1 to C6 (i.e., the reaction center) in the 500 ns MD trajectory, indicating the stability of RCa-1 in the bound state. In contrast, the other three stereoisomeric RCs RCa-2 to RCa-4 show poorer fits to the pocket, which will be discussed later.
Figure 2.
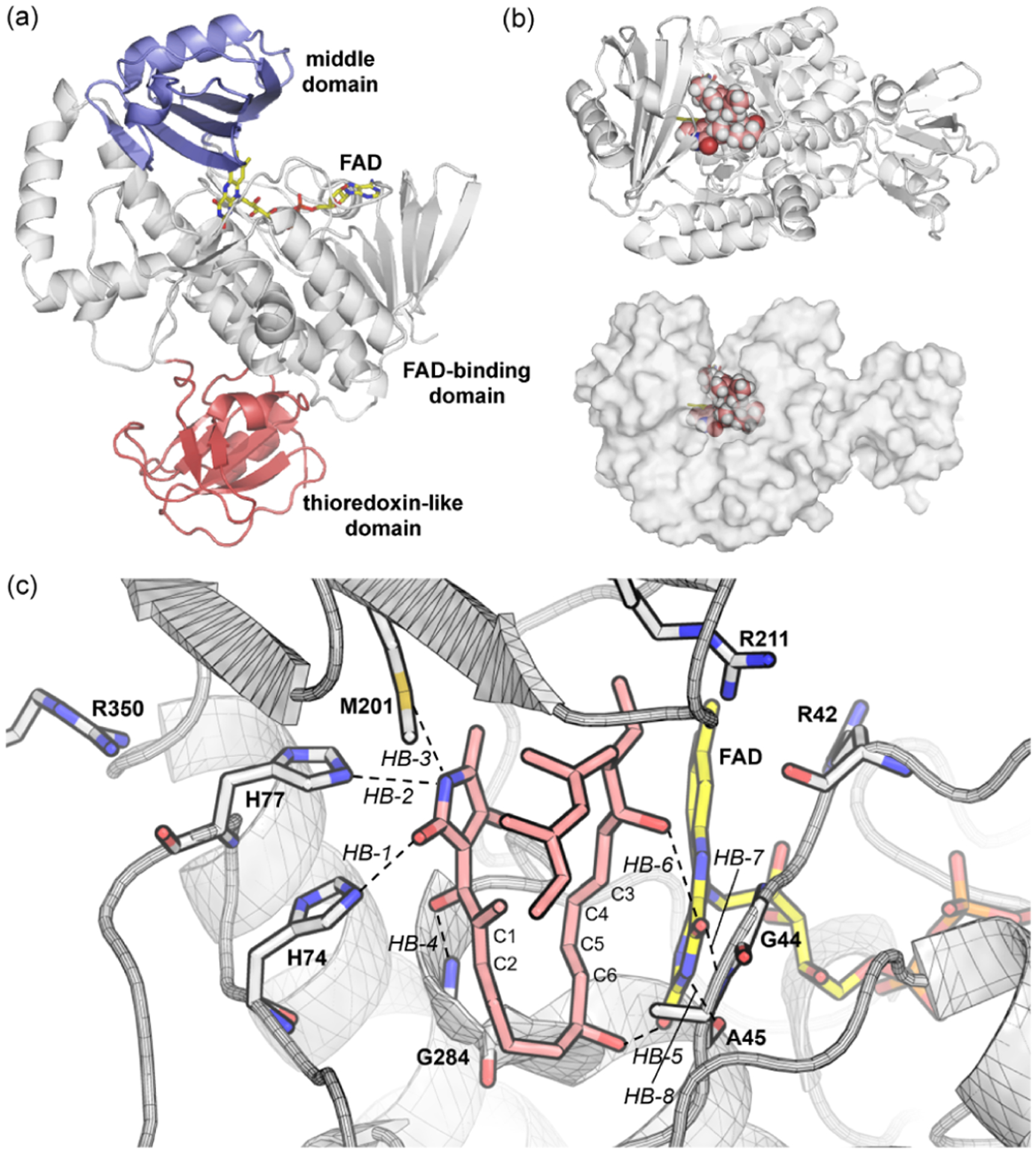
Structure of PyrE3. (a) Apo PyrE3. (b) Substrate–enzyme complex. (c) Substrate binding at endo RC.
2.2.1. Substrate Preorganization.
The folding of the substrate can significantly reduce the activation barrier.15e,20,29–31 In fact, DFT calculations predict that the folding of SMa into RCa-1 requires a free energy of 7.9 kcal/mol (8.9 kcal/mol by enthalpy) in solution, whereas PyrE3 overcomes the folding energy by binding in the active site. The RC is enthalpically higher in energy due to the necessity to introduce a variety of gauche and sterically hindered conformers. In addition, the free substrate SMa can explore many unreactive conformers, as revealed by 500 ns MD simulations in solution (Figure 3). The flexible conformation of SMa implies that a decrease in entropy is required for it to reach TSa-1, in which the rigid trans-decalin ring system is partially formed. This mixture of conformations will lead to a mixed entropy32 that increases the barrier by ca. 2.1 kcal/mol at room temperature (see Supporting Information). PyrE3 can alleviate this negative entropic effect by preorganizing the substrate into one specific pose.33
Figure 3.
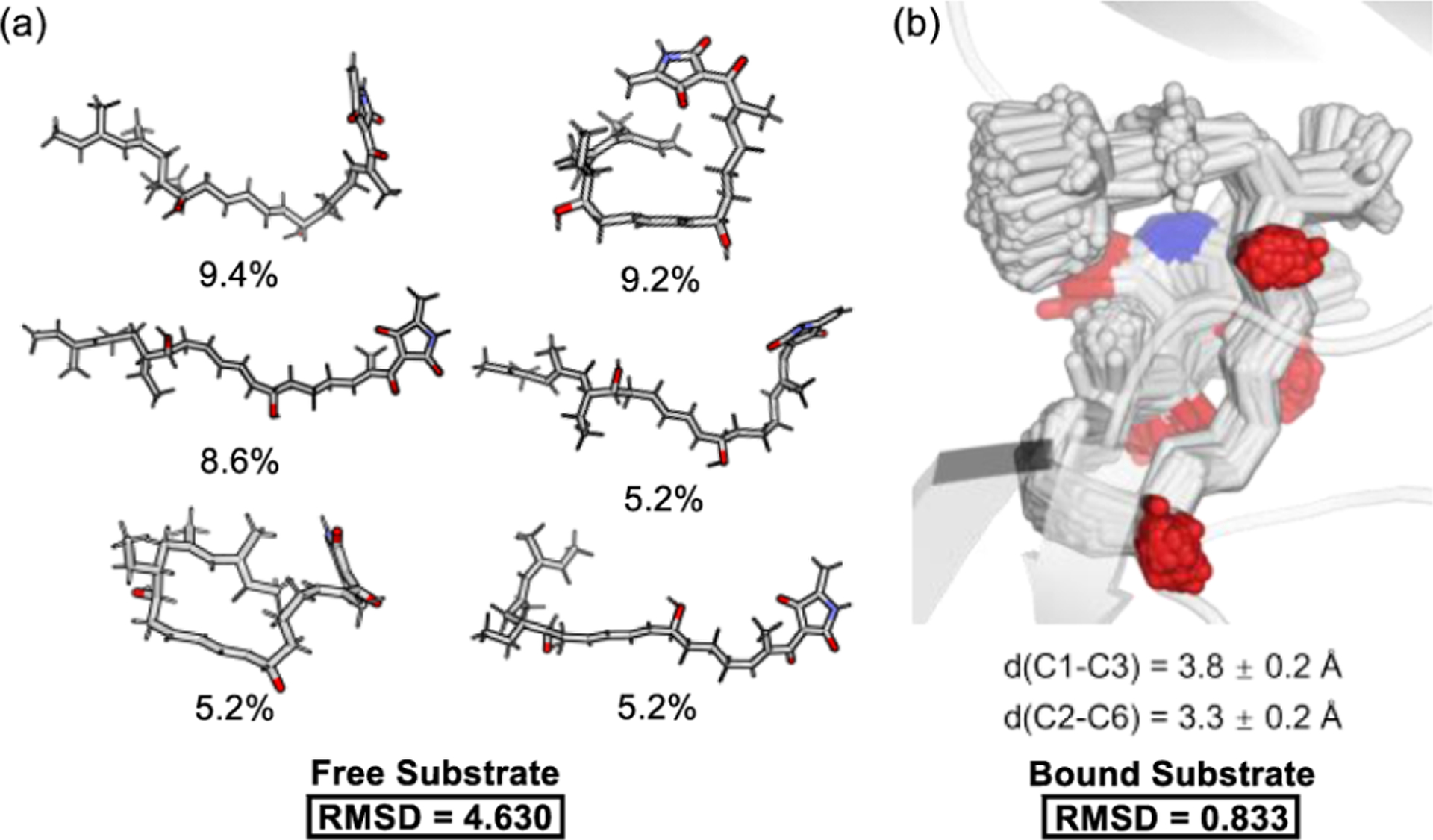
Change of substrate conformation by the PyrE3 active site. (a) Conformation clusters of the solvated substrate. (b) Conformation ensemble in the active site.
Since substrate prearrangement plays a critical role in the catalysis of PyrE3, it becomes important to understand the mode of substrate recognition by the active site. To this end, we carried out MM/GBSA binding energy calculations on the basis of MD simulations. The method combines molecular mechanics energies with generalized Born/surface area continuum solvation, and per-residue contributions can be identified to shed light on their individual roles.34 The MM/GBSA calculations show that the strongest electrostatic attraction results from three arginine residues around the pocket: R211 (−16.7, all in kcal/mol, see Figure 2c for the residues), R350 (−13.0), and R42 (−10.9). Neutral residues H74 (−6.0), H77 (−4.9), L283 (−4.8), and G284 (−3.9) also provide notable electrostatic attractions. Among them, H74, H77, and G284 form H-bonds to anchor and orient the 3-acyltetramate unit. These results explain the increased Km for the R211A (95), H74A (223), H77A (80), and H74A/H77A (477) mutants (52 for wild-type PyrE3).21 Because of the large non-polar interface, 12 residues display strong van der Waals (vdW) interactions with the substrate (larger than −1.0 kcal/mol). Moreover, the FAD co-factor not only acts as a structural motif to maintain the protein backbone and the shape of the active site but also provides non-covalent interactions with the bound substrate to enhance binding. The involvement of FAD and multiple residues was noted earlier in the X-ray crystallographic analysis of the PyrE3 active site.21
To reinforce substrate prearrangement, PyrE3 also possesses a rigid scaffold that can maintain the binding interactions and the shape of the pocket. The backbone of PyrE3 is largely unmoved before and after binding (Figure S3). Also, the substrate–enzyme complex of RCa-1 shows a low average rmsd of 1.63 Å (1.25 Å for the backbone atoms). These are contrasted against PyrI4, for which the entrance of the substrate triggers motions of the lid-like α0-helix to enclose the pocket,20,28 which reflects higher scaffold flexibility.
2.2.2. Enzymatic DA Reaction.
To gain more insights into the enzymatic DA reaction, we constructed a theozyme to carry out quantum mechanical studies at the level of SMD-M06–2X/6–311++G(d,p)//M06–2X/6–31G(d).35 As shown in Figure 4a, the theozyme includes the side chain of H74, the (simplified) flavin group of FAD, and the truncated P282-L283-G284-G285 and G44-A45-I46 loops. All truncation sites were capped with H atom(s). The model construction can reflect the main polar interactions in the pocket. Thermal fluctuations of the enzyme backbone were considered by evenly taking 10 snapshots from a 100 ns MD trajectory as initial geometries, giving rise to an ensemble of 10 theozymes (denoted by T1 to T10, see Figure 4b). Selected atoms were constrained to maintain the positions of the enzyme fragments and better reflect the rigid active pocket (marked with asterisks in Figure 4a). The remaining background of the enzyme is represented by a continuum model of diethyl ether.
Figure 4.
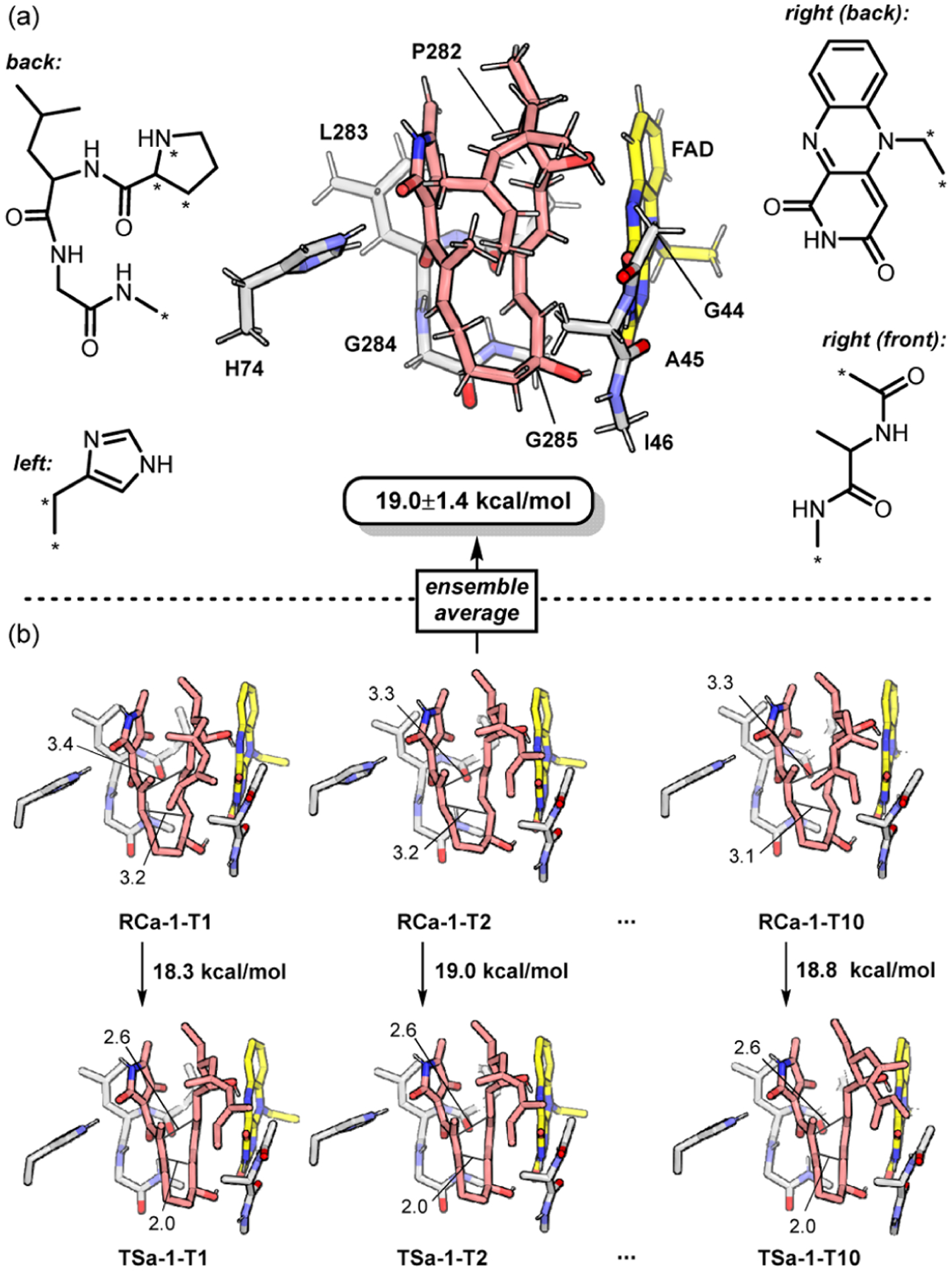
Theozyme model for the PyrE3 active site. (a) Illustration of the theozyme. Constrained atoms are marked with an asterisk. (b) Schematic description for the ensemble of 10 theozymes.
On account of the strong acidity of SM and the positive ESP (Scheme 1b) of the pocket, we propose that the anionic mechanism can be operative with PyrE3.21 This is supported by the agreement between experimental kinetics data (kcat = 223.2 ± 5.4 min−1,17 implying a 17.0 kcal/mol barrier) and the computed barrier (19.0 ± 1.4 kcal/mol, see Table S2 for details). The agreement shows again that PyrE3 differs from PyrI4, as acid catalysis plays an essential role in the latter case.20 The hypothesis thus provides a possible explanation for the distinct electrostatic environments of PyrE3 and PyrI4.
To estimate the catalytic effect of the active site, the barrier at the same pose without the theozyme was computed to be 22.5 kcal/mol, which increased by 3.5 kcal/mol. The theozyme can provide the necessary auxiliary to support shorter C1–C3 and C2–C6 distances in the reactant state (from 4.1 and 3.3 Å in relaxed RC to 3.6 ± 0.3 and 3.2 ± 0.1 Å in the theozyme, respectively). We reasoned that the active site of PyrE3 can result in an optimized diene–dienophile alignment and thereby promote the DA reaction,31 which further emphasizes the importance of substrate prearrangement and destabilization. On the other hand, although the dienophile moiety also forms H-bonds with the H74 side chain and (the N–H unit of) the L283-G284 peptide bond, the theozyme (and likely the PyrE3 enzyme) does not involve H-bond activation of the dienophile.21 In fact, removal of H74 or the L283-G284 peptide bond causes minor changes of 0.0 ± 0.3 and −0.2 ± 0.7 kcal/mol, respectively, to the reaction barrier. The absence of H-bond activation is ascribed to the inverse-electron-demand nature of the anionic DA pathway due to the elevated energy of π-HOMO (dienophile) (Figure 5). The electron-rich dienophile is not likely to accumulate electron density in the transition state, hence no enhancement of H-bonds with the enzyme.
Figure 5.

Frontier orbitals for diene and dienophile fragments.
2.2.3. Discussion.
To summarize, the main mode of catalysis by PyrE3 is the prearrangement of a linear anionic substrate into a near-attack RC with an ideal diene–dienophile alignment. The associated reactant destabilization effect renders an inherently inert inverse-electron-demand DA reaction facile under ambient conditions. The insights explain why most reported mutations undermine enzyme activity by increasing Km rather than decreasing kcat.21 These mutations (e.g., A45L, F70Q, and I213F) might only result in a local shape perturbation that affects substrate binding to some extent. However, when a mutation (e.g., H74A) removes a H-bond with the anionic 3-acyltetramte unit, a significant damage to both substrate binding and diene–dienophile alignment would cause simultaneous changes in Km and kcat (e.g., a 329% increase and a 15% decrease, respectively, for H74A).
2.3. Origin of Stereoselectivity.
With the plausible catalytic mechanism established, we explored the origin of stereoselectivity of PyrE3, which forms the endo-1 trans-decalin product P-1 with a high degree of stereocontrol.17 We remind the reader that four possible stereoisomeric DA adducts, namely P-1 through P-4, can be generated from the corresponding TSs TS-1 through TS-4 (1: endo-1, 2: exo-1, 3: endo-2, and 4: exo-2, see Scheme 2). To probe the intrinsic stereochemical preference, we carried out DFT computations for the uncatalyzed reaction (Figure 6). As presented in Figure 6a, We considered both protonation states because of the uncertainty of pKa calculations. The results suggest that the endo-1 transition states TSa-1 and TSn-1 (27.8 and 29.1 kcal/mol) are not the most favorable in both mechanisms. Instead, TSa-2 (26.0 kcal/mol) and TSn-2/TSn-3 (27.9 and 27.8 kcal/mol) display the lowest barriers, contradicting the stereochemical outcome in pyrroindomycin biosynthesis. These results rely on the lowest-energy conformers in the absence of PyrE3. Since PyrE3 can alter substrate conformation, we also calculated ΔΔG‡ between endo-1 and exo-1 adopting the pose in the bound state (i.e., as shown in Figure 2c) with the enzyme implicitly represented by a continuum model of diethyl ether (Figure 6b). The resulting ΔΔG‡ (0.6 kcal/mol), despite favoring endo-1 over exo-1, is inadequate for explaining the exclusive formation of Pa-1. In sum, these calculations show that Pa-1 is the only product of biosynthesis17 because the enzyme stabilizes the endo-1 pathway. This feature is explored in the following two subsections.
Figure 6.

DFT studies on stereoselectivity. (a) Free-energy profile for the uncatalyzed reaction at different stereochemical relationships. (b) Transition states for the endo-1 and exo-1 pathways at the prearranged pose.
2.3.1. Binding Affinities.
In consideration of the favorable binding interactions and shape complementarity between RCa-1 and the PyrE3 active site, we envisaged that RCa-1 (and thus the structurally similar TSa-1) may exhibit a stronger binding affinity than RCa-2 to RCa-4, thus accounting for the stereopure formation of Pa-1. To assess the total binding affinities of these stereoisomeric RCs, we computed the free-energy cost to reach the RCs (i.e., from SMa to the best docked structures) and the MM/GBSA binding affinities at the resultant RC. Despite overly large numbers (referenced vs gas phase), the MM/GBSA affinities are good indicators for relative affinities.
The results shown in Figure 7a suggest that RCa-1 is in an ideal binding mode with the lowest folding energy (7.9 kcal/mol from SMa to RCa-1) and the strongest binding affinity with the pocket (−55.0 kcal/mol at RCa-1), giving rise to a total affinity of −47.1 kcal/mol. The other three stereoisomeric RCs are docked into somewhat similar poses at which the anionic 3-acyltetramte moiety is positioned downward in the figures (i.e., interfacing with the FAD-binding domain in Figure 2a). The resulting poses of RCa-2 to RCa-4 are substantially less favored, with the folding free energies exceeding 11 kcal/mol and the affinities ranging from −46.1 to −49.5 kcal/mol. Consequently, the total binding affinities are −33.9 to −35.1 kcal/mol, which are notably lower than those of RCa-1. Therefore, the conformation selection of PyrE3 is strongly stereoselective such that the generations of side products Pa-2 to Pa-4 are disfavored. Besides the complementarity of RCa-1, we speculate that the key His and Arg residues on the upper side of the pocket (H74, H77, R211, R350, and R254) can also contribute to the stereo-differentiation. As depicted in Figure 7b and detailed in Table S4, a comparison of MM/GBSA per-residue binding energies discloses that these residues provide stronger electrostatic attractions with RCa-1 than with RCa-2 through RCa-4. This reflects the substrate-His H-bonds and the enhanced Coulomb attractions when the anionic 3-acyltetramate is positioned toward the upper side of the pocket where R211, R350, and R254 exist. These factors can carry over to the respective transition states (which are similar in structure to the RCs), whose energies ultimately determine the stereoselectivity of the catalyzed reaction.
Figure 7.

Assessment of binding affinities. (a) Binding affinities for stereoisomeric RCs RCa-1 through RCa-4. (b) Average differences in MM/GBSA per-residue binding affinities (RCa-1 minus RCa-2-RCa-4).
2.3.2. Stereocontrolling Active Site Environment.
The previous discussion established the energetic differentiation in the favorable binding of RCa-1. Since the transition states of endo-1 and exo-1 share congruent arrangements except for the diene unit (Figure 8a), we envisioned that some feature of the binding site near the reaction center would exert extra control over the orientation of the diene unit. In fact, MD simulations show that the side chain of A45 is complementary to the endo-diene unit of RCa-1 (Figure 8b,c). The corresponding exo-diene, however, will overlay onto A45 and is thus disfavored. This precisely placed steric hindrance must have also enhanced the stability of the cis-1,3-diene, which would otherwise easily turn into the trans conformer (Figure 3a). With details provided in the Supporting Information, we note that the constraint by A45 is stabilized by an FAD-mediated HB network (HB-5 through HB-8 in Figure 2c) as well as favorable vdW interactions.
Figure 8.

Stereocontrolling active site environment. (a) Overlaid geometry of the endo-1 and exo-1 transition states in Figure 6b. (b,c) Stereocontrolling constraint exerted on the 1,3-diene unit.
3. CONCLUSIONS
Pyrroindomycin biosynthesis involves two monofunctional DAases, PyrE3 and PyrI4, that stereoselectively construct the spirotetramate ring system via a cycloaddition cascade. Unlike the acid catalysis by PyrI4, PyrE3 uses a positively charged pocket to promote an inverse-electron-demand DA reaction of the anion with substrate preorganization. The pocket strongly favors an endo RC and maintains the stereochemical information by imposing an exo-disfavoring constraint. The catalysis relies on a synergy of specific binding interactions, electrostatic background, shape complementarity, and scaffold rigidity. Strikingly, the mechanisms of the two DA pathways in pyrroindomycin biosynthesis involve significantly different modes of catalysis. This highlights how nature has evolved multiple ways to catalyze DA reactions.
Supplementary Material
ACKNOWLEDGMENTS
B.L. thanks Prof. Yong Liang, Dr. Wanqing Wei, and Dr. Xin Wang (Nanjing University) for the helpful discussions. We are grateful to the National Institute of General Medical Sciences, the National Institutes of Health (GM 124480), the National Science Foundation (grant CHE-1764328), the National Natural Science Foundation of China (grant 32030002), and the Chinese Academy of Sciences (grant XDB20020200) for financial support of this research. All calculations were performed on the Hoffman2 cluster at the University of California, Los Angeles, and the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation (grant OCI-1053575).
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c00015.
Additional computational details, methods, and supplementary results, including energies of optimized structures, and coordinates of optimized structures (xyz) (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.2c00015
Contributor Information
Bo Li, Department of Chemistry and Biochemistry, University of California, Los Angeles, California 90095-1569, United States.
Xingyi Guan, Department of Chemistry and Biochemistry, University of California, Los Angeles, California 90095-1569, United States.
Song Yang, Department of Chemistry and Biochemistry, University of California, Los Angeles, California 90095-1569, United States.
Yike Zou, Department of Chemistry and Biochemistry, University of California, Los Angeles, California 90095-1569, United States.
Wen Liu, State Key Laboratory of Bioorganic and Natural Products Chemistry, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, Shanghai 200032, China.
K. N. Houk, Department of Chemistry and Biochemistry, University of California, Los Angeles, California 90095-1569, United States.
REFERENCES
- (1).Nicolaou KC; Snyder SA; Montagnon T; Vassilikogiannakis G The Diels-Alder reaction in total synthesis. Angew. Chem., Int. Ed 2002, 41, 1668–1698. [DOI] [PubMed] [Google Scholar]
- (2).Selected reviews for DAases:; (a) Jamieson CS; Ohashi M; Liu F; Tang Y; Houk KN The expanding world of biosynthetic pericyclases: cooperation of experiment and theory for discovery. Nat. Prod. Rep 2019, 36, 698–713. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lichman BR; O’Connor SE; Kries H Biocatalytic Strategies towards [4 + 2] Cycloadditions. Chem.—Eur. J 2019, 25, 6864–6877. [DOI] [PubMed] [Google Scholar]; (c) Walsh CT; Tang Y Recent Advances in Enzymatic Complexity Generation: Cyclization Reactions. Biochem 2018, 57, 3087–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Jeon B-S; Wang S-A; Ruszczycky MW; Liu H-W Natural [4 + 2]-Cyclases. Chem. Rev 2017, 117, 5367–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Minami A; Oikawa H Recent advances of Diels-Alderases involved in natural product biosynthesis. J. Antibiot 2016, 69, 500–506. [DOI] [PubMed] [Google Scholar]; (f) Klas K; Tsukamoto S; Sherman DH; Williams RM Natural Diels-Alderases: Elusive and Irresistable. J. Org. Chem 2015, 80, 11672–11685. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Oikawa H; Tokiwano T Enzymatic catalysis of the Diels-Alder reaction in the biosynthesis of natural products. Nat. Prod. Rep 2004, 21, 321–352. [DOI] [PubMed] [Google Scholar]
- (3).Selected recent discoveries of natural DAases:; (a) Gao L; Su C; Du X; Wang R; Chen S; Zhou Y; Liu C; Liu X; Tian R; Zhang L; Xie K; Chen S; Guo Q; Guo L; Hano Y; Shimazaki M; Minami A; Oikawa H; Huang N; Houk KN; Huang L; Dai J; Lei X FAD-dependent enzyme-catalysed intermolecular [4 + 2] cycloaddition in natural product biosynthesis. Nat. Chem 2020, 12, 620–628. [DOI] [PubMed] [Google Scholar]; (b) Hantke V; Skellam EJ; Cox RJ Evidence for enzyme catalysed intramolecular [4 + 2] Diels–Alder cyclization during the biosynthesis of pyrichalasin H. Chem. Commun 2020, 56, 2925. [DOI] [PubMed] [Google Scholar]; (c) Chen Q; Gao J; Jamieson C; Liu J; Ohashi M; Bai J; Yan D; Liu B; Che Y; Wang Y; Houk KN; Hu Y Enzymatic Intermolecular Hetero-Diels-Alder Reaction in the Biosynthesis of Tropolonic Sesquiterpenes. J. Am. Chem. Soc 2019, 141, 14052–14056. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Tan D; Jamieson CS; Ohashi M; Tang M-C; Houk KN; Tang Y Genome-Mined Diels-Alderase Catalyzes Formation of the cis-Octahydrodecalins of Varicidin A and B. J. Am. Chem. Soc 2019, 141, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Gao L; Zou Y; Liu X; Yang J; Du X; Wang J; Yu X; Fan J; Jiang M; Li Y; Houk KN; Lei X Enzymatic control of endo- and exo-stereoselective Diels–Alder reactions with broad substrate scope. Nat. Catal 2021, 4, 1059–1069. [Google Scholar]; (f) Fujiyama K; Kato N; Re S; Kinugasa K; Watanabe K; Takita R; Nogawa T; Hino T; Osada H; Sugita Y; Takahashi S; Nagano S Molecular Basis for Two Stereoselective Diels–Alderases that Produce Decalin Skeletons. Angew. Chem., Int. Ed 2021, 60, 22401–22410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Selected earlier explorations on catalytic antibodies for Diels–Alder reactions:; (a) Hilvert D; Hill KW; Nared KD; Auditor MTM Antibody catalysis of the Diels-Alder reaction. J. Am. Chem. Soc 1989, 111, 9261–9262. [Google Scholar]; (b) Janda KD; Shevlin CG; Lerner RA Antibody catalysis of a disfavored chemical transformation. Science 1993, 259, 490–493. [DOI] [PubMed] [Google Scholar]; (c) Gouverneur VE; Houk KN; de Pascual-Teresa B; Beno B; Janda KD; Lerner RA Control of the exo and endo Pathways of the Diels Alder Reaction by Antibody Catalysis. Science 1993, 262, 204–208. [DOI] [PubMed] [Google Scholar]; (d) Chen J; Deng Q; Wang R; Houk KN; Hilvert D Shape Complementarity, Binding-Site Dynamics, and Transition State Stabilization: A Theoretical Study of Diels-Alder Catalysis by Antibody 1E9. ChemBioChem 2000, 1, 255–261. [DOI] [PubMed] [Google Scholar]; (e) Xu J; Deng Q; Chen J; Houk KN; Bartek J; Hilvert D; Wilson IA Evolution of shape complementarity and catalytic efficiency from a primordial antibody template. Science 1999, 286, 2345–2348. [DOI] [PubMed] [Google Scholar]
- (5).Selected works for artificial DAases:; (a) Preiswerk N; Beck T; Schulz JD; Milovník P; Mayer C; Siegel JB; Baker D; Hilvert D Impact of scaffold rigidity on the design and evolution of an artificial Diels-Alderase. Proc. Natl. Acad. Sci. U.S.A 2014, 111, 8013–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hilvert D Design of protein catalysts. Annu. Rev. Biochem 2013, 82, 447–470. [DOI] [PubMed] [Google Scholar]; (c) Eiben CB; Siegel JB; Bale JB; Cooper S; Khatib F; Shen BW; Players F; Stoddard BL; Popovic Z; Baker D Increased Diels-Alderase activity through backbone remodeling guided by Foldit players. Nat. Biotechnol 2012, 30, 190–192. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Siegel JB; Zanghellini A; Lovick HM; Kiss G; Lambert AR; Clair JL; Gallaher JL; Hilvert D; Gelb MH; Stoddard BL; Houk KN; Michael FE; Baker D Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science 2010, 329, 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Basler S; Studer S; Zou Y; Mori T; Ota Y; Camus A; Bunzel HA; Helgeson RC; Houk KN; Jiménez-Osés G; Hilvert D Efficient Lewis acid catalysis of an abiological reaction in a de novo protein scaffold. Nat. Chem 2021, 13, 231–235. [DOI] [PubMed] [Google Scholar]; (f) For a relevant review, see Ghattas W; Mahy JP; Réglier M; Simaan AJ Artificial Enzymes for Diels-Alder Reactions. ChemBioChem 2021, 22, 443–459. [DOI] [PubMed] [Google Scholar]
- (6).Selected examples of biomimetic DA catalysts:; (a) Palma A; Artelsmair M; Wu G; Lu X; Barrow SJ; Uddin N; Rosta E; Masson E; Scherman OA Cucurbit[7]uril as a Supramolecular Artificial Enzyme for Diels–Alder Reactions. Angew. Chem., Int. Ed 2017, 56, 15688–15692. [DOI] [PubMed] [Google Scholar]; (b) Martí-Centelles V; Lawrence AL; Lusby PJ High Activity and Efficient Turnover by a Simple, Self-Assembled “Artificial Diels-Alderase”. J. Am. Chem. Soc 2018, 140, 2862–2868. [DOI] [PubMed] [Google Scholar]
- (7).Hashimoto T; Kuzuyama T Mechanistic Insights into Diels-Alder Reactions in Natural Product Biosynthesis. Curr. Opin. Chem. Biol 2016, 35, 117–123. [DOI] [PubMed] [Google Scholar]
- (8).Kim HJ; Ruszczycky MW; Liu H-W Current developments and challenges in the search for a naturally selected Diels-Alderase. Curr. Opin. Chem. Biol 2012, 16, 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Auclair K; Sutherland A; Kennedy J; Witter DJ; Van den Heever JP; Hutchinson CR; Vederas JC Lovastatin Nonaketide Synthase Catalyzes an Intramolecular Diels-Alder Reaction of a Substrate Analogue. J. Am. Chem. Soc 2000, 122, 11519–11520. [Google Scholar]
- (10).(a) Kim R-R; Illarionov B; Joshi M; Cushman M; Lee CY; Eisenreich W; Fischer M; Bacher A Mechanistic Insights on Riboflavin Synthase Inspired by Selective Binding of the 6,7-Dimethyl-8-ribityllumazine Exomethylene Anion. J. Am. Chem. Soc 2010, 132, 2983–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Breugst M; Eschenmoser A; Houk KN Theoretical Exploration of the Mechanism of Riboflavin Formation from 6,7-Dimethyl-8-ribityllumazine: Nucleophilic Catalysis, Hydride Transfer, Hydrogen Atom Transfer, or Nucleophilic Addition? J. Am. Chem. Soc 2013, 135, 6658–6668. [DOI] [PubMed] [Google Scholar]
- (11).(a) Guimarães CRW; Udier-Blagović M; Jorgensen WL Macrophomate Synthase: QM/MM Simulations Address the Diels-Alder versus Michael-Aldol Reaction Mechanism. J. Am. Chem. Soc 2005, 127, 3577–3588. [DOI] [PubMed] [Google Scholar]; (b) Serafimov JM; Gillingham D; Kuster S; Hilvert D The Putative Diels-Alderase Macrophomate Synthase is an Efficient Aldolase. J. Am. Chem. Soc 2008, 130, 7798–7799. [DOI] [PubMed] [Google Scholar]
- (12).Want T; Hoye TR Diels-Alderase-free, bis-pericyclic, [4 +2] dimerization in the biosynthesis of (±)-paracaseolide A. Nat. Chem 2015, 7, 641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Zhang B; Wang KB; Wang W; Wang X; Liu F; Zhu J; Shi J; Li LY; Han H; Xu K; Qiao HY; Zhang X; Jiao RH; Houk KN; Liang Y; Tan RX; Ge HM Enzyme-catalysed [6 +4] cycloadditions in the biosynthesis of natural products. Nature 2019, 568, 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Fage CD; Isiorho EA; Liu Y; Wagner DT; Liu H-W; Keatinge-Clay AT The structure of SpnF, a standalone enzyme that catalyzes [4 + 2] cycloaddition. Nat. Chem. Biol 2015, 11, 256–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).(a) Patel A; Chen Z; Yang Z; Gutiérrez O; Liu H-W; Houk KN; Singleton DA Dynamically Complex [6 + 4] and [4 +2] Cycloadditions in the biosynthesis of spinosyn A. J. Am. Chem. Soc 2016, 138, 3631–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yang Z; Yang S; Yu P; Li Y; Doubleday C; Park J; Patel A; Jeon B-S; Russell WK; Liu H-W; Russell DH; Houk KN Influence of water and enzyme SpnF on the dynamics and energetics of the ambimodal [6 + 4]/[4 + 2] cycloaddition. Proc. Natl. Acad. Sci. U.S.A 2018, 115, No. E848–E855. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Medvedev MG; Zeifman AA; Novikov FN; Bushmarinov IS; Stroganov OV; Titov IY; Chilov GG; Svitanko IV Quantifying Possible Routes for SpnF-Catalyzed Formal Diels-Alder Cycloaddition. J. Am. Chem. Soc 2017, 139, 3942–3945. [DOI] [PubMed] [Google Scholar]; (d) Jeon B-S; Ruszczycky MW; Russell WK; Lin G-M; Kim N; Choi S-H; Wang S-A; Liu Y-N; Patrick JW; Russell DH; Liu H-W Investigation of the mechanism of the SpnF-catalyzed [4 + 2]-cycloaddition reaction in the biosynthesis of spinosyn A. Proc. Natl. Acad. Sci. U.S.A 2017, 114, 10408–10413. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Zheng Y; Thiel W Computational Insights into an Enzyme-Catalyzed [4 + 2] Cycloaddition. J. Org. Chem 2017, 82, 13563–13571. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Chen N; Zhang F; Wu R; Hess BA Jr. Biosynthesis of Spinosyn A: A [4 + 2] or [6 + 4] Cycloaddition? ACS Catal. 2018, 8, 2353–2358. [Google Scholar]; (g) Wang X; Zhang C; Jiang Y; Wang W; Zhou Y; Chen Y; Zhang B; Tan RX; Ge HM; Yang ZJ; Liang Y Influence of Water and Enzyme on the Post-Transition State Bifurcation of NgnD-Catalyzed Ambimodal [6+4]/[4+2] Cycloaddition. J. Am. Chem. Soc 2021, 143, 21003–21009. [DOI] [PubMed] [Google Scholar]
- (16).Kim HJ; Ruszczycky MW; Choi S-H; Liu Y-N; Liu H-W Enzyme-catalysed [4 + 2] cycloaddition is a key step in the biosynthesis of spinosyn A. Nature 2011, 473, 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Tian Z; Sun P; Yan Y; Wu Z; Zheng Q; Zhou S; Zhang H; Yu F; Jia X; Chen D; Mándi A; Kurtán T; Liu W An enzymatic [4 + 2] cyclization cascade creates the pentacyclic core of pyrroindomycins. Nat. Chem. Biol 2015, 11, 259–265. [DOI] [PubMed] [Google Scholar]
- (18).Ding W; Williams DR; Northcote P; Siegel MM; Tsao R; Ashcroft J; Morton GO; Alluri M; Abbanat D; Maiese WM; Ellestad GA Pyrroindomycins, novel antibiotics produced by Streptomyces rugosporus sp. LL-42D005. J. Antibiot 1994, 47, 1250–1257. [DOI] [PubMed] [Google Scholar]
- (19).(a) Li L; Yu P; Tang M-C; Zou Y; Gao S-S; Hung Y-S; Zhao M; Watanabe K; Houk KN; Tang Y Biochemical Characterization of a Eukaryotic Decalin-Forming Diels–Alderase. J. Am. Chem. Soc 2016, 138, 15837–15840. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang Z; Jamieson CS; Zhao Y-L; Li D; Ohashi M; Houk KN; Tang Y Enzyme-Catalyzed Inverse-Electron Demand Diels-Alder Reaction in the Biosynthesis of Antifungal Ilicicolin H. J. Am. Chem. Soc 2019, 141, 5659–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Sato M; Yagishita F; Mino T; Uchiyama N; Patel A; Chooi Y-H; Goda Y; Xu W; Noguchi H; Yamamoto T; Hotta K; Houk KN; Tang Y; Watanabe K Involvement of Lipocalin-Like CghA in Decalin-Forming Stereoselective Intra-molecular [4 + 2] Cycloaddition. ChemBioChem 2015, 16, 2294–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Li L; Yu P; Tang M-C; Zou Y; Gao S-S; Hung Y-S; Zhao M; Watanabe K; Houk KN; Tang Y Biochemical Characterization of a Eukaryotic Decalin-Forming Diels-Alderase. J. Am. Chem. Soc 2016, 138, 15837–15840. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Kato N; Nogawa T; Hirota H; Jang J-H; Takahashi S; Ahn JS; Osada H A New Enzyme Involved in the Control of the Stereochemistry in the Decalin Formation during Equisetin Biosynthesis. Biochem. Biophys. Res. Commun 2015, 460, 210–215. [DOI] [PubMed] [Google Scholar]
- (20).Zou Y; Yang S; Sanders JN; Li W; Yu P; Wang H; Tang Z; Liu W; Houk KN Computational Investigation of the Mechanism of Diels–Alderase PyrI4. J. Am. Chem. Soc 2020, 142, 20232–20239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Zheng Q; Gong Y; Guo Y; Zhao Z; Wu Z; Zhou Z; Chen D; Pan L; Liu W Structural insights into a flavin-dependent [4 + 2] cyclase that catalyzes trans-decalin formation in pyrroindomycin biosynthesis. Cell Chem. Biol 2018, 25, 718–727. [DOI] [PubMed] [Google Scholar]
- (22).Frisch MJ; Trucks GW; Schlegel HB; Scuseria GE; Robb MA; Cheeseman JR; Scalmani G; Barone V; Mennucci B; Petersson GA; Nakatsuji H; Caricato M; Li X; Hratchian HP; Izmaylov AF; Bloino J; Zheng G; Sonnenberg JL; Hada M; Ehara M; Toyota K; Fukuda R; Hasegawa J; Ishida M; Nakajima T; Honda Y; Kitao O; Nakai H; Vreven T; Montgomery JA Jr.; Peralta JE; Ogliaro F; Bearpark M; Heyd JJ; Brothers E; Kudin KN; Staroverov VN; Keith T; Kobayashi R; Normand J; Raghavachari K; Rendell A; Burant JC; Iyengar SS; Tomasi J; Cossi M; Rega N; Millam JM; Klene M; Knox JE; Cross JB; Bakken V; Adamo C; Jaramillo J; Gomperts R; Stratmann RE; Yazyev O; Austin AJ; Cammi R; Pomelli C; Ochterski JW; Martin RL; Morokuma K; Zakrzewski VG; Voth GA; Salvador P; Dannenberg JJ; Dapprich S; Daniels AD; Farkas O; Foresman JB; Ortiz JV; Cioslowski J; Fox DJ Gaussian 09, Revision D.01; Gaussian, Inc.: Wallingford, CT, 2013. [Google Scholar]
- (23).Zhao Y; Truhlar DG The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, non-covalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc 2008, 120, 215–241. [Google Scholar]
- (24).Marenich AV; Cramer CJ; Truhlar DG Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [DOI] [PubMed] [Google Scholar]
- (25).(a) Mulholland TPC; Foster R; Haydock DB Synthesis of pyrrolidine-2,4-diones(tetramic acids) and some derivatives. J. Chem. Soc. Perkin Trans. I 1972, 2121–2128. [Google Scholar]; (b) Stickings CE Studies in the biochemistry of micro-organisms. 106. Metabolites of Alternaria tenuis auct.: the structure of tenuazonic acid. Biochem. J 1959, 72, 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Cherian PT; Deshpande A; Cheramie MN; Bruhn DF; Hurdle JG; Lee RE Design, synthesis and microbiological evaluation of ampicillin–tetramic acid hybrid antibiotics. J. Antibiot 2017, 70, 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Yamaguchi T; Saito K; Tsujimoto T; Yuki H NMR spectroscopic studies on the tautomerism in tenuazonic acid analogs. J. Heterocycl. Chem 1976, 13, 533–537. [Google Scholar]
- (26).(a) Trott O; Olson AJ AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem 2010, 31, 455. [DOI] [PMC free article] [PubMed] [Google Scholar]; Case DA; Ben-Shalom IY; Brozell SR; Cerutti DS; Cheatham TE III; Cruzeiro VWD; Darden TA; Duke RE; Ghoreishi D; Gilson MK; Gohlke H; Goetz AW; Greene D; Harris R; Homeyer N; Izadi S; Kovalenko A; Kurtzman T; Lee TS; LeGrand S; Li P; Lin C; Liu J; Luchko T; Luo R; Mermelstein DJ; Merz KM; Miao Y; Monard G; Nguyen C; Nguyen H; Omelyan I; Onufriev A; Pan F; Qi R; Roe DR; Roitberg A; Sagui C; Schott-Verdugo S; Shen J; Simmerling CL; Smith J; Salomon-Ferrer R; Swails J; Walker RC; Wang J; Wei H; Wolf RM; Wu X; Xiao L; York DM; Kollman PA AMBER 2018; University of California, San Francisco, CA, 2018. [Google Scholar]
- (27).Olsson MHM; Søndergaard CR; Rostkowski M; Jensen JH PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput 2011, 7, 525–537. [DOI] [PubMed] [Google Scholar]
- (28).Zheng Q; Guo Y; Yang L; Zhao Z; Wu Z; Zhang H; Liu J; Cheng X; Wu J; Yang H; Jiang H; Pan L; Liu W Enzyme-dependent [4 + 2] cycloaddition depends on lid-like interaction of the N-terminal sequence with the catalytic core in PyrI4. Cell Chem. Biol 2016, 23, 352–360. [DOI] [PubMed] [Google Scholar]
- (29).Byrne MJ; Lees NR; Han L-C; van der Kamp MW; Mulholland AJ; Stach JEM; Willis CL; Race PR The Catalytic Mechanism of a Natural Diels-Alderase Revealed in Molecular Detail. J. Am. Chem. Soc 2016, 138, 6095–6098. [DOI] [PubMed] [Google Scholar]
- (30).Jencks WP Binding energy, specificity, and enzymic catalysis: The Circe effect. Adv. Enzymol. Relat. Areas Mol. Biol 1975, 43, 219–410. [DOI] [PubMed] [Google Scholar]
- (31).(a) Hur S; Bruice TC The mechanism of catalysis of the chorismate to prephenate reaction by the Escherichia coli mutase enzyme. Proc. Natl. Acad. Sci. U.S.A 2002, 99, 1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hur S; Bruice TC The near attack conformation approach to the study of the chorismate to prephenate reaction. Proc. Natl. Acad. Sci. U.S.A 2003, 100, 12015–12020. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Hur S; Bruice TC Enzymes Do What Is Expected (Chalcone Isomerase versus Chorismate Mutase). J. Am. Chem. Soc 2003, 125, 1472–1473. [DOI] [PubMed] [Google Scholar]; (d) Hur S; Bruice TC Comparison of Formation of Reactive Conformers (NACs) for the Claisen Rearrangement of Chorismate to Prephenate in Water and in the E. coli Mutase: The Efficiency of the Enzyme Catalysis. J. Am. Chem. Soc 2003, 125, 5964–5972. [DOI] [PubMed] [Google Scholar]
- (32).(a) DeTar DF Theoretical ab Initio Calculation of Entropy, Heat Capacity, and Heat Content. J. Phys. Chem. A 1998, 102, 5128–5141. [Google Scholar]; (b) Pracht P; Grimme S Calculation of absolute molecular entropies and heat capacities made simple. Chem. Sci 2021, 12, DOI: DOI: 10.1039/D1SC00621E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Sievers A; Beringer M; Rodnina MV; Wolfenden R The ribosome as an entropy trap. Proc. Natl. Acad. Sci. U.S.A 2004, 101, 7897–7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Genheden S; Ryde U The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expet Opin. Drug Discov 2015, 10, 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).(a) Tantillo DJ; Jiangang C; Houk KN Theozymes and compuzymes: theoretical models for biological catalysis. Curr. Opin. Chem. Biol 1998, 2, 743–750. [DOI] [PubMed] [Google Scholar]; (b) Ujaque G; Tantillo DJ; Hu Y; Houk KN; Hotta K; Hilvert D Catalysis on the coastline: theozyme, molecular dynamics, and free energy perturbation analysis of antibody 21D8 catalysis of the decarboxylation of 5-nitro-3-carboxybenzisoxazole. J. Comput. Chem 2003, 24, 98–110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


