Abstract
Purpose
Low‐field (B0 < 0.1 T) MRI has generated much interest as a means of increased accessibility via reduced cost and improved portability compared to conventional clinical systems (B0 ≥ 1.5 Tesla). Here we measure MR relaxation times at 50 mT and compare results with commonly used models based on both in vivo and ex vivo measurements.
Methods
Using 3D turbo spin echo readouts, T1 and T2 maps of the human brain and lower leg were acquired on a custom‐built 50 mT MRI scanner using inversion‐recovery and multi‐echo–based sequences, respectively. Image segmentation was performed based on a histogram analysis of the relaxation times.
Results
The average T1 times of gray matter, white matter, and cerebrospinal fluid (CSF) were 327 ± 10 ms, 275 ± 5 ms, and 3695 ± 287 ms, respectively. Corresponding values of T2 were 102 ± 6 ms, 102 ± 6 ms, and 1584 ± 124 ms. T1 times in the calf muscle were measured to be 171 ± 11 ms and were 130 ± 5 ms in subcutaneous and bone marrow lipid. Corresponding T2 times were 39 ± 2 ms in muscle and 90 ± 13 ms in lipid.
Conclusions
For tissues except for CSF, the measured T1 times are much shorter than reported at higher fields and generally lie within the range of different models in the literature. As expected, T2 times are similar to those seen at typical clinical field strengths. Analysis of the relaxation maps indicates that segmentation of white and gray matter based purely on T1 or T2 will be quite challenging at low field given the relatively small difference in relaxation times.
Keywords: gray matter, lipid, low‐field MRI, muscle, relaxation times, white matter
1. INTRODUCTION
There is growing interest in low‐field MRI systems as a reduced cost and lower footprint alternative/addition to clinical 1.5 Tesla (T) and 3T systems, particularly as a way of bringing MRI to low resource settings where conventional MR systems are not accessible. 1 , 2 , 3 Various system designs have been proposed: many are based on Halbach and other permanent magnet array systems, either with a homogenous B0 field 4 , 5 or with a built‐in spatial encoding gradient, 6 , 7 , 8 , 9 yoked permanent magnets, 10 , 11 , 12 fast field‐cycling systems, 13 and electromagnets. 14 , 15 Of these systems, a number operate at magnetic field strengths between ~50 and ~80 mT. At low field, there are major advantages in terms of increased implant safety and reduced implant‐induced susceptibility artefacts, 16 as well as reduced specific absorption rate (SAR). 17 The major disadvantage of operating at these field strengths is the significantly reduced signal to noise ratio (SNR) due its power law dependence on the B0 field strength, with a coefficient of ~7/4 at low field where coil noise rather than body noise is the dominant loss‐term. 18
Some of this loss in SNR may, however, be recovered because of favorable relaxation times and fewer limitations on RF power due to the lower specific absorption rate. 19 In vivo T1 relaxation times, dominated by dipole–dipole interactions, are well known to be shorter at lower magnetic fields. 20 , 21 , 22 , 23 , 24 , 25 The T1 value is related to components of the spectral density at the Larmor frequency and twice this frequency. The general relaxation model has 2 components—free and bound water—undergoing exchange that is rapid compared to the MR measurement time scale. The fast exchange 2‐state model shows that the measured relaxation rate is the weighted average of bound and free water. As discussed in Korb and Bryant, 21 the magnetic field dependence arises from the magnetic coupling of water protons to the solid components of the tissue, that is, water molecule exchange between specific binding sites, and to a much lesser degree proton exchange with specific groups (amines, amides, and alcohols) on protein molecules. In lipid tissue, hydrogen nuclei are present mainly in long chain triglycerides, which have relatively slow rotational and molecular motion and therefore generally have a shorter T1 value than tissue water.
T2 times show a much weaker field strength dependence, 22 although tissues with substantial iron concentrations have significantly longer T2 values at lower fields. 24 , 26 The weak dependence is due to the fact that the frequency‐independent static component (0 order) of the spectral density functions for most tissues dominate the first‐ and second‐order terms (which are frequency‐dependent): the greater the degree to which T2 < T1, the lower the frequency dependence.
Whereas in vivo relaxation times have extensively been studied in both healthy subjects and in many pathologies at clinical field strengths, the available literature at low field strength is much sparser. Bottomley 22 collected a large set of relaxation times acquired at different field strengths, primarily from ex vivo samples, including animal tissue. An empirical model for the relaxation time as a function of magnetic field strength was generated from this data and can be used to estimate the T1 relaxation times for various tissues as a function of magnetic field strength, albeit with quite a large standard deviation in the predicted values. Rooney et al 20 performed an in vivo study on brain tissue at several different field strengths and derived a similar empirical formula for T1 relaxation, albeit with different coefficients than those of Bottomley. Fischer et al 23 extended these models to take into account the actual upper and lower bounds of the relaxation time values at very low and very high fields.
In this work, we acquire in vivo T1 and T2 relaxation time maps of the brain and lower leg in healthy subjects on a custom‐built 50 mT permanent magnet‐based MRI scanner. We compare the measured values with those predicted by the models mentioned in the previous paragraph, and also with selected measurements at similar fields. We also discuss the implications for image segmentation at low field.
2. METHODS
2.1. Hardware
All data were acquired on a custom‐built 50 mT (2.15 MHz) Halbach‐based MRI scanner that was previously described in detail. 4 , 5 The magnet is constructed using 2948 12‐mm cuboid N48 neodymium iron boron magnets arranged in a cylindrical Halbach configuration. The magnet is 50.6 cm long and has a 27‐cm diameter bore. The magnetic field homogeneity was optimized over a 20‐cm diameter spherical volume placed at the center of the magnet. A set of 3 linear gradient coils was constructed using an target field method initially proposed by Turner, 27 adapted for the transverse B0 orientation intrinsic to cylindrical Halbach arrays. 28 A Magritek Kea2 spectrometer (Aachen, Germany) was used to generate RF and gradient waveforms and digitize the generated signals. A custom‐built 1 kW RF amplifier with 56 dB gain was used to amplify the RF pulses. 4 The gradient waveforms were amplified using a custom‐built 3‐axis current‐controlled gradient amplifier, powered using 2 Delta Elektronika SM 18‐50 DC power supplies (Zierikzee, the Netherlands). The entire setup is placed inside a Faraday cage constructed from aluminium extrusion and 2‐mm thick aluminium plates. An RF shield is placed inside the inner surface of the bore. During in vivo experiments, the body extends out of the Faraday and couples significant amounts of electromagnetic interference into the RF coil. In order to reduce electromagnetic interference, this the body is placed under a conductive cloth (4711 series, Holland Shielding Systems BV, Dordrecht, the Netherlands).
Brain imaging was performed using an elliptical 20‐cm wide, 25‐cm tall, 15‐cm deep spiral‐solenoid head coil 15 constructed using 0.8 mm copper wire and with a single capacitive segmentation. The bandwidth of the coil was 35 kHz when loaded with a head. Data on the lower leg were acquired using a 15‐turn, 15‐cm long, 15‐cm diameter solenoid constructed using 0.8‐mm diameter copper wire. The bandwidth of the coil was 23 kHz when the coil was loaded with the lower leg. Power optimization for both coils was performed by acquiring 16 spectra with 1 dB increments in the transmitted RF power. The area under the measured signal in the frequency domain was integrated from −1 to 1 kHz around the central frequency for each of the 16 steps, and a sinusoidal curve was fitted to the linearized power. First‐order B0 shimming was performed using the standard automatic shimming algorithm included with the spectrometer, which maximizes the peak of the spectrum of a nonselective FID by applying pseudo‐random gradient offsets to 3 linear gradient coils. Line widths (measured as the full width at half maximum) were less than 1 kHz on all volunteers after shimming. The center frequency of the sample was measured prior to every scan.
2.2. Relaxation mapping
Human studies were conducted with the approval of the institutional review board. Conventional inversion‐recovery and multiple spin‐echo sequences were used to map T1 and T2, respectively. Total imaging times were relatively long and could potentially be shortened considerably by use of more efficient sequences such as Look‐Locker, 29 MR fingerprinting, 30 or compressed sensing. 31 In this preliminary work, however, the choice was made to use conventional sequences for maximum confidence in the reported values. Separate sessions were used for CSF measurements because the long T1 time in particular results in a long imaging session. Our imaging protocol is limited to 1 h maximum; thus, separate measurement sessions were required for each relaxation time.
Data were corrected for any frequency drift induced by heating of the magnet during the acquisition by applying appropriate phase shifts of the k‐space data. The center frequency was measured immediately before and after the scan, and the drift is assumed to be spatially homogeneous over the imaging region and linear over time. Values were typically 500 Hz over the ~45 min imaging time. All data were acquired on healthy volunteers aged 27 to 62 years (6 male, 4 female). Informed consent was obtained from all subjects prior to scanning.
2.3. Brain relaxation time mapping
T1 maps of the gray matter (GM) and white matter (WM) were reconstructed from 6 whole‐brain 3D inversion recovery scans with a turbo spin echo readout acquired with the following scan parameters: resolution: 2.5 × 2.5 × 5 mm3; TR/TE/TEeff: 1250 ms/13 ms/13 ms (center‐out Cartesian k‐space trajectory); no signal averaging; echo train length: 6; and acquisition bandwidth: 20 kHz. Scans were acquired with inversion times of 50, 100, 150, 200, 300, and 500 ms; total scan duration was around 36 min.
T2 maps of the GM and WM were reconstructed from a 3D Carr‐Purcell‐Meiboom‐Gill (CPMG) imaging sequence acquired with the following scan parameters: resolution 2.5 × 2.5 × 5 mm3; TR: 1250 ms; no signal averaging; and acquisition bandwidth: 20 kHz. A total of 10 echoes were acquired with a constant echo spacing of 20 ms; images were reconstructed separately for each of the 10 echo times (TEs).
Separate scans were used to more accurately map the T1 and T2 of CSF due to their much longer values than for WM/GM. For T1: resolution: 2.5 × 2.5 × 5 mm3; TR/TE/TEeff = 12000 ms/11 ms/500 ms (low‐to‐high Cartesian k‐space trajectory); echo train length: 90; and inversion times: 500 ms, 1500 ms, 2000 ms, 2500 ms, 3000 ms, and 4000 ms. Total scan duration for the T1 mapping sequence was around 36 min. Given the relatively coarse spatial resolution, the influence of partial voluming of voxels with CSF and WM/GM could be problematic, which is why a long effective TE using low‐to‐high k‐space coverage (approximately 5 times the T2 of WM/GM) was used to suppress the signal from the WM/GM. T2 maps of the CSF were reconstructed from 7 different turbo spin echo scans: TR = 10,000 ms; echo train length = 60; and TE/TEeff = 15/450, 30/900, 45/1350, 60/1800, 75/2250, 90/2700, 105/3150 ms (low to high Cartesian k‐space coverage). Total acquisition time was around 42 min.
2.4. Lower leg relaxation time mapping
T1 maps: spatial resolution: 2.5 × 2.5 × 5 mm3; TR/TE/TEeff = 850 ms/10 ms/10 ms; echo train length: 4; no signal averaging; imaging bandwidth: 20 kHz. Scans were acquired with 6 different inversion times: 25, 50, 75, 100, 150, and 400 ms. Total scan duration for the T1 mapping sequence was around 36 min. T2 maps: spatial resolution: 2 × 2 × 6 mm3; TR: 800 ms; TE: 10 different TEs 11 to 110 ms in 11 ms steps; and no signal averaging. Scan duration was approximately 28 min.
2.5. Data processing
All images were reconstructed using the Numpy fast Fourier transform implementation, with k‐space filtered using a sine‐bell‐squared filter. Parameter maps were reconstructed from a central slice of the 3D reconstruction on a voxel‐by‐voxel basis using the least squares function in SciPy 1.6.0 running in Python 3.7.3. For T1 mapping the data were fitted to:
| (1) |
The initial guesses for and provided to the least squares function are given by:
| (2) |
| (3) |
For T2 mapping, the data were fitted to:
| (4) |
The initial guesses for and are given by:
| (5) |
| (6) |
Gaussian curves were fit to the histogram, as in Ref. [24] using the curve‐fit fitting function from SciPy 1.6.0. A segmented map was generated for each relaxation time map based on the fitted curves; data points within 2 SDs of the mean were assigned to a curve. If 2 curves overlapped, the crossing point between the 2 curves was set as the value limit for each of the tissue types (eg, WM was limited to less than 300 ms and GM to more than 300 ms in Figure 1).
FIGURE 1.
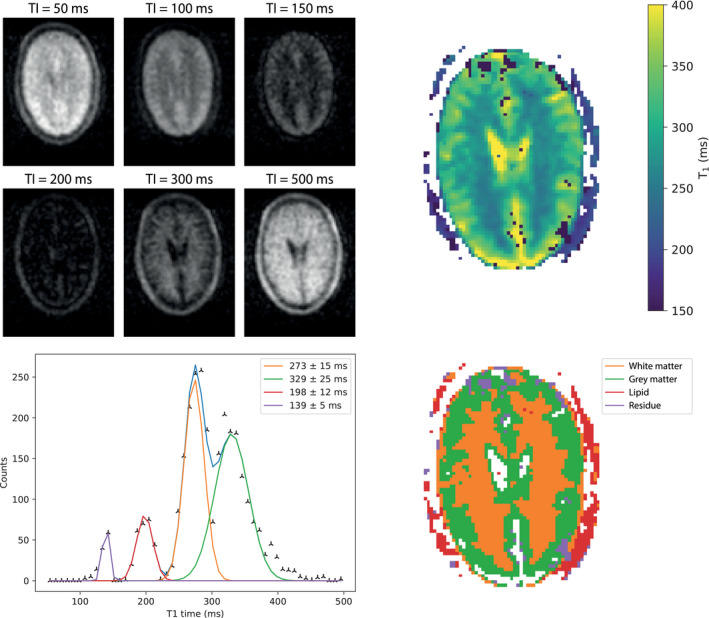
(Top left) Six brain images (central slice) acquired using an inversion‐recovery sequence with different inversion times. (Top right) A T1 map calculated from the acquired images. (Bottom left) A plot of binned T1 values; 4 Gaussian curves are fit to the histogram. (Bottom right) A segmented map of the brain with the color corresponding to the area under each fitted curve: orange is assigned to white matter, green to gray matter, red to lipid, and purple is residual.
3. RESULTS
Figure 1 shows a T1 map acquired from 1 of the healthy volunteers showing T1 contrast between GM and WM as well as lipid. It should be noted that the T1 values obtained in the CSF are inaccurate due to saturation effects, resulting from incomplete longitudinal relaxation due to the short repetition time (1250 ms) relative to the T1 time (around 4 s), which is why a separate scan was performed for accurate quantification. The measured T1 times for each individual are reported in Table 1: the mean T1 times across all 3 subjects are 327 ± 10 ms in the GM and 275 ± 5 ms in the WM.
TABLE 1.
Measured T1 and T2 relaxation times in healthy volunteers
| GM | WM | CSF | GM (Bottomley) | WM (Bottomley) | |
|---|---|---|---|---|---|
| T1 (ms) | 329 ± 25 | 273 ± 15 | 3528 ± 192 | ||
| 335 ± 19 | 280 ± 14 | 4033 ± 325 | 325 ± 55 | 242 ± 41 | |
| 316 ± 19 | 271 ± 7 | 3546 ± 134 | |||
| T2 (ms) | 97 ± 10 | 1687 ± 249 | |||
| 100 ± 8 | 1448 ± 226 | 101 ± 13 | 92 ± 20 | ||
| 108 ± 10 | 1626 ± 253 | ||||
Values (where available) from Bottomley et al 22 are calculated for a field strength of 50 mT.
Abbreviations: GM, gray matter; WM, white matter.
A T2 map of the brain of a different volunteer is shown in Figure 2. The mean T2 measured in both the GM and WM across the 3 subjects is 102 ± 6 ms. Note that the T2 of times of CSF are not accurately represented in this data set due to T1 saturation effects.
FIGURE 2.
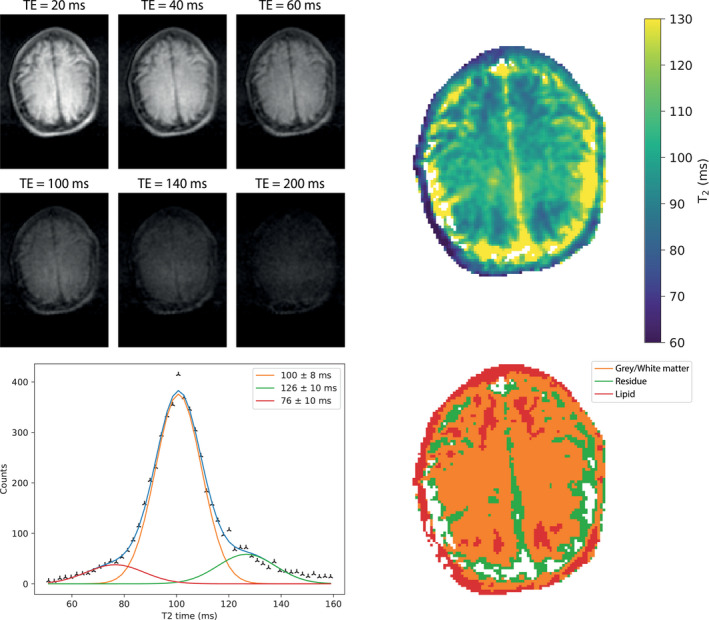
(Top left) images acquired with different TEs using a TSE sequence. (Top right) A T2 map reconstructed from images acquired with 10 different TEs. (Bottom left) A plot of binned T2 times; 3 Gaussian curves are fit to the data; the sum of the 3 curves is shown in blue. (Bottom right) A segmented map of the brain with the color corresponding to the curves of the Gaussian fit: red corresponds to lipid, orange to gray, and white matter (no distinction is possible) and green to incorrect fitting of the T2 of CSF. TSE, turbo spin echo
Figure 3 shows the measured T1 and T2 times in the CSF using a very long repetition time to allow for full longitudinal relaxation. The measured T1 and T2 times in the CSF for each subject are reported in Table 1 (note that T1 and T2 maps were not acquired in the same subjects). The mean T1 in the CSF is measured to be 3695 ± 287 ms, and the mean T2 is 1584 ± 124 ms.
FIGURE 3.
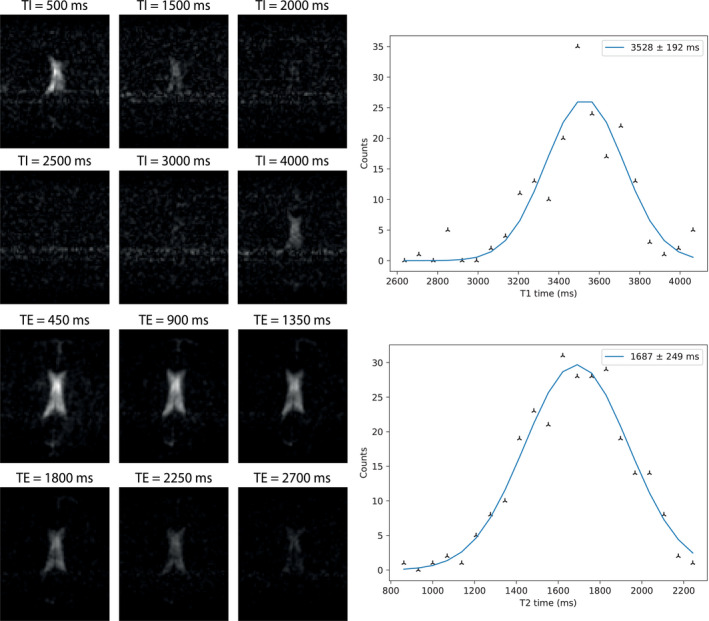
(Top left) Images acquired using an inversion recovery sequence with 6 different inversion times for CSF measurement: the background signal from WM/GM is suppressed by using a very long effective TE. (Top right) A plot of binned T1 times with a single Gaussian curve fitted through the data. (Bottom left) Images acquired using a TSE sequence, with 6 different TEs with (bottom right) a corresponding Gaussian fit
For muscle and lipid measurements in the lower leg, Figure 4 shows a T1 map of 1 of the subjects reconstructed from 6 inversion recovery acquisitions, which are also shown. The mean T1 across all 3 subjects is 171 ± 11 ms in the muscle and 130 ± 5 ms in the lipid. Two areas of much higher T1 values are located at the anterior and posterior tibial arteries and veins and are likely due to flow effects and the relatively short repetition time relative to the expected T1 of blood of around 400 ms 20 : these areas are excluded from the analyses.
FIGURE 4.
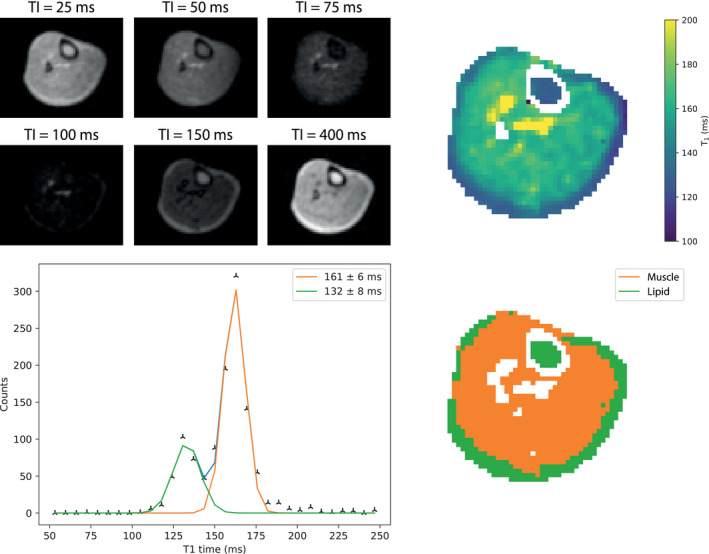
(Top left) Images acquired using an inversion recovery sequence with 6 different inversion times. (Top right) A T1 map reconstructed from the acquired images. (Bottom left) A corresponding histogram plot. Two Gaussian curves are fit to the histogram. (Bottom right) A segmented map of the images with the colors (orange: muscle; green: lipid) corresponding to the area under the fitted curves of the same color
Figure 5 shows a T2 map, together with individual images from a subset of the different TEs. The mean T2 across all 3 subjects for muscle is 39 ± 2 ms and 90 ± 13 ms for lipid. There is no measurable difference in the T1 and T2 between subcutaneous and bone marrow lipid. The measured T1 and T2 times in the muscle and lipid for each individual are reported in Table 2.
FIGURE 5.
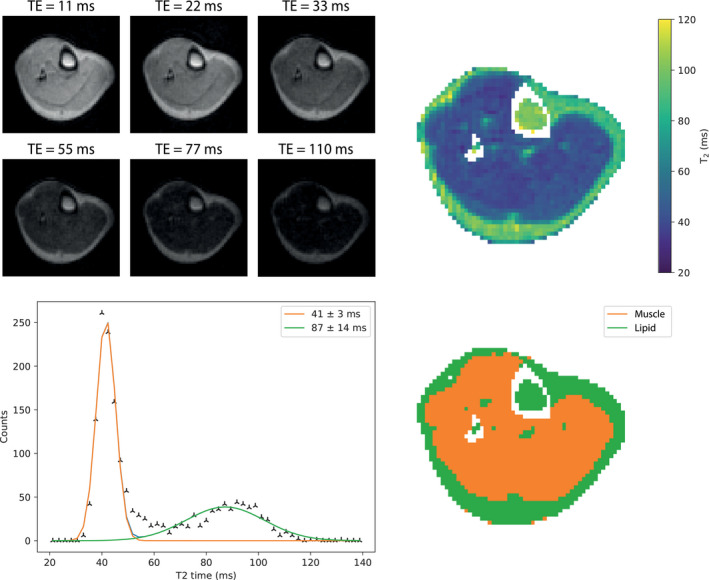
(Top left) Subset of images acquired with different TEs using a TSE readout. (Top right) A T2 map reconstructed from the images; images were acquired with 10 different TEs. (Bottom left) A histogram of the T2 map with 2 Gaussian functions fitted. (Bottom right) A segmented map of the images with the colors (orange: muscle; green: lipid) corresponding to the area under the fitted curves of the same color
TABLE 2.
Measured T1 and T2 times in the lower leg of healthy volunteers
| Muscle | Lipid | Muscle (Bottomley 22 ) | Lipid (Bottomley 22 ) | |
|---|---|---|---|---|
| T1 (ms) | 168 ± 10 | 134 ± 7 | 208 ± 37 | 143 ± 40 |
| 183 ± 10 | 124 ± 11 | |||
| 161 ± 6 | 132 ± 7 | |||
| T2 (ms) | 41 ± 3 | 87 ± 14 | 47 ± 6 | 84 ± 30 |
| 40 ± 4 | 104 ± 10 | |||
| 37 ± 2 | 78 ± 14 |
Values for lipid represent the combination of subcutaneous and bone marrow lipids.
4. DISCUSSION
In this work, we acquired in vivo T1 and T2 maps of the brain (WM, GM, CSF) and lower leg (muscle, subcutaneous lipid, bone marrow) at 50 mT. As expected, T1 times are lower at 50 mT than at clinical field strengths; gray and white matter T1s were measured to be 327 ± 10 ms and 275 ± 5 ms, respectively, compared to 1200 ms and 650 ms at 1.5 T 20 and 1331 ms and 832 ms at 3 T. 32 For the lower leg, T1 times of 171 ± 11 ms for muscle and 130 ± 5 ms for lipid were measured. Again, these are much shorter than at high field, with T1 of the muscle reported as 1130 ms and 1420 ms for 1.5T, and 3T and a T1 of 250 ms and a range 380‐450 ms for subcutaneous lipid at 1.5T 33 and 3T, 32 respectively. The measured T2 times closely match data reported at different field strengths: this is also to be expected given the relative insensitivity of T2 with respect to B0.
In terms of previous work on the field dependence of relaxation times, fundmental work on T1 relaxation times was performed by Koenig and Brown 34 over a range of 0.01 to 20 MHz. Bottomley 22 provided an extensive review of the literature and, based on this collation, proposed an empirical formula of the form:
| (7) |
with different values of A and B for various tissues, for example, A = 0.00362, B = 0.3082, SD = 17% (GM); A = 0.00152, B = 0.3477, SD = 17% (WM); A = 0.000455, B = 0.4203, SD = 18% (skeletal muscle); and A = 0.0113, B = 0.1743, SD = 28% (lipid). At 50 mT, these correspond to T1 values of ~320 ± 55 ms for GM, ~240 ± 40 ms for WM, ~210 ± 38 ms for skeletal muscle, and ~145 ± 40 ms for lipid. Rooney 20 derived a similar formulation with T1 = 0.583(B0)0.382 (WM) and T1 = 0.857(B0)0.376 (GM). These equations correspond to values of 186 ms (WM) and 286 ms (GM). (It should be noted that experimental data with a lower limit of 0.15T were considered in this study; thus, no claim to accuracy below this field strength was made, see page 313 in 20 ). Fischer et al 23 extended the model of Bottomley to incorporate both the “physically plausible and experimentally encountered” low‐field (L) and high‐field (H) limits:
| (8) |
where 1/T1,w is the relaxation rate of pure water (0.23 s−1 at 37°C) ; D is the baseline; A is the height of the dispersion step; fc is the inflection frequency; and β′ is the steepness of the dispersion step. Using a sample of 13 WM and 10 GM samples from 4 different brains, they empirically derive values of D (−1.52 s−1 WM, 0.11 s−1 GM), A (19.07 s−1 WM, 11.08 s−1 GM), fc (0.067 MHz WM, 0.085 MHz GM), and β′ (0.251 WM, 0.438 GM). This gives values of 230 ms for WM and 395 ms for GM.
Placing our experimental results in the context of these different models: T1(GM) of 327 ± 10 ms is right at the center of the range from Bottomley, well above the value from Rooney and below the value from Fischer; T1(WM) of 275 ± 5 ms is at the upper end of the range from Bottomley, well above the value from Rooney and above that from Fischer; T1(skeletal muscle) of 171 ± 11 ms is at the lower end of the Bottomley range; and T1(lipid) of 130 ± 5 ms is in the center of the Bottomley range.
Comparing to previous in vivo low field studies, Agartz et al 35 reported brain relaxation time data at 0.02 T, with T1 values of 200‐220 ms for WM and 185‐200 ms for GM: these values are lower than our measured ones, in line with the lower magnetic field used. Dean et al 36 performed in vivo relaxation time measurements of breast tissue (lipid) at 20 mT. The range of T1 values was 96‐133 ms, which is shorter than those we measured, as expected.
T2 values are generally assumed to be independent of frequency, with Bottomley reporting values of 101 ±13 ms for GM, 92 ± 20 ms for WM, 47 ± 6 ms for skeletal muscle, and 84 ± 36 ms for lipid. Our measured values in vivo agree well with these predictions. T2 values of 92‐98 ms for WM and 81‐87 ms for GM were also reported in the study by Agartz et al at 20 mT, which are very similar to our results. T2 values for lipid in breast at 20 mT were 57‐74 ms in the work by Dean, which is slightly lower than our values.
The relaxation times of CSF are notoriously difficult to measure in vivo, and a wide variety of values have been reported in the literature, in particular for the T1 values 20 , 37 , 38 , 39 , 40 : at 1.5T, reported values range from 3836 ± 470 ms to 4282 ms, and at 3T from 3817 ± 424 to 6873 ms. One report states that the T1 time for CSF shows no significant B0 dependence, with a value of around 4400 ms from 0.15T to 7T. 20 Yamashiro et al 37 showed that the T1 of CSF (measured at 1.5 and 3T) was significantly greater than that of pure water at room temperature. They ascribed this difference to be mainly due to the higher temperature in the body. Tsukiashi 41 showed that the T1 of water increases from approximately 3.2 s at 25°C to 4 s at body temperature, whereas the T2 is much less temperature‐dependent, rising from 2.1 to 2.2 s. Qin 38 mapped both the volume and T2 of CSF, with values at 3T of greater than 2000 ms for ventricle, but lower values (~1600 ms) for frontal cortex and ~1500 ms for both temporal cortex and occipital cortex. The paper attributed the lower T2 values measured in the subarachnoid spaces to be caused by a higher partial pressure of oxygen, which would correlate with the lower T1 values measured by Zaharchuk et al. 42 , 43 Other potential causes included higher protein concentration, although this was not verified or substantiated. T2 values of ~1400 s were measured by Spijkerman et al at 7T. 39 Hopkins 40 performed measurements at a variety of lower field strengths, reporting values of 4360 for the T1 and 1760 for the T2 at 0.15T. Placing our results in context, the average T1 value was ~3700 ms and T2 ~1550 ms, in line with the various literature references.
The major challenges in accurate relaxation time mapping at low field are partial volume effects resulting from the relatively coarse spatial resolution and the low intrinsic SNR. In this work, we used very conventional and somewhat time‐inefficient inversion recovery and multiple spin‐echo sequences. We were only able to measure 1 parameter per imaging session of ~45 min, with separate measurements also necessary for CSF due to its very long T1 and T2 relaxation times. In all the data, there was some evidence of multi‐exponential behavior, although we believe that the source of this is the partial volume effect rather than the tissue itself.
5. CONCLUSION
Practical relaxation time mapping for image segmentation purposes in longitudinal studies, or characterization of pathologies in patients, obviously will require much faster scanning. Reconstruction techniques such as compressive sensing from undersampled k‐space data have already been shown to be applicable to low‐field imaging. 44 The application of techniques such as Look‐Locker for T1 29 and variable tip‐angle turbo spin echo sequences that allow longer echo trains to be acquired 45 would also be valuable to study. Finally, we note that, although strong B0 inhomogeneity is known to cause an underestimation of the T2 times due to diffusion effects, 46 in our case typical line widths over the entire 3D imaging volume were less than 1 kHz and therefore unlikely to introduce significant errors.
The T1 and T2 times measured in this work show that generating inter‐tissue contrast based purely on relaxation times will be more challenging at low field than at higher fields. The absolute difference in T1 relaxation times between tissues becomes smaller at low field, which means conventional methods for generating T1 weighting in images, such as inversion recovery‐based sequences and imaging with a short repetition time, may be suboptimal. It may be that other contrast‐generating techniques such as magnetization transfer 47 or diffusion 48 are required for generating strong tissue contrast at low field. Additionally, lipid suppression becomes challenging at low field because inversion recovery‐based suppression methods such as short tau inversion recovery (STIR) will also largely suppress the muscle signal due to their very similar T1 relaxation times (T1,lipid = 130 ± 5 ms, T1,muscle = 171 ± 11 ms), and the very small chemical shift of less than 10 Hz makes spectrally selective‐based approaches such as spectral presaturation with inversion recovery (SPIR) and DIXON impractical. 49
Finally, in this preliminary work, we did not differentiate between age and gender, both of which have been shown to affect relaxation times. 50 , 51 , 52 Low‐field MRI provides a low(er) cost pathway to understanding how these tissue parameters evolve over time and can be a very useful tool in longitudinal studies. T1 and T2 times are also known to correlate with disease progression 53 , 54 , 55 , 56 ; by making MRI scanners more accessible, there is an opportunity to scan people at a regular interval to gain a more accurate understanding of the current disease state.
ACKNOWLEDGMENT
This work was supported by Horizon 2020 European Research Council (ERC) FET‐OPEN (737180 Histo MRI), Horizon 2020 ERC Advanced (670629 NOMA‐MRI), Dutch research council (NWO) Simon Stevin meester prize, and NWO WOTRO Joint SDG Research Programme (W 07.303.101).
O’Reilly T, Webb AG. In vivo T1 and T2 relaxation time maps of brain tissue, skeletal muscle, and lipid measured in healthy volunteers at 50 mT. Magn Reson Med. 2021;87:884–895. 10.1002/mrm.29009
Funding information
This work was supported by Horizon 2020 European Research Council (ERC) FET‐OPEN grant Histo MRI 737180; Horizon 2020 ERC Advanced grant NOMA‐MRI 670629; Simon Stevin meester prize from the Dutch research council (NWO); and the NWO WOTRO Joint Sustainable Development Goal (SDG) Research Programme (W 07.303.101)
REFERENCES
- 1. Geethanath S, Vaughan JT Jr. Accessible magnetic resonance imaging: a review. J Magn Reson Imaging. 2019;49:e65‐e77. [DOI] [PubMed] [Google Scholar]
- 2. Sarracanie M, Salameh N. Low‐field MRI: how low can we go? A fresh view on an old debate. Front Phys. 2020;8. 10.3389/fphy.2020.00172 [DOI] [Google Scholar]
- 3. Wald LL, McDaniel PC, Witzel T, Stockmann JP, Cooley CZ. Low‐cost and portable MRI. J Magn Reson Imaging. 2020;52:686‐696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. O'Reilly T, Teeuwisse WM, de Gans D, Koolstra K, Webb AG. In vivo 3D brain and extremity MRI at 50 mT using a permanent magnet Halbach array. Magn Reson Med. 2021;85:495‐505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O'Reilly T, Teeuwisse WM, Webb AG. Three‐dimensional MRI in a homogenous 27 cm diameter bore Halbach array magnet. J Magn Reson. 2019;307:106578. [DOI] [PubMed] [Google Scholar]
- 6. Cooley CZ, Haskell MW, Cauley SF, et al. Design of sparse Halbach magnet arrays for portable MRI using a genetic algorithm. IEEE Trans Magn. 2018;54:5100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cooley CZ, McDaniel P, Stockmann J, et al. A portable scanner for magnetic resonance imaging of the brain. Nat Biomed Eng. 2021;5:229–239. 10.1038/s41551-020-00641-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cooley CZ, Stockmann JP, Armstrong BD, et al. Two‐dimensional imaging in a lightweight portable MRI scanner without gradient coils. Magn Reson Med. 2015;73:872‐883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McDaniel PC, Cooley CZ, Stockmann JP, Wald LL. The MR cap: a single‐sided MRI system designed for potential point‐of‐care limited field‐of‐view brain imaging. Magn Reson Med. 2019;82:1946‐1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. He Y, He W, Tan L, et al. Use of 2.1 MHz MRI scanner for brain imaging and its preliminary results in stroke. J Magn Reson. 2020;319:106829. [DOI] [PubMed] [Google Scholar]
- 11. Nakagomi M, Kajiwara M, Matsuzaki J, et al. Development of a small car‐mounted magnetic resonance imaging system for human elbows using a 0.2 T permanent magnet. J Magn Reson. 2019;304:1‐6. [DOI] [PubMed] [Google Scholar]
- 12. Sheth KN, Mazurek MH, Yuen MM, et al. Assessment of brain injury using portable, low‐field magnetic resonance imaging at the bedside of critically Ill patients. JAMA Neurol. 2020;e203263. 10.1001/jamaneurol.2020.3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Broche LM, Ross PJ, Davies GR, Macleod MJ, Lurie DJ. A whole‐body fast field‐cycling scanner for clinical molecular imaging studies. Sci Rep. 2019;9:10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsai LL, Mair RW, Li CH, Rosen MS, Patz S, Walsworth RL. Posture‐dependent human He‐3 lung imaging in an open‐access MRI system: initial results. Acad Radiol. 2008;15:728‐739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sarracanie M, LaPierre CD, Salameh N, Waddington DEJ, Witzel T, Rosen MS. Low‐cost high‐performance MRI. Sci Rep. 2015;5:15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van Speybroeck CDE, O'Reilly T, Teeuwisse W, Arnold PM, Webb AG. Characterization of displacement forces and image artifacts in the presence of passive medical implants in low‐field (<100 mT) permanent magnet‐based MRI systems, and comparisons with clinical MRI systems. Phys Med. 2021;84:116‐124. [DOI] [PubMed] [Google Scholar]
- 17. Bottomley PA, Edelstein WA. Power deposition in whole‐body NMR imaging. Med Phys. 1981;8:510‐512. [DOI] [PubMed] [Google Scholar]
- 18. Hoult DI, Lauterbur PC. Sensitivity of the zeugmatographic experiment involving human samples. J Magn Reson. 1979;34:425‐433. [Google Scholar]
- 19. Marques JP, Simonis FFJ, Webb AG. Low‐field MRI: an MR physics perspective. J Magn Reson Imaging. 2019;49:1528‐1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rooney WD, Johnson G, Li X, et al. Magnetic field and tissue dependencies of human brain longitudinal 1H2O relaxation in vivo. Magn Reson Med. 2007;57:308‐318. [DOI] [PubMed] [Google Scholar]
- 21. Korb JP, Bryant RG. Magnetic field dependence of proton spin‐lattice relaxation times. Magn Reson Med. 2002;48:21‐26. [DOI] [PubMed] [Google Scholar]
- 22. Bottomley PA, Foster TH, Argersinger RE, Pfeifer LM. A review of normal tissue hydrogen NMR relaxation‐times and relaxation mechanisms from 1–100 Mhz—dependence on tissue‐type, NMR frequency, temperature, species, excision, and age. Med Phys. 1984;11:425‐448. [DOI] [PubMed] [Google Scholar]
- 23. Fischer HW, Rinck PA, Van Haverbeke Y, Muller RN. Nuclear relaxation of human brain gray and white matter: analysis of field dependence and implications for MRI. Magn Reson Med. 1990;16:317‐334. [DOI] [PubMed] [Google Scholar]
- 24. Oros‐Peusquens AM, Laurila M, Shah NJ. Magnetic field dependence of the distribution of NMR relaxation times in the living human brain. MAGMA. 2008;21:131‐147. [DOI] [PubMed] [Google Scholar]
- 25. Coletta F, Lacognata C, Tregnaghi A. Frequency‐dependence of T1 and T2 relaxation‐times of water in normal and tumoral lung tissues—T2 relaxation‐time evidence of water different chemical‐shifts and exchange‐rates. Spectrosc Lett. 1994;27:661‐676. [Google Scholar]
- 26. Vymazal J, Brooks RA, Zak O, Mcrill C, Shen C, Dichiro G. T1 and T2 of ferritin at different field strengths—effect on MRI. Magn Reson Med. 1992;27:368‐374. [DOI] [PubMed] [Google Scholar]
- 27. Turner R. A target field approach to optimal coil design. J Phys D Appl Phys. 1986;19:L147‐L151. [Google Scholar]
- 28. de Vos B, Fuchs P, O'Reilly T, Webb A, Remis R. Gradient coil design and realization for a Halbach‐based MRI system. IEEE Trans Magn. 2020;56:1‐8. [Google Scholar]
- 29. Jiang K, Zhu Y, Jia S, Wu Y, Liu X, Chung YC. Fast T1 mapping of the brain at high field using Look‐Locker and fast imaging. Magn Reson Imaging. 2017;36:49‐55. [DOI] [PubMed] [Google Scholar]
- 30. Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature. 2013;495:187‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lustig M, Donoho DL, Santos JM, Pauly JM. Compressed sensing MRI. IEEE Signal Proc Mag. 2008;25(2):72‐82. [Google Scholar]
- 32. Bojorquez JZ, Bricq S, Acquitter C, Brunotte F, Walker PM, Lalande A. What are normal relaxation times of tissues at 3 T? Magn Reson Imaging. 2017;35:69‐80. [DOI] [PubMed] [Google Scholar]
- 33. Pettersson H, Slone R. Magnetic‐resonance relaxation characteristics of muscle, fat and bone‐marrow of the extremities—normal values in a low field‐strength unit. Acta Radiol. 1987;28:363‐364. [PubMed] [Google Scholar]
- 34. Koenig SH, Brown RD. Determinants of proton relaxation rates in tissue. Magn Reson Med. 1984;1:437‐449. [DOI] [PubMed] [Google Scholar]
- 35. Agartz I, Saaf J, Wahlund LO, Wetterberg L. Magnetic‐resonance‐imaging at 0.02‐T in clinical‐practice and research. Magn Reson Imaging. 1987;5:179‐187. [DOI] [PubMed] [Google Scholar]
- 36. Dean KI, Komu M. Breast‐tumor imaging with ultra‐low field MRI. Magn Reson Imaging. 1994;12:395‐401. [DOI] [PubMed] [Google Scholar]
- 37. Yamashiro A, Kobayashi M, Saito T. Cerebrospinal fluid T1 value phantom reproduction at scan room temperature. J Appl Clin Med Phys. 2019;20:166‐175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qin Q. A simple approach for three‐dimensional mapping of baseline cerebrospinal fluid volume fraction. Magn Reson Med. 2011;65:385‐391. [DOI] [PubMed] [Google Scholar]
- 39. Spijkerman JM, Petersen ET, Hendrikse J, Luijten P, Zwanenburg JJM. T2 mapping of cerebrospinal fluid: 3 T versus 7 T. MAGMA. 2018;31:415‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hopkins AL, Yeung HN, Bratton CB. Multiple field‐strength in vivo T1 and T2 for cerebrospinal‐fluid protons. Magn Reson Med. 1986;3:303‐311. [DOI] [PubMed] [Google Scholar]
- 41. Tsukiashi A, Min KS, Kitayama H, et al. Application of spin‐crossover water soluble nanoparticles for use as MRI contrast agents. Sci Rep. 2018;8:14911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zaharchuk G, Busse RF, Rosenthal G, Manley GT, Glenn OA, Dillon WP. Noninvasive oxygen partial pressure measurement of human body fluids in vivo using magnetic resonance imaging. Acad Radiol. 2006;13:1016‐1024. [DOI] [PubMed] [Google Scholar]
- 43. Zaharchuk G, Martin AJ, Rosenthal G, Manley GT, Dillon WP. Measurement of cerebrospinal fluid oxygen partial pressure in humans using MRI. Magn Reson Med. 2005;54:113‐121. [DOI] [PubMed] [Google Scholar]
- 44. Koolstra K, O'Reilly T, Bornert P, Webb A. Image distortion correction for MRI in low field permanent magnet systems with strong B‐0 inhomogeneity and gradient field nonlinearities. MAGMA. 2021;34:631‐642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mugler JP 3rd. Optimized three‐dimensional fast‐spin‐echo MRI. J Magn Reson Imaging. 2014;39:745‐767. [DOI] [PubMed] [Google Scholar]
- 46. Deichmann R, Adolf H, Kuchenbrod E, Noth U, Schwarzbauer C, Haase A. Compensation of diffusion effects in T‐2 measurements. Magn Reson Med. 1995;33:113‐115. [DOI] [PubMed] [Google Scholar]
- 47. Hajnal JV, Baudouin CJ, Oatridge A, Young IR, Bydder GM. Design and implementation of magnetization transfer pulse sequences for clinical use. J Comput Assist Tomogr. 1992;16:7‐18. [DOI] [PubMed] [Google Scholar]
- 48. Yoshiura T, Wu O, Zaheer A, Reese TG, Sorensen AG. Highly diffusion‐sensitized MRI of brain: dissociation of gray and white matter. Magn Reson Med. 2001;45:734‐740. [DOI] [PubMed] [Google Scholar]
- 49. Kaldoudi E, Williams SCR, Barker GJ, Tofts PS. A chemical‐shift selective inversion recovery sequence for fat‐suppressed MRI—theory and experimental validation. Magn Reson Imaging. 1993;11:341‐355. [DOI] [PubMed] [Google Scholar]
- 50. Wansapura JP, Holland SK, Dunn RS, Ball WS. NMR relaxation times in the human brain at 3.0 Tesla. J Magn Reson Imaging. 1999;9:531‐538. [DOI] [PubMed] [Google Scholar]
- 51. Baierl P, Forster C, Fendel H, Naegele M, Fink U, Kenn W. Magnetic‐resonance imaging of normal and pathological white matter maturation. Pediatr Radiol. 1988;18:183‐189. [DOI] [PubMed] [Google Scholar]
- 52. Lee SM, Choi YH, You SK, et al. Age‐related changes in tissue value properties in children: simultaneous quantification of relaxation times and proton density using synthetic magnetic resonance imaging. Invest Radiol. 2018;53:236‐245. [DOI] [PubMed] [Google Scholar]
- 53. Tang X, Cai F, Ding DX, Zhang LL, Cai XY, Fang Q. Magnetic resonance imaging relaxation time in Alzheimer's disease. Brain Res Bull. 2018;140:176‐189. [DOI] [PubMed] [Google Scholar]
- 54. Mosher TJ, Dardzinski BJ. Cartilage MRI T2 relaxation time mapping: overview and applications. Semin Musculoskelet Radiol. 2004;8:355‐368. [DOI] [PubMed] [Google Scholar]
- 55. Willcocks RJ, Arpan IA, Forbes SC, et al. Longitudinal measurements of MRI‐T2 in boys with Duchenne muscular dystrophy: effects of age and disease progression. Neuromuscul Disord. 2014;24:393‐401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Atkinson HF, Birmingham TB, Moyer RF, et al. MRI T2 and T1ρ relaxation in patients at risk for knee osteoarthritis: a systematic review and meta‐analysis. BMC Musculoskelet Disord. 2019;20:182. [DOI] [PMC free article] [PubMed] [Google Scholar]


