Abstract
Recapitulation is a hypothetical concept that assumes embryogenesis of an animal parallels its own phylogenetic history, sequentially developing from more ancestral features to more derived ones. This concept predicts that the earliest developmental stage of various animals should represent the most evolutionarily conserved patterns. Recent transcriptome‐based studies, on the other hand, have reported that mid‐embryonic, organogenetic periods show the highest level of conservation (the developmental hourglass model). This, however, does not rule out the possibility that recapitulation would still be detected after the mid‐embryonic period. In accordance with this, recapitulation‐like morphological features are enriched in late developmental stages. Moreover, our recent chromatin accessibility‐based study provided molecular evidence for recapitulation in the mid‐to‐late embryogenesis of vertebrates, as newly evolved gene regulatory elements tended to be activated at late embryonic stages. In this review, we revisit the recapitulation hypothesis, together with recent molecular‐based studies that support the developmental hourglass model. We contend that the recapitulation hypothesis does not entirely contradict the developmental hourglass model and that these two may even coexist in later embryonic stages of vertebrates. Finally, we review possible mechanisms underlying the recapitulation pattern of chromatin accessibility together with the hourglass‐like evolutionary conservation in vertebrate embryogenesis.
Keywords: chromatin accessibility, developmental hourglass model, evo‐devo, gene expression regulation, parallelism, recapitulation
The developmental hourglass model showing that gene expression profiles and the recapitulation pattern of chromatin accessibility may coincide with each other in later vertebrate embryogenesis. The exact evolutionary mechanism remains largely unclear. However, one possibility would be that the recapitulation pattern in chromatin accessibility of genomic regions (right panel) underlies the diversification of gene expression profiles (left panel) as well as the recapitulation pattern exhibited by some morphological traits during later embryogenesis.

Research Highlights
Recapitulation pattern has been reported for chromatin accessibility during the mid‐to‐late embryogenesis.
The observed recapitulation pattern and the developmental hourglass model may coexist.
The possible evolutionary mechanisms underlying tendencies of embryonic evolution were discussed.
1. INTRODUCTION
Diverse multicellular animals with different body plans originally evolved from a simple unicellular organism. Similarly, animal development starts with a single zygote that develops into a multicellular adult with complex morphological patterns. This tendency of increasing complexity along a time course has inspired scientists to propose parallel relationships between development and evolution (or taxonomic hierarchy) (Gould, 1977; Hall, 1992; Oppenheimer, 1959; Richardson & Keuck, 2002). One of the most famous of these theories is Haeckel's recapitulation theory (Haeckel, 1866), which predicted that development is a rapid replay of the evolutionary trajectory.
Although the theory of recapitulation is no longer accepted in its original form, the essence of the theory led to the early conservation model, which predicts that the earliest stage of development is expected to show the most ancestral, or most evolutionarily conserved features. This early conservation model appears to be reasonable from a developmental perspective, at least intuitively, because changes in earlier developmental processes are thought to have greater and usually harmful impacts on subsequent developmental processes (Arthur, 1997; Garstang, 1922; Riedl, 1978; Wimsatt, 1986), possibly leading to the conservation of earlier stages (Figure 1a). On the contrary, recent transcriptome‐based studies have demonstrated that it is the mid‐embryonic phase, and not the earliest one, that is the most conserved across species of a given phylum (Domazet‐Lošo & Tautz, 2010; Drost et al., 2015; Gildor et al., 2019; Hu et al., 2017; Irie & Kuratani, 2011; Israel et al., 2016; Kalinka et al., 2010; Levin et al., 2012; Malik et al., 2017; Marlétaz et al., 2018; Wang et al., 2013; Yanai et al., 2011). Notably, this tendency was reported in several phyla, indicating that “the developmental hourglass model” (Duboule, 1994) would be one of the most plausible hypotheses that clarifies the apparent relationship between development and evolution in each animal phylum (Figure 1b). While the hourglass‐like conservation of animal embryogenesis appears to rule out the recapitulation theory, there remains a possibility that “recapitulation pattern” may hold true, at least partially, in the diversifying phase of embryogenesis. Furthermore, several morphological features appear to favor the recapitulation theory (see below). In this review, we propose a plausible concept involving the integration of the hourglass model with recapitulation and review possible evolutionary mechanisms underlying the tendency toward embryonic evolution.
Figure 1.
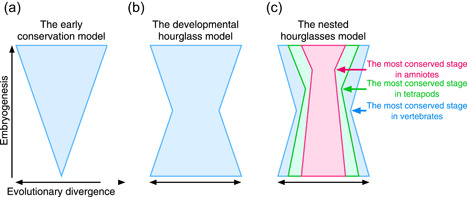
Schematic diagrams that depict evolutionary divergence along embryogenesis. (a) The early conservation model, which assumes that the earliest stage is the most highly conserved. (b) The developmental hourglass model (Duboule, 1994), which assumes the highest conservation across species of the same phylum at the mid‐embryonic period or the phylotypic period. (c) The nested hourglasses model with shifting bottlenecks. The most conserved mid‐embryonic phase shifts to later stages when comparisons were made in smaller phylogenetic groups, which may be regarded as the recapitulation pattern
2. HYPOTHESES ON THE RELATIONSHIP BETWEEN DEVELOPMENT AND EVOLUTION
2.1. Current understanding of the recapitulation theory
The question whether there is any general relationship between development and evolution has attracted much interest for more than a century and has been a central theme in comparative embryology (Gould, 1977; Richardson & Keuck, 2002). A better understanding of this relationship not only helps to infer the ancestral state of the common ancestor, but also provides insights into how animals have diversified by modifying their developmental programs.
A pioneering concept that formulates the relationship between ontogeny and phylogeny was proposed by von Baer (1828); these are currently known as Baer's laws. Baer's third law predicts that basic and widespread traits shared by higher levels of taxa, or larger clades, tend to appear at earlier stages of animal embryogenesis, followed by the more specialized ones shared by lower taxa, or smaller clades (von Baer, 1828). Notochord and pharyngeal arches are well‐known traits that have been argued to fit Baer's law, as they are chordate‐shared features and develop at the early stages of vertebrates (Abzhanov, 2013; Gould, 1977; Richardson & Keuck, 2002). While Baer did not accept the idea of evolution, Ernst Haeckel (1866) explicitly related the embryogenetic process to evolutionary processes. Haeckel's recapitulation theory assumes that animal embryos pass through developmental stages representing adult forms of ancestors. However, this scheme is no longer accepted (Richardson & Keuck, 2002), because embryos do not completely resemble the adult morphologies of ancestors (Garstang, 1922; Morgan, 1916; Sedgwick, 1894). For example, Richardson and Keuck (2002) argued that although human embryos do display many ancestral features, such as pharyngeal arches and heart, they do not show features that resemble those of adult ancestral fish. Moreover, different features that appear at different stages of development may often evolve independently (Garstang, 1922; Richardson & Keuck, 2002), thereby contradicting predictions of the original recapitulation theory. Considered together, it may be more appropriate to regard the recapitulation as a tendency that evolutionarily more derived features tend to appear sequentially at later developmental stages, in the same order of their evolutionary origins. The current review aims to follow this definition.
2.2. The developmental hourglass model
According to the re‐defined recapitulation mentioned above, earlier embryonic stages may be expected to display more ancestral features than later embryonic stages. This early conservation model (Figure 1a) has been upheld by several theoretical studies (Arthur, 1997; Garstang, 1922; Riedl, 1978; Wimsatt, 1986). However, no consensus has been reached as to whether the earliest stage is evolutionarily the most conserved. For example, morphologically diverse features observed during early developmental stages, such as variable cell cleavage patterns, gastrulation, and axis formation, are often referred to as typical examples of evidence that counter the early conservation model (Kalinka & Tomancak, 2012; Raff, 1996; Schierenberg, 2006; Slack et al., 1993). Moreover, the situation has become far more complicated as several counter models have been proposed (Bininda‐Emonds et al., 2003; Duboule, 1994; Poe & Wake, 2004; Raff, 1996; Richardson, 1999; Slack et al., 1993). The developmental hourglass model, for instance, assumes the highest conservation within each phylum at the mid‐embryonic period (Duboule, 1994; Raff, 1996). This originates from morphological similarities that were apparent at vertebrate organogenesis stages, which have been pointed out so far by many biologists (von Baer, 1828; Ballard, 1981; Duboule, 1994; Haeckel, 1874; Raff, 1996; Slack et al., 1993). For example, the vertebrate embryos around pharyngula period share a variety of primordial organs, such as notochord, somite, dorsal nerve cord, the pharyngeal arch, rhombomere, and optic anlagen. Despite these apparent similarities, another morphological study with detailed quantitative analyses showed that the highest conservation did not occur at the mid‐embryonic phase, but rather proposed that the mid‐embryonic phase was the most divergent stage (Bininda‐Emonds et al., 2003). Furthermore, others proposed the ontogenetic adjacency model, which predicts that there is no global tendency in the evolutionary conservation of embryonic stages, except for small changes (Poe & Wake, 2004).
While comparative morphological approaches were unable to resolve the controversy (Bininda‐Emonds et al., 2003; Poe, 2006; Poe & Wake, 2004; Richardson et al., 1997), the situation changed with the advent of molecular‐based studies focusing on gene expression profiles during embryogenesis (Domazet‐Lošo & Tautz, 2010; Hazkani Covo et al., 2005; Irie & Kuratani, 2011; Irie & Sehara‐Fujisawa, 2007; Kalinka et al., 2010; Roux & Robinson‐Rechavi, 2008). Specifically, transcriptomic technologies, such as microarray and RNA sequencing, provided quantitative approaches that allowed conserved developmental stages between different species to be evaluated. The findings of most studies, including ours, have supported mid‐embryonic conservation rather than early conservation in various animal groups, including chordates (Domazet‐Lošo & Tautz, 2010; Hu et al., 2017; Irie & Kuratani, 2011; Irie & Sehara‐Fujisawa, 2007; Marlétaz et al., 2018; Wang et al., 2013), echinoderms (Gildor et al., 2019; Li et al., 2020; Malik et al., 2017), arthropods (Domazet‐Lošo & Tautz, 2010; Kalinka et al., 2010), nematodes (Levin et al., 2012; Schiffer et al., 2018), and mollusks (Xu et al., 2016). Thus, accumulating evidence suggests that the mid‐embryonic period reflects the most highly conserved expression profiles in a phylum or less inclusive groups, thereby contradicting the early conservation model. However, this does not necessarily refute the possibility that the recapitulation pattern square with the developmental hourglass model at later developmental stages with increasing diversity (Abzhanov, 2013; Kuratani, 2004). Specifically, if the most conserved stage of the lower taxa shifts sequentially to later periods, these two ideas may coexist with each other at the latter part of embryogenesis (Figure 1c; the nested hourglasses model with shifting bottlenecks).
With regard to this viewpoint, the nested hourglasses model, which was tested at the transcriptomic‐level using several vertebrate species, demonstrated that the target of the mid‐embryonic conservation remained mostly the same, irrespective of the phylogenetic scales analyzed (Hu et al., 2017; Wang et al., 2013). For example, the highly conserved mid‐embryonic stage identified in chicken‐turtle comparison (around stage HH16 in chicken) largely overlapped those identified in other tetrapod comparisons (Wang et al., 2013). Similarly, Hu et al. (2017) examined eight chordates covering different taxa and showed that conserved developmental stages in different phylogenetic groups (i.e., Osteichthyes, Tetrapoda, Aminiota, Xenopus, and Archelosauria) largely overlapped each other (Figure 2a). Furthermore, concerning the higher level of taxa, mid‐embryonic conservation was observed up to Vertebrata, but was only weakly supported in Chordata and Olfactores (Figure 1a; Hu et al., 2017). This does not contradict the study by Marlétaz et al. (2018) which suggested that the most conserved stages are not comparable between vertebrates and chordates. Considered together, these comparative transcriptomic studies imply that the mid‐embryonic period may have been the target of conservation persistently through the vertebrate evolution (persistent conservation; Irie & Kuratani, 2014), and the recapitulation pattern does not seem to coexist even partially during later developmental stages.
Figure 2.

The persistently conserved mid‐embryonic period at the transcriptomic level and recapitulation pattern observed in chromatin accessibility in the late embryogenesis. In both panels, the results of chicken development are shown. (a) Conserved developmental stages in different taxonomic groups are estimated by ortholog‐group‐based comparative transcriptome (Hu et al., 2017). The percentages of chicken developmental stages included in the most similar combinations of staged embryos from five different taxonomic groups are shown as Ptop in different colored bar graphs (for detail, see Methods in Hu et al., 2017). Higher values in Ptop represent higher levels of whole‐embryonic transcriptomic conservation for the species compared. Note that stages that show the body plan for vertebrates (potential phylotypic period, highlighted in gray) largely overlap each other, except for the bottom graph showing conservation for chordate‐conserved stages in chicken. Meanwhile, mid‐embryonic conservations up to Chordata were observed in the analysis with strictly conserved 1‐to‐1 orthologs (see Supplementary Information in Hu et al., 2017). Essentially the same results were observed for the other vertebrate species (see Hu et al., 2017). (b) Evolutionarily categorized chromatin accessibility during chicken development (Uesaka et al., 2019). For each developmental stage, the percentage on the vertical axis represents the relative chromatin accessibilities of the genomic regions categorized by the evolutionary ages of their sequences (see color and phylogenetic tree in the right for the category). In each graph after the conserved mid‐embryonic period, the developmental stages with the highest signal (highlighted in the corresponding colors) shift sequentially to later stages by their evolutionary category. While earlier stages before the conserved stages show high signals of ancestral genomic regions (e.g., those conserved in vertebrates and olfactores), newly acquired genomic regions (e.g., those acquired since the chicken‐alligator split) also show higher chromatin accessibilities. Essentially the same patterns were observed for the other species (see Uesaka et al., 2019), and these indicate that embryogenesis before the conserved stages does not show the recapitulation pattern with the unknown evolutionary mechanism. The figures are adapted and modified from Hu et al. (2017) and Uesaka et al. (2019)
3. POSSIBLE COEXISTENCE OF THE DEVELOPMENTAL HOURGLASS MODEL AND THE RECAPITULATION PATTERN IN VERTEBRATE EMBRYOGENESIS
3.1. Morphological examples showing the recapitulation pattern
In addition to the denial of the recapitulation pattern at transcriptomic level in later developmental stages, previous studies further supported the idea that the body plan of each animal phylum is defined by the most conserved mid‐embryonic stage (phylotype hypothesis; Duboule, 1994; Hu et al., 2017; Irie et al., 2018; Raff, 1996; Wang et al., 2013). This implies that embryos develop the body plan of their phylum first and then develop lineage‐specific features afterward, indicating that no recapitulation pattern emerges after the conserved mid‐embryonic phase. However, similarities between whole‐embryonic gene expression profiles do not necessarily indicate morphological conservation (Irie & Kuratani, 2014), although these presumably reflect the composition of homologous cells. It is thus worth overviewing studies based on comparative morphological approach.
In accordance with the transcriptomic studies, clear counter examples against the recapitulation pattern in the mid‐to‐late embryogenesis has also been reported for morphological features (Bininda‐Emonds et al., 2003; Koyabu & Son, 2014; Poe, 2006; Prum, 1999; Prum & Brush, 2015; Richardson et al., 2009; Weisbecker et al., 2008; Ziermann & Diogo, 2014). Nonetheless, a reasonable number of morphological traits display recapitulation patterns during development, especially in vertebrates (Gould, 1977; Richardson & Keuck, 2002). One example is the developmental pattern of ribs and scapulae during turtle embryogenesis (Nagashima et al., 2009). Unlike other amniotes, adult turtles are exceptional in that they have scapulae (shoulder blades) beneath the ribs, or the carapace. Despite the unique anatomy of adult turtles, they retain a typical amniote pattern early in their embryogenesis as follows: primordia of scapulae emerge at the outside of rib primordia and then tilt to locate at the ventral side of the ribs, which in intermediate stage resemble that of an ancestral fossil turtle, Odontochelys (Li et al., 2008), seemingly recapitulating the ancestral state. In addition, there are more examples that seem to display the recapitulation‐like pattern, such as the caudal morphology of teleost fishes (Metscher & Ahlberg, 2001; see also Sallan, 2016), the elongated jaw of some needlefishes (Lovejoy et al., 2004; Lovejoy, 2000), the hind limb buds that become regressed during embryogenesis in snakes and cetaceans (Bejder & Hall, 2002; Cohn & Tickle, 1999; Thewissen et al., 2006), the opposable hallux of birds (Botelho et al., 2015), the position of the hallux in the foot of birds (Botelho et al., 2017), the position of the external naris of cetaceans (Thewissen, 2018), and the three‐digit hind limb of jerboas and camels (Cooper et al., 2014). Diogo et al. (2015) also argued that the majority of head muscles of zebrafish, salamanders, and turtles tend to develop sequentially in the order of their evolutionary origins. Importantly, all these examples that seem to show the recapitulation pattern were observed in later embryogenesis following the organogenesis phase.
3.2. Testing the recapitulation pattern in the context of chromatin accessibility
Considering that previous comparative studies did not support coexistence between the hourglass model and recapitulation at the level of the whole embryonic transcriptome (Hu et al., 2017, Wang et al., 2013), how could we reconcile the recapitulation pattern observed for a significant number of morphological traits with the hourglass model (Bejder & Hall, 2002; Botelho et al., 2015, 2017; Cohn & Tickle, 1999; Cooper et al., 2014; Diogo et al., 2015; Gould, 1977; Lovejoy et al., 2004; Metscher & Ahlberg, 2001; Nagashima et al., 2009; Richardson & Keuck, 2002; Thewissen et al., 2006). One possibility would be that, although some traits display recapitulation patterns during development both at morphologic and transcriptomic levels, the overall recapitulation pattern in developing whole embryos disappears when all traits of the embryos are taken into account. Briefly, if only a minority of traits follow recapitulation, then the recapitulation pattern would not be detected at whole embryo level. In this case, no reconciliation is needed. Another possibility is that the transcriptomic approach lacks sufficient resolution to detect the recapitulation pattern. This concern arises from the fact that a cross‐species comparative transcriptomic approach mainly focuses on the expression of orthologous coding genes, whereas many protein‐coding genes are known to be repeatedly recruited at different developmental stages (Hu et al., 2017). In addition, most genomic mutations that accumulated during the evolution of major vertebrate groups have been identified in noncoding regions rather than in protein‐coding regions (Mikkelsen et al., 2007; Seki et al., 2017). Thus, it would be helpful to measure the activities of noncoding regulatory regions to test recapitulation patterns during development. One of a proxy of the regulatory region activity at various developmental stages is chromatin accessibility in embryos. Accessible chromatin marks active regulatory regions, including enhancers, promoters, and silencers (Gross & Garrard, 1988; Klemm et al., 2019), because accessible chromatin regions become physically accessible to transcription factors for the execution of their functions. ATAC‐seq (an assay for transposase‐accessible chromatin using sequencing; Buenrostro et al., 2013) is widely utilized to profile chromatin accessibility on a genome‐wide scale. Thus, with this method, we measured chromatin accessibilities of developing embryos in a recent study of ours (Uesaka et al., 2019). The chromatin accessibilities of whole embryos are expected to reflect the number of cells in which the chromatin state of the examined region is accessible, which would be consistent with the recently confirmed similarities between ATAC‐seq profiles of bulk samples and aggregate profiles of single‐cell ATAC‐seq (Lareau et al., 2019; Satpathy et al., 2019). Along with this postulate, we quantitatively examined chromatin accessibilities of genomic regions during development, and found that these showed a recapitulation pattern (Uesaka et al., 2019). In brief, we estimated the activities of gene regulatory elements through embryogenesis of three vertebrates (chicken, mouse, and medaka) using ATAC‐seq, and analyzed whether sequence conservation of these accessible chromatin regions showed any evolutionary tendencies along with the development of each species. The results indicated that genomic regions conserved at the sequence level in lower taxa tended to show the highest chromatin accessibilities at later developmental stages (Figure 2b for chicken; for mice and medaka, see Uesaka et al., 2019). In other words, evolutionarily newer genomic regions tend to become accessible at later developmental stages, suggesting a parallel relationship between the evolutionary age of genomic regions, and the timing of their accessibility along the development. Unlike the embryogenesis from the conserved mid‐embryonic period of vertebrates onwards, early‐to‐mid embryogenesis did not show a recapitulation pattern in chromatin accessibility (Figure 2b), indicating that a chromatin‐level recapitulation pattern can only be observed after the conserved mid‐embryonic period. The recapitulation pattern observed in whole embryonic chromatin accessibility (Uesaka et al., 2019) coincides well with morphological traits that show recapitulation patterns, at least in terms of timing in development, as both take place mainly after the conserved mid‐embryonic phase of the hourglass model.
3.3. The relationship between the recapitulation pattern of chromatin accessibility and the hourglass‐like embryonic divergence
Both transcriptome and chromatin accessibility‐based studies indicated that the mid‐embryonic phase retained the most ancestral features, followed by derived features that developed afterward. According to this perspective, the recapitulation pattern of chromatin accessibility and transcriptomic hourglass‐like divergence may coexist during mid‐to‐late embryogenesis (Figure 3; coexistence model). Although there is less empirical evidence, several assumptions can be made for possible mechanisms underlying this model. One is that evolutionarily newer gene regulatory elements could be the mechanism behind diversified gene expression profiles, as well as the morphological traits showing recapitulation patterns that appear in the later embryonic stages. Another assumption is that evolutionarily newer genomic regions activated at earlier stages lead to divergence in early embryogenesis at both the transcriptomic level (Hu et al., 2017; Irie & Kuratani, 2011; Wang et al., 2013) and the morphological level, such as the size of eggs, forms of blastula, and developing gastrula (Kalinka & Tomancak, 2012; Raff, 1996; Schierenberg, 2006; Slack et al., 1993). Meanwhile, higher chromatin accessibility of conserved genomic regions at early stages, such as the genomic regions conserved in vertebrates and in Olfactores, could be associated with deeply conserved embryonic patterns, such as egg cleavage, gastrulation, and neurulation (Figure 2b).
Figure 3.

Possible coexistence of the hourglass model and the recapitulation pattern of chromatin accessibility. While no recapitulation pattern was observed for transcriptomic divergencies during vertebrate embryogenesis (a), chromatin accessibility displayed the recapitulation pattern only after the conserved mid‐embryonic phase (b). The mechanism underlying this discrepancy remains unclear; however, it is tempting to know if activation of newly acquired regulatory regions is the underlying mechanism behind the diversifying gene expression profiles observed in the mid‐to‐late embryogenesis and some of the morphological traits that show the recapitulation pattern. (a) The hourglass‐like evolutionary divergence was observed for gene expression profiles during embryogenesis. Color lines represent transcriptome divergences among species of vertebrates and less inclusive taxonomic groups, respectively (e.g., red for amniotes, green for tetrapods, and blue for vertebrates). This nested hourglass model with non‐shifting bottlenecks represents that the same mid‐embryonic period was always the target of conservation throughout the vertebrate evolution (Hu et al., 2017; Irie & Kuratani, 2014). (b) Schematic graphs showing relative chromatin accessibilities (horizontal axis) of four different categories based on the evolutionary ages of genomic regions at different developmental stages (vertical axis; Uesaka et al., 2019). In the graphs, the evolutionary ages of genomic regions represent the levels of taxa the corresponding sequences are conserved in. For example, the graph of evolutionarily newest genomic regions (the rightmost, red one) schematically show the chromatin accessibility patterns of genomic regions specific to the chicken and those shared only in chicken and turkey. (c) Examples of morphological features in different periods of embryogenesis
The above‐stated assumptions require detailed investigation, mainly because the association between developmental patterns in chromatin accessibility and hourglass‐like gene expression divergence as well as morphological traits that show recapitulation patterns remain unclear. Even the relationship between the recapitulation pattern of chromatin accessibility and hourglass‐like transcriptomic divergence requires clarification on several points. For example, genomic regions with conserved sequences do not necessarily show a conserved pattern in chromatin accessibility (Stergachis et al., 2014; Vierstra et al., 2014), implying that such conserved genomic regions may not be linked with evolutionarily conserved gene expression. Another issue is raised by the fact that not all accessible chromatin regions contribute to the regulation of gene expression (The ENCODE Project Consortium, 2012). Therefore, identification of regulatory sequences, such as enhancers and repressors that promote hourglass‐like divergence of gene expression is needed. More detailed analyses of single‐cell‐based genome‐wide chromatin accessibility and transcriptomes in the same cell may help elucidate the relationship between the recapitulation pattern of chromatin accessibilities and hourglass‐like gene expression divergence (Cao et al., 2018; Chen et al., 2019; Zhu et al., 2019). In addition, the previous study conducted by us (Uesaka et al., 2019) did not include a cross‐species comparison of developmental chromatin accessibility to confirm whether evolutionarily newer chromatin accessibility displays a higher diversity to match with the latter part of the hourglass model.
Furthermore, the relationship between the chromatin accessibility and morphological features that display recapitulation patterns also remains unclear. Based on the reports indicating that the evolution of lineage‐specific morphological traits in vertebrates is often associated with evolutionary acquisitions of gene regulatory elements (Carroll, 2005; Nishihara et al., 2016; Seki et al., 2017; Shim et al., 2012; Tashiro et al., 2011), one might expect that morphological development showing a recapitulation pattern could be attributed to the sequential activation of gene regulatory elements in a recapitulation manner. However, it is not clear whether this relationship holds true for the overall tendency of morphological traits. In certain cases, newly acquired gene regulatory elements may replace ancestral regulatory elements during evolution, while still maintaining their function (Odom et al., 2007; Weirauch & Hughes, 2010; Wilson & Odom, 2009). Finally, the recapitulation pattern of developmental chromatin accessibility has only been observed in a limited number of vertebrates, and thus it is unclear how widely this pattern can be observed in the animal kingdom. Therefore, further studies involving both comparative transcriptomic analyses as well as whole‐embryonic chromatin accessibility analyses would be needed to test the generality of the possible coexistence.
4. POTENTIAL EVOLUTIONARY MECHANISMS UNDERLYING THE RECAPITULATION PATTERN OBSERVED IN VERTEBRATE EMBRYOGENESIS
Based on the findings that developmental chromatin accessibility at the whole embryo level shows the recapitulation pattern only after the conserved mid‐embryonic phase of the vertebrates and that many morphological features that show the recapitulation pattern appear during the later embryonic stages, we have proposed the potential coexistence of the recapitulation and the hourglass model. Meanwhile, the evolutionary mechanisms underlying this relationship remain largely unknown. In this last section, we discuss the possible mechanisms that may underlie the evolutionary tendency of vertebrate embryogenesis.
4.1. Possible mechanisms underlying the evolutionary tendency of chromatin accessibility
Why do the chromatin accessibilities of more evolutionarily derived genomic regions tend to become higher at later stages to show a recapitulation pattern? One attractive concept to explain this would be “developmental burden” (Riedl, 1978). According to this concept, since preceding developmental processes serve as a prerequisite for later processes, earlier developmental processes are less prone to change, leading to the evolutionary conservation of earlier processes. Similar mechanisms, such as the stepping stone model (Garstang, 1922), somatic program (Mayr, 1994), epigenetic characters (Løvtrup, 1978), and generative entrenchment (Wimsatt, 1986) have been proposed by other scientists.
Developmental burden has often been used to explain the conservation of certain morphological traits, such as the pharyngeal arches and notochord of vertebrate embryos. For example, while adult terrestrial vertebrates do not have gills, they still develop pharyngeal arches during embryogenesis, and this could be attributed to its responsibility for the development of other organs, such as the thymus. Similarly, a possible scenario based on the concept of the developmental burden would be that gene regulatory activities at earlier stage exert a greater impact on the following developmental processes when mutated, and thus are more difficult to change during evolution. This could have led to the conservation of these gene regulatory elements in both nucleotide sequences and their activities during early stages, which furthered the recapitulation pattern.
However, our recent report measured only sequence‐level conservation of potential regulatory regions, and whether or not the activities of these regions show cross‐species conservation remains unknown. Critically, the recapitulation pattern of chromatin accessibility was observed only up to the conserved mid‐embryonic phase, indicating that simple application of developmental burden throughout embryogenesis may not be justified. This problem will be discussed further in the next section.
4.2. Why is the recapitulation pattern restricted to the later part of embryogenesis?
Our recent findings indicated that the most evolutionarily new genomic regions showed higher chromatin accessibilities not only in the latest developmental stages, but also during the stages before the transcriptomically conserved mid‐embryonic period (Figure 2b). Considered together with transcriptomic divergence observed during the earlier stages, these appear to debunk the assumption that developmental burden acts on all stages and proceeds throughout embryogenesis.
In interpreting the developmental burden concept using these observations, one assumption would be that the developmental burden acts upon all stages of embryogenesis, but its effect is masked by stronger diversifying effects from other evolutionary mechanisms such as natural selection. For example, some studies contend that strong positive selective pressure is exerted on early embryonic stages during diversification and adaptation to a variety of environments (Kalinka & Tomancak, 2012; Slack et al., 1993). This could have led to high chromatin accessibilities in recently acquired genomic regions during early stages, as reported for transcriptomic divergency (Hu et al., 2017; Irie & Kuratani, 2011; Wang et al., 2013) and some lineage‐specific morphological traits (Kalinka & Tomancak, 2012; Raff, 1996; Schierenberg, 2006; Slack et al., 1993). In addition, selective pressures on maternal reproductive strategies could also exert similar effects at the beginning of embryogenesis, as developmental features that emerge during initial stages, such as egg size and yolk amount, are established and regulated by maternal phenotypes (Abzhanov, 2013; Kalinka & Tomancak, 2012). To further expand this burden‐oriented perspective, the activation of highly ancestral chromatin accessibility during earlier stages also requires further explanation (Figure 2b), as these may be the remnants of a strong conservation effect due to developmental burden. Unfortunately, the above‐mentioned burden‐oriented assumptions have no experimental basis, and thus far, no study has succeeded in measuring the force of developmental burden to substantiate its existence. Thus, potential evolutionary mechanisms that underlie the later‐stage recapitulation patterns of chromatin accessibility remain largely unknown.
4.3. Gene regulatory background for the mid‐embryonic conservation of gene expression
The question remains as to why developmental stages only after organogenesis exhibit the recapitulation pattern in chromatin accessibility. As observed so far, even the most attractive hypothesis, such as developmental burden, fails to fully explain these findings. It is tempting to speculate that the conserved mid‐embryonic phase acts as a singular point, or boundary that inhibits the recapitulation pattern before this phase. With this perspective, a classic argument is that the mid‐embryonic phase is the target of the strongest negative selection due to embryonic lethality (Raff, 1996). This concept has been referred to as the conservation mechanism for the bottleneck part of the hourglass model; however, it was not supported by our recent study (Uchida et al., 2018). Namely, embryonic lethality of vertebrate embryos under normal and perturbation conditions suggested that it was the early embryonic phase, rather than the mid‐embryonic phase, that was targeted by negative selection pressure owing to lethality (Uchida et al., 2018). Another feature unique to the conserved mid‐embryonic phase is the enrichment of pleiotropically expressed genes (Hu et al., 2017; Li et al., 2020; Liu & Robinson‐Rechavi, 2018). Based on this, Hu et al. suggested that pleiotropic constraint (Duboule & Wilkins, 1998; Galis, 1999) at the gene regulation level could be a contributing factor for mid‐embryonic conservation. Although the exact molecular mechanism of pleiotropic constraint at the gene regulation level remains to be clarified, it would be of interest to investigate the relevance of regulatory elements, such as pleiotropic enhancers (Sabarís et al., 2019) or chromatin accessibility, in this process.
These factors, however, do not provide any reasonable mechanisms for the boundary of the recapitulation pattern at the mid‐embryonic phase. Furthermore, it is also problematic that the recapitulation pattern has been observed only for chromatin accessibility, and not for the transcriptome (Hu et al., 2017; Irie & Kuratani, 2011; Uesaka et al., 2019; Wang et al., 2013; see Figure 1c; the nested hourglasses model with multiple bottlenecks). Notably, a theoretical study suggested that gene activations showing the recapitulation pattern arise with a computer‐based simulation that evolved linearly aligned artificial cells with simple gene regulatory networks (Kohsokabe & Kaneko, 2016). However, the discrepancies observed between this and the experimental studies would be due to the different conditions of development and evolution between artificial and real organisms. Further analyses utilizing the epigenome and transcriptome data from multiple developing tissues, including public databases, such as the ENCODE database (Davis et al., 2018), would provide a basis for understanding the relationship between the gene regulatory elements and hourglass‐like transcriptomic divergency.
5. CONCLUSIONS AND FUTURE ISSUES
While the recapitulation pattern was not observed for whole‐embryonic transcriptomes, it was observed for developmental chromatin accessibility during vertebrate embryogenesis after the most conserved mid‐embryonic period. This discrepancy remains to be clarified. However, it is possible that the recapitulation and the developmental hourglass model coincide with each other during later vertebrate embryogenesis, as both imply that more derived features appear after the most conserved mid‐embryonic period. The evolutionary mechanism underlying such coexistence at later stages remains largely unknown. However, it may be useful to clarify whether the recapitulation pattern of chromatin accessibility underlies the diversifying gene expression profiles and the recapitulation pattern in the morphological development of some traits. It also has to be noted that the coexistence model has not been validated enough even in vertebrates, and thus the generality of this model remains unclear. Thus, further studies focusing on not only various vertebrates but also species from different phyla are awaited. Clarifying these remaining questions will further advance our understanding not only of the laws on the evolution of animal embryogenesis but also of how animal embryogenesis has evolved and would evolve.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
ACKNOWLEDGMENTS
We thank Dr. Takahiro Kohsokabe, Dr. Shiro Egawa, and Dr. Tatsuya Hirasawa for their inspiring comments. This study was supported in part by JSPS KAKENHI Grant JP18K14711 (Early‐Career Scientists) to Masahiro Uesaka; JSPS KAKENHI Grants JP15H05603 (Grant‐in‐Aid for Young Scientists (A)) and JP17H06387 (Grant‐in‐Aid for Scientific Research on Innovative Areas) and the Platform for Dynamic Approaches to Living System from MEXT, Japan, to Naoki Irie.
Uesaka M, Kuratani S, Irie N. The developmental hourglass model and recapitulation: An attempt to integrate the two models. J Exp Zool B (Mol Dev Evol). 2022;338:76–86. 10.1002/jezb.23027
DATA AVAILABILITY STATEMENT
Original data are available from the authors upon request.
REFERENCES
- Abzhanov, A. (2013). von Baer's law for the ages: Lost and found principles of developmental evolution. Trends in Genetics, 29, 712–722. [DOI] [PubMed] [Google Scholar]
- Arthur, W. (1997). The origin of animal body plans: A study in evolutionary developmental biology. Cambridge University Press. [Google Scholar]
- von Baer, K. E. (1828). Über Entwickelungsgeschichte der Thiere: Beobachtung und Reflektion. Gebrüder Borntraeger. [Google Scholar]
- Ballard, W. W. (1981). Morphogenetic movements and fate maps of vertebrates. American Zoologist, 21, 391–399. [Google Scholar]
- Bejder, L. , & Hall, B. K. (2002). Limbs in whales and limblessness in other vertebrates: Mechanisms of evolutionary and developmental transformation and loss. Evolution & Development, 4, 445–458. [DOI] [PubMed] [Google Scholar]
- Bininda‐Emonds, O. R. P. , Jeffery, J. E. , & Richardson, M. K. (2003). Inverting the hourglass: Quantitative evidence against the phylotypic stage in vertebrate development. Proceedings of the Royal Society B: Biological Sciences, 270, 341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho, J. F. , Smith‐Paredes, D. , Soto‐Acuña, S. , Mpodozis, J. , Palma, V. , & Vargas, A. O. (2015). Skeletal plasticity in response to embryonic muscular activity underlies the development and evolution of the perching digit of birds. Scientific Reports, 5, 9840–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho, J. F. , Smith‐Paredes, D. , Soto‐Acuña, S. , Núñez‐León, D. , Palma, V. , & Vargas, A. O. (2017). Greater growth of proximal metatarsals in bird embryos and the evolution of hallux position in the grasping foot. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution, 328, 106–118. [DOI] [PubMed] [Google Scholar]
- Buenrostro, J. D. , Giresi, P. G. , Zaba, L. C. , Chang, H. Y. , & Greenleaf, W. J. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA‐binding proteins and nucleosome position. Nature Methods, 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J. , Cusanovich, D. A. , Ramani, V. , Aghamirzaie, D. , Pliner, H. A. , Hill, A. J. , Daza, R. M. , McFaline‐Figueroa, J. L. , Packer, J. S. , Christiansen, L. , Steemers, F. J. , Adey, A. C. , Trapnell, C. , & Shendure, J. (2018). Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science, 361, 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, S. B. (2005). Evolution at two levels: On genes and form. PLOS Biology, 3, e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Lake, B. B. , & Zhang, K. (2019). High‐throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nature Biotechnology, 37, 1452–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn, M. J. , & Tickle, C. (1999). Developmental basis of limblessness and axial patterning in snakes. Nature, 399, 474–479. [DOI] [PubMed] [Google Scholar]
- Cooper, K. L. , Sears, K. E. , Uygur, A. , Maier, J. , Baczkowski, K.‐S. , Brosnahan, M. , Antczak, D. , Skidmore, J. A. , & Tabin, C. J. (2014). Patterning and post‐patterning modes of evolutionary digit loss in mammals. Nature, 511, 41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, C. A. , Hitz, B. C. , Sloan, C. A. , Chan, E. T. , Davidson, J. M. , Gabdank, I. , Hilton, J. A. , Jain, K. , Baymuradov, U. K. , Narayanan, A. K. , Onate, K. C. , Graham, K. , Miyasato, S. R. , Dreszer, T. R. , Strattan, J. S. , Jolanki, O. , Tanaka, F. Y. , & Cherry, J. M. (2018). The encyclopedia of DNA elements (ENCODE): Data portal update. Nucleic Acids Research, 46, D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diogo, R. , Smith, C. M. , & Ziermann, J. M. (2015). Evolutionary developmental pathology and anthropology: A new field linking development, comparative anatomy, human evolution, morphological variations and defects, and medicine. Developmental Dynamics, 244, 1357–1374. [DOI] [PubMed] [Google Scholar]
- Domazet‐Lošo, T. , & Tautz, D. (2010). A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature, 468, 815–818. [DOI] [PubMed] [Google Scholar]
- Drost, H.‐G. , Gabel, A. , Grosse, I. , & Quint, M. (2015). Evidence for active maintenance of phylotranscriptomic hourglass patterns in animal and plant embryogenesis. Molecular Biology and Evolution, 32, 1221–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule, D. (1994). Temporal colinearity and the phylotypic progression: A basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Development (Supplement), 1994, 135–142. [PubMed] [Google Scholar]
- Duboule, D. , & Wilkins, A. S. (1998). The evolution of 'bricolage'. Trends in Genetics, 14, 54–59. [DOI] [PubMed] [Google Scholar]
- Galis, F. (1999). Why do almost all mammals have seven cervical vertebrae? Developmental constraints, Hox genes, and cancer. Journal of Experimental Zoology, 285, 19–26. [PubMed] [Google Scholar]
- Garstang, W. (1922). The theory of recapitulation: A critical re‐statement of the biogenetic law. Zoological Journal of the Linnean Society, 35, 81–101. [Google Scholar]
- Gildor, T. , Cary, G. A. , Lalzar, M. , Hinman, V. F. , Ben‐Tabou, & de‐Leon, S. (2019). Developmental transcriptomes of the sea star, Patiria miniata, illuminate how gene expression changes with evolutionary distance. Scientific Reports, 9, 16201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, S. J. (1977). Ontogeny and phylogeny. Harvard University Press. [Google Scholar]
- Gross, D. S. , & Garrard, W. T. (1988). Nuclease hypersensitive sites in chromatin. Annual Review of Biochemistry, 57, 159–197. [DOI] [PubMed] [Google Scholar]
- Haeckel, E. (1866). Generelle Morphologie der Organismen: Allgemeine Grundzuge der organischen Formen‐Wissenschaft, mechanisch begrundet durch die von Charles Darwin reformirte Descendenz‐Theorie. Georg Reimer. [Google Scholar]
- Haeckel, E. (1874). Anthropogenie oder entwickelungsgeschichte des menschen. Leipzig: W. Engelmann.
- Hall, B. K. (1992). Evolutionary developmental biology. Chapman & Hall. [Google Scholar]
- Hazkani Covo, E. , Wool, D. , & Graur, D. (2005). In search of the vertebrate phylotypic stage: A molecular examination of the developmental hourglass model and von Baer's third law. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution, 304B, 150–158. [DOI] [PubMed] [Google Scholar]
- Hu, H. , Uesaka, M. , Guo, S. , Shimai, K. , Lu, T.‐M. , Li, F. , Fujimoto, S. , Ishikawa, M. , Liu, S. , Sasagawa, Y. , Zhang, G. , Kuratani, S. , Yu, J.‐K. , Kusakabe, T. G. , Khaitovich, P. , & Irie, N. (2017). Constrained vertebrate evolution by pleiotropic genes. Nature Ecology and Evolution, 1(11), 1722–1730. [DOI] [PubMed] [Google Scholar]
- Irie, N. , & Kuratani, S. (2011). Comparative transcriptome analysis reveals vertebrate phylotypic period during organogenesis. Nature Communications, 2, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie, N. , & Kuratani, S. (2014). The developmental hourglass model: A predictor of the basic body plan? Development, 141, 4649–4655. [DOI] [PubMed] [Google Scholar]
- Irie, N. , Satoh, N. , & Kuratani, S. (2018). The phylum Vertebrata: A case for zoological recognition. Zoological Letters, 4, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie, N. , & Sehara‐Fujisawa, A. (2007). The vertebrate phylotypic stage and an early bilaterian‐related stage in mouse embryogenesis defined by genomic information. BMC Biology, 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel, J. W. , Martik, M. L. , Byrne, M. , Raff, E. C. , Raff, R. A. , McClay, D. R. , & Wray, G. A. (2016). Comparative developmental transcriptomics reveals rewiring of a highly conserved gene regulatory network during a major life history switch in the sea urchin genus heliocidaris. PLOS Biology, 14, e1002391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinka, A. T. , & Tomancak, P. (2012). The evolution of early animal embryos: Conservation or divergence? Trends in Ecology & Evolution, 27, 385–393. [DOI] [PubMed] [Google Scholar]
- Kalinka, A. T. , Varga, K. M. , Gerrard, D. T. , Gerrard, D. T. , Preibisch, S. , Corcoran, D. L. , Corcoran, D. L. , Jarrells, J. , Jarrells, J. , Ohler, U. , Bergman, C. M. , Bergman, C. M. , & Tomancak, P. (2010). Gene expression divergence recapitulates the developmental hourglass model. Nature, 468, 811–814. [DOI] [PubMed] [Google Scholar]
- Klemm, S. L. , Shipony, Z. , & Greenleaf, W. J. (2019). Chromatin accessibility and the regulatory epigenome. Nature Reviews Genetics, 20, 207–220. [DOI] [PubMed] [Google Scholar]
- Kohsokabe, T. , & Kaneko, K. (2016). Evolution‐development congruence in pattern formation dynamics: Bifurcations in gene expression and regulation of networks structures. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution, 326, 61–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyabu, D. , & Son, N. T. (2014). Patterns of postcranial ossification and sequence heterochrony in bats: Life histories and developmental trade‐offs. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution, 322, 607–618. [DOI] [PubMed] [Google Scholar]
- Kuratani, S. (2004). Evolutionary morphology: Bauplan and embryonic development of vertebrates. Tokyo Daigaku Shuppankai. [Google Scholar]
- Lareau, C. A. , Duarte, F. M. , Chew, J. G. , Kartha, V. K. , Burkett, Z. D. , Kohlway, A. S. , Pokholok, D. , Aryee, M. J. , Steemers, F. J. , Lebofsky, R. , & Buenrostro, J. D. (2019). Droplet‐based combinatorial indexing for massive‐scale single‐cell chromatin accessibility. Nature Biotechnology, 37, 916–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, M. , Hashimshony, T. , Wagner, F. , & Yanai, I. (2012). Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Developmental Cell, 22, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Li, C. , Wu, X.‐C. , Rieppel, O. , Wang, L.‐T. , & Zhao, L.‐J. (2008). An ancestral turtle from the Late Triassic of southwestern China. Nature, 456, 497–501. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Omori, A. , Flores, R. L. , Satterfield, S. , Nguyen, C. , Ota, T. , Tsurugaya, T. , Ikuta, T. , Ikeo, K. , Kikuchi, M. , Leong, J. C. K. , Reich, A. , Hao, M. , Wan, W. , Dong, Y. , Ren, Y. , Zhang, S. , Zeng, T. , Uesaka, M. , … Irie, N. (2020). Genomic insights of body plan transitions from bilateral to pentameral symmetry in Echinoderms. Commununications Biology, 3, 371–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , & Robinson‐Rechavi, M. (2018). Developmental constraints on genome evolution in four bilaterian model species. Genome Biology and Evolution, 10, 2266–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy, N. R. (2000). Reinterpreting recapitulation: Systematics of needlefishes and their allies (Teleostei: Beloniformes). Evolution, 54, 1349–1362. [DOI] [PubMed] [Google Scholar]
- Lovejoy, N. R. , Iranpour, M. , & Collette, B. B. (2004). Phylogeny and jaw ontogeny of beloniform fishes. American Zoologist, 44, 366–377. [DOI] [PubMed] [Google Scholar]
- Løvtrup, S. (1978). On von Baerian and Haeckelian recapitulation. Systematic Zoology, 27, 348. [Google Scholar]
- Malik, A. , Gildor, T. , Sher, N. , Layous, M. , & Ben‐Tabou de‐Leon, S. (2017). Parallel embryonic transcriptional programs evolve under distinct constraints and may enable morphological conservation amidst adaptation. Developmental Biology, 430, 202–213. [DOI] [PubMed] [Google Scholar]
- Marlétaz, F. , Firbas, P. N. , Maeso, I. , Tena, J. J. , Bogdanović, O. , Perry, M. , Wyatt, C. D. R. , Calle‐Mustienes, E. , Bertrand, S. , Burguera, D. , Acemel, R. D. , Heeringen, S. J. , Naranjo, S. , Herrera‐Ubeda, C. , Skvortsova, K. , Jimenez‐Gancedo, S. , Aldea, D. , Marquez, Y. , Buono, L. , … Irimia, M. (2018). Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature, 138, 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr, E. (1994). Recapitulation reinterpreted: The somatic program. Quarterly Review of Biology, 69, 223–232. [Google Scholar]
- Metscher, B. D. , & Ahlberg, P. E. (2001). Origin of the teleost tail: Phylogenetic frameworks for developmental studies. In Ahlberg P. E. (Ed.), Major events in early vertebrate evolution. CRC Press. [Google Scholar]
- Mikkelsen, T. S. , Wakefield, M. J. , Aken, B. , Amemiya, C. T. , Chang, J. L. , Duke, S. , Garber, M. , Gentles, A. J. , Goodstadt, L. , Heger, A. , Jurka, J. , Kamal, M. , Mauceli, E. , Searle, S. M. J. , Sharpe, T. , Baker, M. L. , Batzer, M. A. , Benos, P. V. , Belov, K. , … Lindblad‐Toh, K. (2007). Genome of the marsupial Monodelphis domestica reveals innovation in non‐coding sequences. Nature, 447, 167–177. [DOI] [PubMed] [Google Scholar]
- Morgan, T. H. (1916). A critique of the theory of evolution. Oxford University Press. [Google Scholar]
- Nagashima, H. , Sugahara, F. , Takechi, M. , Ericsson, R. , Kawashima‐Ohya, Y. , Narita, Y. , & Kuratani, S. (2009). Evolution of the turtle body plan by the folding and creation of new muscle connections. Science, 325, 193–196. [DOI] [PubMed] [Google Scholar]
- Nishihara, H. , Kobayashi, N. , Kimura‐Yoshida, C. , Yan, K. , Bormuth, O. , Ding, Q. , Nakanishi, A. , Sasaki, T. , Hirakawa, M. , Sumiyama, K. , Furuta, Y. , Tarabykin, V. , Matsuo, I. , & Okada, N. (2016). Coordinately co‐opted multiple transposable elements constitute an enhancer for wnt5a expression in the mammalian secondary palate. PLOS Genetics, 12, e1006380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom, D. T. , Dowell, R. D. , Jacobsen, E. S. , Gordon, W. , Danford, T. W. , MacIsaac, K. D. , Rolfe, P. A. , Conboy, C. M. , Gifford, D. K. , & Fraenkel, E. (2007). Tissue‐specific transcriptional regulation has diverged significantly between human and mouse. Nature Genetics, 39, 730–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer, J. M. (1959). Embryology and evolution: Nineteenth century hopes and twentieth century realities. Quarterly Review of Biology, 34, 271–277. [DOI] [PubMed] [Google Scholar]
- Poe, S. (2006). Test of von Baer's law of the conservation of early development. Evolution, 60(11), 2239–2245. [PubMed] [Google Scholar]
- Poe, S. , & Wake, M. H. (2004). Quantitative tests of general models for the evolution of development. American Naturalist, 164, 415–422. [DOI] [PubMed] [Google Scholar]
- Prum, R. O. (1999). Development and evolutionary origin of feathers. Journal of Experimental Zoology, 285, 291–306. [PubMed] [Google Scholar]
- Prum, R. O. , & Brush, A. H. (2015). The evolutionary origin and diversification of feathers. Quarterly Review of Biology, 77, 261–295. [DOI] [PubMed] [Google Scholar]
- Raff, R. A. (1996). The shape of life: Genes, development, and the evolution of animal form. University of Chicago Press. [Google Scholar]
- Richardson, M. K. (1999). Vertebrate evolution: The developmental origins of adult variation. BioEssays, 21, 604–613. [DOI] [PubMed] [Google Scholar]
- Richardson, M. K. , Gobes, S. M. H. , van Leeuwen, A. C. , Polman, J. A. E. , Pieau, C. , & Sánchez‐Villagra, M. R. (2009). Heterochrony in limb evolution: Developmental mechanisms and natural selection. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution, 312(6), 639–664. [DOI] [PubMed] [Google Scholar]
- Richardson, M. K. , Hanken, J. , Gooneratne, M. L. , Pieau, C. , Raynaud, A. , Selwood, L. , & Wright, G. M. (1997). There is no highly conserved embryonic stage in the vertebrates: Implications for current theories of evolution and development. Anatomy and Embryology, 196, 91–106. [DOI] [PubMed] [Google Scholar]
- Richardson, M. K. , & Keuck, G. (2002). Haeckel's ABC of evolution and development. Biological Reviews of the Cambridge Philosophical Society, 77, 495–528. [DOI] [PubMed] [Google Scholar]
- Riedl, R. (1978). Order in living organisms: A systems analysis of evolution. John Wiley & Sons. [Google Scholar]
- Roux, J. , & Robinson‐Rechavi, M. (2008). Developmental constraints on vertebrate genome evolution. PLOS Genetics, 4, e1000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabarís, G. , Laiker, I. , Preger‐Ben Noon, E. , & Frankel, N. (2019). Actors with multiple roles: Pleiotropic enhancers and the paradigm of enhancer modularity. Trends in Genetics, 35, 423–433. [DOI] [PubMed] [Google Scholar]
- Sallan, L. (2016). Fish “tails” result from outgrowth and reduction of two separate ancestral tails. Current Biology, 26, R1224–R1225. [DOI] [PubMed] [Google Scholar]
- Satpathy, A. T. , Granja, J. M. , Yost, K. E. , Qi, Y. , Meschi, F. , McDermott, G. P. , Olsen, B. N. , Mumbach, M. R. , Pierce, S. E. , Corces, M. R. , Shah, P. , Bell, J. C. , Jhutty, D. , Nemec, C. M. , Wang, J. , Wang, L. , Yin, Y. , Giresi, P. G. , Chang, A. L. S. , … Chang, H. Y. (2019). Massively parallel single‐cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nature Biotechnology, 37, 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierenberg, E. (2006). Embryological variation during nematode development, WormBook: The online review of C. elegans biology (pp. 1–13). WormBook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer, P. H. , Polsky, A. L. , Cole, A. G. , Camps, J. I. R. , Kroiher, M. , Silver, D. H. , Grishkevich, V. , Anavy, L. , Koutsovoulos, G. , Hashimshony, T. , & Yanai, I. (2018). The gene regulatory program of Acrobeloides nanus reveals conservation of phylum‐specific expression. Proceedings ofthe National Academy of Sciences of the United States of America, 115, 4459–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick, A. (1894). On the law of development commonly known as von Baer's law; and on the significance of ancestral rudiments in embryonic development. The Journal of Cell Science, s2–36, 35–52. https://jcs.biologists.org/content/s2-36/141/35 [Google Scholar]
- Seki, R. , Li, C. , Fang, Q. , Hayashi, S. , Egawa, S. , Hu, J. , Xu, L. , Pan, H. , Kondo, M. , Sato, T. , Matsubara, H. , Kamiyama, N. , Kitajima, K. , Saito, D. , Liu, Y. , Gilbert, M. T. P. , Zhou, Q. , Xu, X. , Shiroishi, T. , … Zhang, G. (2017). Functional roles of Aves class‐specific cis‐regulatory elements on macroevolution of bird‐specific features. Nature Communications, 8, 14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim, S. , Kwan, K. Y. , Li, M. , Lefebvre, V. , & Šestan, N. (2012). Cis‐regulatory control of corticospinal system development and evolution. Nature, 486, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack, J. M. , Holland, P. W. , & Graham, C. F. (1993). The zootype and the phylotypic stage. Nature, 361, 490–492. [DOI] [PubMed] [Google Scholar]
- Stergachis, A. B. , Sandstrom, R. , Reynolds, A. P. , Zhang, M. , Stelhing‐Sun, S. , Stelhing‐Sun, S. , Thurman, R. E. , Vong, S. , Vierstra, J. , Sabo, P. J. , Wilken, M. S. , Reh, T. A. , Treuting, P. M. , & Stamatoyannopoulos, J. A. (2014). Conservation of trans‐acting circuitry during mammalian regulatory evolution. Nature, 515, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro, K. , Teissier, A. , Kobayashi, N. , Nakanishi, A. , Sasaki, T. , Yan, K. , Tarabykin, V. , Vigier, L. , Sumiyama, K. , Hirakawa, M. , Nishihara, H. , Pierani, A. , & Okada, N. (2011). A mammalian conserved element derived from SINE displays enhancer properties recapitulating Satb2 expression in early‐born callosal projection neurons. PLOS One, 6, e28497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium . (2012). An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. http://www.nature.com/nature/journal/v489/n7414/full/nature11247.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thewissen, J. G. M. (2018). Highlights of cetacean embryology. Aquatic Mammals, 44, 591–602. 10.1578/AM.44.6.2018.591 [DOI] [Google Scholar]
- Thewissen, J. G. M. , Cohn, M. J. , Stevens, L. S. , Bajpai, S. , Heyning, J. , & Horton, W. E. (2006). Developmental basis for hind‐limb loss in dolphins and origin of the cetacean bodyplan. Proceedings of the National Academy of Sciences of the United States of America, 103, 8414–8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, Y. , Uesaka, M. , Yamamoto, T. , Takeda, H. , & Irie, N. (2018). Embryonic lethality is not sufficient to explain hourglass‐like conservation of vertebrate embryos. EvoDevo, 9, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesaka, M. , Kuratani, S. , Takeda, H. , & Irie, N. (2019). Recapitulation‐like developmental transitions of chromatin accessibility in vertebrates. Zoological Letters, 5, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra, J. , Rynes, E. , Sandstrom, R. , Zhang, M. , Canfield, T. , Hansen, R. S. , Stehling‐Sun, S. , Sabo, P. J. , Byron, R. , Humbert, R. , Thurman, R. E. , Johnson, A. K. , Vong, S. , Lee, K. , Bates, D. , Neri, F. , Diegel, M. , Giste, E. , Haugen, E. , … Stamatoyannopoulos, J. A. (2014). Mouse regulatory DNA landscapes reveal global principles of cis‐regulatory evolution. Science, 346, 1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Pascual‐Anaya, J. , Zadissa, A. , Niimura, Y. , White, S. , Xiong, Z. , Wang, B. , Zheng, Y. , Pignatelli, M. , Nozawa, M. , Wang, J. , Zhang, H. , Yu, L. , Shigenobu, S. , Wang, J. , Searle, S. , Wang, J. , Yin, Y. , Zhang, G. , & Irie, N. (2013). The draft genomes of soft‐shell turtle and green sea turtle yield insights into the development and evolution of the turtle‐specific body plan. Nature Genetics, 45, 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch, M. T. , & Hughes, T. R. (2010). Conserved expression without conserved regulatory sequence: The more things change, the more they stay the same. Trends in Genetics, 26, 66–74. [DOI] [PubMed] [Google Scholar]
- Weisbecker, V. , Goswami, A. , Wroe, S. , & Sánchez‐Villagra, M. R. (2008). Ossification heterochrony in the therian postcranial skeleton and the marsupial‐placental dichotomy. Evolution, 62, 2027–2041. [DOI] [PubMed] [Google Scholar]
- Wilson, M. D. , & Odom, D. T. (2009). Evolution of transcriptional control in mammals. Current Opinion in Genetics & Development, 19, 579–585. [DOI] [PubMed] [Google Scholar]
- Wimsatt, W. C. (1986). Developmental constraints, generative entrenchment, and the innate‐acquired distinction. In Bechtel W. (Ed.), Integrating scientific disciplines. Science and philosophy. Springer. [Google Scholar]
- Xu, F. , Domazet‐Lošo, T. , Fan, D. , Dunwell, T. L. , Li, L. , Fang, X. , & Zhang, G. (2016). High expression of new genes in trochophore enlightening the ontogeny and evolution of trochozoans. Scientific Reports, 6, 34664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai, I. , Peshkin, L. , Jorgensen, P. , & Kirschner, M. W. (2011). Mapping gene expression in two Xenopus species: Evolutionary constraints and developmental flexibility. Developmental Cell, 20, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, C. , Yu, M. , Huang, H. , Juric, I. , Abnousi, A. , Hu, R. , Lucero, J. , Behrens, M. M. , Hu, M. , & Ren, B. (2019). An ultra high‐throughput method for single‐cell joint analysis of open chromatin and transcriptome. Nature Structural & Molecular Biology, 26, 1063–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziermann, J. M. , & Diogo, R. (2014). Cranial muscle development in frogs with different developmental modes: Direct development versus biphasic development. Journal of Morphology, 275, 398–413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Original data are available from the authors upon request.


