Abstract
The evolution of mass raiding has allowed army ants to become dominant arthropod predators in the tropics. Although a century of research has led to many discoveries about behavioural, morphological and physiological adaptations in army ants, almost nothing is known about the molecular basis of army ant biology. Here we report the genome of the iconic New World army ant Eciton burchellii, and show that it is unusually compact, with a reduced gene complement relative to other ants. In contrast to this overall reduction, a particular gene subfamily (9‐exon ORs) expressed predominantly in female antennae is expanded. This subfamily has previously been linked to the recognition of hydrocarbons, key olfactory cues used in insect communication and prey discrimination. Confocal microscopy of the brain showed a corresponding expansion in a putative hydrocarbon response centre within the antennal lobe, while scanning electron microscopy of the antenna revealed a particularly high density of hydrocarbon‐sensitive sensory hairs. E. burchellii shares these features with its predatory and more cryptic relative, the clonal raider ant. By integrating genomic, transcriptomic and anatomical analyses in a comparative context, our work thus provides evidence that army ants and their relatives possess a suite of modifications in the chemosensory system that may be involved in behavioural coordination and prey selection during social predation. It also lays the groundwork for future studies of army ant biology at the molecular level.
Keywords: chemosensation, ecological adaptation, evolution, evolutionary genomics, gene family, genome evolution, genomics/proteomics
1. INTRODUCTION
Army ants are dominant arthropod predators in tropical ecosystems worldwide (Brady, 2003; Kronauer, 2009). It has been estimated that a given square metre of rain forest is subject to an army ant raid more than once a day on average (Kaspari & O’Donnell, 2003), with some species depleting ~25% of invertebrate biomass in such events (Kaspari et al., 2011). The key to the ecological success of army ants is mass raiding, a form of obligate collective foraging in which colonies attack and overwhelm live prey (Gotwald, 1995; Kronauer, 2020; Rettenmeyer, 1963; Schneirla, 1971). Army ant mass raids contain up to hundreds of thousands of individuals collectively searching for and exploiting food resources (Brady, 2003; Gotwald, 1995; Kronauer, 2009; Schneirla, 1971). This behaviour has evolved at least twice within the ant subfamily Dorylinae, giving rise to separate Old World and New World army ant lineages that display striking convergence in behaviour and morphology (Borowiec, 2017). A suite of other traits have evolved in parallel with mass raiding, including nomadism; morphologically specialized, wingless queens; and reproduction via colony fission (Brady, 2003; Gotwald, 1995; Kronauer, 2009; Schneirla, 1971). Together, these traits are referred to as the “army ant adaptive syndrome” (Gotwald, 1982). Both army ant lineages display elevated rates of diversification (Borowiec, 2017), demonstrating the evolutionary success of this lifestyle. Army ants have proven excellent models for studying social predation, and several behavioural and physiological adaptations for this behaviour have been described (Brady, 2003; Bulova et al., 2016; Gotwald, 1995; Kronauer, 2009; Rettenmeyer, 1963; Schneirla, 1971). However, molecular adaptations facilitating social predation—a premise for the ecological dominance of army ants in tropical rainforests—remain poorly understood, and no genomic studies of army ants have been conducted.
Like most other ants (Wilson, 1965), army ants have evolved a sophisticated pheromone communication system to coordinate social behaviour, including navigation, recruitment of nestmates to prey sources, alerting nestmates of danger, and coordinating defence of the queen and care for the brood (Brown, 1959; Chadab & Rettenmeyer, 1975; Schneirla, 1971; Torgerson & Akre, 1970). The chemosensory system is responsible for recognizing these pheromones (Dumpert, 1972; Ozaki et al., 2005; Pask et al., 2017; Sharma et al., 2015; Slone et al., 2017; Trible et al., 2017; Yan et al., 2017). Cuticular hydrocarbons (CHCs), a diverse class of chemicals that are displayed on the surface of arthropods, play a particularly prominent role in ant communication by conveying species, colony and caste identity (Liebig, 2010; van Zweden & D’Ettorre, 2010). Chemosensation is also important for ant foraging by facilitating prey detection and discrimination (Lang & Menzel, 2011; Manubay & Powell, 2020; Mirenda et al., 1980; Witte et al., 2010), and here again, CHCs play an important role (Lang & Menzel, 2011; Manubay & Powell, 2020; Witte et al., 2010). While CHCs and other army ant pheromones have been studied in some detail (e.g., Bagneres et al., 1991; von Beeren et al., 2018; Brückner et al., 2018; Keegans et al., 1993), nothing is known about the army ant chemosensory system and how it compares with that of other ants. Genomic and neuroanatomical studies in the nonarmy ant doryline Ooceraea biroi, which possesses a subset of army ant traits (Kronauer, 2009), have revealed an especially elaborate chemosensory system, with several features that have so far not been observed in other ants (McKenzie et al., 2016; McKenzie & Kronauer, 2018; Oxley et al., 2014; Trible et al., 2017). Chemosensory receptor genes in the 9‐exon odorant receptor (OR) gene family, which detect CHCs, are particularly expanded in O. biroi, as are associated neural structures in the brain (T6 glomeruli in the antennal lobe; McKenzie et al., 2016). Furthermore, workers of this species possess a unique arrangement of CHC‐detecting sense hairs (basiconic sensilla; McKenzie et al., 2016). Specifically, in O. biroi, basiconic sensilla occur in an unusually high density along the ventral (with respect to the body axis while antennae are held naturally) surface of the female antennal club, but nowhere else on the antenna. Given the paucity of molecular or neurobiological data within the Dorylinae, it is unknown whether any of these features are conserved across dorylines and whether they might be linked to the evolution of army ant‐like lifestyles.
The conspicuous Eciton burchellii (Figure 1a)—an abundant and ecologically impactful species in Neotropical forests (Kaspari et al., 2011; O’Donnell et al., 2007)—is perhaps the best studied army ant (Gotwald, 1995; Rettenmeyer, 1963; Schneirla, 1971). Like other army ants, E. burchellii is nomadic, reproduces via colony fission, possesses morphologically specialized queens and forages via mass raiding (Gotwald, 1995; Rettenmeyer, 1963; Schneirla, 1971). E. burchellii stands out among New World army ants in being an epigaeic swarm raider, meaning that foragers fan out across the surface of the forest floor into a large “raiding carpet” (Schneirla, 1971). This species consumes more nonant prey than most other New World army ant species: ~50% by mass according to Franks (1982), 7% of prey items according to Hoenle et al. (2019). However, the majority of E. burchellii prey items belong to a few species of Camponotus carpenter ants, indicating that, like most other army ants, E. burchellii is still a specialized ant predator (Hoenle et al., 2019).
FIGURE 1.
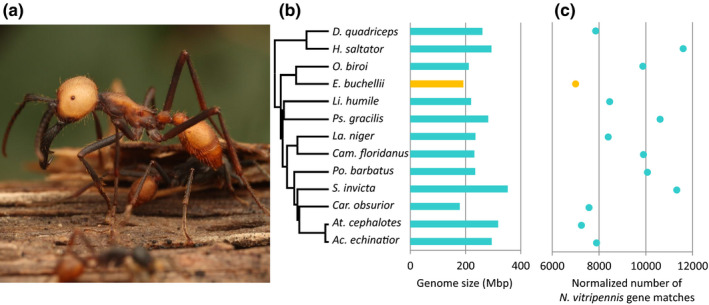
The genome of Eciton burchellii. (a) An E. burchellii major (photo by D.J.C.K.). (b) Phylogeny and genome assembly sizes of representative ant species. Phylogeny after Moreau and Bell (2013). Genus abbreviations are as follows: D. = Dinoponera, H. = Harpegnathos, O. = Ooceraea, E. = Eciton, Li. = Linepithema, Ps. = Pseudomyrmex, La. = Lasius, Cam. = Camponotus, Po. = Pogonomyrmex, S. = Solenopsis, Car. = Cardiocondyla, At. = Atta, Ac. = Acromyrmex. (c) Normalized number of N. vitripennis gene matches in the genomes of representative ant species, as calculated by the Blat algorithm with a 50% alignment coverage cutoff, and relativized to genome completeness as estimated by busco. Data from E. burchellii are in orange
To investigate the genomic and chemosensory basis of predation in E. burchellii, we sequenced the species’ genome and worker and male antennal and whole‐body transcriptomes, and investigated worker and male antennal and antennal lobe anatomy. Compared with other ants, we found that E. burchellii has a compact genome, with a particular paucity of interspersed repeats. E. burchellii also shows a general reduction in gene number, but no large, significant contractions in any specific gene family. Correspondingly, E. burchellii possesses relatively few chemosensory genes, with reduced repertoires of most OR gene subfamilies. However, as in O. biroi, the hydrocarbon‐sensitive 9‐exon OR subfamily is expanded. Gene evolution analysis revealed that these expansions happened independently in both species, indicating parallel selection pressures on OR genes. In addition, we detected an unusually high number of macroglomeruli in the antennal lobes, which may play a role in the recognition of nonhydrocarbon pheromones. Finally, we observed striking similarities in the peripheral and central chemosensory systems of E. burchellii and O. biroi. Integrating all data, our study suggests that E. burchellii has evolved specific adaptations in the olfactory system, potentially for processing semiochemicals in the contexts of predation and social information transfer. This suggests that the chemosensory system has played an important role in the evolution of army ants and other dorylines, and lays a foundation for studying the molecular basis for social predation.
2. METHODS
2.1. Genome sequencing and assembly
From a single colony, Eciton burchellii (subspecies E. burchellii foreli) workers and males were collected at La Selva Biological Station in Heredia Province, Costa Rica, frozen alive at −80°C, and split into two packages containing dry ice (see RNA sequencing below). No queens were collected due to the difficulty of sampling army ant queens and the fact that queen removal destroys the whole colony. One package contained only males and was transported to the University of Wisconsin‐Madison, Madison, WI, USA. Genomic DNA was extracted from one male using a modified version of the Genomic‐tip DNA extraction protocol for mosquitoes and other insects (Qiagen) as previously described (Suen et al., 2011). Three libraries for Illumina sequencing were constructed, including a 3‐ and an 8‐kbp insertion mate‐pair library, and sequenced at the University of Wisconsin‐Madison Biotechnology Center on two independent 2×300‐bp paired‐end Illumina MiSeq runs, yielding 10 and 16 million reads, respectively. A third shotgun library was sequenced on an Illumina HiSeq 2500 with 2×150‐bp paired‐end reads to yield 157 million reads. Reads were trimmed and prepared with nextclip (Leggett et al., 2014), and assembled using allpaths‐lg (Gnerre et al., 2011). The final assembly contained 190,515,515 bp on 2,492 contigs, scaffolded onto 449 scaffolds for a final length of 192,351,860 bp including gaps. The sequencing data and genome assemblies are available at the NCBI SRA and genome databases (Bioproject PRJNA632625).
2.2. RNA sequencing
The second package contained workers and males and was transported to the Rockefeller University. The antennae of 10 workers, spanning a range of worker sizes including representatives of each size class (minims, media, submajors and majors), were separated from bodies at 4°C. All antennae and all remaining bodies were then transferred separately into two vials containing Trizol reagent (Thermo Fisher Scientific), and subsequently the two samples were homogenized using a TissueLyser II (Qiagen). Five males were processed in the same way, except that only one of the antennae‐less bodies was used due to the exceptionally large size of E. burchellii males. Chloroform was then added to the Trizol, and the aqueous phase was retrieved using Phase Lock columns (5Prime) and cleaned using Qiagen RNeasy columns following the manufacturer's protocol. Illumina cDNA libraries were prepared at the Rockefeller University Genome Resource Center and sequenced on an Illumina HiSeq 2500. Reads were aligned to the assembled genome with star (Dobin et al., 2013), and gene expression was calculated using cufflinks (Trapnell et al., 2010).
2.3. Manual annotation of chemosensory genes
Odorant receptor genes, ionotropic receptor genes, gustatory receptor genes, odorant binding protein genes and chemosensory protein genes were annotated manually, largely following Oxley et al. (2014). Briefly, gene loci were located using tblastn (Altschul et al., 1990) and exonerate (Slater & Birney, 2005), with queries from Ooceraea biroi, Solenopsis invicta and Pogonomyrmex barbatus. Alignments from tblastn (Altschul et al., 1990) and exonerate (Slater & Birney, 2005) were then pieced together manually in the Apollo genome annotation editor (Lee et al., 2009), using RNA‐sequencing (RNA‐seq) data and muscle (Edgar, 2004) alignment to query proteins to generate complete annotations in cases where blast and exonerate alignments were insufficient. Gene models encoding over 200 amino acids with premature stop codons, frame shifts, missing splice sites, or truncated or missing exons (which could not be explained by an assembly gap) were designated as pseudogenes, while gene models encoding over 200 amino acids with no such issues were designated as putatively functional.
2.4. Automated gene annotation
The ab‐initio gene predictors augustus (Stanke et al., 2006) and genemark (Lomsadze et al., 2005) were trained within the BRAKER1 pipeline (Hoff et al., 2016) using all RNA‐seq reads aligned to the genome with tophat. trinity (Haas et al., 2013) was used to assemble RNA‐seq reads into putative transcripts, using the “jaccard clip” option to avoid gene fusions from overlapping untranslated regions (UTRs). The ab‐initio gene predictor snap (Korf, 2004) was trained with genes predicted by the CEGMA pipeline (Parra et al., 2007) as well as with trinity‐assembled transcripts which met all of the following criteria: (i) complete open reading frames, (ii) a blastp alignment to a Drosophila melanogaster gene (FlyBase release 6) which was at least 95% of the length of both the target and query proteins, and (iii) exonerate alignment to the genome with at least 95% of the possible alignment score. augustus, genemark and snap were then run in the MAKER2 pipeline (Holt & Yandell, 2011). Protein evidence for MAKER2 consisted of official gene sets for D. melanogaster and Apis mellifera, and NCBI refseq predictions for Acromyrmex echinatior and Ooceraea biroi. blastx evidence was excluded as it led to numerous fusions in tandem arrays. “EST” evidence consisted of trinity‐assembled RNA‐seq reads as well as tophat junctions, and the “avoid EST fusion” option was used because many UTRs overlapped. maker repeat masking (repeat masker and repeat runner programs, www.repeatmasker.org) was used with custom‐trained repeat libraries constructed by repeat modeler (www.repeatmasker.org). Final gene models from MAKER2, as well as all ab‐initio predictions from both MAKER2 and BRAKER1 and alignment‐based models from CEGMA, were then input into evidence modeler (Haas et al., 2008), along with exonerate‐aligned proteins and trinity‐assembled RNA‐seq evidence (as used in MAKER2) and PASA‐assembled transcript evidence from unguided and genome‐guided trinity RNA‐seq assemblies. Gene models with no significant blast hits in D. melanogaster or O. biroi, no expression in our RNA‐seq data, and no PFAM‐A (Finn et al., 2010) nonretrotransposon domain (as determined by interpro scan; Zdobnov & Apweiler, 2001) were then excluded, manual annotations of chemosensory genes were added, and exons from the evidence modeler gene set that overlapped manual annotations were deleted.
2.5. Comparative genomics
Genomes from 12 additional ant species (Bonasio et al., 2010; Konorov et al., 2017; Nygaard et al., 2011; Oxley et al., 2014; Patalano et al., 2015; Rubin & Moreau, 2016; Schrader et al., 2014; C.D. Smith et al., 2011a; Smith et al., 2011b; Suen et al., 2011; Wurm et al., 2011) were downloaded from the Hymenoptera Genomes Database (Elsik et al., 2016) or the NCBI Assembly database. Genome assembly statistics were calculated using quast (Gurevich et al., 2013). Genome repeat content, including transposable element content, was calculated for each genome using the repeatmodeler2 pipeline (Flynn et al., 2020). To obtain an approximation of gene complement independent of annotation effort/quality, we used blat (Kent, 2002) to align the NCBI RefSeq gene set (version 101) from Nasonia vitripennis to each ant genome, using a 50% alignment coverage cutoff to ensure that genes were not counted twice. This gene set was chosen because N. vitripennis is an outgroup to all ants, and therefore equally related to all ants, and because the N. vitripennis genome is exceptionally well annotated. Apis mellifera also matches these criteria and is more closely related to ants. However, it possesses a slightly reduced gene complement (Elsik et al., 2014). The busco program (version 3.0.2) (Simão et al., 2015) was used to determine genome completeness for all ant species using the Hymenopteran ODB9 gene set, and the number of N. vitripennis gene blat hits was relativized to the percentage of complete genes recovered by busco to control for genome completeness effects on estimated gene content. For analyses of protein family and pathway evolution, protein sequences for all species were functionally annotated with GO terms, PFAM domains and KEGG pathways using eggnog‐mapper (Huerta‐Cepas et al., 2017). ℓ1ou (Khabbazian, Kriebel, Rohe, & An′e, 2016) was used to detect significant shifts in gene repertoire evolution for total gene number, GO Slim terms, PFAM domains, KEGG pathways and OR subfamilies (see below) in an explicitly phylogenetic framework. ℓ1ou estimates the number and position of shifts in phenotypic optima of quantitative traits (e.g., gene number) along phylogenetic trees using Ornstein–Uhlenbeck (OU) models. Statistical support for proposed shifts was calculated using likelihood ratio tests against models where the phenotypic optimum was not allowed to shift along a specified branch (using the “candid.edges” option of the “estimate_shift_configuration” function).
2.6. Odorant receptor evolutionary analyses
Amino acid sequences for ORs from eight additional ant species and two outgroup hymenopterans were obtained from other studies (McKenzie et al., 2016; Oxley et al., 2014; Robertson et al., 2010; C.D. Smith et al., 2011a; Smith et al., 2011b; Zhou et al., 2012) and aligned using mafft with the linsi parameter set (Katoh et al., 2005). raxml (Stamatakis, 2014) was used to reconstruct a maximum‐likelihood phylogeny for these genes under the LG +gamma model of protein evolution (Le & Gascuel, 2008), and the raxml rapid bootstrap algorithm (Stamatakis et al., 2008) was used to assess node support. The LG +gamma model was chosen as it was consistently recovered as the best or second best (after JTT +gamma) model in our previous OR phylogenetic analyses (McKenzie et al., 2016; McKenzie & Kronauer, 2018). For gene birth and death analysis, this tree was imported into notung (Chen et al., 2000) and reconciled with a species tree following the topology of Moreau et al. (2006), Brady et al. (2006) and Borowiec et al. (2017), and the divergence times from Borowiec et al. (2017), with an edge weight threshold of 70% bootstrap support (i.e., notung was allowed to rearrange nodes with lower than 70% bootstrap support when it led to a more parsimonious gene‐tree/species‐tree reconciliation). notung was then used to count gene duplications and losses along each branch of the species tree. We calculated birth and death rates on a per‐gene basis along each branch in the species tree as Log2((ancestral repertoire +duplication events or loss events)/ancestral repertoire)/branch length, which represents the number of times a gene family doubled or halved per unit time. Net expansion was calculated similarly as Log2((ancestral repertoire +duplication ‐ loss events)/ancestral repertoire)/branch length. Positive selection analysis was performed with the codeml algorithm of the paml package (Yang, 2007), with coding sequences aligned based on their translated peptide sequences using the mafft aligner with the ginsi parameter set (Katoh et al., 2005). codeml site models 1a and 2a, 7 and 8, and 8a and 8 were compared using likelihood ratio tests.
2.7. Neuroanatomical analyses
Additional E. burchellii workers and males were collected at La Selva Biological Station and preserved in 70% ethanol. Brains were later dissected from one male and one medium‐sized worker in 70% ethanol and further dehydrated in 80%, 90%, 95% and 3 × 100% ethanol. Brains were then cleared and mounted in methyl salicylate. Imaging was performed on a Zeiss LSM 880 laser scanning confocal microscope with 488‐nm laser stimulation used to image tissue autofluorescence. We found that ethanol fixation and preservation was sufficient to preserve antennal lobe morphology, and in fact, this method performed better than immersing whole ants in 4% PFA for autofluorescence imaging (data not shown). Glomeruli were reconstructed manually using the segmentation editor plugin in the Fiji distribution of imagej (Schindelin et al., 2012), and assigned to input tracts following Zube et al. (2008). Potentially due to the general decrease in glomerulus number outside of the T6 cluster, glomeruli innervated by tracts T1, T2 and T4 could not be unambiguously distinguished. These input tracts were therefore not separated in our analysis. Glomerulus volume was calculated using the 3D analysis plugin in the 3D imagej suite (Ollion et al., 2013). Glomerulus radius was calculated following Kelber et al. (2009) as the radius of a sphere with equal volume to the glomerulus in question. We found that this transformation (essentially a cube‐root transformation) yielded more normal distributions compared to raw volume measurements. Putative macroglomeruli were identified using two criteria. First, following Kelber et al. (2009), we identified glomeruli with radii at least five standard deviations larger than the median glomerulus radius. Because this cutoff is arbitrary, we also tested whether larger glomeruli belonged to a size distribution separate from the remaining glomeruli by fitting statistical distributions to our measures of glomerular radii using finite Gaussian mixture modelling with an expectation maximization algorithm implemented in the R package mclust (Scrucca et al., 2016). The Bayesian information criterion (BIC) was used to determine the best fitting model parameters, including the number of distribution components/clusters. A Shapiro–Wilk's test was used to assess whether the clusters identified by mclust matched the Gaussian distribution assumed by the model.
2.8. Scanning electron microscopy
Eciton burchellii workers were fixed immediately after collection in Karnovsky solution (2.5% glutaraldehyde and 2% paraformaldehyde, 0.1 m phosphate buffer, pH 7.4) and post‐fixed in 1% osmium tetraoxide for 1 hr. Samples were dehydrated in a gradient of ethanol (30, 50, 70, 80, 90, 95, 100%), critical point dried with CO2 (Leica EM CDP300, critical point dryer), and coated with gold (Eiko IB5 ion coater). Male specimens stored at −80°C were washed in 50% DMSO overnight, and then washed in distilled water at least 10 times prior to fixation. All samples were imaged with an Hitachi S‐3700N scanning electron microscope.
2.9. Statistics
All other statistics were calculated in R or with the stats package from SciPy (R Core Team, 2013; Virtanen et al., 2019). For Pearson's correlation analysis, normality of residuals was assessed using the Shapiro–Wilk's test for normality.
3. RESULTS
Assembly of 157 million paired‐end 150‐bp Illumina HiSeq shotgun reads and 26 million 300‐bp Illumina MiSeq mate‐pair reads (with 3‐ and 8‐kbp inserts) yielded a 192‐Mbp draft genome. Contig N50 was 187 kbp, while scaffold N50 was 2,433 kbp. This genome is relatively small compared to other ants, but within the range of variation (Figure 1b; Table S1). busco (Simão et al., 2015) analysis of genome completeness showed that 97.1% of all widely conserved hymenopteran genes were present in this draft assembly, making this one of the most complete ant genomes reported to date (Table S1).
Our annotation procedure yielded a final set of 12,670 genes, including 499 manually annotated chemosensory genes. Of these genes, 6,941 had a blastp alignment to a Drosophila melanogaster protein (FlyBase release 5) that spanned >50% of the length of the D. melanogaster sequence. The total number of identified genes is small relative to the other ant genomes we examined with similar annotation strategies (Lasius niger and Cardiocondyla obscurior), and the number of D. melanogaster blastp hits is likewise small compared with most of the other ant genomes we examined (Table S1). Eciton burchellii also had the fewest Nasonia vitripennis blat matches against the genome itself relative to busco completeness (Figure 1c; Table S1), indicating that the reduced gene number we observed is due to a true reduction in gene content, rather than an annotation artefact. However, phylogenetic analysis of trait shifts using OU models implemented in ℓ1ou (Khabbazian et al., 2016) did not detect a statistically significant shift in total gene repertoire size along the branch leading to E. burchellii (Figure S1a). Gene count as approximated by our methods showed only a weak correlation with genome size (Pearson's correlation, R 2 =.32, p =.045), even though E. burchellii is on the low end for both. To elucidate the evolutionary forces leading to gene repertoire reduction in E. burchellii, we used ℓ1ou to test for significant repertoire size shifts for gene GO terms, PFAM domains and KEGG pathways along the phylogenetic tree branch leading to E. burchellii. Two GO terms showed significant expansion in the E. burchellii lineage: GO:0030705 (cytoskeleton‐dependent intracellular transport; false discovery rate [FDR]‐adjusted p =.042, Figure S1b) and GO:0003729 (mRNA binding; FDR‐adjusted p =.042, Figure S1c). However, both of these GO terms also show expansion in the L. niger and C. obscurior lineages (Figure S1b,c). All three of these species were annotated with in‐house pipelines using similar ab‐initio predictors (Table S1), while all other annotations used were from the NCBI RefSeq pipeline. We therefore suspect that these expansions are technical artefacts rather than biologically meaningful. One PFAM domain (PF00412; LIM domain) also showed trait deviation in the three maker‐annotated lineages (expanded in E. burchellii [FDR‐adjusted p =.019] and in C. obscurior, contracted in L. niger; Figure S1d), while another (PF07524; Bromodomain associated) showed significant contraction in E. burchellii (FDR‐adjusted p =.0011) potentially unrelated to annotation method (Figure S1e). The contraction observed in PF07524 was rather small, however, from a median of 11 genes in most ants to seven in E. burchellii. No significant shifts in the E. burchellii lineage were found for any KEGG pathway. Together, our analyses failed to recover any reduction in gene functional categories which might be driving the overall trend of reduced gene numbers in E. burchellii.
Analysis of repeat content showed fairly typical amounts of LINE and LTR element transposons and all types of clustered repeats in E. burchellii, but notable reductions in SINE and DNA element transposons as well as unclassified interspersed repeats (Table S1). As a consequence, E. burchellii has the second lowest repeat content of any ant examined in this study, second only to the only ant with a smaller genome, C. obscurior (Table S1). In fact, we found that total repeat content was highly correlated with genome size across all species (R 2 =.95, p <.00001), suggesting that this is a major driver of genome size evolution in ants. This correlation held for every subtype of repeat except for simple repeats and low‐complexity sequences (FDR‐adjusted p <.05 for SINEs, LINEs, LTR elements, DNA elements, unclassified interspersed repeats and satellites). This contrasts with the findings of Schrader et al. (2014), who found the opposite pattern in their analysis of eight species using a custom TE annotation pipeline.
Our manual annotation of chemosensory genes in E. burchellii yielded 318 intact, putatively functional OR gene models, as well as 12 CSPs, 13 OBPs, 20 GRs and 23 IRs (Dataset S1). We also recovered 109, two and two putative pseudogenes in the OR, GR and IR families, respectively. The numbers of CSPs, OBPs and IRs all fall into the typical ranges for ants (Table S1). The number of GRs is lower than in many ants, but it is similar to the repertoire size seen in Harpegnathos saltator and O. biroi, which along with E. burchellii belong to phylogenetic lineages (the poneroids [H. saltator] and dorylines [O. biroi and E. burchellii]) that branched off before the other ants with high‐quality chemosensory gene annotations (Borowiec et al., 2017).
Strikingly, E. burchellii possesses one of the smallest putatively functional OR repertoires known for an ant (Figure 2a,b). This repertoire reduction is primarily due to high rates of gene death in the ancestral lineage leading to the two sequenced dorylines, E. burchellii and O. biroi, followed by relatively high rates of gene death and lower rates of gene birth in the E. burchellii lineage compared to the O. biroi lineage (Figure 2a; Figure S2a,b, Datasets S2–S4). In contrast to this overall trend, the 9‐exon subfamily of ORs is expanded in E. burchellii compared to most other ants (Figure 2b). Phylostratigraphy showed elevated rates of gene duplication in 9‐exon ORs in the ancestral lineage leading to E. burchellii and O. biroi, as well as along the branches leading to each species (Figure 2c; Figure S2c). The high rate of 9‐exon OR gene death in the ancestral lineage indicates that most extant gene expansions are specific to either E. burchellii or O. biroi (Figure S2d, Dataset S5). The overall net reduction in OR repertoire in the common ancestor of E. burchellii and O. biroi is driven by a stark net reduction in non‐9‐exon ORs, due to high rates of gene death (Figure 2d; Figure S2e,f). To determine whether OR repertoire size evolution in E. burchellii and O. biroi relative to other ants represented a significant evolutionary shift vs. random trait drift, we tested for trait optimality shifts along the tree using ℓ1ou for all ORs and for the five largest ant OR subfamilies (9‐exon, L, V, U and E). We found a significant expansion in total OR number along the branch leading to the ants, but none within the ants (Figure S3a). Along the branch leading to E. burchellii and O. biroi, we observed a significant expansion in 9‐exon ORs and a significant contraction in V ORs (p =.02 for both subfamilies; Figure S3b,c), with no significant changes in the other large OR subfamilies.
FIGURE 2.
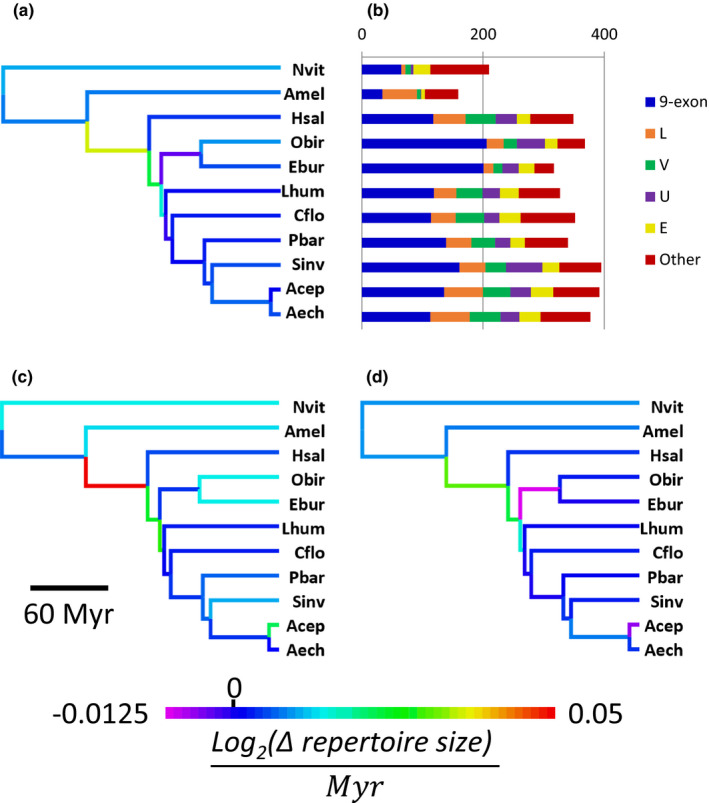
Evolution of odorant receptor gene repertoires in Hymenoptera. (a) Phylogeny of 11 species of Hymenoptera with branches coloured according to the rate of net change in OR gene repertoire size. Species abbreviations are as follows: Nvit: Nasonia vitripennis, Amel: Apis mellifera, Hsal: Harpegnathos saltator, Obir: Ooceraea biroi, Ebur: Eciton burchellii, Lhum: Linepithema humile, Cflo: Camponotus floridanus, Pbar: Pogonomyrmex barbatus, Sinv: Solenopsis invicta, Acep: Atta cephalotes, Aech: Acromyrmex echinatior. (b) Number of intact ORs in 11 species of Hymenoptera. OR numbers are broken down by subfamily for the five largest odorant receptor subfamilies. (c) Hymenoptera phylogeny with branches coloured according to the rate of net change in 9‐exon OR gene repertoire size. (d) Hymenoptera phylogeny with branches coloured according to the rate of net change in OR gene repertoire size excluding 9‐exon ORs
To determine whether misannotation of functional genes as pseudogenes due to sequencing errors or stop codon readthrough (e.g., Prieto‐Godino et al., 2016) could be influencing our results, we characterized the expression, OR subfamily and number of putative inactivating features (frame shifts, splice site mutations or premature stop codons) of all putative pseudogenes (Table S2). Fifteen pseudogene models had a single putative inactivating feature, five pseudogene models had two putative inactivating features and two pseudogene models had three putative inactivating features. The remaining 87 pseudogene models were classified as “extensively pseudogenized” based on having four or more putative inactivating features or else having missing exons that could not be explained by an assembly gap. Of the 22 genes with three or fewer putative inactivating features, only 11 showed expression at >2 FPKM in any sex/tissue. Ten of these 11 potential false‐pseudogene models were 9‐exon subfamily ORs.
To investigate evolutionary forces acting on the recent E. burchellii expansions, we examined site‐specific positive selection in the six species‐specific 9‐exon OR expansions in E. burchellii which contained more than four genes, following Engsontia et al. (2015). Site model tests revealed that one of these expansions showed statistically significant signs of positive selection at some sites under the most robust model test (M1a vs. M2a; LRT, p =.009), and one additional expansion was significant under the more powerful tests (M7 vs. M8 and M8a vs. M8; LRT, p =.007 and 0.02, respectively) (Table S3).
Most E. burchellii 9‐exon ORs belong to a single clade (9e‐α ORs sensu McKenzie et al., 2016), and genes in this clade comprise over half of all intact ORs in the E. burchellii genome (Figure 3a). RNA‐seq of pooled antennae from workers and males, pooled bodies (without antennae) from workers and a body (without antennae) from a single male showed that all but one of the 9e‐α ORs are expressed specifically in worker antennae (Figure 3b; Table S4). Most of the remaining ORs showed a mixture of weak male‐ and worker‐biased expression, although 21 additional ORs (mostly in the E and U subfamilies) were expressed specifically in worker antennae, and five ORs were expressed specifically in male antennae. Eleven ORs were expressed in the bodies of workers and/or the male (four and two in workers and the male, respectively, and five in both). Of the ORs classified as expressed in bodies (at a cutoff of 2.0 FPKM), the median expression within those bodies was 10.0 FPKM, while the median expression in the antennae of all ORs classified as expressed in the antennae was 11.6 FPKM (Mann–Whitney U test, p =.26). These findings indicate genuine (though not necessarily functional) expression of a small number of ORs in nonantennal tissues.
FIGURE 3.
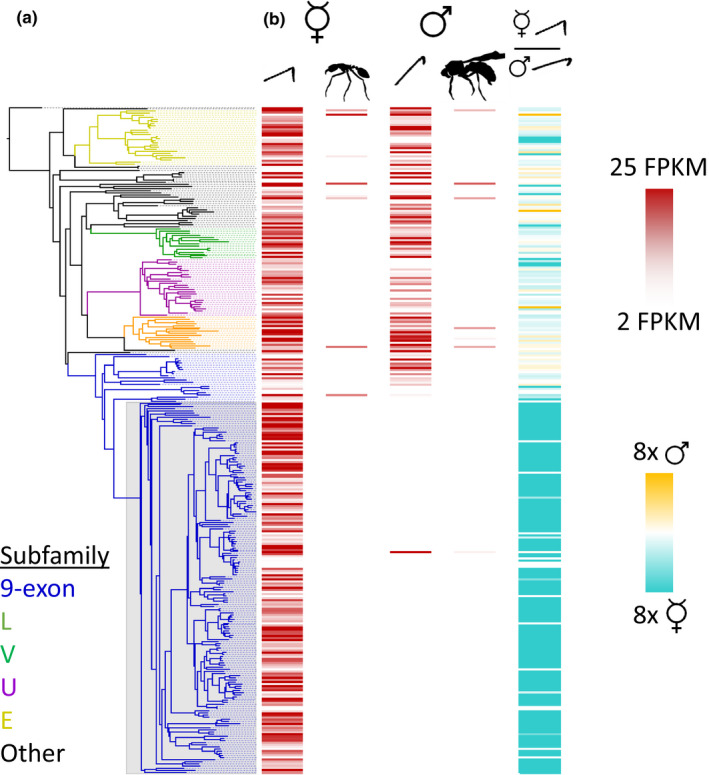
Tissue‐ and sex‐specific expression of Eciton burchellii odorant receptors. (a) Phylogeny of E. burchellii ORs, with branches belonging to the five largest ant OR subfamilies coloured by subfamily. 9e‐α ORs are designated by a grey box. (b) Heat maps of gene expression in antennae and antennae‐less bodies of E. burchellii workers and males. The first four columns represent absolute gene expression in each tissue, while the last represents fold enrichment in male vs. worker antennae. Expression data should be interpreted cautiously given the lack of biological replicates
To test whether the expansion of 9‐exon ORs was mirrored by an expansion of T6 glomeruli as seen in O. biroi (McKenzie et al., 2016), we next examined the olfactory neuroanatomy of E. burchellii workers and males. Three‐dimensional reconstruction of antennal lobe glomeruli from confocal micrograph stacks of E. burchellii worker and male brains confirmed that E. burchellii workers do indeed have an expanded T6 cluster of glomeruli compared with nondoryline ants with published antennal lobe reconstructions (Camponotus floridanus, C. japonicus and Atta vollenweideri [Kelber et al., 2010; Nakanishi et al., 2010; Zube et al., 2008]) (Figure 4; Table S5). Consistent with the lack of expression of 9‐exon ORs in male antennae, males lacked an expansion of T6 glomeruli (Figure 4; Table S5). E. burchellii also exhibits a corresponding reduction in non‐T6 glomeruli, possessing fewer non‐T6 glomeruli than any other ant yet examined (Table S5).
FIGURE 4.
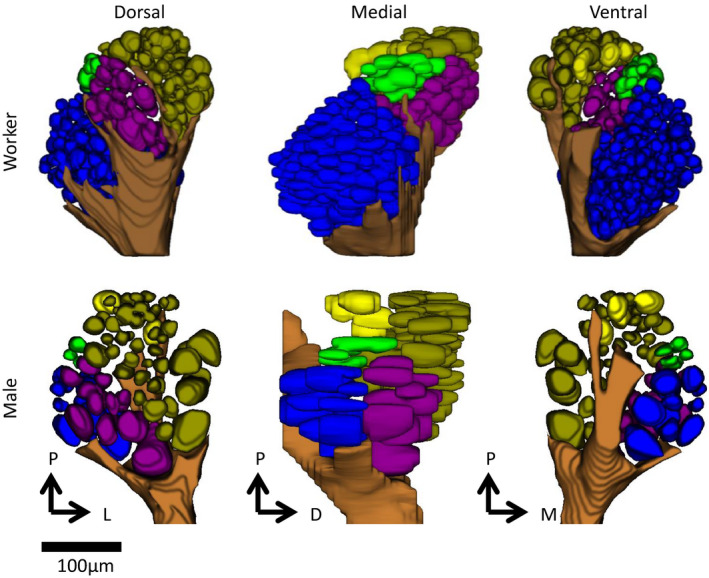
Reconstructed antennal lobes of an Eciton burchellii worker and male, with glomeruli coloured according to input tract. Clusters T1, T2 and T4 could not be reliably distinguished and are all coloured gold. T3 is coloured purple, T5 green, T6 blue and T7 yellow. The antennal nerve is shown in brown. P = posterior (n‐anterior), D = dorsal (n‐ventral), M = medial, L = lateral
Apart from the general reduction in glomerulus number, we observed many exceptionally large glomeruli in clusters T1–4 in workers (Figure 5), which resembled pheromone‐responsive “macroglomeruli” observed in other insect species (Boeckh & Selsam, 1984; Kuebler et al., 2010; Matsumoto & Hildebrand, 1981). Two E. burchellii glomeruli passed the threshold proposed by Kelber et al. (2009) for defining macroglomeruli, and Gaussian mixture modelling showed that 16 glomeruli were significantly larger than all other glomeruli (Figure 5). None of the larger glomeruli belonged to the T6 cluster of glomeruli. We also observed many relatively large glomeruli in E. burchellii males (Figure S4). However, none qualified as macroglomeruli according to the definition of Kelber et al. (2009). Although mclust also found two distinct populations of glomeruli in E. burchellii males (Figure S4), the distribution of glomerular radii in the larger cluster deviated significantly from a Gaussian distribution (Shapiro–Wilk test, p =.004), suggesting that the models tested by mclust did not fit our male data well.
FIGURE 5.
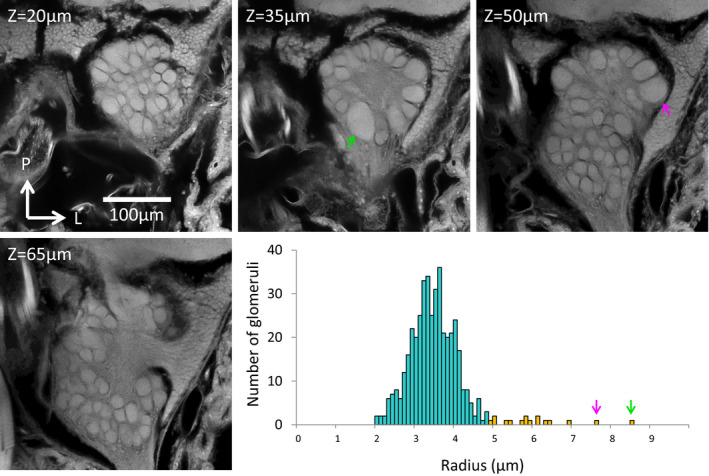
Optical slices from a confocal micrograph stack of the Eciton burchellii worker antennal lobe (top and lower left) and a plot of the size distribution of worker glomeruli (lower right). The two largest glomeruli, which pass Kelber et al.’s (2009) threshold for “macroglomeruli,” are indicated by green and magenta arrows. Bars in the size distribution plot are coloured according to their corresponding mclust size distribution cluster classification
Scanning electron microscopy of E. burchellii worker antennae (Figure 6) revealed that basiconic sensilla were present at high density along the ventral surface of the six terminal antennal segments (segments 7–12; directionality is relative to body axis with antennae held in the resting position). This pattern is reminiscent of that observed in O. biroi (McKenzie et al., 2016). A small number of additional basiconic sensilla were detected on the ventral surface of antennal segment 6, while segments 1–5 had no basiconic sensilla. As in other ants, males lacked basiconic sensilla entirely (Figure S5).
FIGURE 6.
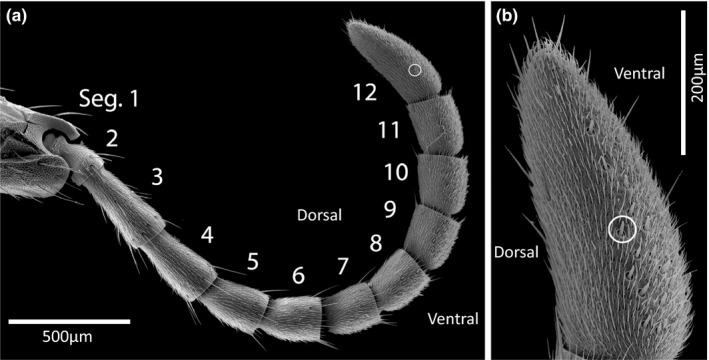
Scanning electron micrographs of Eciton burchellii worker antennae showing basiconic sensilla restricted to the ventral surface (as defined relative to the body axis while antennae are held naturally) of the seven terminal antennal segments. (a) Whole antennal flagellum with segments numbered (1–12). (b) Magnified terminal antennal segment. A representative basiconic sensillum is circled in both images
4. DISCUSSION
The New World army ant Eciton burchellii is an extensively studied, prevalent and ecologically important arthropod predator in Neotropical ecosystems. By providing genomic resources for this species, our study will facilitate future investigations into the molecular basis of social predation. We found that E. burchellii possesses a relatively compact genome, with a general reduction in gene complement compared to other ant species. Examination of evolutionary dynamics in functional subgroups of genes did not show accelerated loss of any specific functional subgroup, suggesting relatively even gene loss across the E. burchellii genome. Analysis of repeat content across ants shows that it is tightly correlated with genome size, indicating that repeat content expansion and contraction probably plays a major role in shaping genome size in ants—as previously observed for many different types of organisms (Lynch, 2007). A paucity of interspersed repeats in particular is probably largely responsible for the compact genome size of E. burchellii.
Corresponding to the general reduction in gene complement, we found that E. burchellii possesses one of the smallest intact odorant receptor gene repertoires among the ants we examined in this study. However, the 9‐exon subfamily of ORs is expanded in E. burchellii, and in fact this species possesses the second largest 9‐exon OR repertoire of any ant with high‐quality OR annotations examined in this study. We also found that E. burchellii has an unusually large number of T6 glomeruli in the antennal lobe, and an unusually dense region of basiconic sensilla along the ventral surface of the antenna. Almost all 9‐exon ORs are expressed specifically in worker antennae in E. burchellii. While our lack of replicates prevents us from drawing statistical conclusions about any given OR, this pattern is consistent with observations from Ooceraea biroi, Camponotus floridanus and Harpegnathos saltator (McKenzie et al., 2016; Zhou et al., 2012). All of these data are consistent with previous work which indicated that 9‐exon ORs are expressed in OSNs that are housed in basiconic sensilla and innervate T6 glomeruli (McKenzie et al., 2016). Intriguingly, the clonal raider ant O. biroi, which together with E. burchellii belongs to the ant subfamily Dorylinae, also possesses an expanded 9‐exon OR repertoire and T6 glomerulus cluster, as well as an unusually dense region of basiconic sensilla on the ventral surface of the antennal club (McKenzie et al., 2016). We also observed signatures of positive selection in two of six E. burchellii‐specific 9‐exon OR expansions. This mirrors findings of Engsontia et al. (2015) in O. biroi, where four of six species‐specific 9‐exon OR expansions also showed signatures of positive selection. Engsontia et al. (2015) found only one other positively selected species‐specific clade, a 9‐exon OR expansion in C. floridanus, suggesting that doryline 9‐exon ORs are under unusually high selective pressure.
Both 9‐exon ORs and basiconic sensilla have been shown to respond to CHCs, an important class of ant pheromones used in nest‐mate recognition, fertility signalling and division of labour (Holman et al., 2010; Liebig, 2010; Ozaki et al., 2005; Pask et al., 2017; Sharma et al., 2015; Sturgis & Gordon, 2012; van Zweden & D’Ettorre, 2010). Hydrocarbons have also been shown to function in species recognition and prey selection in ants (Kidokoro‐Kobayashi et al., 2012; Lang & Menzel, 2011; Manubay & Powell, 2020), and basiconic sensilla are specifically implicated in species recognition in the ant Formica yessensis and prey discrimination in the wasp Liris niger (Anton & Gnatzy, 1998; Kidokoro‐Kobayashi et al., 2012). Because both O. biroi and E. burchellii are believed to be specialist ant predators (Franks, 1982; Hoenle et al., 2019; Rettenmeyer, 1963; Tsuji & Yamauchi, 1995), the expansion of the hydrocarbon detection system in O. biroi and E. burchellii may represent adaptations for prey discrimination, particularly for discriminating between prey and nonprey ant species. Unfortunately, O. biroi and E. burchellii are the only doryline ants with sequenced genomes. Additionally, these species share further life history traits not seen in the other ant species in our study, namely nomadism and cyclical reproduction (Oxley et al., 2014; Rettenmeyer, 1963; Schneirla, 1944; Tsuji & Yamauchi, 1995). Future comparative genomics studies with increased phylogenetic and ecological resolution will therefore be necessary to conclusively determine the evolutionary significance, if any, of 9‐exon OR gene expansions.
Recently, two studies have been published examining the genomic consequences of ecological specialization in socially parasitic inquiline (Schrader et al., 2021) and kidnapping (Jongepier et al., 2021) ant species. While these were published too recently to be included in our in‐depth comparative analyses, it is worth noting some striking similarities and differences in these ant groups relative to E. burchellii. Specifically, both studies found large contractions in the OR family in social parasites compared with their free‐living relatives (Schrader et al., 2021, Jongepier et al., 2021). This suggests that olfactory system simplification is a common theme of ecological specialization in ants. However, both studies also showed the strongest contractions in the 9‐exon ORs, while we find a major expansion in this gene subfamily in E. burchellii. This suggests that there may be fundamental differences in the olfactory processes related to finding host colonies to parasitize vs. distinguishing between prey species during raids. Of note, most inquilines and kidnapping ants parasitize closely related ant species (Schrader et al., 2021, Jongepier et al., 2021), and thus they may be ancestrally able to detect volatile host pheromones.
Our neuroanatomical investigation of the E. burchellii AL also revealed many exceptionally large glomeruli, all located outside of the T6 cluster and thus probably not involved in hydrocarbon perception. Enlarged glomeruli, termed macroglomeruli, are often involved in the detection of ecologically particularly important odours, such as sex pheromones in male moths, cockroaches and bees (Boeckh & Selsam, 1984; Matsumoto & Hildebrand, 1981; Sandoz, 2006), and food odours in specialist flies (Ibba et al., 2010). Notably, in the leaf‐cutting ant Atta vollenweideri, large workers (majors) possess a single macroglomerulus that responds to trail pheromones (Kuebler et al., 2010). Workers in C. floridanus also possess a single enlarged glomerulus, although its function is unknown (Zube et al., 2008). Most insects with macroglomeruli possess one or two such structures. However, our analysis of glomerulus size distribution suggested that 16 glomeruli in E. burchellii workers belong to a distinctly larger size class. As they are located outside of the T6 cluster, we hypothesize that these glomeruli are involved in volatile pheromone detection, such as alarm, trail and recruitment pheromones. Functional studies will be necessary to confirm pheromone responses in these glomeruli. While our study was restricted to males and medium‐sized workers, future studies on neuroanatomical differences between worker subcastes may also prove informative, given the above‐mentioned major‐restricted macroglomeruli observed in Atta. Intriguingly, previous work on AL size in Eciton has shown a reduction in AL size in Eciton majors relative to the brain as a whole (O’Donnell et al., 2018). Future work should also examine differences in neuroanatomy and genomics of different E. burchellii populations, as recent work has found striking genetic differentiation between E. burchellii subspecies (Winston et al., 2017).
Overall, our findings suggest that doryline ants possess a suite of chemosensory system traits that have so far not been observed in other ants. These include an expanded repertoire of putative hydrocarbon sensors, and a dense clustering of hydrocarbon‐sensitive sensilla along the ventral surface of the antennae. These traits may represent adaptations for specialized predation and myrmecophagy. Moving forward, it will be important to investigate the chemosensory systems of other army ant‐like species with separate evolutionary origins, such as the ponerine genera Simopelta and Leptogenys, the amblyoponine genus Onychomyrmex, and the army ant‐like members of the myrmicine genus Carebara (previously Pheidologeton). The E. burchellii genome will also provide an invaluable resource for future studies of the genomics of social predation and army ant biology in general.
4.1. Data Accessibility
Sequencing data and genomic assemblies and annotations have been deposited in the NCBI SRA and genome databases under the BioProject accession no. PRJNA632625. All other data are available as supplemental material accompanying this paper.
CONFLICT OF INTEREST
N/A.
AUTHOR CONTRIBUTION
N/A.
Supporting information
Fig S1‐5
Table S1
Table S2
Table S3
Table S4
Table S5
ACKNOWLEDGMENTS
The genetic resources were accessed in Costa Rica in compliance with the Biodiversity Law # 7788 and the Convention on Biological Diversity (CBD). Biodiversity access permits were granted by the Institutional Biodiversity Committee, University of Costa Rica, Resolution Nos. 023 and 094; and collecting permits were authorized by La Selva Biological Station, Organization for Tropical Studies. We thank Robert L. Kerby for assistance in DNA extraction for whole‐genome sequencing and Alexander Rodríguez for his valuable contribution to improving our electron microscopy images. Open Access Funding provided by Universite de Lausanne.
McKenzie, S. K. , Winston, M. E. , Grewe, F. , Vargas Asensio, G. , Rodríguez‐Hernández, N. , Rubin, B. E. R. , Murillo‐Cruz, C. , von Beeren, C. , Moreau, C. S. , Suen, G. , Pinto‐Tomás, A. A. , & Kronauer, D. (2021). The genomic basis of army ant chemosensory adaptations. Molecular Ecology, 30, 6627–6641. 10.1111/mec.16198
Contributor Information
Sean K. McKenzie, Email: sean.mckenzie@nanoporetech.com.
Adrian A. Pinto‐Tomás, Email: adrian.pinto@ucr.ac.cr.
Daniel J. C. Kronauer, Email: dkronauer@rockefeller.edu.
Data Availability Statement
McKenzie, S. K., Winston, M. W., Grewe, F., Vargas Asensio, G., Rodríguez‐Hernández, N., Rubin, B.E.R., Murillo‐Cruz, C., von Beeren, C., Moreau, C.S., Suen, G., Pinto‐Tomás, A.A., & Kronauer, D.J.C. (2020) Sequencing and genome assembly of the army ant Eciton burchellii. NCBI SRA and Genome Databases, BioProject. PRJNA632625.
REFERENCES
- Altschul, S. F. , Gish, W. , Miller, W. , Myers, E. W. , & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Anton, S. , & Gnatzy, W. (1998). Prey specificity and the importance of close‐range chemical cues in prey recognition in the digger wasp, Liris niger . Journal of Insect Behavior, 11(5), 671–690. 10.1023/A:1022346825811 [DOI] [Google Scholar]
- Bagneres, A.‐G. , Billen, J. , & Morgan, E. D. (1991). Volatile secretion of Dufour gland of workers of an army ant, Dorylus (Anomma) molestus. Journal of Chemical Ecology, 17(8), 1633–1639. 10.1007/BF00984694 [DOI] [PubMed] [Google Scholar]
- Boeckh, J. , & Selsam, P. (1984). Quantitative investigation of the odour specificity of central olfactory neurones in the American cockroach. Chemical Senses, 9(4), 369–380. 10.1093/chemse/9.4.369 [DOI] [Google Scholar]
- Bonasio, R. , Zhang, G. , Ye, C. , Mutti, N. S. , Fang, X. , Qin, N. , Donahue, G. , Yang, P. , Li, Q. , Li, C. , Zhang, P. , Huang, Z. , Berger, S. l , Reinberg, D. , Wang, J. , & Liebig, J. (2010). Genomic Comparison of the Ants Camponotus floridanus and Harpegnathos saltator. Science, 329(5995), 1068–1071. 10.1126/science.1192428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec, M. L. (2017). Convergent evolution of the army ant syndrome and congruence in big‐data phylogenetics. BioRxiv, 1–61, 10.1101/134064 [DOI] [PubMed] [Google Scholar]
- Borowiec, M. L. , Rabeling, C. , Brady, S. G. , Fisher, B. L. , Schultz, T. R. , & Ward, P. S. (2017). Compositional heterogeneity and outgroup choice influence the internal phylogeny of the ants. BioRxiv, 173393, 10.1101/173393 [DOI] [PubMed] [Google Scholar]
- Brady, S. G. (2003). Evolution of the army ant syndrome: the origin and long‐term evolutionary stasis of a complex of behavioral and reproductive adaptations. Proceedings of the National Academy of Sciences of the United States of America, 100(11), 6575–6579. 10.1073/pnas.1137809100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady, S. G. , Schultz, T. R. , Fisher, B. L. , & Ward, P. S. (2006). Evaluating alternative hypotheses for the early evolution and diversification of ants. Proceedings of the National Academy of Sciences of the United States of America, 103(48), 18172–18177. 10.1073/pnas.0605858103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, W. L. (1959). The Release of Alarm and Attack Behavior in Some New world Army Ants. Psyche, 66, 25–27. 10.1155/1959/71808 [DOI] [Google Scholar]
- Brückner, A. , Hoenle, P. O. , & von Beeren, C. (2018). Comparative chemical analysis of army ant mandibular gland volatiles (Formicidae: Dorylinae). PeerJ, 2018(7), 1–13. 10.7717/peerj.5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulova, S. , Purce, K. , Khodak, P. , Sulger, E. , & O’Donnell, S. (2016). Into the black and back: The ecology of brain investment in Neotropical army ants (formicidae: Dorylinae). Science of . Nature, 103(3–4), 10.1007/s00114-016-1353-4 [DOI] [PubMed] [Google Scholar]
- Chadab, R. , & Rettenmeyer, C. W. (1975). Mass recruitment by army ants. Science, 188(4193), 1124–1125. [DOI] [PubMed] [Google Scholar]
- Chen, K. , Durand, D. , & Farach‐Colton, M. (2000). NOTUNG: a program for dating gene duplications and optimizing gene family trees. Journal of Computational Biology : A Journal of Computational Molecular Cell Biology, 7(3–4), 429–447. 10.1089/106652700750050871 [DOI] [PubMed] [Google Scholar]
- Dobin, A. , Davis, C. A. , Schlesinger, F. , Drenkow, J. , Zaleski, C. , Jha, S. , Batut, P. , Chaisson, M. , & Gingeras, T. R. (2013). STAR: ultrafast universal RNA‐seq aligner. Bioinformatics, 29(1), 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumpert, K. (1972). Alarmstoffrezeptorcn auf der Antenne yon Lasius fuliginosus (Latr.) (Hymenoptera, Formieidae). Z. Vergl. Physiologie, 425, 403–425. [Google Scholar]
- Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5), 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik, C. G. , Tayal, A. , Diesh, C. M. , Unni, D. R. , Emery, M. L. , Nguyen, H. N. , & Hagen, D. E. (2016). Hymenoptera Genome Database: Integrating genome annotations in HymenopteraMine. Nucleic Acids Research, 44(D1), D793–D800. 10.1093/nar/gkv1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik, C. G. , Worley, K. C. , Bennett, A. K. , Beye, M. , Camara, F. , Childers, C. P. , de Graaf, D. C. , Debyser, G. , Deng, J. , Devreese, B. , Elhaik, E. , Evans, J. D. , Foster, L. J. , Graur, D. , Guigo, R. , Hoff, K. , Holder, M. E. , Hudson, M. E. , Hunt, G. J. , Jiang, H. , Joshi, V. , Khetani, R. S. , Kosarev, P. , Kovar, C. L. , Ma, J. , Maleszka, R. , Moritz, R. F. A. , Munoz‐Torres, M. C. , Murphy, T. D. , Muzny, D. M. , Newsham, I. F. , Reese, J. T. , Robertson, H. M. , Robinson, G. E. , Rueppell, O. , Solovyev, V. , Stanke, M. , Stolle, E. , Tsuruda, J. M. , Vaerenbergh, M. , Waterhouse, R. M. , Weaver, D. B. , Whitfield, C. W. , Wu, Y. , Zdobnov, E. M. , Zhang, L. , Zhu, D. , & Gibbs, R. A. (2014). Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics, 15(1), 86. 10.1186/1471-2164-15-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engsontia, P. , Sangket, U. , Robertson, H. M. , & Satasook, C. (2015). Diversification of the ant odorant receptor gene family and positive selection on candidate cuticular hydrocarbon receptors. BMC Research Notes, 8, 380. 10.1186/s13104-015-1371-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R. D. , Mistry, J. , Tate, J. , Coggill, P. , Heger, A. , Pollington, J. E. , Gavin, O. L. , Gunasekaran, P. , Ceric, G. , Forslund, K. , Holm, L. , Sonnhammer, E. L. L. , Eddy, S. R. , & Bateman, A. (2010). The Pfam protein families database. Nucleic Acids Research, 38(suppl_1), D211–D222. 10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn, J. M. , Hubley, R. , Goubert, C. , Rosen, J. , Clark, A. G. , Feschotte, C. , & Smit, A. F. (2020). RepeatModeler2 for automated genomic discovery of transposable element families. Proceedings of the National Academy of Sciences of the United States of America, 117(17), 9451–9457. 10.1073/pnas.1921046117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks, N. R. (1982). Ecology and population regulation in the army ant Eciton burchelli. In Leigh E. G., Rand A. S., & Windsor D. M. (Eds.), The Ecology of a Tropical Forest (pp. 389–395). Smithsonian Institution Press. [Google Scholar]
- Gnerre, S. , MacCallum, I. , Przybylski, D. , Ribeiro, F. J. , Burton, J. N. , Walker, B. J. , Sharpe, T. , Hall, G. , Shea, T. P. , Sykes, S. , Berlin, A. M. , Aird, D. , Costello, M. , Daza, R. , Williams, L. , Nicol, R. , Gnirke, A. , Nusbaum, C. , Lander, E. S. , & Jaffe, D. B. (2011). High‐quality draft assemblies of mammalian genomes from massively parallel sequence data. Proceedings of the National Academy of Sciences, 108(4), 1513–1518. 10.1073/pnas.1017351108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotwald, W. H. (1982). Army ants. In Herman H. R. (Ed.), Social insects (pp. 157–254). Academic Press. [Google Scholar]
- Gotwald, W. H. (1995). Army Ants: The Biology of Social Predation. Ithaca, NY, USA: Cornell University Press. [Google Scholar]
- Gurevich, A. , Saveliev, V. , Vyahhi, N. , & Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics (Oxford, England), 29(8), 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B. J. , Papanicolaou, A. , Yassour, M. , Grabherr, M. , Blood, P. D. , Bowden, J. , Couger, M. B. , Eccles, D. , Li, B. , Lieber, Matthias , MacManes, M. D. , Ott, M. , Orvis, J. , Pochet, N. , Strozzi, F. , Weeks, N. , Westerman, R. , William, T. , Dewey, C. N. , Henschel, R. , LeDuc, R. D. , Friedman, N. , & Regev, A. (2013). De novo transcript sequence reconstruction from RNA‐seq using the Trinity platform for reference generation and analysis. Nature Protocols, 8(8), 1494–1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B. J. , Salzberg, S. L. , Zhu, W. , Pertea, M. , Allen, J. E. , Orvis, J. , White, O. , Buell, C. R. , & Wortman, J. R. (2008). Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biology, 9(1), 1–22. 10.1186/gb-2008-9-1-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenle, P. O. , Blüthgen, N. , Brückner, A. , Fiala, B. , Donoso, D. A. , Smith, M. A. , Ospina Jara, B. , & Beeren, C. (2019). Species ‐ level predation network uncovers high prey specificity in a Neotropical army ant community. Molecular Ecology, March, 1–18. 10.1111/mec.15078 [DOI] [PubMed] [Google Scholar]
- Hoff, K. J. , Lange, S. , Lomsadze, A. , Borodovsky, M. , & Stanke, M. (2016). BRAKER1: Unsupervised RNA‐Seq‐Based Genome Annotation with GeneMark‐ET and AUGUSTUS. Bioinformatics, 32(5), 767–769. 10.1093/bioinformatics/btv661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman, L. , Jørgensen, C. G. , Nielsen, J. , & d'Ettorre, P. (2010). Identification of an ant queen pheromone regulating worker sterility. Proceedings of the Royal Society B: Biological Sciences, 277(1701), 3793–3800. 10.1098/rspb.2010.0984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, C. , & Yandell, M. (2011). MAKER2: an annotation pipeline and genome‐database management tool for second‐generation genome projects. BMC Bioinformatics, 12(1), 491. 10.1186/1471-2105-12-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta‐Cepas, J. , Forslund, K. , Coelho, L. P. , Szklarczyk, D. , Jensen, L. J. , von Mering, C. , & Bork, P. (2017). Fast Genome‐Wide Functional Annotation through Orthology Assignment by eggNOG‐Mapper. Molecular Biology and Evolution, 34(8), 2115–2122. 10.1093/molbev/msx148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba, I. , Angioy, A. M. , Hansson, B. S. , & Dekker, T. (2010). Macroglomeruli for fruit odors change blend preference in Drosophila. Naturwissenschaften, 97(12), 1059–1066. 10.1007/s00114-010-0727-2 [DOI] [PubMed] [Google Scholar]
- Jongepier, E. , Séguret, A. , Labutin, A. , Feldmeyer, B. , Gstöttl, C. , Foitzik, S. , Bornberg‐Bauer, E. (2021). Convergent loss of chemoreceptors across independent origins of slave‐making in ants. BioRxiv, 10.1101/2021.05.11.443570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspari, M. , & O’Donnell, S. (2003). High rates of army ant raids in the Neotropics and implications for ant colony and community structure. Evolutionary Ecology Research, 5(6), 933–939. [Google Scholar]
- Kaspari, M. , Powell, S. , Lattke, J. , & O’Donnell, S. (2011). Predation and patchiness in the tropical litter: Do swarm‐raiding army ants skim the cream or drain the bottle? Journal of Animal Ecology, 80(4), 818–823. 10.1111/j.1365-2656.2011.01826.x [DOI] [PubMed] [Google Scholar]
- Katoh, K. , Kuma, K. , Toh, H. , & Miyata, T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Research, 33(2), 511–518. 10.1093/nar/gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegans, S. J. , Billen, J. , Morgan, E. D. , & Gökcen, O. A. (1993). Volatile glandular secretions of three species of new world army ants, Eciton burchelli, Labidus coecus, andLabidus praedator. Journal of Chemical Ecology, 19(11), 2705–2719. 10.1007/BF00980702 [DOI] [PubMed] [Google Scholar]
- Kelber, C. , Rössler, W. , & Kleineidam, C. J. (2010). Phenotypic plasticity in number of glomeruli and sensory innervation of the antennal lobe in leaf‐cutting ant workers (A. vollenweideri). Developmental Neurobiology, 70(4), 222–234. 10.1002/dneu.20782 [DOI] [PubMed] [Google Scholar]
- Kelber, C. , Rössler, W. , Roces, F. , & Kleineidam, C. J. (2009). The antennal lobes of fungus‐growing ants (Attini): Neuroanatomical traits and evolutionary trends. Brain, Behavior and Evolution, 73(4), 273–284. 10.1159/000230672 [DOI] [PubMed] [Google Scholar]
- Kent, W. J. (2002). BLAT–‐The BLAST‐Like Alignment Tool. Genome Research, 12(4), 656–664. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabbazian, M. , Kriebel, R. , Rohe, K. , & Ané, C. (2016). Fast and accurate detection of evolutionary shifts in Ornstein–Uhlenbeck models. Methods in Ecology and Evolution, 7(7), 811–824. 10.1111/2041-210X.12534 [DOI] [Google Scholar]
- Kidokoro‐Kobayashi, M. , Iwakura, M. , Fujiwara‐Tsujii, N. , Fujiwara, S. , Sakura, M. , Sakamoto, H. , Higashi, S. , Hefetz, A. , & Ozaki, M. (2012). Chemical discrimination and aggressiveness via cuticular hydrocarbons in a supercolony‐forming ant, formica yessensis. PLoS ONE, 7(10), e46840. 10.1371/journal.pone.0046840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konorov, E. A. , Nikitin, M. A. , Mikhailov, K. V. , Lysenkov, S. N. , Belenky, M. , Chang, P. L. , Nuzhdin, S. V. , & Scobeyeva, V. A. (2017). Genomic exaptation enables Lasius niger adaptation to urban environments. BMC Evolutionary Biology, 17(S1), 39. 10.1186/s12862-016-0867-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf, I. (2004). Gene finding in novel genomes. BMC Bioinformatics, 5(1), 59. 10.1186/1471-2105-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronauer, D. J. C. (2009). Recent advances in army ant biology. Myrmecological News, 12, 51–65. [Google Scholar]
- Kronauer, D. J. C. (2020). Army ants. In Starr C. K. (Ed.), Encyclopedia of Social Insects. Berlin, DE: Springer International. [Google Scholar]
- Kuebler, L. S. , Kelber, C. , & Kleineidam, C. J. (2010). Distinct antennal lobe phenotypes in the leaf‐cutting ant (Atta vollenweideri). Journal of Comparative Neurology, 518(3), 352–365. 10.1002/cne.22217 [DOI] [PubMed] [Google Scholar]
- Lang, C. , & Menzel, F. (2011). Lasius niger ants discriminate aphids based on their cuticular hydrocarbons. Animal Behaviour, 82(6), 1245–1254. 10.1016/j.anbehav.2011.08.020 [DOI] [Google Scholar]
- Le, S. Q. , & Gascuel, O. (2008). An improved general amino acid replacement matrix. Molecular Biology and Evolution, 25(7), 1307–1320. 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- Lee, E. , Harris, N. , Gibson, M. , Chetty, R. , & Lewis, S. (2009). Apollo: A community resource for genome annotation editing. Bioinformatics (Oxford, England), 25(14), 1836–1837. 10.1093/bioinformatics/btp314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett, R. M. , Clavijo, B. J. , Clissold, L. , Clark, M. D. , & Caccamo, M. (2014). NextClip: an analysis and read preparation tool for Nextera Long Mate Pair libraries. Bioinformatics (Oxford, England), 30(4), 566–568. 10.1093/bioinformatics/btt702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebig, J. (2010). Hydrocarbon profiles indicate fertility and dominance status in ant, bee, and wasp colonies. In Blomquist G. J. & Bagneres A. G. (Eds.), Insect Hydrocarbons. Cambridge, UK: Cambridge University Press. 10.1017/CBO9780511711909 [DOI] [Google Scholar]
- Lomsadze, A. , Ter‐Hovhannisyan, V. , Chernoff, Y. O. , & Borodovsky, M. (2005). Gene identification in novel eukaryotic genomes by self‐training algorithm. Nucleic Acids Research, 33(20), 6494–6506. 10.1093/nar/gki937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M. (2007). The Origins of Genome Architecture. Sinaur Associates. [Google Scholar]
- Manubay, J. A. , & Powell, S. (2020). Detection of prey odors underpins dietary specialization in a Neotropical top‐predator: How army ants find their ant prey. Journal of Animal Ecology, 89(5), 1165–1174. [DOI] [PubMed] [Google Scholar]
- Matsumoto, S. G. , & Hildebrand, J. G. (1981). Olfactory Mechanisms in the Moth Manduca sexta: Response Characteristics and Morphology of Central Neurons in the Antennal Lobes. Proceedings of the Royal Society B: Biological Sciences, 213(1192), 249–277. 10.1098/rspb.1981.0066 [DOI] [Google Scholar]
- McKenzie, S. K. , Fetter‐Pruneda, I. , Ruta, V. , & Kronauer, D. J. C. (2016). Transcriptomics and neuroanatomy of the clonal raider ant implicate an expanded clade of odorant receptors in chemical communication. Proceedings of the National Academy of Sciences USA, 113(49), 14091–14096. 10.1073/pnas.1610800113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie, S. K. , & Kronauer, D. J. C. (2018). The genomic architecture and molecular evolution of ant odorant receptors. Genome Research, 28, 1757–1765. 10.1101/gr.237123.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirenda, J. T. , Eakins, D. G. , Gravelle, K. , & Topoff, H. (1980). Predatory Behavior and Prey Selection by Army Ants in a Desert‐ Grassland Habitat. Behavioral Ecology and Sociobiology, 7(2), 119–127. 10.1007/BF00299517 [DOI] [Google Scholar]
- Moreau, C. S. , & Bell, C. D. (2013). Testing the museum versus cradle tropical biological diversity hypothesis: Phylogeny, diversification, and ancestral biogeographic range evolution of the ants. Evolution, 67(8), 2240–2257. 10.1111/evo.12105 [DOI] [PubMed] [Google Scholar]
- Moreau, C. S. , Bell, C. D. , Vila, R. , Archibald, S. B. , & Pierce, N. E. (2006). Phylogeny of the ants: Diversification in the age of angiosperms. Science, 312(5770), 101–104. 10.1126/science.1124891 [DOI] [PubMed] [Google Scholar]
- Nakanishi, A. , Nishino, H. , Watanabe, H. , Yokohari, F. , & Nishikawa, M. (2010). Sex‐specific antennal sensory system in the ant Camponotus japonicus: Glomerular organizations of antennal lobes. Journal of Comparative Neurology, 518(12), 2186–2201. 10.1002/cne.22326 [DOI] [PubMed] [Google Scholar]
- Nygaard, S. , Zhang, G. , Schiøtt, M. , Li, C. , Wurm, Y. , Hu, H. , & Boomsma, J. J. (2011). The genome of the leaf‐cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming. Genome Research, 21(8), 1339–1348. 10.1101/gr.121392.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell, S. , Bulova, S. , Barrett, M. , & von Beeren, C. (2018). Brain investment under colony‐level selection: Soldier specialization in Eciton army ants (Formicidae: Dorylinae). BMC Zoology, 3(1), 1–6. 10.1186/s40850-018-0028-3 [DOI] [Google Scholar]
- O’Donnell, S. , Lattke, J. , Powell, S. , & Kaspari, M. (2007). Army ants in four forests: Geographic variation in raid rates and species composition. Journal of Animal Ecology, 76(3), 580–589. 10.1111/j.1365-2656.2007.01221.x [DOI] [PubMed] [Google Scholar]
- Ollion, J. , Cochennec, J. , Loll, F. , Escudé, C. , & Boudier, T. (2013). TANGO: a generic tool for high‐throughput 3D image analysis for studying nuclear organization. Bioinformatics (Oxford, England), 29(14), 1840–1841. 10.1093/bioinformatics/btt276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxley, P. R. , Ji, L. , Fetter‐Pruneda, I. , McKenzie, S. K. , Li, C. , Hu, H. , Zhang, G. , & Kronauer, D. J. C. (2014). The genome of the clonal raider ant Cerapachys biroi . Current Biology, 24(4), 451–458. 10.1016/j.cub.2014.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki, M. , Wada‐Katsumata, A. , Fujikawa, K. , Iwasaki, M. , Yokohari, F. , Satoji, Y. , & Yamaoka, R. (2005). Ant nestmate and non‐nestmate discrimination by a chemosensory sensillum. Science, 309(5732), 311–314. 10.1126/science.1105244 [DOI] [PubMed] [Google Scholar]
- Parra, G. , Bradnam, K. , & Korf, I. (2007). CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics, 23(9), 1061–1067. 10.1093/bioinformatics/btm071 [DOI] [PubMed] [Google Scholar]
- Pask, G. M. , Slone, J. D. , Millar, J. G. , Das, P. , Moreira, J. A. , Zhou, X. , Bello, J. , Berger, S. L. , Bonasio, R. , Desplan, C. , Reinberg, D. , Liebig, J. , Zwiebel, L. J. , & Ray, A. (2017). Specialized odorant receptors in social insects that detect cuticular hydrocarbon cues and candidate pheromones. Nature Communications, 8(1), 297. 10.1038/s41467-017-00099-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patalano, S. , Vlasova, A. , Wyatt, C. , Ewels, P. , Camara, F. , Ferreira, P. G. , Asher, C. L. , Jurkowski, T. P. , Segonds‐Pichon, A. , Bachman, M. , González‐Navarrete, I. , Minoche, A. E. , Krueger, F. , Lowy, E. , Marcet‐Houben, M. , Rodriguez‐Ales, J. L. , Nascimento, F. S. , Balasubramanian, S. , Gabaldon, T. , Tarver, J. E. , Andrews, S. , Himmelbauer, H. , Hughes, W. O. H. , Guigó, R. , Reik, W. , & Sumner, S. (2015). Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. Proceedings of the National Academy of Sciences of the United States of America, 112(45), 13970–13975. 10.1073/pnas.1515937112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto‐Godino, L. L. , Rytz, R. , Bargeton, B. , Abuin, L. , Arguello, J. R. , Peraro, M. D. , & Benton, R. (2016). Olfactory receptor pseudo‐pseudogenes. Nature, 539(7627), 93–97. 10.1038/nature19824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2013). R: A Language and Environment for Statistical Computing. Austria. [Google Scholar]
- Rettenmeyer, C. W. (1963). Behavioral studies of army ants. University of Kansas Science Bulletin, 44(9), 281–465. [Google Scholar]
- Robertson, H. M. , Gadau, J. , & Wanner, K. W. (2010). The insect chemoreceptor superfamily of the parasitoid jewel wasp Nasonia vitripennis . Insect Molecular Biology, 19(s1), 121–136. 10.1111/j.1365-2583.2009.00979.x [DOI] [PubMed] [Google Scholar]
- Rubin, B. E. R. , & Moreau, C. S. (2016). Comparative genomics reveals convergent rates of evolution in ant–plant mutualisms. Nature Communications, 7, 12679. 10.1038/ncomms12679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoz, J.‐C. (2006). Odour‐evoked responses to queen pheromone components and to plant odours using optical imaging in the antennal lobe of the honey bee drone Apis mellifera L. The Journal of Experimental Biology, 209(Pt 18), 3587–3598. 10.1242/jeb.02423 [DOI] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , Preibisch, S. , Rueden, C. , Saalfeld, S. , Schmid, B. , Tinevez, J‐Y. , White, D. J. , Hartenstein, V. , Eliceiri, K. , Tomancak, P. , & Cardona, A. (2012). Fiji: an open‐source platform for biological‐image analysis. Nature Methods, 9(7), 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneirla, T. C. (1944). Studies on the Army‐Ant Behavior Pattern. ‐Nomadism in the Swarm‐Raider Eciton burchelli. Proceedings of the American Philosophical Society, 87(5), 438–457. [Google Scholar]
- Schneirla, T. C. (1971). Army ants; a study in social organization. W.H.Freeman & Co Ltd. [Google Scholar]
- Schrader, L. , Kim, J. W. , Ence, D. , Zimin, A. , Klein, A. , Wyschetzki, K. , Weichselgartner, T. , Kemena, C. , Stökl, J. , Schultner, E. , Wurm, Y. , Smith, C. D. , Yandell, M. , Heinze, J. , Gadau, J. , & Oettler, J. (2014). Transposable element islands facilitate adaptation to novel environments in an invasive species. Nature Communications, 5, 5495. 10.1038/ncomms6495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader, L. , Pan, H. , Bollazzi, M. , Schiøtt, M. , Larabee, F. J. , Bi, X. , Deng, Y. , Zhang, G. , Boomsma, J. J. , & Rabeling, C. (2021). Relaxed selection underlies genome erosion in socially parasitic ant species. Nature Communications, 12(1), 1–13. 10.1038/s41467-021-23178-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrucca, L. , Fop, M. , Murphy, T. B. , & Raftery, A. E. (2016). mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. The R Journal, 8(1), 289–317. 10.32614/RJ-2016-021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, K. R. , Enzmann, B. L. , Schmidt, Y. , Moore, D. , Jones, G. R. , Parker, J. , Berger, S. L. , Reinberg, D. , Zwiebel, L. J. , Breit, B. , Liebig, J. , & Ray, A. (2015). Cuticular hydrocarbon pheromones for social behavior and their coding in the ant antenna. Cell Reports, 12(8), 1261–1271. 10.1016/j.celrep.2015.07.031 [DOI] [PubMed] [Google Scholar]
- Simão, F. A. , Waterhouse, R. M. , Ioannidis, P. , Kriventseva, E. V. , & Zdobnov, E. M. (2015). BUSCO: Assessing genome assembly and annotation completeness with single‐copy orthologs. Bioinformatics, 31(19), 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Slater, G. S. C. , & Birney, E. (2005). Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics, 6(1), 31. 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slone, J. D. , Pask, G. M. , Ferguson, S. T. , Millar, J. G. , Berger, S. L. , Reinberg, D. , & Zwiebel, L. J. (2017). Functional characterization of odorant receptors in the ponerine ant, Harpegnathos saltator. Proceedings of the National Academy of Sciences of the United States of America, 114(32), 8586–8591. 10.1073/pnas.1704647114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C. R. , Smith, C. D. , Robertson, H. M. , Helmkampf, M. , Zimin, A. , Zimin, A. , Yandell, M. , Holt, C. , Hu, H. , Abouheif, E. , Benton, R. , Cash, E. , Croset, V. , Currie, C. R. , Elhaik, E. , Elsik, C. G. , Fave, M. J. , Fernandes, V. , Gibson, J. D. , Graur, D. , Gronenberg, W. , Grubbs, K. J. , Hagen, D. E. , Viniegra, A. S. I. , Johnson, B. R. , Johnson, R. M. , Khila, A. , Kim, J. W. , Mathis, K. A. , Munoz‐Torres, M. C. , Murphy, M. C. , Mustard, J. A. , Nakamura, R. , Niehuis, O. , Nigam, S. , Overson, R. P. , Placek, J. E. , Rajakumar, R. , Reese, J. T. , Suen, G. , Tao, S. , Torres, C. W. , Tsutsui, N. D. , Viljakainen, L. , Wolschin, F. , & Gadau, J. (2011). Draft genome of the red harvester ant Pogonomyrmex barbatus. Proceedings of the National Academy of Sciences of the United States of America, 108(14), 5667–5672. 10.1073/pnas.1007901108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C. D. , Zimin, A. , Holt, C. , Abouheif, E. , Benton, R. , Cash, E. , Croset, V. , Currie, C. R. , Elhaik, E. , Elsik, C. G. , Fave, M‐J. , Fernandes, V. , Gadau, J. , Gibson, J. D. , Graur, D. , Grubbs, K. J. , Hagen, D. E. , Helmkampf, M. , Holley, J‐A. , Hu, H. , Viniegra, A. S. I. , Johnson, B. R. , Johnson, R. M. , Khila, A. , Kim, J. W. , Laird, J. , Mathis, K. A. , Moeller, J. A. , Munoz‐Torres, M. C. , Murphy, M. C. , Nakamura, R. , Nigam, S. , Overson, R. P. , Placek, J. E. , Rajakumar, R. , Reese, J. T. , Robertson, H. M. , Smith, C. R. , Suarez, A. V. , Suen, G. , Suhr, E. l , Tao, S. , Torres, C. W. , van Wilgenburg, E. , Viljakainen, L. , Walden, K. K. O. , Wild, A. l , Yandell, M. , Yorke, J. A. , & Tsutsui, N. D. (2011). Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proceedings of the National Academy of Sciences of the United States of America, 108(14), 5673–5678. 10.1073/pnas.1008617108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics (Oxford, England), 30(9), 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis, A. , Hoover, P. , & Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology, 57(5), 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Stanke, M. , Schöffmann, O. , Morgenstern, B. , & Waack, S. (2006). Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics, 7, 62. 10.1186/1471-2105-4-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgis, S. J. , & Gordon, D. M. (2012). Nestmate recognition in ants (Hymenoptera: Formicidae): A review. Myrmecological News, 16, 101–110. [Google Scholar]
- Suen, G. , Teiling, C. , Li, L. , Holt, C. , Abouheif, E. , Bornberg‐Bauer, E. , Bouffard, P. , Caldera, E. J. , Cash, E. , Cavanaugh, A. , Denas, O. , Elhaik, E. , Favé, M.‐J. , Gadau, J. , Gibson, J. D. , Graur, D. , Grubbs, K. J. , Hagen, D. E. , Harkins, T. T. , Helmkampf, M. , Hu, H. , Johnson, B. R. , Kim, J. , Marsh, S. E. , Moeller, J. A. , Muñoz‐Torres, M. C. , Murphy, M. C. , Naughton, M. C. , Nigam, S. , Overson, R. , Rajakumar, R. , Reese, J. T. , Scott, J. J. , Smith, C. R. , Tao, S. , Tsutsui, N. D. , Viljakainen, L. , Wissler, L. , Yandell, M. D. , Zimmer, F. , Taylor, J. , Slater, S. C. , Clifton, S. W. , Warren, W. C. , Elsik, C. G. , Smith, C. D. , Weinstock, G. M. , Gerardo, N. M. , & Currie, C. R. (2011). The genome sequence of the leaf‐cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLoS Genetics, 7(2), 10.1371/journal.pgen.1002007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson, R. L. , & Akre, R. D. (1970). Interspecific Responses to Trail and Alarm Pheromones by New World Army Ants. Journal of the Kansas Entomological Society, 43(4), 395–404. [Google Scholar]
- Trapnell, C. , Williams, B. A. , Pertea, G. , Mortazavi, A. , Kwan, G. , van Baren, M. J. , Salzberg, S. L. , Wold, B. J. , & Pachter, L. (2010). Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology, 28(5), 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trible, W. , Olivos‐Cisneros, L. , McKenzie, S. K. , Saragosti, J. , Chang, N.‐C. , Matthews, B. J. , Oxley, P. R. , & Kronauer, D. J. C. (2017). orco mutagenesis causes loss of antennal lobe glomeruli and impaired social behavior in ants. Cell, 170(4), 727–735. 10.1016/j.cell.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, K. , & Yamauchi, K. (1995). Production of females by parthenogenesis in the ant. Cerapachys Biroi. Insectes Sociaux, 42(3), 333–336. 10.1007/BF01240430 [DOI] [Google Scholar]
- van Zweden, J. S. , & D’Ettorre, P. (2010). Nestmate recognition in social insects and the role of hydrocarbons. In Blomquist G. J. & Bagneres A. G. (Eds.), Insect Hydrocarbons. Cambridge, UK: Cambridge University Press. 10.1017/CBO9780511711909 [DOI] [Google Scholar]
- Virtanen, P. , Gommers, R. , Oliphant, T. E. , Haberland, M. , Reddy, T. , Cournapeau, D. , Contributors, S. (2019). SciPy 1.0‐‐Fundamental Algorithms for Scientific Computing in Python. ArXiv E‐Prints, arXiv:1907.10121.
- von Beeren, C. , Brückner, A. , Maruyama, M. , Burke, G. , Wieschollek, J. , & Kronauer, D. J. C. (2018). Chemical and behavioral integration of army ant‐associated rove beetles–a comparison between specialists and generalists. Frontiers in Zoology, 15(1), 8. 10.1186/s12983-018-0249-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, E. O. (1965). Chemical Communication in the Social Insects. Science, 149(3688), 1064–1071. 10.1126/science.149.3688.1064 [DOI] [PubMed] [Google Scholar]
- Winston, M. E. , Kronauer, D. J. C. , & Moreau, C. S. (2017). Early and dynamic colonization of Central America drives speciation in Neotropical army ants. Molecular Ecology, 26(3), 859–870. 10.1111/MEC.13846 [DOI] [PubMed] [Google Scholar]
- Witte, V. , Schliessmann, D. , & Hashim, R. (2010). Attack or call for help? Rapid individual decisions in a group‐hunting ant. Behavioral Ecology, 21(5), 1040–1047. 10.1093/beheco/arq100 [DOI] [Google Scholar]
- Wurm, Y. , Wang, J. , Riba‐Grognuz, O. , Corona, M. , Nygaard, S. , Hunt, B. G. , Ingram, K. K , Falquet, L. , Nipitwattanaphon, M. , Gotzek, D. , Dijkstra, M. B , Oettler, J. , Comtesse, F. , Shih, C‐J , Wu, W‐J , Yang, C‐C , Thomas, J. , Beaudoing, E. , Pradervand, S. , Flegel, V. , Cook, E. D , Fabbretti, R. , Stockinger, H. , Long, L. , Farmerie, W. G , Oakey, J. , Boomsma, J. J , Pamilo, P. , Yi, S. V , Heinze, J. , Goodisman, M. A. D. , Farinelli, L. , Harshman, K. , Hulo, N. , Cerutti, L. , Xenarios, I. , Shoemaker, D. , & Keller, L. (2011). The genome of the fire ant Solenopsis invicta. Proceedings of the National Academy of Sciences of the United States of America, 108(14), 5679–5684. 10.1073/pnas.1009690108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H. , Opachaloemphan, C. , Mancini, G. , Yang, H. , Gallitto, M. , Mlejnek, J. , & Desplan, C. (2017). An engineered orco mutation produces aberrant social behaviour and defective neural development in ants. Cell, 170(4), 736–747. 10.1016/j.cell.2017.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution, 24(8), 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zdobnov, E. M. , & Apweiler, R. (2001). InterProScan ‐ an integration platform for the signature‐recognition methods in InterPro. Bioinformatics, 17(9), 847–848. 10.1093/bioinformatics/17.9.847 [DOI] [PubMed] [Google Scholar]
- Zhou, X. , Slone, J. D. , Rokas, A. , Berger, S. L. , Liebig, J. , Ray, A. , & Zwiebel, L. J. (2012). Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex‐specific signatures of odor coding. PLoS Genetics, 8(8), e1002930. 10.1371/journal.pgen.1002930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zube, C. , Kleineidam, C. J. , Kirschner, S. , Neef, J. , & Rössler, W. (2008). Organization of the olfactory pathway and odor processing in the antennal lobe of the ant Camponotus floridanus. The Journal of Comparative Neurology, 506(3), 425–441. 10.1002/cne.21548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐5
Table S1
Table S2
Table S3
Table S4
Table S5
Data Availability Statement
Sequencing data and genomic assemblies and annotations have been deposited in the NCBI SRA and genome databases under the BioProject accession no. PRJNA632625. All other data are available as supplemental material accompanying this paper.
McKenzie, S. K., Winston, M. W., Grewe, F., Vargas Asensio, G., Rodríguez‐Hernández, N., Rubin, B.E.R., Murillo‐Cruz, C., von Beeren, C., Moreau, C.S., Suen, G., Pinto‐Tomás, A.A., & Kronauer, D.J.C. (2020) Sequencing and genome assembly of the army ant Eciton burchellii. NCBI SRA and Genome Databases, BioProject. PRJNA632625.


