Abstract
Methyl iodide (CH3I) plays an important role in the natural iodine cycle and participates in atmospheric ozone destruction. However, the main source of this compound in nature is still unclear. Here we report that a wide variety of bacteria including terrestrial and marine bacteria are capable of methylating the environmental level of iodide (0.1 μM). Of the strains tested, Rhizobium sp. strain MRCD 19 was chosen for further analysis, and it was found that the cell extract catalyzed the methylation of iodide with S-adenosyl-l-methionine as the methyl donor. These results strongly indicate that bacteria contribute to iodine transfer from the terrestrial and marine ecosystems into the atmosphere.
In the geochemical cycling of iodine, methyl iodide (CH3I) plays a significant role serving as an effective carrier of iodine from the biosphere into the atmosphere (4, 12, 14). CH3I is emitted into the atmosphere mainly as a result of biological methylation of iodine (13, 14) and is readily photolysed to produce iodine atoms, which may influence the atmospheric ozone budget (4, 5, 33). Subsequent precipitation of atmospheric iodine satisfies the needs of humans and animals for this essential element. The methylation of iodine should also be considered from the viewpoint of the hazard posed by anthropogenic 129I (half life, 1.6 × 107 years), which has been released into the environment from nuclear facilities (3, 24). Once it is methylated, 129I can spread far from a contaminated area and may accumulate in the human thyroid gland. Therefore, it is very important to elucidate the mechanisms of iodine methylation in the environment.
The predominant source of atmospheric CH3I has been considered to be marine organisms (13, 14, 20, 27, 28). Considerable evidence has supported the notion that macroalgae (seaweed such as kelp) are CH3I sources, but many laboratory culture experiments have indicated that this production (107 to 108 g year−1) (7, 16, 26) is insignificant compared with the global CH3I flux (1 × 1011 to 4 × 1011 g year−1) (21, 32). This appears to be due to the limited distribution of macroalgae only in coastal regions and hence the limited biomass available. Microalgae (phytoplankton) are widely distributed in the ocean and have a greater biomass than macroalgae. However, several laboratory studies have shown that microalgal production (109 to 1010 g year−1) also seems to be insufficient to account for the global flux (10, 11, 15). Therefore, the involvement of other organisms such as bacteria has been inferred (17), but no direct evidence has yet been reported.
In this report, we provide evidence that a wide variety of bacteria, including both laboratory strains and natural isolates, have capacities for CH3I production. They are also able to methylate environmental level of iodide (I−) under oligotrophic conditions.
MATERIALS AND METHODS
Organisms.
Bacterial and archaeal strains were obtained from the National Collection of Industrial and Marine Bacteria Ltd. (NCMB), Aberdeen, Scotland; the Institute of Molecular and Cellular Biosciences (IAM), University of Tokyo, Tokyo, Japan; the Japan Collection of Microorganisms (JCM), Wako, Japan; and the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSM), Braunschweig, Germany. Most of other strains were from our own culture collection (MRCD). MRCD strains were isolated from soil environments in Ibaraki Prefecture, Japan.
Culture conditions for CH3I production by growing cultures.
Methylosinus trichosporium OB3b (ATCC 35070) and Methylomonas sp. strain KSWIII were grown in NMS medium (35) under a methane-air (20:80) atmosphere. Methanosarcina mazei DSM 9195 was grown in medium 120 as described in the DSM 1993 catalog. The other three methanogens were grown in medium 141 under an atmosphere of H2-CO2 (80:20), but final NaCl concentrations were modified to 0.4 g per liter for Methanobacterium formicicum DSM 1535 and Methanospirillum hungateii DSM 864 and to 6 g per liter for Methanoculleus olentangyi DSM 2772. Clostridium novyi ACR was grown under a nitrogen (N2) atmosphere in medium 334 from which sodium acetate had been removed and to which 0.1 g of yeast extract per liter and 0.9 g of glucose per liter were added. All the other strains were grown aerobically in 0.1× PTYG medium. (1× PTYG medium contains 2.5 g of tryptone per liter, 2.5 g of peptone per liter, 5.0 g of yeast extract per liter, 5.0 g of glucose per liter, 0.3 g of MgSO4 per liter, and 0.035 g of CaCl2 per liter.) The growth temperature was 30°C except for methanogens (which were grown at 37°C).
All strains were grown in 120-ml serum bottles containing 20 ml of culture medium that included sodium iodide (NaI) at a final concentration of 0.1 mM. The bottles were sealed with butyl rubber stoppers which were sterilized with 70% ethanol. We avoided autoclaving the butyl rubber stoppers, because this resulted in the formation of an unidentified substance which strongly interfered with the CH3I peak in gas chromatography (GC) analysis. For each strain, a culture without NaI was prepared as a control. For each medium, an uninoculated medium was incubated with NaI as another control. From these control samples, no CH3I was detected in GC analysis.
Detection and identification of CH3I.
CH3I emitted into the headspace was detected with a Shimadzu 14A gas chromatograph equipped with an electron capture detector (ECD; 63Ni source operated at 200°C). After the cells reached the stationary growth phase, 50 to 500 μl of headspace gas was directly injected into the gas chromatograph (injector port, 100°C). The stainless steel column (length, 3 m; inner diameter, 2.6 mm) was packed with 20% silicone DC-550. N2 was used as the carrier gas at a flow rate of 40 ml min−1. The column temperature was 50°C. Under these conditions, not only CH3I but iodoethane (C2H5I), 1-iodopropane (1-C3H7I), 2-iodopropane (2-C3H7I), and chloroiodomethane (CH2ClI) could be detected. The detection limits were less than 2 pmol/ml of headspace gas for all of the compounds. CH3I and CH2ClI showed the highest response to the ECD and therefore the lowest detection limit of approximately 0.05 pmol/ml of headspace gas. Each compound (Aldrich Chemical Co. Inc.) was diluted in methanol and used as a standard. For CH3I, a gaseous standard prepared by dilution of 45 nmol of CH3I/ml of helium (Takachiho Trading Co. Ltd., Tokyo, Japan) was also used.
Biogenic CH3I was identified using a Hewlett-Packard 5890A series II gas chromatograph equipped with a PoraPLOT Q-HT capillary column (length, 25 m; inner diameter, 0.32 mm; film thickness, 10 μm) and coupled to a Hewlett-Packard 5972A mass-selective detector. Headspace samples (2.5 ml) were cryogenically concentrated (Chrompack CP4010) at −130°C before being injected into the gas chromatograph. The temperature program started at 40°C and rose to 220°C at a rate of 10°C min−1. The temperature was subsequently kept at 220°C for 20 min. The target ions for CH3I were 142 and 127 (m/z).
Radioanalytical determination of CH3I using resting cells.
Terrestrial bacterial strains were grown to the mid-exponential phase in 1× PTYG medium at 30°C. Cells were harvested by centrifugation (10,000 × g at 4°C), washed with 20 mM potassium phosphate buffer (pH 7.0), and resuspended in the same buffer to give final cell concentrations of approximately 108 to 109 ml−1 (the cell concentration was determined by plating the suspensions on agar medium). Marine bacterial strains were grown in marine broth (Difco Laboratories) at 20°C and resuspended in 20 mM potassium phosphate–340 mM NaCl–1 mM MgCl2 buffer (pH 7.0) in the same manner. Each cell suspension (19 ml) was dispensed into a sterile 120-ml serum bottle. Potassium iodide (KI) (final concentration, 0.1 μM) containing approximately 30 kBq of K125I (Dupont NEN Products) were added to each suspension. To determine the effect of the I− concentration on CH3I formation, we chose Rhizobium sp. strain MRCD 19 and Alteromonas macleodii IAM 12920 as representative organisms, and stable I− at final concentrations ranging from 0.1 μM to 5 mM was added to the suspensions. Serum bottles were sealed with butyl rubber stoppers and then incubated in the dark with gentle shaking at 30°C (terrestrial bacteria) or 20°C (marine bacteria).
After 24 h of incubation, CH3I produced was collected in an activated charcoal trap by sweeping under nitrogen gas flushing. Nitrogen gas was introduced into the vial at 250 ml min−1 from a needle. The charcoal trap was constructed in a plastic syringe with a needle and contained 0.4 g of activated charcoal (Merck; 35 to 50 mesh) supplemented with 10% (wt/wt) 1,4-diazabicyclo-[2,2,2]-octane (Wako Pure Chemical Industries, Ltd.) to enhance the efficiency of organic iodine collection (22). During this sampling, the vial was heated in a hot bath (100°C) to expel the dissolved CH3I into the gas phase. In this method, all of the CH3I present in the vial could be collected in the trap within 10 min. Volatile inorganic iodine (I2 or HOI) was also determined by using silver wool traps (22), but no production was observed.
The activity of 125I collected in the trap was measured with an NaI scintillation counter (Aloka ARC-380) for 20 min, and counting errors were 0.5 to 20% (in most cases they were less than 10%). Quintuplet bottles per bacterial strain were prepared, and two of these bottles were used at zero time for background determination. CH3I production (femtomoles day−1 1010 cells−1) was calculated when the average counting value of the triplicates at 24 h of incubation was significantly higher (P < 0.05) than that time zero (Student's t test). The detection limit was about 0.03 pmol of CH3I/bottle, which is approximately 300 times higher than the sensitivity of GC-ECD. Preliminary experiments confirmed that the viabilities of resting cells did not change significantly after 24 h of incubation.
By using GC-ECD, it was also confirmed that resting cells of Rhizobium sp. strain MRCD 19 and A. macleodii IAM 12920 did not produce alkyl iodides other than CH3I under 1 mM I−. Assuming that the Henry's law constant of CH3I under our experimental conditions was approximately 0.2 (19), the CH3I production normalized to the cell number (nanomoles day−1 1010 cells−1) quantified by GC-ECD (2.3 for Rhizobium, 0.065 for A. macleodii) showed good agreement with that quantified by the radiotracer experiment (2.1 for Rhizobium, 0.11 for A. macleodii). This indicated that our radiotracer experiment was sufficiently quantitative.
Preparation of cell extract for the enzyme assay.
Rhizobium sp. strain MRCD 19 was grown in 1× PTYG medium. Cells were harvested in the late exponential phase by centrifugation (10,000 × g at 4°C), and washed with 100 mM potassium phosphate buffer (pH 7.0). Washed cells were resuspended in the same buffer to give approximately 0.2 g (wet weight) of cells ml−1. The cell suspension was then passed through a French press (SLM-Aminco) at 1,300 lb/in2. After the cell debris was removed by centrifugation (30,000 × g for 10 min), the supernatant was used as the crude enzyme.
Enzyme assay.
Enzyme reactions were carried out in 60-ml serum bottles sealed with butyl rubber stoppers. The reaction mixture (10 ml) contained 100 mM potassium phosphate (pH 7.0), 10 mM KI, approximately 30 kBq of K125I, and 50 to 100 mg of enzyme preparation. In some cases, S-adenosyl-l-methionine (SAM; Sigma Chemical Co.) and/or S-adenosyl-l-homocysteine (SAHC; Sigma) was added to the reaction mixture at final concentrations of 0.5 mM each. In the presence of SAM, CH3I production was linear for the first 20 h. After 20 h of incubation at 30°C with gentle shaking, the CH3I produced was collected and quantified as described above. The protein concentration was determined by the method of Bradford (2) with bovine serum albumin as the standard. In the preliminary experiment, it was confirmed by GC-ECD that the enzyme preparation does not produce alkyl iodides other than CH3I under the reaction conditions.
The Km values for I− and SAM were determined by varying the concentration of one substrate while keeping that of the other constant (I−, 10 mM; SAM, 0.5 mM).
RESULTS
Distribution of CH3I-producing ability among microorganisms.
We first examined whether bacteria are capable of producing CH3I from I− by using growing cultures. Various bacterial strains maintained in our laboratory were chosen randomly and grown in media containing 0.1 mM I−. The phylogenetic affiliations and the number of selected strains were as follows: gram-positive high-GC group, six strains (Arthrobacter oxydans, Cellulomonas sp., Curtobacterium citreum, Microbacterium schleiferi, Rhodococcus equi, and Streptomyces fradiae); gram-positive low-GC group, three strains (Bacillus subtilis, Bacillus thuringiensis, and Clostridium novyi); Thermus-Deinococcus group, one strain (Deinococcus grandis); Cytophaga-Bacteroides group, two strains (Flexibacter sp. and Sphingobacterium sp.); Proteobacteria α subgroup, three strains (Rhizobium sp., Methylobacterium sp., and Methylosinus trichosporium); Proteobacteria β subgroup, two strains (Variovorax sp. and Zoogloea sp.); Proteobacteria γ subgroup, five strains (Beggiatoa sp., Escherichia coli, Methylomonas sp., Pseudomonas straminae, and Xanthomonas sp.); and Archaea (methanogens), four strains (Methanobacterium formicicum, Methanoculleus olentangyi, Methanosarcina mazei, and Methanospirillum hungatei). Of these 26 microorganisms, 14 strains produced more than 0.05 pmol of CH3I/ml of headspace, the detection limit of GC-ECD. CH3I formation was verified by mass spectroscopy. The aerobic bacteria Rhizobium sp. strain MRCD 19 and Methylosinus trichosporium OB3b showed much higher production than the other strains (3.8 and 3.7 pmol/ml of headspace, respectively). On the other hand, the anaerobic microorganisms (Clostridium and methanogens) did not show CH3I production. Production of other alkyl iodides such as C2H5I, 1-C3H7I, 2-C3H7I, and CH2ClI was not observed in any microorganisms.
Bacterial CH3I production at environmental levels of iodide.
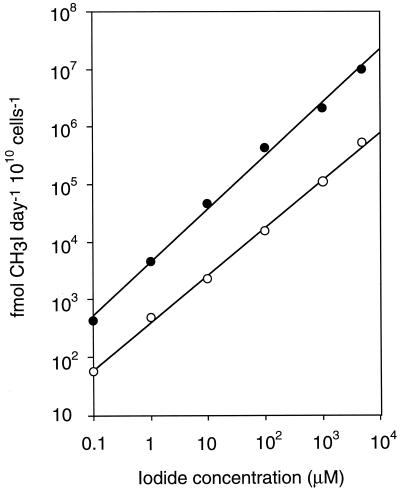
Microorganisms do not grow as fast in natural environments such as soils and seawater, as in the laboratory because of the oligotrophic conditions, and the natural iodine levels are usually much lower than the concentration in the experiment described above. To examine bacterial CH3I production under natural conditions, we measured CH3I production in the presence of an environmental concentration of I− (0.1 μM) (36) using a radioactive iodine tracer. This method allowed us to detect much smaller amounts of CH3I than by GC-ECD. The cells were suspended in phosphate buffer and were not supplied with any nutrients that support bacterial growth. From the bacterial strains investigated above, we chose 10 strains which did not lose viability during the incubation period and used them as terrestrial bacteria. Ten marine bacterial strains were also used to determine CH3I production. Of the 20 bacterial strains, 13 showed CH3I formation under the experimental conditions (Table 1). Although Variovorax sp. strain MRCD 30 produced the largest amount of CH3I per cell mass among terrestrial strains (909 ± 121 fmol day−1 1010 cells−1 [Table 1]), it could grow only very slowly in our medium (1× PTYG). Therefore, we chose Rhizobium sp. strain MRCD 19 as a representative terrestrial bacterium for further investigation. By using resting cells of Rhizobium sp. strain MRCD 19 and A. macleodii IAM 12920, we measured CH3I production at I− concentrations of 0.1 μM to 5 mM. These strains showed increased CH3I production in the presence of increased I− concentrations (Fig. 1), indicating that bacterial CH3I production depends greatly on the surrounding iodine levels. Heat-treated (80°C) or autoclaved cells of Rhizobium sp. strain MRCD 19 did not show any CH3I production, suggesting that the methylation was mediated enzymatically.
TABLE 1.
Bacterial methyl iodide production in the presence of 0.1 μM iodide
| Organism | CH3I production (fmol day−1 1010 cells−1) (mean ± SD, n = 3) |
|---|---|
| Terrestrial bacteria | |
| Arthrobacter oxydans JCM 2521 | 0 |
| Curtobacterium citreum JCM 1345 | 0 |
| Deinococcus grandis JCM 6269 | 0 |
| Escherichia coli DH5α | 0 |
| Methylobacterium sp. strain MRCD 18 | 9.0 ± 1.0 |
| Pseudomonas straminae JCM 2783 | 13.2 ± 3.3 |
| Rhizobium sp. strain MRCD 19 | 428.1 ± 6.1 |
| Rhodococcus equi JCM 1311 | 5.0 ± 1.0 |
| Variovorax sp. strain MRCD 30 | 908.8 ± 121.3 |
| Zoogloea sp. strain MRCD 32 | 11.0 ± 0.3 |
| Marine bacteria | |
| Alteromonas macleodii IAM 12920 | 56.8 ± 0.8 |
| Deleya aquamarina IAM 12550 | 0 |
| Deleya marina IAM 14107 | 5.4 ± 0.4 |
| Oceanospirillum comune IAM 12914 | 0 |
| Photobacterium leiognathi NCIMB 2193 | 40.4 ± 1.0 |
| Photobacterium phosphoreum IAM 14401 | 22.2 ± 2.6 |
| Pseudoalteromonas haloplanktis IAM 12915 | 6.0 ± 0.7 |
| Shewanella putrefaciens IAM 12079 | 2.2 ± 0.3 |
| Vibrio alginolyticus NCIMB 1903 | 0 |
| Vibrio splendidus NCIMB 1 | 42.2 ± 10.0 |
FIG. 1.
CH3I production by resting cells of Rhizobium sp. strain MRCD 19 (●) and A. macleodii IAM 12920 (○) at various concentrations of iodide. Means and standard deviations of triplicate samples are shown, but the error bars are hidden under the symbols. Of the 12 data points depicted, only 1 point showed a standard deviation of more than 10%.
Kinetics of methylation activity.
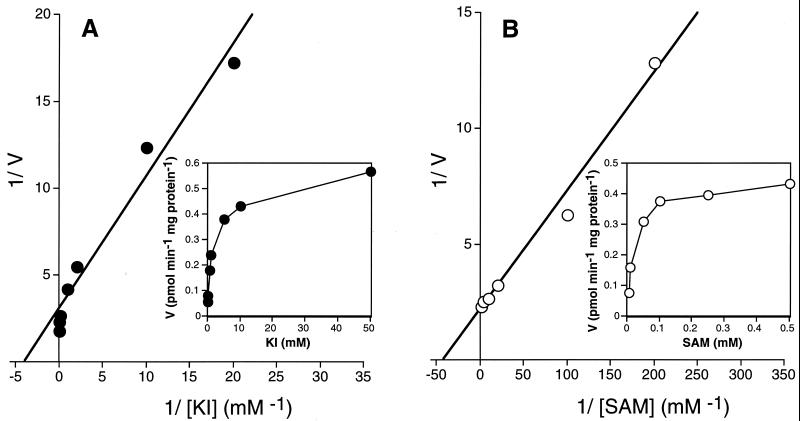
We measured the I−-methylating activity by using cell extracts of Rhizobium sp. strain MRCD 19. We found that SAM could serve as a methyl donor for methylation of I− (the specific activity was 0.547 pmol of CH3I min−1 mg of protein−1). Only very low methylating activity (0.014 pmol of CH3I min−1 mg of protein−1) was detected in the absence of SAM. We also found that SAHC, an inhibitor of SAM-dependent enzymes (1, 31), completely inhibited this background level of activity, suggesting that SAM functions as a methyl donor in vivo, at least in this strain. The inhibition was approximately 65% at equimolar concentrations of SAM and SAHC (0.189 pmol of CH3I min−1 mg of protein−1). The Km values for I− and SAM, calculated from Lineweaver-Burk plots, were 0.26 and 0.024 mM, respectively (Fig. 2).
FIG. 2.
The iodide-methylating reaction by cell extracts of Rhizobium sp. strain MRCD 19 is dependent on the concentrations of KI (A) and SAM (B). I−-methylating rates (picomoles per minute per milligram of protein) and their reciprocals were plotted against KI concentrations and their reciprocals (A) and SAM concentrations and their reciprocals (B). The reactions were carried out in potassium phosphate buffer (pH 7.0) containing KI, SAM, approximately 30 kBq of K125I, and 50 mg of cell extract. CH3I was collected and quantified as described in Materials and Methods. The concentration of KI was varied from 0.05 to 50 mM in the presence of 0.5 mM SAM (●), and the concentration of SAM was varied from 0.005 to 0.5 mM in the presence of 10 mM KI (○). Means of triplicate samples are shown.
DISCUSSION
Since atmospheric iodine is thought to originate mainly from the ocean (6, 18, 34), much attention has been paid to the methylation of iodine in marine ecosystems. However, several experimental results have shown that iodine is emitted from terrestrial ecosystems such as rice paddy fields (22, 29). In addition, several terrestrial organisms such as plants and wood-rotting fungi have been reported to possess halide-methylating abilities (8, 9, 30, 37). In this study, we have found that a wide variety of terrestrial bacteria have capacities for methylating I−. Considering their enormous biomass and the ubiquity of methylation activity regardless of their taxonomic positions, it would not be surprising if bacteria contributed to iodine transfer from terrestrial environments. The extrapolation of our results to natural soil environments is, however, difficult because of the dependence of iodine availability on the properties of soils, as discussed below, and the large variations in iodine levels among different soils (6).
As shown in Fig. 1, bacterial CH3I production depends greatly on the surrounding iodine levels. Since the iodine content in soils is sometimes very high (more than 50 mg kg−1) (6, 24, 34), a large amount of iodine emission should occur if iodine availability is high enough for bacterial methylation. However, iodine is usually sorbed by soil components such as soil organic matter (38). It can be desorbed and leaches into soil water when the soil redox potential (Eh) decreases to −100 mV and below (23). Therefore, bacterial methylation of iodine may preferentially occur at oxic-anoxic interfaces of soil environments such as flooded rice fields, peat bogs, and swamps. On the other hand, methylation would not occur in strictly anoxic sediments or soil subsurface, because anaerobic microorganisms (Clostridium and methanogens) did not show CH3I production in this study. In addition, we have previously measured CH3I production in four types of soil water supplemented with 1 mM I−. Little CH3I was detected by GC-ECD in samples incubated under an N2 atmosphere, whereas significant production (0.3 to 1.6 pmol/ml of headspace) was observed in aerobically incubated samples (S. Amachi, Y. Kamagata, and Y. Muramatsu, unpublished data). These results suggest that anaerobic methylation of iodine is not common or occurs very slowly.
Our findings will provide another insight into the migration of long-lived 129I. Brauer and Strebin (3) have reported interesting observations on gaseous radioiodine concentrations near the Hanford fuel-reprocessing plant in Washington State. They found that concentrations of 129I in air samples were 2 orders of magnitude higher than in the samples from a remote place, over 7 years after the fuel-reprocessing operations discontinued. Most of 129I was considered to be in organic forms, probably as a result of emission from the contaminated soil surface (3). These observations, together with our findings, possibly indicate that soil microorganisms are involved in the methylation and subsequent emission of radioiodine. They also indicate that if iodine were released from nuclear facilities, weapons testing, or ground storage of nuclear waste, the pathway of methylation by bacteria should be considered in the assessment of its environmental migration.
Regarding marine ecosystems, macro- and microalgae have been the most significant candidates for primary CH3I producers in the ocean. However, the discrepancy between the annual global flux (1× 1011 to 4 × 1011 g) (21, 32) and the estimated annual algal production (107 to 1010 g) (7, 10, 11, 15, 16, 26) suggests that other organisms should be involved in the production. Manley and Dastoor (17) have reported weak CH3I production by undefined microbial populations obtained from decaying kelp tissue. However, there has been no direct evidence that marine bacteria are involved in CH3I production in the ocean. In this study, we have found that CH3I-producing ability is widespread among representative marine bacteria such as A. macleodii, Photobacterium spp., and V. splendidus (Table 1). It should be noted that these bacteria showed significant CH3I production from low levels of I− (0.1 μM), corresponding to the average I− level in surface sea water (36). Thus, it is possible that they actually produce CH3I in the marine environments, together with macro- and microalgae. However, accurate global significance of marine bacteria cannot be discussed here because of the difference in physiological conditions between laboratory and sea water.
Cell extract of Rhizobium sp. strain MRCD 19 showed SAM-dependent I−-methylating activity. In several eucaryotic organisms such as marine algae, wood-rotting fungi, and terrestrial higher plants, similar enzymes catalyzing the methylation of halides have been characterized (1, 11, 25, 31, 37). Further studies to determine the properties of bacterial enzyme such as substrate specificity are required. In addition, it would be of great interest if we could confirm that this type of enzyme is widely distributed in bacteria.
ACKNOWLEDGMENTS
We thank C. H. Nakatsu (University of Purdue) and T. Hamilton (Lawrence Livermore National Laboratory) for critical reading of the manuscript and S. Yoshida (KAKEN, Mito, Japan) for his technical support.
REFERENCES
- 1.Attieh J M, Hanson A D, Saini H S. Purification and characterization of a novel methyltransferase responsible for biosynthesis of halomethanes and methanethiol in Brassica oleracea. J Biol Chem. 1995;270:9250–9257. doi: 10.1074/jbc.270.16.9250. [DOI] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Brauer F P, Strebin R S., Jr . Environmental migration of long-lived radionuclides. Proceedings of an International Symposium. Vienna, Austria: International Atomic Energy Agency; 1982. Environmental concentration and migration of 129I; pp. 465–480. [Google Scholar]
- 4.Chameides W L, Davis D D. Iodine: its possible role in tropospheric photochemistry. J Geophys Res. 1980;85:7383–7398. [Google Scholar]
- 5.Davis D, Crawford J, Liu S, McKeen S, Brandy A, Thornton D, Rowland F, Blake D. Potential impact of iodine on tropospheric levels of ozone and other critical oxidants. J Geophys Res. 1996;101:2135–2147. [Google Scholar]
- 6.Fuge R, Johnson C C. The geochemistry of iodine. Environ Geochem Health. 1986;8:31–54. doi: 10.1007/BF02311063. [DOI] [PubMed] [Google Scholar]
- 7.Giese B, Laturnus F, Adams F C, Wiencke C. Release of volatile iodinated C1–C4 hydrocarbons by marine macroalgae from various climate zones. Environ Sci Technol. 1999;33:2432–2439. [Google Scholar]
- 8.Harper D B. Halomethane from halide ion-a highly efficient fungal conversion of environmental significance. Nature. 1985;315:55–57. [Google Scholar]
- 9.Harper D B, Kennedy J T. Effect of growth conditions on halomethane production by Phellinus species: biological and environmental implications. J Gen Microbiol. 1986;132:1231–1246. [Google Scholar]
- 10.Itoh N. Volatile halogenated compounds from marine algae; their formation mechanisms and geochemical aspects. Recent Res Dev Phytochem. 1997;1:309–327. [Google Scholar]
- 11.Itoh N, Tsujita M, Ando T, Hisatomi G, Higashi T. Formation and emission of monohalomethanes from marine algae. Phytochemistry. 1997;45:67–73. [Google Scholar]
- 12.Liss P S, Slater P G. Flux of gases across the air-sea interface. Nature. 1974;247:181–184. [Google Scholar]
- 13.Lovelock J E. Natural halocarbons in the air and in the sea. Nature. 1975;256:193–194. doi: 10.1038/256193a0. [DOI] [PubMed] [Google Scholar]
- 14.Lovelock J E, Maggs R J, Wade R J. Halogenated hydrocarbons in and over the Atlantic. Nature. 1973;241:194–196. [Google Scholar]
- 15.Manley S L, de la Cuesta J L. Methyl iodide production from marine phytoplankton cultures. Limnol Oceanogr. 1997;42:142–147. [Google Scholar]
- 16.Manley S L, Goodwin K, North W J. Laboratory production of bromoform, methylene bromide, and methyl iodide by macroalgae and distribution in nearshore southern California waters. Limnol Oceanogr. 1992;37:1652–1659. [Google Scholar]
- 17.Manley S L, Dastoor M N. Methyl iodide (CH3I) production by kelp and associated microbes. Mar Biol. 1988;98:477–482. [Google Scholar]
- 18.Miyake Y, Tsunogai S. Evaporation of iodine from the ocean. J Geophys Res. 1963;68:3989–3993. [Google Scholar]
- 19.Moore R M, Geen C E, Tait V K. Determination of Henry's law constants for a suite of naturally occurring halogenated methanes in seawater. Chemosphere. 1995;30:1183–1191. [Google Scholar]
- 20.Moore R M, Tokarczyk R. Volatile biogenic halocarbons in the northwest Atlantic. Global Biogeochem Cycles. 1993;7:195–210. [Google Scholar]
- 21.Moore R M, Groszko W. Methyl iodide distribution in the ocean and fluxes to the atmosphere. J Geophys Res. 1999;104:11163–11171. [Google Scholar]
- 22.Muramatsu Y, Yoshida S. Volatilization of methyl iodide from the soil-plant system. Atmos Environ. 1995;29:21–25. [Google Scholar]
- 23.Muramatsu Y, Yoshida S, Uchida S, Hasebe A. Iodine desorption from rice paddy soil. Water Air Soil Pollut. 1996;86:359–371. [Google Scholar]
- 24.Muramatsu Y, Ohmomo Y. Iodine-129 and iodine-127 in environmental samples collected from Tokaimura/Ibaraki, Japan. Sci Total Environ. 1986;48:33–43. doi: 10.1016/0048-9697(87)90207-5. [DOI] [PubMed] [Google Scholar]
- 25.Ni X, Harger L P. cDNA cloning of Batis maritima methyl chloride transferase and purification of the enzyme. Proc Natl Acad Sci USA. 1998;95:12866–12871. doi: 10.1073/pnas.95.22.12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nightingale P D, Malin G, Liss P S. Production of chloroform and other low-molecular-weight halocarbons by some species of macroalgae. Limnol Oceanogr. 1995;40:680–689. [Google Scholar]
- 27.Oram D E, Penkett S A. Observation in eastern England of elevated methyl iodide concentration in air of Atlantic origin. Atmos Environ. 1994;28:1159–1174. [Google Scholar]
- 28.Rasmussen R A, Khalil M A K, Gunawardena R, Hoyt S D. Atmospheric methyl iodide (CH3I) J Geophys Res. 1982;87:3086–3090. [Google Scholar]
- 29.Redeker K R, Wang N-Y, Low J C, McMillan A, Tyler S C, Cicerone R J. Emission of methyl halides and methane from rice paddies. Science. 2000;290:966–969. doi: 10.1126/science.290.5493.966. [DOI] [PubMed] [Google Scholar]
- 30.Saini H S, Attieh J M, Hanson A D. Biosynthesis of halomethanes and methanethiol by higher plants via a novel methyltransferase reaction. Plant Cell Environ. 1995;18:1027–1033. [Google Scholar]
- 31.Saxena D, Aouad S, Attieh J, Saini H S. Biochemical characterization of chloromethane emission from the wood-rotting fungus Phellinus pomaceus. Appl Environ Microbiol. 1998;64:2831–2835. doi: 10.1128/aem.64.8.2831-2835.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh H B, Salas L J, Stiles R E. Methyl halides in and over the eastern Pacific (40°N–32°S) J Geophys Res. 1983;88:3684–3690. [Google Scholar]
- 33.Solomon S, Garcia R R, Ravishankara A R. On the role of iodine in ozone depletion. J Geophys Res. 1994;99:20491–20499. [Google Scholar]
- 34.Whitehead D C. The distribution and transformations of iodine in the environment. Environ Int. 1984;10:321–339. [Google Scholar]
- 35.Whittenbury R, Phillips K C, Wilkinson J F. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol. 1970;61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 36.Wong G T F. The marine geochemistry of iodine. Rev Aquat Sci. 1991;4:45–73. [Google Scholar]
- 37.Wuosmaa A M, Hager L P. Methyl chloride transferase: a carbocation route for biosynthesis of halometabolites. Science. 1990;249:160–162. doi: 10.1126/science.2371563. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida S, Muramatsu Y, Uchida S. Studies on the sorption of I− (iodide) and IO3− (iodate) onto andosols. Water Air Soil Pollut. 1992;63:321–329. [Google Scholar]