Abstract
Small‐for‐size syndrome (SFSS) is a common complication following partial liver transplantation and extended hepatectomy. SFSS is characterized by postoperative liver dysfunction caused by insufficient regenerative capacity and portal hyperperfusion and is more frequent in patients with preexisting liver disease. We explored the effect of the Mesenchymal‐epithelial transition factor (MET)‐agonistic antibody 71D6 on liver regeneration and functional recovery in a mouse model of SFSS. Male C57/BL6 mice were exposed to repeated carbon tetrachloride injections for 10 weeks and then randomized into 2 arms receiving 3 mg/kg 71D6 or a control immunoglobulin G (IgG). At 2 days after the randomization, the mice were subjected to 70% hepatectomy. Mouse survival was recorded up to 28 days after hepatectomy. Satellite animals were euthanized at different time points to analyze liver regeneration, fibrosis, and inflammation. Serum 71D6 administration significantly decreased mouse mortality consequent to insufficient regeneration of the cirrhotic liver. Analysis of liver specimens in satellite animals revealed that 71D6 promoted powerful activation of the extracellular signal‐regulated kinase pathway and accelerated liver regeneration, characterized by increased liver‐to‐body weight, augmented mitotic index, and higher serum albumin levels. Moreover, 71D6 accelerated the resolution of hepatic fibrosis as measured by picrosirius red, desmin, and α–smooth muscle actin staining, and suppressed liver infiltration by macrophages as measured by CD68 and F4/80 staining. Analysis of gene expression by reverse‐transcription polymerase chain reaction confirmed that 71D6 administration suppressed the expression of key profibrotic genes, including platelet‐derived growth factor, tissue inhibitor of metalloproteinase 3, and transforming growth factor‐β1, and of key proinflammatory genes, including tumor necrosis factor‐α, interleukin‐1β, chemokine (C‐C motif) ligand 3, and chemokine (C‐C motif) ligand 5. These results suggest that activating the MET pathway via an hepatocyte growth factor–mimetic antibody may be beneficial in patients with SFSS and possibly other types of acute and chronic liver disorders.
Short abstract
Abbreviations
- agomAbs
agonistic monoclonal antibodies
- α‐SMA
α–smooth muscle actin
- a.u.
arbitrary units
- CCL3
chemokine (C‐C motif) ligand 3
- CCl4
carbon tetrachloride
- CCL5
chemokine (C‐C motif) ligand 5
- ELISA
enzyme‐linked immunosorbent assay
- ERK
extracellular signal‐regulated kinase
- H & E
hematoxylin‐eosin
- HGF
hepatocyte growth factor
- hMET
human mesenchymal‐epithelial transition factor
- HSCs
hepatic stellate cells
- IgG
immunoglobulin G
- IgG1
immunoglobulin G1
- IL‐1β
interleukin‐1β
- IP
intraperitoneally
- IV
intravenously
- mMET
mouse mesenchymal‐epithelial transition factor
- mRNA
messenger RNA
- N/A
Not applicable
- PCNA
proliferating cell nuclear antigen
- PDGF
platelet‐derived growth factor
- PSR
picrosirius red
- qRT‐PCR
quantitative reverse‐transcription polymerase chain reaction
- RT‐PCR
reverse‐transcription polymerase chain reaction
- SD
standard deviation
- SFSS
small‐for‐size syndrome
- TIMP3
tissue inhibitor of metalloproteinase 3
- TNF‐ α
tumor necrosis factor‐α
Small‐for‐size syndrome (SFSS) is a common yet underrecognized complication following partial liver transplantation and extended hepatectomy.( 1 ) It is characterized by postoperative liver dysfunction caused by insufficient regenerative capacity and portal hyperperfusion and is more frequent in patients with preexisting liver disease.( 2 ) Approximately one‐third of liver transplant recipients who develop early graft failure qualify for SFSS. Small‐for‐size liver grafts show delayed and impaired regeneration( 3 ) and have greater risks of failure including microcirculatory damage, inflammatory injury, and accelerated acute rejection, leading to liver failure with associated coagulopathy, ascites, prolonged cholestasis, and encephalopathy. Because of the persistent organ shortage, living donor liver transplantation is becoming the most viable option for patients with end‐stage liver disease. Donor safety always comes first in living donor liver transplantation, and there is a growing momentum for the increased use of small‐for‐size grafts in association with hepato‐regenerative therapies.( 4 , 5 )
Hepatocyte growth factor (HGF) is a pleiotropic cytokine of mesenchymal origin that plays a key role in organ regeneration.( 6 ) Its high‐affinity receptor, the MET tyrosine kinase, is mainly expressed by epithelial and endothelial cells, but it is also present on some immune cells as well as in various types of myofibroblasts.( 7 ) In the liver, HGF is typically secreted by hepatic stellate cells (HSCs) and plays a key role in hepatic regeneration. Following injury, increased HGF secretion initiates a repair program that limits cell damage, ensures hepatocyte regeneration, inhibits myofibroblast hyperproliferation, and suppresses inflammation, restoring liver function.( 8 )
Despite the broad therapeutic potential of HGF in liver diseases, its translation to the clinic has been challenging. In fact, HGF does not display ideal drug‐like properties: its very short plasma half‐life (a few minutes) causes an unfavorable pharmacokinetics because of its high avidity for the extracellular matrix, it has a poor biodistribution; it needs proteolytic activation to acquire biological activity, and once activated it is unstable; lastly, its industrial manufacture is difficult and costly. To overcome the limitations of HGF and to generate a drug that could effectively promote liver regeneration in patients, we generated a series of anti‐MET agonistic monoclonal antibodies (“agomAbs”) that bind to MET at high affinity and determine MET activation, mimicking the biochemical and biological activity of HGF. AgomAbs combine the powerful therapeutic potential of HGF with the excellent pharmacokinetic, pharmacodynamic, and manufacturing properties of antibodies.
In this study, we explored the therapeutic potential of 71D6, a fully agonistic anti‐MET antibody that cross‐reacts with rodent, nonhuman primate, and human MET in a mouse model of SFSS.
Materials and Methods
Serum 71D6 Antibody Generation and Characterization
Serum 71D6 was generated by immunization of Llama glama using the SIMPLE antibody platform.( 9 ) A detailed description of its generation and characterization has been published previously.( 10 ) Binding of 71D6 to MET and HGF‐mediated and 71D6‐mediated MET autophosphorylation and analysis of MET downstream signaling were performed as described.( 10 )
Animal Model
Mouse procedures were authorized by the National Research Institute for Child Health and Development (permission no. A2014‐010‐C06). The 8‐week‐old male C57BL/6JJmsSLc mice (Shizuoka Laboratory Animal Center, Shizuoka, Japan) were subjected to repeated subcutaneous injection of 10% carbon tetrachloride (CCl4; Wako) dissolved in olive oil (100 μL/mouse) twice a week for 10 weeks. On the day of the last CCl4 administration, the mice were randomized into 2 arms receiving 3 mg/kg of 71D6 or a control antibody against the F glycoprotein of respiratory syncytial virus( 11 ) (both in the mouse immunoglobulin G1 [IgG1] format). At 2 days after randomization, all of the mice were subjected to 70% hepatectomy by removal of the anterior 2 lobes and posterior left lobe. The 71D6 or control IgG were administered intraperitoneally (IP) at a dose of 3 mg/kg 2 days before hepatectomy and at days 0, 2, 4, 6, 8, 10, and 12. Mice recruited in the trial included main study animals and satellite animals. The spontaneous survival rate of the main study animals was recorded from days 0 to 28. Satellite animals (3 mice per group) were euthanized at 3, 7, and 28 days after hepatectomy. Serum and liver samples were stored at −80℃.
Serum Biochemical Measurements
Serum was collected from whole‐blood samples after standing for 30 minutes at 37℃ and then centrifuged at 1800g for 25 minutes at 4℃. Serum samples were then analyzed for serum albumin concentrations using a commercially available kit (Fujifilm) and an automatic biochemical analyzer (DRI‐CHEM 3500i; Fujifilm) according to the manufacturer’s instructions. Serum HGF concentration was measured using the mouse/rat HGF Quantikine enzyme‐linked immunosorbent assay (ELISA) kit (R&D Systems). Serum 71D6 concentration was determined by ELISA as described.( 10 )
Histopathological and Immunohistochemical Examination
Both the liver portion extracted at the time of hepatectomy and that collected at autopsy were processed for histopathological examination. Liver tissues were fixed in 10% formalin for 48 hours, routinely processed, and sliced into sections of 4 μm in thickness. For detection of liver fibrosis, sections were stained with picrosirius red (PSR; Sigma‐Aldrich), anti–α‐smooth muscle actin (α‐SMA) antibodies (AbCam) and anti‐desmin antibodies (Boehringer). For detection of macrophage infiltration, sections were stained with antibodies against CD68 (AbCam) and F4/80 (AbCam). For detection of liver proliferation, sections were stained with anti–proliferating cell nuclear antigen (PCNA) antibodies (Dako). Sections were also stained with hematoxylin‐eosin (H & E) and periodic acid–Schiff (both from Sigma‐Aldrich). After staining, specimens were photographed under a microscope (Olympus). Histological and immunohistochemical results were quantified using WinRoof 7.4 software (Mitani Corporation).
Total Messenger RNA Preparation and Quantitative Reverse‐Transcription Polymerase Chain Reaction Analysis
Total messenger RNA (mRNA) was extracted from frozen liver tissues using RNeasy Mini Kit (Qiagen). Each 0.8 µg aliquot of mRNA was reverse transcribed to complementary DNA using a Prime Script reverse‐transcription (RT) reagent kit (RR037A; Takara). Quantitative reverse‐transcription polymerase chain reaction (qRT‐PCR) was performed using the SYBR Green system or the primer/probe set system (primer sequences are listed in Table 1) the Applied Biosystem PRISM7900 apparatus (Thermo Fisher Scientific) is a machine which is used to do RT‐PCR. The PCR cycle conditions for the SYBR Green system were 95℃ for 3 minutes, 45 cycles of 95℃ for 3 seconds, and 60℃ for 30 seconds. The PCR cycle conditions for the primer/probe set system were 50℃ for 2 minutes, 95℃ for 15 minutes, 40 cycles of 95℃ for 30 seconds, 60℃ for 1 minute, and 25℃ for 2 minutes. The comparative threshold cycle (ΔΔCt) method was used for determining relative gene expression, and the results of target genes (including fibrosis‐related genes and inflammation‐related genes) were normalized by subtracting 18S expression values.
TABLE 1.
Primers and Probes Used in This Study
| Genes (PCR) | Forward (5′–3′) | Reverse (5′–3′) | Probe |
|---|---|---|---|
| TIMP3 (SYBR Green) | CACAAAGTTGCACAGTCCTG | TTTGTCGCCTCAAGCTAGA | N/A |
| PDGF (SYBR Green) | TACAGTTGCACTCCCAGGAAT | CTTCCAGTTGACAGTTCCGCA | N/A |
| TGF‐β (SYBR Green) | ATCCTGTCCAAACTAAGGCTCG | ACCTCTTTAGCATAGTAGTCCGC | N/A |
| TNF‐α (SYBR Green) | AAGCCTGTAGCCCACGTCGTA | GGCACCACTAGTTGGTTGTCTTTG | N/A |
| CCL5 (SYBR Green) | TGCCCTCACCATCATCCTCACT | GGCGGTTCCTTCGAGTGACA | N/A |
| IL‐1β (SYBR Green) | ACCTTCCAGGATGAGGACATGA | AACGTCACACACCAGCAGGTTA | N/A |
| 18S (SYBR Green) | ATGAGTCCACTTTAAATCCTTTAACGA | CTTTAATATACGCTATTGGAGCTGGAA | N/A |
| CCL3 (Taqman) | ACCCAGGTCTCTTTGGAGTCAGCGCA | TCCCAGCCAGGTGTCATTTTC | AGGCATTCAGTTCCAGGTCAG |
| 18S (Taqman) | ATCCATTGGAGGGCAAGTCTGGTGC | ATGAGTCCACTTTAAATCCTTTAACGA | CTTTAATATACGCTATTGGAGGCTGGAA |
Liver‐specific expression of profibrotic and proinflammatory genes determined by SYBR green PCR or Taqman PCR as indicated in the table. N/A, Not applicable.
In Vivo Analysis of Extracellular Signal‐Regulated Kinase Activation
Frozen liver tissue was homogenized in radio immunoprecipitation assay buffer containing 1% protease inhibitor cocktail‐1 and 1% protease inhibitor cocktail‐2 (Sigma‐Aldrich) followed by centrifugation in a microfuge at top speed for 30 minutes. Protein concentrations were assayed using a Protein Assay kit (Bio‐Rad). Samples were separated by electrophoresis on 10% polyacrylamide gels and transferred to Immobilon (Bio‐Rad) polyvinylidene fluoride. After brief incubation with 5% nonfat milk to block nonspecific binding, membranes were exposed overnight at 4℃ to specific phosphorylated anti‐p44/p42 extracellular signal‐regulated kinase (ERK) antibodies (Cell Signaling Technology). Membranes were washed and exposed to alkaline phosphatase‐conjugated secondary antibodies and visualized by incubation in 5% nonfat milk. Phosphorylated p44/p42 ERK activity was quantified by laser densitometric analysis of the radiographic film using ImageJ software (National Institutes of Health, Bethesda, MD).
Statistical Analysis
Prism7 software (GraphPad, San Diego, CA) was used to calculate statistical significance. A 2‐way analysis of variance method and Student t test method were used for comparisons between groups. Survival rate analysis was performed using a log‐rank (Cox‐Mantel) test. Data are expressed as mean ± standard deviation (SD). A value of P < 0.05 was considered to be statistically significant.
Results
Serum 71D6 Binds to MET at High Affinity and Promotes MET Activation, Mimicking HGF
HGF‐mimetic, agonistic anti‐MET antibodies were generated by immunization of L. glama using the SIMPLE antibody platform.( 9 ) Their biochemical and biological characterizations have been published previously.( 10 ) Among these molecules, which include both partial and full agonists of MET, serum 71D6 represents the most potent fully agonistic antibody. Serum 71D6 was produced as a chimera between variable llama regions and human or mouse IgG1/λ constant regions. Serum 71D6 bound to either the human MET (hMET) or mouse MET (mMET) extracellular domain with the concentration for 50% of maximal effect in the picomolar range (Fig. 1A). Stimulation of immortalized mouse liver cells with 71D6 or HGF resulted in a similar dose‐dependent activation of MET and of its downstream signaling (Fig. 1B). An overlapping pattern of MET activation and signaling was observed in MET‐expressing human epithelial cells of various origin (not shown).
FIG. 1.
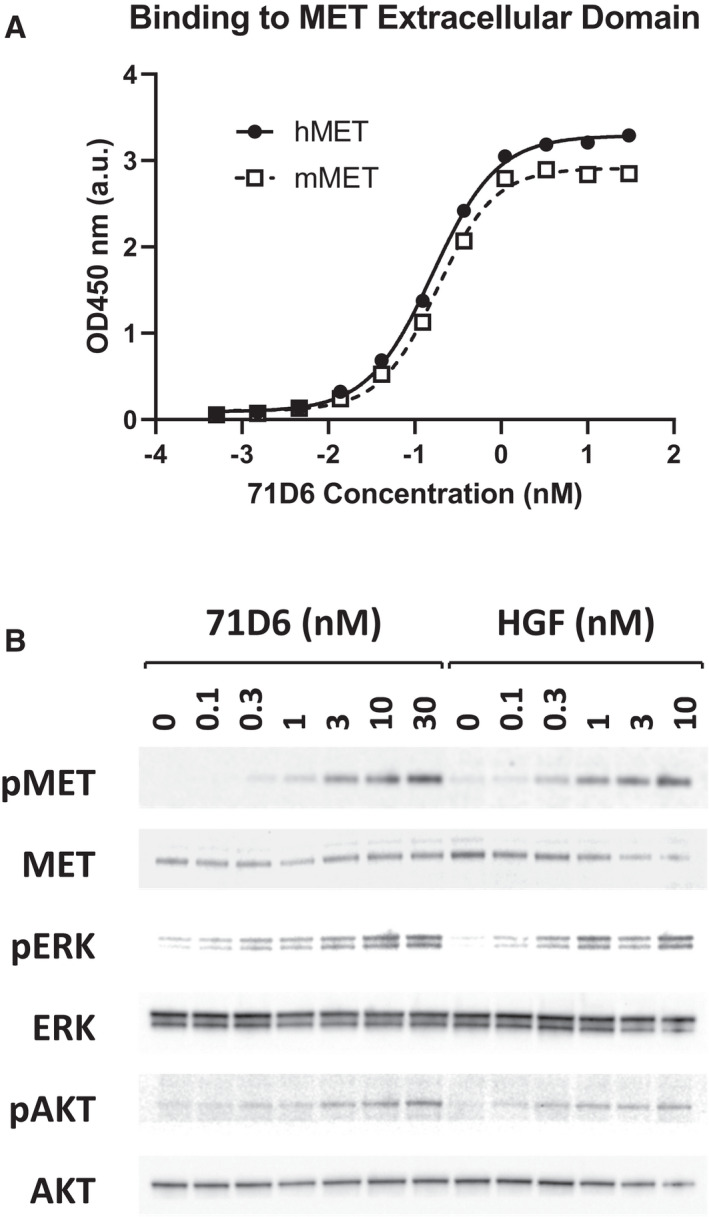
71D6 binds to MET at high affinity and elicits MET activation and downstream signaling, mimicking HGF function. (A) 71D6 binding to hMET or mMET extracellular domain was analyzed by ELISA (a.u.). (B) MET activation and downstream signaling induced by HGF or 71D6 was studied in human mouse MLP29 liver precursor cells. Cell lysates were analyzed by Western blotting using antibodies specific for the phosphorylated forms of MET, ERK, and AKT (pMET, pERK and pAKT) as well as antibodies against total MET, ERK, and AKT.
Systemic 71D6 Administration Results in Biologically Significant Plasma Levels
The pharmacokinetic properties of 71D6 were tested in various mouse strains using various routes of administration. In all studies, increasing doses of the antibody were delivered as a single bolus, and antibody levels in plasma were determined at different time points using a MET‐based ELISA assay. Table 2 shows the results obtained in a representative study conducted by IP injection. Serum 71D6 concentration reached a peak 8 hours after injection (65 nM at 1 mg/kg and 2638 nM at 30 mg/kg). After 2 days, plasma 71D6 levels showed only a minor deflection (38 nM at 1 mg/kg and 2466 nM at 30 mg/kg). After 8 days, all dose levels except the lowest were still detectable and well in the nanomolar range. Based on these data, the mean plasma half‐life of 71D6 corresponded to approximately 5 days (note that recombinant HGF has a half‐life of a few minutes in both rodents( 12 ) and humans).( 13 ) Considering that in normal, healthy mice endogenous HGF plasma levels range from 1 to 10 pM( 14 ) and that in the MET phosphorylation assays shown in Fig. 1B both HGF and 71D6 reached saturation at about 10 nM, the concentrations reached following IP injections of 71D6 shown in Table 2 are certainly relevant from a biologic viewpoint.
TABLE 2.
Pharmacokinetic Properties of 71D6
| Dose (mg/kg) | Maximum Concentration (nM) | Day 2 Concentration (nM) | Day 8 Concentration (nM) |
|---|---|---|---|
| 1 | 65 ± 3 | 38 ± 6 | 0 ± 0 |
| 3 | 271 ± 56 | 249 ± 70 | 17 ± 12 |
| 10 | 1265 ± 270 | 1062 ± 114 | 257 ± 27 |
| 30 | 2638 ± 327 | 2466 ± 494 | 446 ± 45 |
71D6 concentration in plasma was determined at different time points following IP injection of different dose levels of antibody as a single bolus. Values represent the mean ± standard deviation of at least 3 biological replicates.
Serum 71D6 Displays a Potent Hepatotrophic Activity in Mice
The biological effects of 71D6 administration were compared with those of recombinant HGF in vivo. In a first experiment, a single bolus of 1 mg/kg of either 71D6 or HGF was administered intravenously (IV) to adult Balb‐c mice. Liver‐to‐body weight ratio and serum albumin levels were measured 4 and 10 days later. As shown in Fig. 2A, 71D6 administration resulted in a marked increase in liver weight both at day 4 (91%) and at day 10 (42%). In contrast, recombinant HGF administration promoted only a minor increase in liver weight (20% at day 4 and 0% at day 10). Serum albumin levels, expression of the synthetic activity of the liver, were invariably higher in the 71D6‐treated mice, confirming the superior potency of the antibody. In a second experiment, we aimed at compensating the shorter half‐life of HGF with more frequent administration. Because of the challenge of IV injecting the same animals multiple times, IP injection was preferred. Mice were injected IP with either a single bolus of 1 mg/kg 71D6 or with 1 mg/kg recombinant HGF every 12 hours for 5 days. Mice were euthanized at day 6 and subjected to the same analysis as noted previously. Even when administered more frequently, HGF could not match the potent hepatotrophic activity of 71D6. In fact, mice injected with a single bolus of 71D6 displayed a 70% larger liver compared with controls, whereas animals injected repeatedly with HGF showed a modest 10% increase in liver weight (Fig. 2B). Similarly, 71D6 injection resulted in 121% higher albumin levels compared with controls, whereas HGF injection increased serum albumin secretion by 52%. Therefore, 71D6 elicits a significantly more potent hepatotrophic effect in mice compared with recombinant HGF.
FIG. 2.
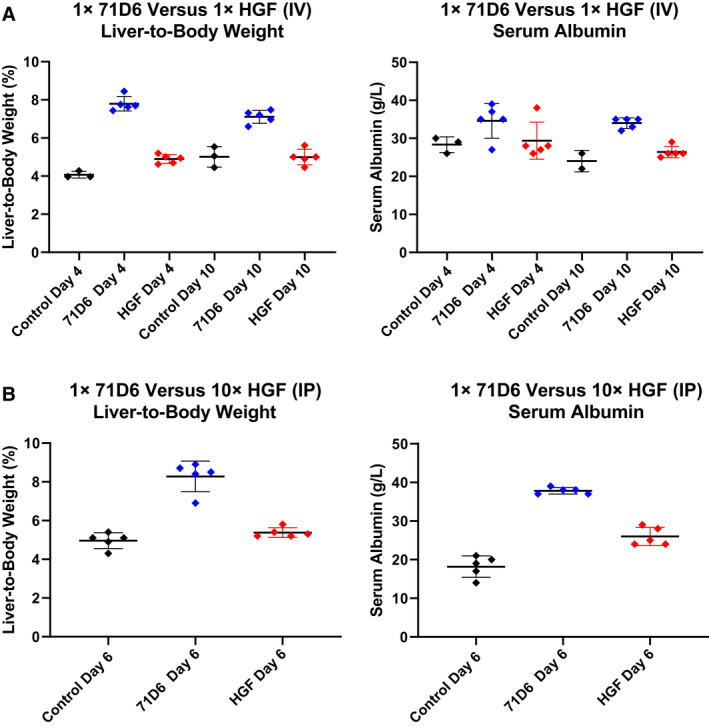
71D6 displays a potent hepatotrophic activity in mice. (A) A single 1 mg/kg bolus of 71D6 or human recombinant HGF was injected IV into adult Balb‐c mice, and the liver‐to‐body weight and serum albumin levels were determined 4 and 8 days afterward. (B) A single 1 mg/kg bolus of 71D6 or multiple doses (10 over 5 days) of 1 mg/kg recombinant human HGF were injected IP into adult Balb‐c mice. Liver‐to‐body weight and serum albumin levels were determined 6 days after the first injection. Black represents control group. Red represents HGF group. Blue represents 71D6 group.
Serum 71D6 Promotes Liver Regeneration and Increases Survival in Mice Undergoing CCl4 Injury and Partial Hepatectomy
Prompted by the previous results, we evaluated the therapeutic effect of 71D6 on liver regeneration in a mouse model of SFSS. This model reproduces the impaired regenerative capacity of the liver in patients with cirrhosis or other hepatic diseases. Mice were subjected to chronic CCl4 intoxication for 10 weeks and then randomized into 2 arms receiving either 3 mg/kg 71D6 or a control IgG1. At 3 days after randomization, all mice were subjected to 70% hepatectomy. Mouse survival was recorded up to 28 days after hepatectomy. Satellite animals were euthanized at different time points to analyze liver regeneration, fibrosis, and inflammation. This hepatectomy model typically presents high mortality within 4 days after surgery. Remarkably, the survival rate observed in the 71D6‐treated group was significantly higher than that observed in the control group (P = 0.03). Although only 5 of 24 mice (21%) in the control group survived for 28 days after hepatectomy, 7 of 13 71D6‐treated mice (54%) were alive at the end of the experiment (Fig. 3A). As shown in Fig. 3B, 71D6 also accelerated body weight recovery following surgery compared with controls (P < 0.001). Liver weight was also evaluated upon autopsy of satellite animals at various time intervals. As shown in Fig. 3C, 71D6 significantly increased the liver‐to‐body weight ratio at most time points analyzed (days 1 and 3, P < 0.05; day 7, nonsignificant; day 28, P < 0.001). Consistent with these findings, serum albumin levels in 71D6‐treated animals increased (Fig. 3D) and showed a significant difference with the control group starting on day 7 (P < 0.01) and reaching a peak on day 28 (P < 0.01). These data indicate that 71D6 treatment promotes liver regeneration and accelerates recovery of liver function after partial hepatectomy.
FIG. 3.
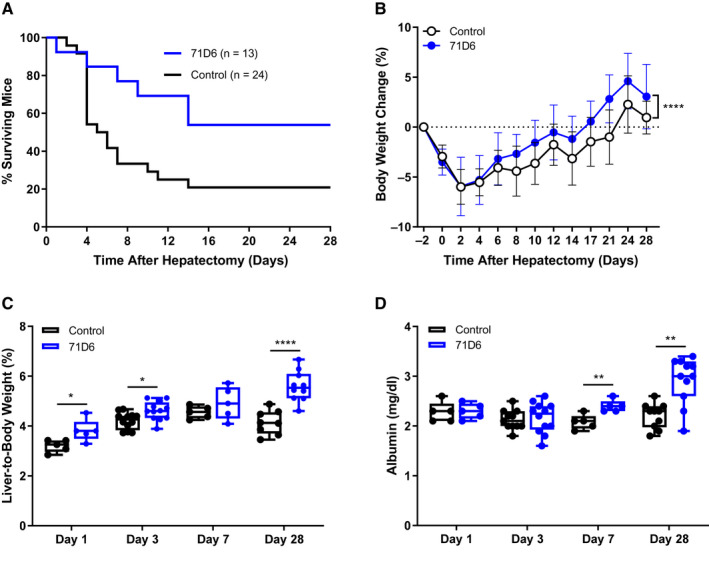
71D6 promotes liver regeneration and increases survival in mice undergoing CCl4 injury and partial hepatectomy. (A) Kaplan‐Meier curve analysis of mouse survival (P = 0.03). (B) Body weight change over time (mean ± SD) (P < 0.001). (C) Liver‐to‐body weight over time (days 1 and 3, P < 0.05; day 7, nonsignificant; day 28, P < 0.001). (D) Serum albumin levels over time (days 1 and 3, nonsignificant; day 7, P < 0.01; day 28, P < 0.01).
Serum 71D6 Promotes Hepatic Proliferation Following Hepatectomy Through Activation of the ERK Signaling Pathway
Hepatocyte proliferation following hepatectomy was assessed by staining liver sections with anti‐PCNA antibodies. As shown in Fig. 4A, the number of PCNA‐positive cells was higher in the 71D6‐treated group starting at day 3 and peaking at day 28. To further characterize 71D6‐induced liver proliferation, we analyzed the expression and activation of p44 ERK1 and p42 ERK2, a key event in the postinjury liver regeneration program.( 15 ) This analysis revealed that 71D6 significantly increased the levels of phosphorylated (activated) ERK1/2 on both day 3 and day 28 following hepatectomy (Fig. 4B). Activation of ERK1/2 directly correlated with PCNA expression, suggesting that 71D6 stimulates activation of ERK1/2 in the remnant liver, resulting in accelerated hepatic regeneration. Notably, however, no ERK1/2 activation was detected in control animals despite hepatectomy typically inducing spontaneous liver regeneration within a few days. This could be attributed to, and is consistent with, the impaired regenerative capacity of a cirrhotic liver. To cast light onto the mechanism underlying this regenerative impairment, we analyzed serum HGF levels in posthepatectomy mice as well as in healthy, naïve animals. This analysis revealed that posthepatectomy mice treated with the control IgG protein displayed only a marginal increase in HGF levels compared with naïve mice (Fig. 3C). Interestingly, serum HGF levels were slightly higher in the 71D6 group, probably because of larger liver mass and/or increased hepatic function. In any case, serum HGF levels were confined to the picomolar range, whereas 71D6 levels measured in the same samples remained well above MET saturating levels for the entire duration of the study (full activation of MET is reached with 10 nM 71D6). These results explain the absence of ERK activation and the poor liver regeneration observed in the control group on one hand and justify the superior regenerative ability of the animals treated with 71D6 on the other hand.
FIG. 4.
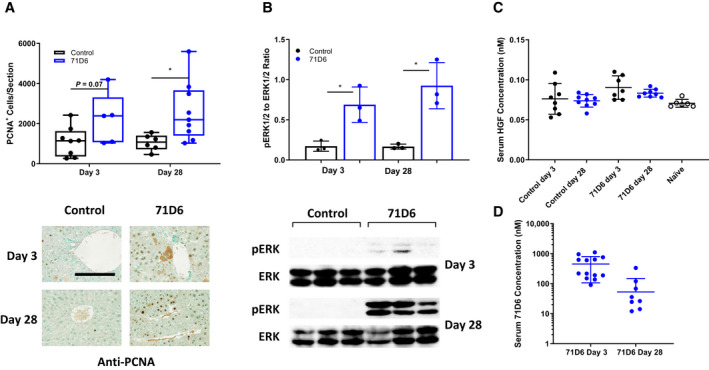
71D6 promotes hepatic proliferation following hepatectomy of cirrhotic mice through activation of the ERK signaling pathway. (A) Hepatic proliferation is expressed as the number of PCNA‐positive cells per liver section analyzed. Representative images are shown below the graph. The bar is 250 µM. (B) The levels of phosphorylated and total ERK1/2 (p44/p42) were determined by Western blot analysis of liver samples. The ratio between phosphorylated ERK1/2 and total ERK1/2 was determined by densitometric analysis. (C) Serum HGF levels in mice of the control and 71D6 groups as well as in healthy, naïve animals were determined by ELISA. (D) Serum 71D6 levels in mice of the 71D6 group were determined by ELISA.
Serum 71D6 Ameliorates Hepatic Fibrosis After Partial Hepatectomy on Cirrhotic Background
Sections of livers harvested from satellite animals were stained with H & E, PSR, and antibodies against desmin or α‐SMA. Liver specimens of the control group showed consistent formation of thin fibrotic septa at the portal and central areas on day 3. Although milder, fibrosis was still present at day 28. Control liver sections also displayed high density of myofibroblasts. In contrast, liver specimens of the 71D6 group showed significantly milder signs of fibrosis and a reduced presence of myofibroblasts already at day 3. Liver fibrosis was quantified and expressed as the percentage of PSR‐positive area (Fig. 5A), number of desmin‐positive cells (Fig. 5B), and percentage of α‐SMA–positive area (Fig. 5C). To further strengthen these results, the expression of fibrosis‐related genes was analyzed by quantitative RT‐PCR. This analysis revealed that 71D6 significantly reduced mRNA expression of platelet‐derived growth factor (PDGF), tissue inhibitor of metalloproteinase 3 (TIMP3), and transforming growth factor β1 (TGF‐β1), which are involved in collagen deposition and activation of HSCs (Fig. 5D). Together, these results suggest that in mice subjected to CCl4 treatment and partial hepatectomy, the agonistic anti‐MET antibody 71D6 effectively accelerates the regression of hepatic fibrosis by inhibiting the activation of multiple fibrogenic pathways.
FIG. 5.
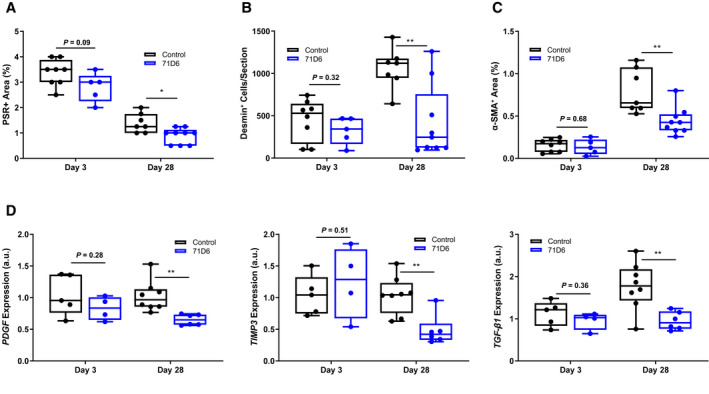
71D6 ameliorates hepatic fibrosis after partial hepatectomy on cirrhotic background. (A) Liver sections were stained with PSR. Data are expressed as percentage of PSR‐positive area. (B) Liver sections were stained with anti‐desmin antibodies. Data are expressed as number of desmin‐positive cells per section analyzed. (C) Liver sections were stained with anti–α‐SMA antibodies. Data are expressed as percentage of α‐SMA–positive area. (D) Liver specimens were analyzed by RT‐PCR to determine the levels of PDGF, TGF‐β1, and TIMP3 expression. Data are expressed as a.u.
Serum 71D6 Reduces Macrophage Infiltration in the Regenerating Liver
Liver specimens extracted at autopsy were analyzed by immunohistochemistry using antibodies against F4/80 and CD68. The number of cells positive for F4/80 (P < 0.05; Fig. 6A) and CD68 (P < 0.0001; Fig. 6B) was significantly lower in the 71D6 group compared with the control group on day 28. Consistent with histological findings, mRNA levels of inflammatory cytokine genes (tumor necrosis factor α [TNF‐α], interleukin 1β [IL1β], chemokine [C‐C motif] ligand 3 [CCL3], and chemokine [C‐C motif] ligand 5 [CCL5]) were significantly lower in the liver homogenates of 71D6‐treated mice compared with control mice on day 28 (P < 0.05; Fig. 6C).
FIG. 6.
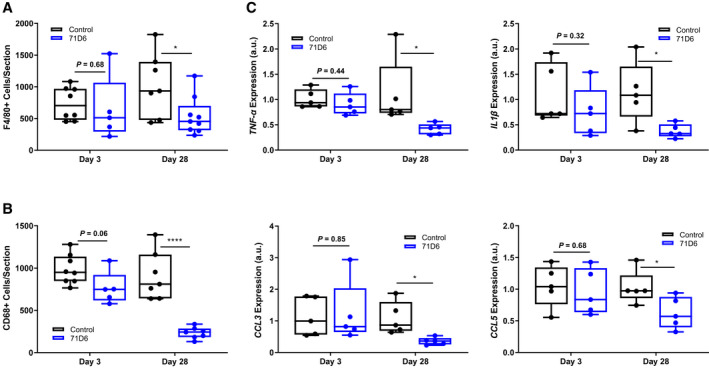
71D6 reduces macrophage infiltration in the regenerating liver. (A) Liver sections were stained with anti‐F4/80 antibodies. Data are expressed as number of F4/80‐positive cells per section analyzed. (B) Liver sections were stained with anti‐CD68 antibodies. Data are expressed as number of CD68‐positive cells per section analyzed. (C) Liver specimens were analyzed by RT‐PCR to determine the levels of TNF‐α, IL1β, CCL3, and CCL5 expression. Data are expressed as a.u.
Discussion
Liver failure is prone to occur after liver transplantation or extended resection when the size and function of the remnant liver is unable to meet the metabolic demand of the patient. SFSS limits the use of living donor and split‐liver transplants, and lifesaving large resections of tumors or nonmalignant lesions may be limited by concerns of postsurgical liver failure. Furthermore, SFSS is analogous to end‐stage liver disease where the functional liver mass no longer meets metabolic demand. Therefore, there is a major need for effective therapies capable of enhancing and accelerating liver regeneration.
Most studies evaluating regenerative therapies take advantage of the 70% hepatectomy model in mice or rats. In this model, 70% of the liver, usually the median and left lateral lobes, is surgically removed. In response to this, the remnant liver enlarges until it restores normal mass and function. Although the liver has a remarkable potential for regeneration, this regenerative capacity becomes impaired with serious liver fibrosis.( 16 ) The transition of quiescent HSCs to activated, scar‐forming, myofibroblast‐like cells leads to excessive extracellular matrix synthesis.( 17 , 18 ) This abnormal scar formation in the liver has been shown to hold back hepatocyte proliferation. In the present study, we studied a newly developed agonistic anti‐MET antibody that increased hepatocyte proliferation after partial hepatectomy in mice with CCl4‐induced liver disease.
Following 70% hepatectomy, both hepatocytes and nonparenchymal cells are activated and integrate multiple signals originating from immune, hormonal, and metabolic networks to induce liver regeneration.( 19 ) Within this process, the activation of HGF/MET signaling pathway has been demonstrated to be 1 of the essential mechanisms that lead hepatocytes into the cell cycle after hepatectomy.( 20 ) Following resection, HGF protein levels in plasma typically rise,( 21 ) but liver fibrosis is known to prevent this process. The lack of HGF induction on a cirrhotic background was fully confirmed in our study (Fig. 4C). To circumvent defective endogenous HGF activation, we employed the 71D6 agonistic anti‐MET antibody. When bound by 71D6, the MET receptor dimerizes and becomes phosphorylated on tyrosine residues to initiate MET downstream signaling (Fig. 1).
The results presented here indicate that liver‐to‐body weight ratio increased very slowly in the control group, whereas it increased markedly and constantly in the 71D6‐treated group (Fig. 3). Quantitative analysis of PCNA immunostaining also confirmed that the agonistic anti‐MET antibody potently promotes hepatocyte proliferation (Fig. 4A). We also clarified the involvement of the ERK1/2 signaling pathway in this hepato‐proliferative effect. It is known that the Ras/Raf/MEK/ERK cascade has the ability to lead to cellular responses, including proliferation. The ERK1/2 signaling pathway can also regulate the hepatocyte proliferative response during the regeneration of normal liver.( 22 ) Our Western blot results showed that 71D6 strongly activated ERK p44/p42, closely reflecting the high levels of PCNA expression (Fig. 4B). Our results clearly show that 71D6 has the ability to mimic HGF signaling in vivo, including activation of downstream kinases and promotion of hepatocyte proliferation. However, in contrast to HGF, 71D6 is very stable in vivo (Table 2) and displays superior pharmacodynamic properties (Fig. 2). These results highlight the therapeutic potential of 71D6 in liver regeneration.
Liver fibrosis is an inadequate wound‐healing response to chronic liver injury and is characterized by the excessive deposition and reduced degradation of the extracellular matrix. Excessive accumulation of extracellular matrix alters the hepatic architecture to progress to liver fibrosis, and if not prevented it may eventually lead to cirrhosis and even liver cancer. Our study provides evidence that 71D6 treatment leads to a significantly lower degree of collagen, desmin, and α‐SMA compared with controls (Fig. 5A,B). This is explained by the direct inhibition of multiple fibrogenic pathways including those controlled by PDGF, TIMP3, and TGF‐β1 (Fig. 5C). These data support the hypothesis that 71D6 has the ability to antagonize TGF‐β1 directly so as to reduce liver fibrosis and improve liver regeneration.( 23 )
Furthermore, our study showed that 71D6 also inhibited the infiltration of inflammatory cells into the liver. In fact, the number of F4/80‐positive and CD68‐positive cells in the 71D6 group was significantly lower than that observed in the control group (Fig. 6A,B). Consistent with these findings, qRT‐PCR analysis revealed that the expressions of proinflammatory cytokines and chemokines such as TNF‐α, IL1β, CCL3, and CCL5 were much lower in the liver homogenates of 71D6‐treated mice compared with control animals (Fig. 6C).
Thus, not only does 71D6 overcome the inability of a cirrhotic liver to regenerate following hepatectomy, but it also achieves faster resolution of fibrosis and the effective suppression of inflammation. Together, these results suggest that activating the MET pathway via an HGF‐mimetic antibody may be beneficial in patients with SFSS and possibly other types of acute and chronic liver disorders.
Acknowledgments
The authors thank Philippe Wiesel for critically reading the manuscript, Luca Rossi for in vivo work, Damiana Sattanino for assay setup, and the entire AgomAb team for continuous support.
This study was supported in part by the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grants‐in‐Aid 16K11064, 24/17H04277, and 18K08558), the National Center for Child Health and Development (Grant 29‐09), and Science and Technology Innovation Talents in Henan Universities (no. 19HASTIT003).
Manuela Cazzanti, Paolo Michieli, and Virginia Morello consult for, own stock in, and are employed by AgomAb Therapeutics NV.
Earn MOC for this article: www.wileyhealthlearning.com/aasld.aspx
SEE EDITORIAL ON PAGE 749
Contributor Information
Paolo Michieli, Email: paolo.michieli@unito.it.
Terumi Takahara, Email: terutaka-tym@umin.ac.jp.
Xiao‐Kang Li, Email: ri-k@ncchd.go.jp.
REFERENCES
- 1. Riddiough GE, Christophi C, Jones RM, Muralidharan V, Perini MV. A systematic review of small for size syndrome after major hepatectomy and liver transplantation. HPB 2020;22:487‐496. [DOI] [PubMed] [Google Scholar]
- 2. Masuda Y, Yoshizawa K, Ohno Y, Mita A, Shimizu A, Soejima Y. Small‐for‐size syndrome in liver transplantation: definition, pathophysiology and management. Hepatobiliary Pancreat Dis Int 2020;19:334‐341. [DOI] [PubMed] [Google Scholar]
- 3. Zhong Z, Schwabe RF, Kai Y, He L, Yang L, Bunzendahl H, et al. Liver regeneration is suppressed in small‐for‐size liver grafts after transplantation: involvement of c‐Jun N‐terminal kinase, cyclin D1, and defective energy supply. Transplantation 2006;82:241‐250. [DOI] [PubMed] [Google Scholar]
- 4. Greenbaum LE, Ukomadu C, Tchorz JS. Clinical translation of liver regeneration therapies: a conceptual road map. Biochem Pharmacol 2020;175:113847. [DOI] [PubMed] [Google Scholar]
- 5. Forbes SJ, Newsome PN. Liver regeneration—mechanisms and models to clinical application. Nat Rev Gastroenterol Hepatol 2016;13:473‐485. [DOI] [PubMed] [Google Scholar]
- 6. Nakamura T, Mizuno S. The discovery of hepatocyte growth factor (HGF) and its significance for cell biology, life sciences and clinical medicine. Proc Jpn Acad Ser B Phys Biol Sci 2010;86:588‐610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Matsumoto K, Funakoshi H, Takahashi H, Sakai K. HGF‐Met pathway in regeneration and drug discovery. Biomedicines 2014;2:275‐300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakamura T, Sakai K, Nakamura T, Matsumoto K. Hepatocyte growth factor twenty years on: much more than a growth factor. J Gastroenterol Hepatol 2011;26(suppl 1):188‐202. [DOI] [PubMed] [Google Scholar]
- 9. Klarenbeek A, Blanchetot C, Schragel G, Sadi AS, Ongenae N, Hemrika W, et al. Combining somatic mutations present in different in vivo affinity‐matured antibodies isolated from immunized Lama glama yields ultra‐potent antibody therapeutics. Protein Eng Des Sel 2016;29:123‐133. [DOI] [PubMed] [Google Scholar]
- 10. Michieli P. Anti‐Met antibodies and uses thereof. European patent application EP 3475302 A1 2019. https://www.lens.org/lens/patent/062‐815‐545‐734‐095. [Google Scholar]
- 11. Cingoz O. Motavizumab. mAbs 2009;1:439‐442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ido A, Moriuchi A, Kim I, Numata M, Nagata‐Tsubouchi Y, Hasuike S, et al. Pharmacokinetic study of recombinant human hepatocyte growth factor administered in a bolus intravenously or via portal vein. Hepatol Res 2004;30:175‐181. [DOI] [PubMed] [Google Scholar]
- 13. Ido A, Moriuchi A, Numata M, Murayama T, Teramukai S, Marusawa H, et al. Safety and pharmacokinetics of recombinant human hepatocyte growth factor (rh‐HGF) in patients with fulminant hepatitis: a phase I/II clinical trial, following preclinical studies to ensure safety. J Transl Med 2011;9:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xue F, Takahara T, Yata Y, Minemura M, Morioka CY, Takahara S, et al. Attenuated acute liver injury in mice by naked hepatocyte growth factor gene transfer into skeletal muscle with electroporation. Gut 2002;50:558‐562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tao Y, Wang M, Chen E, Tang H. Liver regeneration: analysis of the main relevant signaling molecules. Mediators Inflamm 2017;2017:4256352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chan A, Kow A, Hibi T, Di Benedetto F, Serrablo A. Liver resection in cirrhotic liver: are there any limits? Int J Surg 2020;82S:109‐114. [DOI] [PubMed] [Google Scholar]
- 17. Issa R, Zhou X, Trim N, Millward‐Sadler H, Krane S, Benyon C, Iredale J. Mutation in collagen‐1 that confers resistance to the action of collagenase results in failure of recovery from CCl4‐induced liver fibrosis, persistence of activated hepatic stellate cells, and diminished hepatocyte regeneration. FASEB J 2003;17:47‐49. [DOI] [PubMed] [Google Scholar]
- 18. Henderson NC, Forbes SJ. Hepatic fibrogenesis: from within and outwith. Toxicology 2008;254:130‐135. [DOI] [PubMed] [Google Scholar]
- 19. Yagi S, Hirata M, Miyachi Y, Uemoto S. Liver regeneration after hepatectomy and partial liver transplantation. Int J Mol Sci 2020;21:8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thorgeirsson SS. The central role of the c‐Met pathway in rebuilding the liver. Gut 2012;61:1105‐1106. [DOI] [PubMed] [Google Scholar]
- 21. Eguchi S, Yanaga K, Okudaira S, Sugiyama N, Miyamoto S, Furui J, Kanematsu T. Changes in serum levels of hepatocyte growth factor in patients undergoing adult‐to‐adult living‐donor liver transplantation. Transplantation 2003;76:1769‐1770. [DOI] [PubMed] [Google Scholar]
- 22. Chen XG, Xu CS, Liu YM. Involvement of ERK1/2 signaling in proliferation of eight liver cell types during hepatic regeneration in rats. Genet Mol Res 2013;12:665‐677. [DOI] [PubMed] [Google Scholar]
- 23. Fabregat I, Moreno‐Càceres J, Sánchez A, Dooley S, Dewidar B, Giannelli G, ten Dijke P. TGF‐β signalling and liver disease. FEBS J 2016;283:2219‐2232. [DOI] [PubMed] [Google Scholar]


