Abstract
Methylation of host‐cell deoxyribonucleic acid (DNA) has been proposed as a promising biomarker for triage of high‐risk (hr) human papillomavirus (HPV) positive women at screening. Our study aims to validate recently identified host‐cell DNA methylation markers for triage in an hrHPV‐positive cohort derived from primary HPV‐based cervical screening in The Netherlands. Methylation markers ASCL1, LHX8, ST6GALNAC5, GHSR, ZIC1 and SST were evaluated relative to the ACTB reference gene by multiplex quantitative methylation‐specific PCR (qMSP) in clinician‐collected cervical samples (n = 715) from hrHPV‐positive women (age 29‐60 years), who were enrolled in the Dutch IMPROVE screening trial (NTR5078). Primary clinical end‐point was cervical intraepithelial neoplasia grade 3 (CIN3) or cancer (CIN3+). The single‐marker and bi‐marker methylation classifiers developed for CIN3 detection in a previous series of hrHPV‐positive clinician‐collected cervical samples were applied. The diagnostic accuracy was visualised using receiver operating characteristic (ROC) curves and assessed through area under the ROC curve (AUC). The performance of the methylation markers to detect CIN3+ was determined using the predefined threshold calibrated at 70% clinical specificity. Individual methylation makers showed good performance for CIN3+ detection, with highest AUC for ASCL1 (0.844) and LHX8 (0.830). Combined as a bi‐marker panel with predefined threshold, ASCL1/LHX8 yielded a CIN3+ sensitivity of 76.9% (70/91; 95% CI 68.3‐85.6%) at a specificity of 74.5% (465/624; 95% CI 71.1‐77.9%). In conclusion, our study shows that the individual host‐cell DNA methylation classifiers and the bi‐marker panel ASCL1/LHX8 have clinical utility for the detection of CIN3+ in hrHPV‐positive women invited for routine screening.
Keywords: cervical intraepithelial neoplasia, clinical performance, cytology, diagnostic accuracy, epigenetic, genotyping, primary HPV screening
What's new?
As cervical screening transitions from cytology to primary human papillomavirus (HPV) testing worldwide, effective triage tests are increasingly needed. Here, the authors report on the performance of host‐cell DNA methylation biomarkers ASCL1, LHX8, ST6GALNAC5, GHSR, ZIC1, and SST in an HPV‐positive cohort derived from primary HPV‐based screening in The Netherlands. All markers exhibited significant differences in methylation levels between cervical intraepithelial neoplasia grade 3 or worse (CIN3/CIN3+) and CIN1, CIN2, and women with normal histology. The robust triage performance for CIN3+ as compared to cytology and HPV16/18 genotyping highlights the potential of methylation biomarker‐based triage for HPV‐positive women.
Abbreviations
- ACTB
β‐actin
- AIS
adenocarcinoma in situ
- ASCL1
achaete‐scute family BHLH transcription factor 1
- ASC‐US
atypical squamous cells of undetermined significance
- AUC
area under the curve
- BMD
borderline or mild dyskaryosis
- CI
confidence interval
- CIN
cervical intraepithelial neoplasia
- CIS
carcinoma in situ
- CISOE‐A
composition, inflammation, squamous, other and endometrium, and endocervical cylindrical epithelium, and adequacy
- Cq
quantification cycle
- DNA
deoxyribonucleic acid
- GHSR
growth hormone secretagogue receptor
- H&E
haematoxylin‐eosin
- HPV
human papillomavirus
- hr
high risk
- ICC
intraclass coefficient
- LHX8
LIM homeobox 8
- LSIL
low‐grade squamous intraepithelial lesions
- N
group size
- n
number of
- NILM
negative for intraepithelial lesion or malignancy
- NPV
negative predictive value
- PALGA
nationwide network and registry of histo‐ and cytopathology in the Netherlands
- PPV
positive predictive value
- qMSP
quantitative methylation‐specific PCR
- ROC
receiver operating characteristic
- RR
relative risk
- SST
somatostatin
- ST6GALNAC5
ST6 N‐acetylgalactosaminide alpha‐2,6‐sialyltransferase 5
- ZIC1
zinc finger of the cerebellum family member 1
1. INTRODUCTION
Since a negative high‐risk (hr) human papillomavirus (HPV) test provides high reassurance against cervical cancer, many countries have switched from cytology to primary HPV‐based screening. 1 Women with a positive hrHPV test require additional triage testing as many hrHPV infections are transient and harmless without causing any pre‐malignancy. Cytology is widely used as a triage test, but a variety of molecular tests are presently under investigation as promising alternative, because they can offer an objective, non‐morphological modality. 2 , 3 , 4 Particularly, the analysis of deoxyribonucleic acid (DNA) hypermethylation of host‐cell genes involved in cervical carcinogenesis seems to have great potential. 5 Methylation levels of various genes show an increase with cervical intraepithelial neoplasia (CIN) grade and are almost universally high in cervical cancer. 6 Currently available data support the capability of DNA methylation tests for triage of hrHPV‐positive women in cervical screening. 6 , 7
Using epigenome‐wide screens, we recently identified new host‐cell DNA methylation markers that may hold promise for full‐molecular screening. 8 , 9 The utility of these methylation markers ASCL1, LHX8, ST6GALNAC5, GHSR, SST and ZIC1 for the detection of cervical intraepithelial neoplasia grade 3 (CIN3) and cancer (CIN3+) in clinician‐collected cervical samples of hrHPV‐positive women was verified in a large training series. The six methylation markers showed good triage performance, with the bi‐marker panel ASCL1/LHX8 detecting 89.1% of CIN3 and all 50 cervical cancers at a specificity of 70%, 10 providing an optimised sensitivity over reported methylation markers 6 that warrants further investigation.
Therefore, our study was set out to externally validate these methylation markers for the detection of CIN3+ in a large, independent hrHPV‐positive cohort derived from primary HPV‐based screening. We made use of cervical samples of hrHPV‐positive women participating in the IMPROVE study that was carried out within the organised population‐based screening programme in the Netherlands. To evaluate triage performance of the methylation markers in a routine cervical screening setting, CIN diagnosis by pathologists made in daily practice was used. In addition, centralised reviewed diagnosis was included to verify the accuracy of the methylation markers.
2. METHODS
2.1. Study population
This is a post hoc analysis of the IMPROVE trial (NTR5078), a randomised non‐inferiority trial, which was performed to evaluate the clinical accuracy of hrHPV testing on self‐collected and clinician‐collected samples within the setting of the Dutch cervical screening programme. The IMPROVE study was approved by the Dutch Ministry of Health Welfare, and Sport (2014/32). A detailed description of the trial has been published before. 11 In brief, 16 410 women were enrolled and randomised (1:1) to the intervention group (self‐sampling) and the control group (clinician‐based sampling). HrHPV‐positive women (n = 1 020) were re‐tested using the other collection method. In accordance with the current guidelines of the Dutch primary HPV screening programme, women with a positive hrHPV test were triaged by cytology. Cytology results were classified according to the reporting on composition, inflammation, squamous, other and endometrium, and endocervical cylindrical epithelium, and adequacy (CISOE‐A) used in the Netherlands. In the IMPROVE trial, hrHPV‐positive women with cytology triage result of borderline or mild dyskaryosis (BMD) or worse (>BMD) at baseline were immediately referred to the gynaecologist for colposcopy. HrHPV‐positive women with normal cytology (ie, negative for intraepithelial lesion of malignancy [NILM]) at baseline were advised to undergo repeat cytological testing after 6 months, and referred to colposcopy when repeat cytology was ≥BMD.
For our study, we included all women who had an hrHPV‐positive clinician‐collected cervical sample and provided consent for follow‐up research (n = 739; 72.5%). Data on cytology and hrHPV were retrieved from the study database. 11 For analysis, the CISOE‐A classification was translated into the Bethesda nomenclature. 12 HPV genotype was categorised as HPV16, HPV18 or other hrHPV types (ie, HPV31, ‐33, ‐35, ‐39, ‐45, ‐51, ‐52, ‐56, ‐58, ‐59, ‐66 and ‐68).
2.2. Histology
2.2.1. Original histology
The original histology results as reported during regular care were retrieved from the pathology laboratories through the nationwide network and registry of histo‐ and cytopathology in the Netherlands (PALGA). 13 Histology was categorised as normal (no dysplasia; CIN0), LSIL/CIN1, HSIL/CIN2, HSIL/CIN3 (further referred to as CIN1, CIN2 and CIN3, respectively) or invasive cervical cancer according to the latest WHO classifications. 14 Adenocarcinoma in situ (AIS) and carcinoma in situ (CIS) were classified as CIN3.
2.2.2. Revised histology
A revision diagnosis based on morphologic features combined with the interpretation of Ki‐67 and p16INK4A immunostainings was rendered centrally by an expert pathologist, blinded to the original diagnosis. For this, the original haematoxylin‐eosin (H&E)‐stained slides and formalin‐fixed paraffin‐embedded tissue blocks were retrieved from the local pathology laboratories. Ki‐67 and p16INK4A immunohistochemistry was performed as described before. 15 In case no tissue was left in the blocks or lesions had an uninterpretable immunohistochemical staining, the revision diagnosis was based solely on expert review of the original H&E‐stained slide.
2.3. Host‐cell DNA methylation analysis
DNA from cervical screening samples was isolated using the NucleoMag 96 (Macherey‐Nagel, Düren, Germany) and a Microlab Star robotic system (Hamilton, Gräfelfing, Germany) according to the recommendations of the manufacturer. DNA concentrations were quantified using a Qubit fluorometer (ThermoFisher Scientific, Waltham, MA). Sodium bisulphite treatment of DNA and multiplex qMSPs were performed as described previously for markers GHSR/SST/ZIC1 8 and ASCL1/LHX8/ST6GALNAC5. 9 Samples with a quantification cycle (Cq) value for ACTB above 30 in one or both qMSP assays were considered to have an inadequate sample quality and excluded from further analyses. Methylation levels were normalised to the reference gene ACTB using the Cq values (2−ΔCq × 100) to obtain ΔCq ratios. 16 All methylation testing was performed blinded for cyto‐ and histopathology outcomes.
2.4. Data and statistical analysis
All histology until 2 years after baseline was included. To calculate the agreement between original and revision diagnosis, the intraclass correlation coefficient (ICC) with 95% confidence intervals (95% CI) was used.
To assess differences in DNA methylation levels across disease categories, the Kruskal‐Wallis omnibus test was performed on square‐root transformed ΔCq ratios. After a significant result, post hoc testing was performed using the Wilcoxon rank‐sum test. Square‐root transformed ΔCq ratios were visualised in boxplots.
In an earlier study, single‐marker classifiers using univariable logistic regression and a multi‐marker classifier using a LASSO regression model, which yielded a bi‐marker panel consisting of ASCL1 and LHX8, were developed for triage of hrHPV‐positive women on clinician‐collected cervical samples. 10 All classifiers were trained to discriminate between CIN3 and controls. Predicted probabilities (value range 0‐1), representing the risk of underlying CIN3, were calculated and thresholds calibrated at 70% clinical specificity among hrHPV‐positive women, were defined for all classifiers. 16 In our study, these classifiers and corresponding thresholds are applied.
The diagnostic accuracy was evaluated and visualised by receiver operating characteristic (ROC) curves and evaluated by area under the curve (AUC). Estimates of cytology with threshold atypical squamous cells of undetermined significance (ASCUS, ie, threshold BMD) and HPV16/18 genotyping were included in the ROC curve.
Sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and relative risk (RR) were determined with Wald 95% CI for the following triage strategies: (I) cytology, (II) HPV16/18 genotyping, (III) methylation analysis, (IV) HPV16/18 genotyping combined with cytology and (V) HPV16/18 genotyping combined with methylation analysis. Strategy (I) was labelled positive if cytology was ≥ASCUS (ie, ≥BMD). Strategy (II) was labelled positive if HPV16/18 was present. Strategy (III) was labelled positive if predicted probability was above the predefined threshold of the ASCL1/LHX8 marker panel. Strategy (IV) was labelled positive if HPV16/18 was present or the cytology result was positive. Strategy (V) was labelled positive if HPV16/18 was present or the methylation result was positive.
Primary clinical end‐point was CIN3+. Additional analysis with CIN2 or worse (CIN2+) as endpoint was performed. Controls were defined as ≤CIN1, including women with no histology, normal histology and CIN1. All statistical analyses were performed using SPSS software for Windows (version 26.0; IBM SPSS Inc., Chicago, IL), STATA (version 14.1; StataCorp, College Station, TX) and R open source software version 3.6.1. 17
3. RESULTS
3.1. Study cohort
A total of 739 women who participated in the IMPROVE study had an hrHPV‐positive clinician‐collected cervical sample available for methylation analysis. Twenty‐four samples (24/739; 3.2%) with invalid results based on an ACTB Cq value above 30 were excluded, leaving 715 hrHPV‐positive women in the final analysis. Median age was 40.0 years (IQR 34‐49; range 29‐60). Genotyping results were available for 650/715 (84.9%) women, of whom 183/650 (25.6%) were positive for HPV16 and 62/650 (8.7%) for HPV18. Three‐hundred and sixteen (44.2%) women had abnormal cytology at baseline, of whom 81 (25.6%) women had CIN3+, including one adenocarcinoma and two squamous cell carcinomas, and 58 (18.4%) had CIN2. Three‐hundred and ninety‐nine (55.8%) women had normal cytology at baseline, of whom 359 (90.0%) underwent repeat cytology at 6 months. A total of 281 (78.3%) women had a normal repeat cytology result and 78 (21.7%) an abnormal repeat cytology result, of whom 10 (12.8%) women had CIN3 and 14 (17.9%) CIN2 (Figure 1).
FIGURE 1.
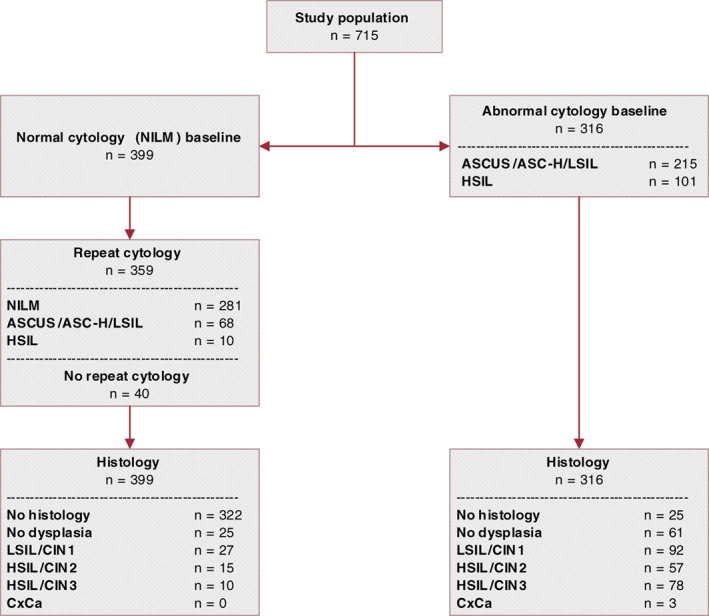
Flowchart of the study cohort. Low‐grade cytology: BMD equalling ASC‐US/ASC‐H/LSIL. High‐grade cytology: >BMD equalling HSIL. ASC‐H, atypical squamous cells cannot rule out high‐grade squamous intraepithelial lesion; ASC‐US, atypical squamous cells of undetermined significance; BMD, borderline or mild dyskaryosis; CIN, cervical intraepithelial neoplasia; HSIL, high‐grade squamous intraepithelial lesions; LSIL, low‐grade squamous intraepithelial lesions; n, number of; NILM, negative for intraepithelial lesion or malignancy [Color figure can be viewed at wileyonlinelibrary.com]
The data on revision diagnosis, available for 696 women (97.3%). Pathology review using biomarkers Ki‐67 and p16INK4A demonstrated an excellent correlation with the original diagnosis (ICC 0.942; 95% CI 0.933‐0.950%; Table S1). In the analyses presented below the original histology scores obtained during regular care are used; results using revision diagnosis are presented in Figures S1 and S2 and Tables S2 and S3.
3.2. Single marker classifiers
The methylation level distributions of ASCL1, LHX8, ST6GALNAC5, GHSR, SST and ZIC1 in hrHPV‐positive cervical screening samples across histological subgroups are shown in Figure 2, displaying an increase with the severity of the underlying cervical disease (all P‐values <3.8 × 10−10). All markers demonstrated a significant difference between methylation levels in CIN3 and subgroups no CIN (i.e., women with no histology or normal histology), CIN1 and CIN2 (all P‐values <.0001). The performance for CIN3+ was evaluated by ROC curves (Figure 3A) and quantified by AUCs ranging from 0.720 to 0.844. Two markers had AUC above 0.800, with highest AUC achieved by ASCL1 (AUC = 0.844), followed by LHX8 (AUC = 0.830).
FIGURE 2.
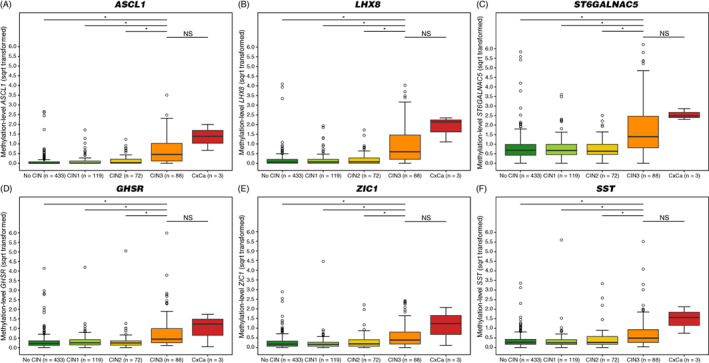
Methylation levels increase with severity of underlying cervical disease. DNA methylation levels of (A) ASCL1, (B) LHX8, (C) ST6GALNAC5, (D) GHSR, (E) ZIC1 and (F) SST represented by the square‐root transformed Cq‐ratios (y‐axis) in the different histology categories (x‐axis). Differences between histological categories upon Kruskal‐Wallis omnibus test, followed by post hoc testing using the Wilcoxon rank‐sum test: *P‐value <.05; **P‐value <.01; ***P‐value <.001; NS, not significant. (○) Outlier sample. CIN, cervical intraepithelial neoplasia; CxCa, cervical carcinoma; no CIN, women with no histology or normal histology [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 3.
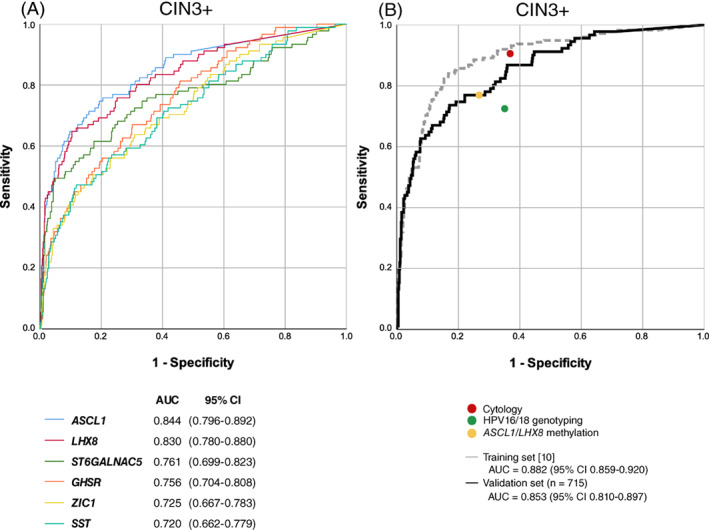
Diagnostic accuracy. ROC curves and corresponding AUC for CIN3+ detection for (A) single marker classifiers ASCL1, LHX8, ST6GALNAC5, GHSR, ZIC1 and SST and (B) bi‐marker panel ASCL1/LHX8 (black line), including point estimates of cytology (red circle), HPV16/18 genotyping (green circle) and bi‐marker panel ASCL1/LHX8 with the predefined threshold calibrated at 70% clinical specificity (yellow circle). For comparison, the cross‐validated ROC curve and corresponding AUC, as obtained in the hrHPV‐positive training series, are projected (grey line). 10 AUC, area under the curve; CIN, cervical intraepithelial neoplasia; ROC, receiver operating characteristic [Color figure can be viewed at wileyonlinelibrary.com]
3.3. Validation of the predefined bi‐marker panel ASCL1 / LHX8
The bi‐marker panel ASCL1/LHX8 was found most discriminative for CIN3 and cancer among hrHPV‐positive women in our earlier study. 10 The performance of this predefined panel for CIN3+ is depicted in Figure 3B, with inclusion of the point estimates for cytology and HPV16/18 genotyping. The bi‐marker panel achieved an AUC of 0.853 for CIN3+. At the predefined threshold 10 , the panel yielded a CIN3+ sensitivity of 76.9% (95% CI 68.3‐85.6%) and a specificity of 74.5% (95% CI 71.1‐77.9%; Table 1). All cervical carcinomas (n = 3) were classified as methylation positive. Cytology with threshold ASCUS had a CIN3+ sensitivity of 89.0% (95% CI 82.6‐95.4%) and specificity 62.3% (95% CI 58.5‐66.1%), and HPV16/18 genotyping a CIN3+ sensitivity 75.3% (95% CI 65.9‐84.7%) and specificity of 65.0% (95% CI 60.9‐69.1%). Combinations of HPV16/18 genotyping with cytology or methylation resulted in a CIN3+ sensitivity of 95.2% (95% CI 90.6‐99.8%) and 86.7% (95% CI 79.5‐94.0%), and a specificity of 43.7% (95% CI 39.7‐47.8%) and 53.3% (95% CI 49.2‐57.4%), respectively. The clinical performance for CIN2+ is shown in Table 1.
TABLE 1.
Sensitivity and specificity of baseline cytology, genotyping and ASCL1/LHX8 methylation for detection of CIN3+ and CIN2+
| Sensitivity | Specificity | PPV | NPV | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n/N | % | 95% CI | n/N | % | 95% CI | % | 95% CI | % | 95% CI | |
| (A) Cytology | ||||||||||
| CIN3+ | 81/91 | 89.0% | (82.6‐95.4%) | 389/624 | 62.3% | (58.5‐66.1%) | 25.6% | (20.8‐30.4%) | 97.5% | (96.0‐99.0%) |
| CIN2+ | 138/163 | 84.7% | (79.1‐90.2%) | 374/552 | 67.8% | (63.9‐71.7%) | 43.7% | (38.2‐49.1%) | 93.7% | (91.4‐96.1%) |
| (B) HPV16/18 genotyping | ||||||||||
| CIN3+ | 61/83 | 73.5% | (64.0‐83.0%) | 383/567 | 67.5% | (63.7‐71.4%) | 24.9% | (19.5‐30.3%) | 94.6% | (92.4‐96.8%) |
| CIN2+ | 76/146 | 68.7% | (51.6‐67.5%) | 346/504 | 68.7% | (64.6‐72.7%) | 35.5% | (29.5‐41.5%) | 85.4% | (82.0‐88.9%) |
| (C) ASCL1/LHX8 methylation a | ||||||||||
| CIN3+ | 70/91 | 76.9% | (68.3‐85.6%) | 465/624 | 74.5% | (71.1‐77.9%) | 30.6% | (24.6‐36.5%) | 95.7% | (93.9‐97.5%) |
| CIN2+ | 97/163 | 59.5% | (52.0‐67.0%) | 420/552 | 76.1% | (72.5‐79.6%) | 42.4% | (36.0‐48.8%) | 86.4% | (83.4‐89.5%) |
| (D) Cytology + HPV16/18 genotyping | ||||||||||
| CIN3+ | 79/83 | 95.2% | (90.6‐99.8%) | 248/567 | 43.7% | (39.7‐47.8%) | 19.8% | (15.9‐23.8%) | 98.4% | (96.9‐100.0%) |
| CIN2+ | 130/146 | 89.0% | (84.0‐94.1%) | 236/504 | 46.8% | (42.5‐51.2%) | 32.7% | (28.1‐37.3%) | 93.7% | (90.6‐96.7%) |
| (E) ASCL1/LHX8 methylation a + HPV16/18 genotyping | ||||||||||
| CIN3+ | 72/83 | 86.7% | (79.5‐94.0%) | 302/567 | 53.3% | (49.2‐57.4%) | 21.4% | (17.0‐25.7%) | 96.5% | (94.4‐98.5%) |
| CIN2+ | 112/146 | 76.7% | (69.9‐83.6%) | 279/504 | 55.4% | (51.0‐59.7%) | 33.2% | (28.2‐38.3%) | 89.1% | (85.7‐92.6%) |
Abbreviations: CI, confidence interval; CIN, cervical intraepithelial neoplasia; n, number of; N, group size; NPV, negative predicted value; PPV, positive predicted value.
Using a predefined threshold (70% specificity).
When methylation data were stratified by cytology, the sensitivity for CIN3+ was 86.4% (95% CI 77.7‐95.2%), and specificity was 52.4% (95% CI 37.3‐67.5%; Table 2) among hrHPV‐positive women with high‐grade cytology. For hrHPV‐positive women with low‐grade cytology, CIN3+ sensitivity was 63.6% (95% CI 43.5‐83.7%), and specificity was 75.1% (95% CI 69.0‐81.2%). For hrHPV‐positive women with normal cytology, CIN3+ sensitivity was 50.0% (95% CI 19.0‐81.0%), and specificity was 76.6% (95% CI 72.4‐80.8%). Methylation demonstrated a particularly good discriminatory power among hrHPV‐positive women with low‐grade cytology (RR CIN3+ 4.32; 95% CI 1.91‐9.78%, P‐value <.0001). HPV16/18 genotyping had an RR for CIN3+ of 2.53 (95% CI 1.14‐5.62%, P‐value = .0188) in this subgroup (Table 2). Data on the combinations of HPV16/18 genotyping and methylation stratified by cytology as well as estimates for endpoint CIN2+ can be found in Table 2.
TABLE 2.
Sensitivity and specificity of ASCL1/LHX8 methylation and HPV16/18 genotyping for detection of CIN3+ and CIN2+, stratified by cytology class
| Sensitivity | Specificity | PPV | NPV | Relative risk | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n/N | % | 95% CI | n/N | % | 95% CI | % | 95% CI | % | 95% CI | 95% CI | |||
| (A) ASCL1/LHX8 methylation a | |||||||||||||
| CIN3+ | NILM | 5/10 | 50.0% | (19.0‐81.0%) | 298/389 | 76.6% | (72.4‐80.8%) | 5.2% | (0.8‐9.7%) | 98.3% | (96.9‐99.8%) | 3.15 | (0.93‐10.67%) |
| Low‐grade cytology | 14/22 | 63.6% | (43.5‐83.7%) | 145/193 | 75.1% | (69.0‐81.2%) | 22.6% | (12.2‐33.0%) | 94.8% | (91.2‐98.3%) | 4.32 | (1.91‐9.78%) | |
| High‐grade cytology | 51/59 | 86.4% | (77.7‐95.2%) | 22/42 | 52.5% | (37.3‐67.5%) | 71.8% | (61.4‐82.3%) | 73.3% | (57.5‐89.2%) | 2.69 | (1.46‐4.96%) | |
| CIN2+ | NILM | 9/25 | 36.0% | (17.2‐54.8%) | 287/374 | 76.7% | (72.5‐81.0%) | 9.4% | (3.5‐15.2%) | 94.7% | (92.2‐97.2%) | 1.78 | (0.81‐3.87%) |
| Low‐grade cytology | 25/55 | 45.5% | (32.3‐58.6%) | 123/160 | 76.9% | (70.3‐83.4%) | 40.3% | (28.1‐52.5%) | 80.4% | (74.1‐86.7% | 2.06 | (1.32‐3.20%) | |
| High‐grade cytology | 63/83 | 75.9% | (66.7‐85.1%) | 10/18 | 55.6% | (32.6‐78.5%) | 88.7% | (81.4‐96.1%) | 33.3% | (16.5‐50.2%) | 1.33 | (1.01‐1.74%) | |
| (B) HPV16/18 genotyping | |||||||||||||
| CIN3+ | NILM | 8/10 | 80.0% | (55.2‐104.8%) | 247/354 | 69.8% | (65.0‐74.6%) | 7.0% | (2.3‐11.6%) | 99.2% | (98.1‐100.3%) | 8.66 | (1.87‐40.14%) |
| Low‐grade cytology | 13/22 | 59.1% | (38.5‐79.6%) | 117/176 | 66.5% | (59.5‐73.5%) | 18.1% | (9.2‐26.9%) | 92.9% | (88.4‐97.4%) | 2.53 | (1.14‐5.62%) | |
| High‐grade cytology | 40/51 | 78.4% | (67.1‐89.7%) | 19/37 | 51.4% | (35.2‐67.5%) | 69.0% | (57.1‐80.9%) | 63.3% | (46.1‐80.6%) | 1.88 | (1.14‐3.10%) | |
| CIN2+ | NILM | 10/23 | 43.5% | (23.2‐63.7%) | 236/341 | 69.2% | (64.3‐74.1%) | 8.7% | (3.5‐13.8%) | 94.8% | (92.0‐97.5%) | 1.67 | (0.75‐3.68%) |
| Low‐grade cytology | 26/52 | 50.0% | (36.4‐63.6%) | 100/146 | 68.5% | (61.0‐76.0%) | 36.1% | (25.0‐47.2%) | 79.4% | (72.3‐86.4%) | 1.75 | (1.10‐2.77%) | |
| High‐grade cytology | 51/71 | 71.8% | (61.4‐82.3%) | 10/17 | 58.8% | (35.4‐82.2%) | 87.9% | (79.5‐96.3%) | 33.3% | (16.5‐50.2%) | 1.32 | (1.01‐1.73%) | |
| (C) ASCL1/LHX8 methylation a + HPV16/18 genotyping | |||||||||||||
| CIN3+ | NILM | 8/10 | 80.0% | (55.2‐104.8%) | 200/354 | 56.5% | (51.3‐61.7%) | 4.9% | (1.6‐8.3%) | 99.0% | (97.6‐100.4%) | 4.42 | (0.95‐20.51%) |
| Low‐grade cytology | 17/22 | 77.3% | (59.8‐94.8%) | 92/176 | 52.3% | (44.9‐59.7%) | 16.8% | (9.5‐24.1%) | 94.8% | (90.4‐99.2%) | 5.61 | (1.71‐18.35%) | |
| High‐grade cytology | 47/51 | 92.2% | (84.8‐99.5%) | 10/37 | 27.0% | (12.7‐41.3%) | 63.5% | (52.5‐74.5%) | 71.4% | (47.8‐95.1%) | 2.22 | (0.95‐5.18%) | |
| CIN2+ | NILM | 13/23 | 56.5% | (36.3‐76.8%) | 192/341 | 56.3% | (51.0‐61.6%) | 8.0% | (3.8‐12.2%) | 95.0% | (92.1‐98.0%) | 1.43 | (0.65‐3.19%) |
| Low‐grade cytology | 36/52 | 69.2% | (56.7‐81.8%) | 81/146 | 55.5% | (47.4‐63.5%) | 35.6% | (26.3‐45.0%) | 83.5% | (76.1‐90.9%) | 2.66 | (1.51‐4.66%) | |
| High‐grade cytology | 63/71 | 88.7% | (81.4‐96.1%) | 6/17 | 35.3% | (12.6‐58.0%) | 85.1% | (77.0‐93.2%) | 42.9% | (16.9‐68.8%) | 1.49 | (0.94‐2.37%) | |
Note: Low‐grade cytology: BMD equalling ASC‐US/ASC‐H/LSIL. High‐grade cytology: >BMD equalling HSIL.
Abbreviations: ASC‐H, atypical squamous cells that cannot exclude high‐grade squamous intraepithelial lesions; ASCUS, atypical squamous cells of undetermined significance; CI, confidence interval; CIN, cervical intraepthelial neoplasia; HPV, human papillomavirus; HSIL, high‐grade squamous intraepithelial lesions; LSIL, low‐grade squamous intraepithelial lesions; N, group size; n, number of; NILM, negative for intraepthelial lesion or malignancy; NPV, negative predicted value; PPV, positive predicted value.
Using a predefined threshold (70% specificity).
4. DISCUSSION
The most important outcome of our study is the validation of host‐cell DNA methylation markers ASCL1, LHX8, ST6GALNAC5, GHSR, ZIC1, SST and the bi‐marker panel ASCL1/LHX8, as defined in our previous study, 10 in a large and independent hrHPV‐positive screening cohort, confirming their good and robust triage performance for CIN3+. Current findings underscore the potential of ASCL1/LHX8 methylation analysis as a useful alternative to cytology as a triage test in primary HPV‐based screening with the advantage of being objective, less prone to training and interpretational errors, automatable and offering a full‐molecular approach.
In our study, the bi‐marker panel ASCL1/LHX8 had a similar sensitivity as HPV16/18 genotyping but showed a markedly higher specificity. Though small in numbers, the ASCL1/LHX8 marker panel was able to detect all cervical cancer cases (n = 3), in line with Dick et al. 10 The HPV genotypes in these cancers were HPV16 (n = 1), HPV18 (n = 1) and HPV35 (n = 1). A triage strategy comprising a combination of HPV16/18 genotyping and methylation resulted in an increase in sensitivity over either triage test alone, but at a marked drop in specificity. Comparison with cytology, either or not combined with HVP16/18 genotyping, must be done with caution due to the fact that the hrHPV‐positive women in the IMPROVE study were managed based on cytology.
The CIN3+ sensitivity (76.9%) and specificity (74.5%) achieved by the bi‐marker panel ASCL1/LHX8 are highly reassuring when compared to a meta‐analysis of methylation markers for the detection of CIN3+. 6 This meta‐analysis reported, when restricting to studies in which specificity was set at 70%, a pooled sensitivity for CIN3+ of 71.1%. It is assumed that methylation analysis identifies the subset of CIN2/3 lesions with a high progression risk to cancer that are in need of direct treatment. 5 Methylation levels of several genes were found significantly higher in CIN2/3 lesions associated with a long‐standing (>5 years) hrHPV infection (ie, so‐called advanced lesions, with high copy number aberrations), compared to CIN2/3 with a more recently acquired (≤5 years) hrHPV infection (early or incident lesions, with lower copy number aberrations). 5 , 18 The high sensitivity for advanced CIN as well as cervical cancer makes methylation marker analysis an attractive triage tool.
As cytology may remain a triage method of choice and hrHPV‐positive women with low‐grade cytology generate a substantial increase in referral rate while relatively few CIN3+ are detected, it is noteworthy that additional triage of hrHPV‐positive women with low‐grade cytology by ASCL1/LHX8 methylation showed good performance. Secondary triage of hrHPV‐positive women with low‐grade cytology by ASCL1/LHX8 methylation might reduce the number of women referred for colposcopy with 70%, while ensuring that most women with CIN3+ receive colposcopy (CIN3+ sensitivity of 63.6%; Table 2). Even though ASCL1/LHX8 methylation showed good performance data in hrHPV‐positive women with high‐grade cytology, the diagnosis of high‐grade cytology among hrHPV‐positives carries high risk of CIN3+ making that these women should be sent to colposcopy regardless of methylation status. The ASCL1/LHX8 methylation data stratified by cytology presented here are in line with a multi‐centre study by Bonde et al., which reported on another methylation marker panel consisting of FAM19A4 and miR124‐2. 19
The methylation levels of ASCL1, LHX8, ST6GALNAC5, GHSR, SST and ZIC1 in hrHPV‐positive cervical screening samples displayed an increase with the severity of the underlying cervical disease, confirming previous findings. 10 Slight variations in methylation level range between the six markers may be explained technologically by target‐to‐target differences in the qMSP or may be related to biology as some of the markers are known to reside on chromosomal regions that can be affected by copy number alterations. In earlier work, we observed that the methylation levels of GHSR, SST and ZIC1 (located at 3q) in cervical scrapes were significantly higher in the presence of a 3q gain in the corresponding CIN2/3 lesions compared to the absence of a 3q gain. 8 The markers used in our study were identified by genome‐wide methylation profiling of cervical samples 8 , 9 and have been reported in other cancer types as well. For example, ASCL1 was found to be methylated in among others colorectal, oral and anal cancer, 20 , 21 , 22 and LHX8 in breast and lung cancer. 23 , 24 ASCL1 is a proneural, oncogenic transcription factor, 25 , 26 and LHX8 is a highly conserved transcription factor involved in cell fate in neurogenesis, tooth morphogenesis and oogenesis. 24 The functional involvement and methylation‐dependent expression regulation of these genes in cervical carcinogenesis remain to be clarified.
The major strength of the current study is the evaluation of a large and independent series of hrHPV‐positive clinician‐collected cervical samples derived from primary HPV‐based screening. The IMPROVE study was nested within a population‐based screening setting and intended to reflect the new routine primary HPV‐based screening programme that was introduced in 2017 in the Netherlands, suggesting that results could be scalable to the screening population level. Many studies evaluating host‐cell DNA methylation markers so far have been performed in referral populations or case‐control settings. Histology endpoints were retrieved from the nationwide network and registry of histo‐ and cytopathology (PALGA), which covers all pathology labs in the Netherlands. The use of the original pathology diagnoses represents routine setting, which underscores the value of our findings for potential future implementation in screening practice. The use of revision diagnoses in the analyses did not change our conclusions, verifying the good performance of methylation analysis as triage tool. A limitation of our study is that our results could possibly be biased, because no histology endpoint was available in a number of hrHPV‐positive women. This could potentially lead to an underestimation of CIN3+. It is not likely that this would have a marked influence on the outcome given that of all women without a histological endpoint, the far majority was documented with two times normal cytology.
In conclusion, our study shows that the individual markers ASCL1, LHX8, ST6GALNAC5, GHSR, ZIC1 and SST and the bi‐marker panel ASCL1/LHX8 have clinical utility for the detection of CIN3+ in hrHPV‐positive women at screening. ASCL1/LHX8 methylation test on cervical screening samples has potential as triage test to identify hrHPV‐infected women in need of colposcopy.
CONFLICT OF INTEREST
Daniëlle A. M. Heideman, Renske D. M. Steenbergen and Chris J. L. M. Meijer are minority shareholders of Self‐screen B.V., a spin‐off company of VUmc; Self‐screen B.V. develops, manufactures and licences high‐risk HPV and methylation marker assays for cervical cancer screening and holds patents on these tests. Daniëlle A. M. Heideman has been on the speaker's bureau of Qiagen and serves occasionally on the scientific advisory boards of Pfizer and Bristol‐Myers Squibb. Chris J. L. M. Meijer is part‐time CEO of Self‐screen B.V. and has a very small number of shares of MDXHealth and previously of QIAGEN, has received speakers fees from GSK, QIAGEN and SPMSD/Merck, has served occasionally on the scientific advisory boards (expert meeting) of these companies and is co‐investigator of an SPMSD sponsored trial of which his research institute received research funding. Lisanne Verhoef, Maaike C. G. Bleeker, Nicole Polman, Willem J. G. Melchers, Ruud L. Bekkers, Anco C. Molijn, Wim G. Quint and Folkert J. van Kemenade declare no conflicts of interest.
ETHICS STATEMENT
The work in our study was approved by the Medical Ethics Committee of Amsterdam UMC, Vrije Universiteit Amsterdam (Amsterdam, The Netherlands; METC 2018/09, TcB 2018.106). All participants provided written informed consent.
Supporting information
TABLE S1 Original diagnosis vs revision diagnosis.
TABLE S2 Sensitivity and specificity of baseline cytology, genotyping and ASCL1/LHX8 methylation for detection of CIN3+ and CIN2+—subgroup with revision diagnosis.
TABLE S3 Sensitivity and specificity of ASCL1/LHX8 and HPV16/18 genotyping for detection of CIN3+, stratified by cytology class—subgroup with revision diagnosis.
FIGURE S1. Methylation levels increase with severity of underlying cervical disease—subgroup with revision diagnosis. DNA methylation levels of (A) ASCL1, (B) LHX8, (C) ST6GALNAC5 22, (D) GHSR, (E) ZIC1 and (F) SST represented by the square‐root transformed Cq‐ratios (y‐axis) in the different histology categories (x‐axis). Differences between histological categories upon Kruskal‐Wallis omnibus test, followed by post hoc testing using the Wilcoxon rank‐sum test and Bonferroni multiple testing correction: *P‐value <.05;**P‐value <.01; ***P‐value <.001; NS, not significant. ○, outlier sample. Abbreviations: CIN, cervical intraepithelial neoplasia; CxCa, cervical carcinoma; no CIN, women with no histology or normal histology.
FIGURE S2. Diagnostic accuracy—subgroup with revision diagnosis. ROC curves and corresponding AUC for CIN3+ detection for (A) single marker classifiers ASCL1, LHX8, ST6GALNAC5, GHSR, ZIC1 and SST and (B) bi‐marker panel ASCL1/LHX8 (black line), including point estimates of cytology (red circle), HPV16/18 genotyping (green circle) and bi‐marker panel ASCL1/LHX8 with the predefined threshold calibrated at 70% clinical specificity (yellow circle). For comparison, the cross‐validated ROC curve and corresponding AUC, as obtained in the hrHPV‐positive training series, are projected (grey line) [10]. Abbreviations: AUC, area under the curve; CIN, cervical intraepithelial neoplasia; ROC, receiver operating characteristic.
ACKNOWLEDGEMENTS
This work is dedicated to our colleague Prof. Dr. P.J.F. Snijders, who passed away on May 27, 2018. The authors thank all women who participated in the IMPROVE study; the screening organisations Midden‐West, Zuid‐West, and Oost; the National Institute for Public Health and the Environment; and the physicians, gynaecologists, and pathologists in the study regions. They also thank Daisy Hoek for technical assistance and Mark van de Wiel and Ivonne Martin for statistical support. The authors would like to acknowledge the nationwide network and registry of histo‐ and cytopathology in the Netherlands (PALGA) for providing data and the Amsterdam UMC Biobank for their high‐quality storage of collected samples. The graphical abstract accompanying this article was created with BioRender.com. J.B. had financial support from the European Commission (RISCC, grant number 847845).
Verhoef L, Bleeker MCG, Polman N, et al. Performance of DNA methylation analysis of ASCL1, LHX8, ST6GALNAC5, GHSR, ZIC1 and SST for the triage of HPV‐positive women: Results from a Dutch primary HPV‐based screening cohort. Int. J. Cancer. 2022;150(3):440-449. doi: 10.1002/ijc.33820
Funding information KWF Kankerbestrijding, Grant/Award Number: 11337; European Commission, Grant/Award Number: 847845
DATA AVAILABILITY STATEMENT
The data that support the findings of our study are in anonymised form available from the corresponding author upon reasonable request and after the data protection regulations.
REFERENCES
- 1. Maver PJ, Poljak M. Primary HPV‐based cervical cancer screening in Europe: implementation status, challenges, and future plans. Clin Microbiol Infect. 2020;26:579‐583. [DOI] [PubMed] [Google Scholar]
- 2. Shiraz A, Crawford R, Egawa N, Griffin H, Doorbar J. The early detection of cervical cancer. The current and changing landscape of cervical disease detection. Cytopathology. 2020;31:258‐270. [DOI] [PubMed] [Google Scholar]
- 3. Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225‐3229. [PubMed] [Google Scholar]
- 4. Feng Q, Balasubramanian A, Hawes SE, et al. Detection of hypermethylated genes in women with and without cervical neoplasia. J Natl Cancer Inst. 2005;97:273‐282. [DOI] [PubMed] [Google Scholar]
- 5. Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV‐induced cervical precancerous lesions. Nat Rev Cancer. 2014;14:395‐405. [DOI] [PubMed] [Google Scholar]
- 6. Kelly H, Benavente Y, Pavon MA, De Sanjose S, Mayaud P, Lorincz AT. Performance of DNA methylation assays for detection of high‐grade cervical intraepithelial neoplasia (CIN2+): a systematic review and meta‐analysis. Br J Cancer. 2019;121:954‐965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kremer WW, Steenbergen R, Heideman D, Kenter GG, Meijer C. The use of host cell DNA methylation analysis in the detection and management of women with advanced cervical intraepithelial neoplasia: a review. BJOG. 2021;128:504‐514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Verlaat W, Snijders PJF, Novianti PW, et al. Genome‐wide DNA methylation profiling reveals methylation markers associated with 3q gain for detection of cervical precancer and cancer. Clin Cancer Res. 2017;23:3813‐3822. [DOI] [PubMed] [Google Scholar]
- 9. Verlaat W, Snoek BC, Heideman DAM, et al. Identification and validation of a 3‐gene methylation classifier for HPV‐based cervical screening on self‐samples. Clin Cancer Res. 2018;24:3456‐3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dick S, Verhoef L, De Strooper LM, et al. Evaluation of six methylation markers derived from genome‐wide screens for detection of cervical precancer and cancer. Epigenomics. 2020;12:1569‐1578. [DOI] [PubMed] [Google Scholar]
- 11. Polman NJ, Ebisch RMF, Heideman DAM, et al. Performance of human papillomavirus testing on self‐collected versus clinician‐collected samples for the detection of cervical intraepithelial neoplasia of grade 2 or worse: a randomised, paired screen‐positive, non‐inferiority trial. Lancet Oncol. 2019;20:229‐238. [DOI] [PubMed] [Google Scholar]
- 12. Bulk S, Van Kemenade FJ, Rozendaal L, Meijer CJ. The Dutch CISOE‐A framework for cytology reporting increases efficacy of screening upon standardisation since 1996. J Clin Pathol. 2004;57:388‐393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Casparie M, Tiebosch AT, Burger G, et al. Pathology databanking and biobanking in The Netherlands, a central role for PALGA, the nationwide histopathology and cytopathology data network and archive. Cell Oncol. 2007;29:19‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Editorial Board WCoT . WHO Classification of Tumours Female Genital Tumours. Vol 4. 5th ed. International Agency for Research on Cancer; 2020. [Google Scholar]
- 15. Vink FJ, Dick S, Heideman DAM, et al. Classification of high‐grade CIN by p16(ink4a), Ki‐67, HPV E4 and FAM19A4/miR124‐2 methylation status demonstrates considerable heterogeneity with potential consequences for management. Int J Cancer. 2021;149(3):707‐716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmittgen TD, Livak KJ. Analyzing real‐time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101‐1108. [DOI] [PubMed] [Google Scholar]
- 17. Robin X, Turck N, Hainard A, et al. pROC: an open‐source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bierkens M, Wilting SM, van Wieringen WN, et al. Chromosomal profiles of high‐grade cervical intraepithelial neoplasia relate to duration of preceding high‐risk human papillomavirus infection. Int J Cancer. 2012;131:E579‐E585. [DOI] [PubMed] [Google Scholar]
- 19. Bonde J, Floore A, Ejegod D, et al. Methylation markers FAM19A4 and miR124‐2 as triage strategy for primary HPV screen positive women; a large European multi‐center study. Int J Cancer. 2020;148(2):396‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jin B, Yao B, Li JL, et al. DNMT1 and DNMT3B modulate distinct polycomb‐mediated histone modifications in colon cancer. Cancer Res. 2009;69:7412‐7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li YF, Hsiao YH, Lai YH, et al. DNA methylation profiles and biomarkers of oral squamous cell carcinoma. Epigenetics. 2015;10:229‐236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van der Zee RP, Richel O, van Noesel CJM, et al. Host cell deoxyribonucleic acid methylation markers for the detection of high‐grade anal intraepithelial neoplasia and anal cancer. Clin Infect Dis. 2019;68:1110‐1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tommasi S, Karm DL, Wu X, Yen Y, Pfeifer GP. Methylation of homeobox genes is a frequent and early epigenetic event in breast cancer. Breast Cancer Res. 2009;11:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou C, Yang G, Chen M, et al. Lhx6 and Lhx8: cell fate regulators and beyond. FASEB J. 2015;29:4083‐4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vasconcelos FF, Castro DS. Transcriptional control of vertebrate neurogenesis by the proneural factor ASCL1. Front Cell Neurosci. 2014;8:412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lenhart R, Kirov S, Desilva H, et al. Sensitivity of small cell lung cancer to BET inhibition is mediated by regulation of ASCL1 gene expression. Mol Cancer Ther. 2015;14:2167‐2174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE S1 Original diagnosis vs revision diagnosis.
TABLE S2 Sensitivity and specificity of baseline cytology, genotyping and ASCL1/LHX8 methylation for detection of CIN3+ and CIN2+—subgroup with revision diagnosis.
TABLE S3 Sensitivity and specificity of ASCL1/LHX8 and HPV16/18 genotyping for detection of CIN3+, stratified by cytology class—subgroup with revision diagnosis.
FIGURE S1. Methylation levels increase with severity of underlying cervical disease—subgroup with revision diagnosis. DNA methylation levels of (A) ASCL1, (B) LHX8, (C) ST6GALNAC5 22, (D) GHSR, (E) ZIC1 and (F) SST represented by the square‐root transformed Cq‐ratios (y‐axis) in the different histology categories (x‐axis). Differences between histological categories upon Kruskal‐Wallis omnibus test, followed by post hoc testing using the Wilcoxon rank‐sum test and Bonferroni multiple testing correction: *P‐value <.05;**P‐value <.01; ***P‐value <.001; NS, not significant. ○, outlier sample. Abbreviations: CIN, cervical intraepithelial neoplasia; CxCa, cervical carcinoma; no CIN, women with no histology or normal histology.
FIGURE S2. Diagnostic accuracy—subgroup with revision diagnosis. ROC curves and corresponding AUC for CIN3+ detection for (A) single marker classifiers ASCL1, LHX8, ST6GALNAC5, GHSR, ZIC1 and SST and (B) bi‐marker panel ASCL1/LHX8 (black line), including point estimates of cytology (red circle), HPV16/18 genotyping (green circle) and bi‐marker panel ASCL1/LHX8 with the predefined threshold calibrated at 70% clinical specificity (yellow circle). For comparison, the cross‐validated ROC curve and corresponding AUC, as obtained in the hrHPV‐positive training series, are projected (grey line) [10]. Abbreviations: AUC, area under the curve; CIN, cervical intraepithelial neoplasia; ROC, receiver operating characteristic.
Data Availability Statement
The data that support the findings of our study are in anonymised form available from the corresponding author upon reasonable request and after the data protection regulations.


