Abstract
Bacterioplankton from a meso-eutrophic dam reservoir was size fractionated to reduce (<0.8-μm treatment) or enhance (<5-μm treatment) protistan grazing and then incubated in situ for 96 h in dialysis bags. Time course samples were taken from the bags and the reservoir to estimate bacterial abundance, mean cell volume, production, protistan grazing, viral abundance, and frequency of visibly infected cells. Shifts in bacterial community composition (BCC) were examined by denaturing gradient gel electrophoresis (DGGE), cloning and sequencing of 16S rDNA genes from the different treatments, and fluorescence in situ hybridization (FISH) with previously employed and newly designed oligonucleotide probes. Changes in bacterioplankton characteristics were clearly linked to changes in mortality rates. In the reservoir, where bacterial production about equaled protist grazing and viral mortality, community characteristics were nearly invariant. In the “grazer-free” (0.8-μm-filtered) treatment, subject only to a relatively low mortality rate (∼17% day−1) from viral lysis, bacteria increased markedly in concentration. While the mean bacterial cell volume was invariant, DGGE indicated a shift in BCC and FISH revealed an increase in the proportion of one lineage within the beta proteobacteria. In the grazing-enhanced treatment (5-μm filtrate), grazing mortality was ∼200% and viral lysis resulted in mortality of 30% of daily production. Cell concentrations declined, and grazing-resistant flocs and filaments eventually dominated the biomass, together accounting for >80% of the total bacteria by the end of the experiment. Once again, BCC changed strongly and a significant fraction of the large filaments was detected using a FISH probe targeted to members of the Flectobacillus lineage. Shifts of BCC were also reflected in DGGE patterns and in the increases in the relative importance of both beta proteobacteria and members of the Cytophaga-Flavobacterium cluster, which consistently formed different parts of the bacterial flocs. Viral concentrations and frequencies of infected cells were highly significantly correlated with grazing rates, suggesting that protistan grazing may stimulate viral activity.
Many experimental studies have shown that protistan predation yields distinct changes in bacterioplankton communities. The phenomena typically reported include changes in average physiological rates, as well as average cell morphology. For example, protistan bacterivory is associated with increases in per-bacterium production (e.g., thymidine incorporation rate) and decreases in average cell size or, perhaps more commonly, development of larger, “grazing-resistant” filament or floc forms (see e.g., references 15, 18, 29, and 49). Recent studies of grazing effects on natural or mixed communities (see, e.g., references 19, 26, 38, 40, 45, and 48) have shown that protistan bacterivory can also affect bacterial community composition (BCC). Interestingly, some similar morphological changes associated with increases in grazing pressure, i.e., development of filaments and flocs, can be due to quite variable shifts in the taxonomic composition of the community (19, 38). Filament formation is apparently a growth rate-controlled defense mechanism, which is widespread among bacteria (16). For example, manipulating grazing mortality among natural communities during different seasons has shown that filaments can be formed by bacteria of the alpha subclass of Proteobacteria in spring and by species of the Cytophaga-Flavobacterium (CF) group in the summer community (38).
While there is little doubt that protist-induced mortality yields changes in bacterial communities, protists are not the sole source of bacterial mortality. Virus-induced mortality is probably responsible for 5 to 50% of bacterial mortality, depending on the system (see reference 8 for a review). Surprisingly few attempts have been made to make simultaneous estimates of virus-induced and grazer-induced bacterial mortality (4, 9, 13, 43, 53). Furthermore, the significance of experimentally induced changes in natural bacterioplankton, whether virus or protist related, is difficult to assess. While there are data on the spatial variability of natural bacterioplankton communities on a large variety of scales (see e.g., references 1, 14, 17, and 51), data on temporal variability are limited to seasonal changes (see, e.g., references 25 and 47). Remarkably, over the timescales used in most experimental studies (hours to days), even data on simple morphological variability of natural bacterial communities are rare (41). Thus, protist grazing no doubt affects bacterioplankton, but the significance of differential mortality effects remains obscure since the background variability in BCC over short timescales has not been examined. Here we present the results of an investigation which addresses these problems.
We conducted a field study employing communities from a meso-eutrophic freshwater reservoir during the late clear-water phase, a period when relatively low protistan grazing pressure is exerted on the in situ bacterioplankton (38). Using a size fractionation approach to manipulate natural populations, we monitored changes in bacterioplankton subjected to negligible or to high levels of protistan bacterivory. Communities were incubated in situ in dialysis bags to minimize experimental artifacts. Building on our previous study (38), viral abundances and virus-induced mortality were also assessed and the natural variability of bacterioplankton characteristics was examined through simultaneous sampling of the reservoir population.
Data were gathered on the abundance and size structure of the bacterial community, on biomass production rates, and on community composition. Taxonomic shifts were monitored by fluorescent in situ hybridization (FISH) with oligonucleotide probes. Changes in bacterial community composition were also examined by using denaturing gradient gel electrophoresis (DGGE) and by cloning and sequencing of 16S rDNA genes retrieved from the different treatments and reservoir water. Specific oligonucleotide probes were designed based on sequence information from cloning. We intended to (i) confirm previous results which suggested that increasing bacterivory on populations naturally subjected to low levels of bacterivory yields large changes; (ii) examine viral dynamics and, based on the frequency of visibly infected bacterial cells, estimate bacterial mortality; and (iii) assess the short-term in situ variability of BCC. Furthermore, we provide a more detailed analysis of the “grazing (or mortality)-resistant” community than is found in similar previous studies on grazing in natural communities (19, 38), especially with respect to the structure of bacterial flocs and the identity of dominant filament-forming bacteria. We also provide preliminary evidence that protistan grazing may increase viral lysis, suggesting a synergy between grazing- and virus-induced mortality.
MATERIALS AND METHODS
Study site and experimental design.
The experiment was conducted in the meso-eutrophic Římov Reservoir (South Bohemia) (for details, see reference 36). The sampling site was located above the former river valley (which is 30 m deep), about 250 m from the reservoir dam. On 28 May 1999, water samples were collected with a 2-liter Friedinger sampler from a depth of 0.5 m (15 samples) and a final volume of 30 liters was mixed in a 50-liter plastic container. The experiment was run during the late clear-water phase (28 May to 1 June 1999; water temperature, 18 to 20°C; chlorophyll a concentration, 5 to 9 μg liter−1). The experimental design and protocol were similar to those described in detail by Šimek et al. (38), partly simplified since two treatments were employed, but the experiments were done in triplicate.
Treatments represented nominally (i) “bacterivore free,” via filtration through 0.8-μm-pore-size filters, which remove most bacteriovorous protists, and (ii) “increased bacterivory,” via filtration through 5.0-μm-pore size filters, which remove the predators (thus allowing rapid growth) of bacterivorous heterotrophic nanoflagellates (HNF). Thus, water samples were sequentially size fractionated through 5- and 0.8-μm-pore-size Poretics filters (diameter, 47 mm; OSMONIC Inc., Livermore, Calif.) into the fractions <5 μm (bacteria and HNF only) and <0.8 μm (bacteria only), which were used along with unfiltered samples taken directly from the surrounding reservoir water (all bacterivores and HNF predators present); these samples are designated throughout the text as <5 μm, <0.8 μm, and reservoir treatments, respectively. The last <0.8-μm filtration step was conducted in sterilized glass Poretics filter holders to minimize HNF contamination. The water fractions were placed in ∼2-liter pretreated (distilled water rinsed and boiled) dialysis tubes (diameter, 75 mm; molecular mass cutoff, 12,000 to 16,000 Da [Poly Labo]). The dialysis bags were incubated in the reservoir at a depth of 0.5 m, oriented horizontally in open Plexiglas holders. Samples (∼250 to 300 ml) were taken from each dialysis bag and reservoir water at 0, 12, 24, 48, 72, and 96 h (t0, t12, t24, t48, t72, and t96).
Bacterial abundance and biomass.
Samples were fixed with formaldehyde (2% [vol/vol] final concentration) stained with 4′-6-diamidino-2-phenylindole (DAPI) (final concentration, 0.1 μg ml−1), and enumerated by epifluorescence microscopy (Olympus BX 60 microscope). Between 450 and 700 DAPI-stained bacterial cells were recorded at a magnification of ×1,000 with an analog monochrome charge-coupled device camera (Cohu Inc., San Diego, Calif.) mounted on the microscope and processed with the semiautomatic image analysis systems LUCIA D (Lucia 3.52; resolution, 750 by 520 pixels; 256 grey levels [Laboratory Imaging, Prague, Czech Republic]). Details of the image processing (gray transformation and edge finding) are described by Posch et al. (28). Bacterial biomass was calculated from the allometric relationship between cell volume and carbon content as described by Simon and Azam (42) and modified by Norland (24). Since the cell width showed very little variability, the cell length was the most important factor in determining cell volume. Cells longer than 4 μm were assumed to be ungrazable by most of the bacterivorous HNF (15, 40). Correspondingly, we used this criterion to classify bacteria as biomass in the form of small cells (<4 μm) that were susceptible to grazing, as opposed to biomass in the form of grazing-resistant, bacterial filamentous cells (>4 μm). Since some of the filaments in the <5-μm treatments were even longer than 100 μm, they were recorded at a magnification of ×400 and processed separately.
Bacterial production.
Bacterial production was measured by a thymidine incorporation method modified from that of Riemann and Søndergaard (31). Duplicate 5-ml subsamples were incubated for 30 min at in situ temperature with 10 nmol of [methyl-3H]thymidine (Amersham) per liter, preserved with neutral buffered formaldehyde (2% [vol/vol] final concentration) filtered through 0.2-μm membrane filters (Poretics), and extracted 10 times with 1 ml of ice-cold 5% trichloroacetic acid (see reference 36 for details). Replicate blanks prefixed by 2% formaldehyde were processed in parallel. An empirical conversion factor between the thymidine incorporation rate and the bacterial cell production rate was determined using data from the triplicate <0.8-μm treatments. The cell production rate was calculated from the slope of the increase of the natural logarithm of bacterial abundance over time (0, 24, and 48 h). Our determined empirical conversion factor (2.3 × 1018 cells mol of thymidine−1) was used for calculations.
DGGE, clone libraries, phylogenetic tree, and probe design.
DGGE of 16S rRNA gene segments (21) amplified by PCR was performed for samples from two dialysis bags of each treatment and from reservoir water at t0, t48, and t72. Oligonucleotide primers 341F and 907R (21) were used to directly amplify an approximately 550-nucleotide segment of the 16S rDNA genes from cells concentrated on polycarbonate filters (7). DGGE was performed by the method of Muyzer et al. (21), and banding patterns were visualized by ethidium bromide staining. Two gels were run for each of the 0.8- and 5 μm-filtered treatments, and samples from the reservoir served as references on each of the gels.
Three clone libraries were constructed from a reservoir sample at t0, a dialysis bag containing 0.8-μm-filtered water at t72, and a bag with 5-μm-filtered water at t72. From cells concentrated on membrane filters, PCR with bacterial 16S rRNA primers 8F and 1492R (6) was used to amplify almost full-length 16S rDNAs under the amplification conditions described by Glöckner et al. (11). The amplified rDNA was inserted into the pGEM-T vector (Promega Corp., Madison, Wis.) and cloned into competent Escherichia coli cells as specified by the manufacturer. Plasmid DNA inserts of 25 clones from each library were digested with 7.5 U of Sau3a-1 (Promega) for 3 h at 37°C. The fragments were analyzed by agarose gel electrophoresis, and restriction patterns were compared visually.
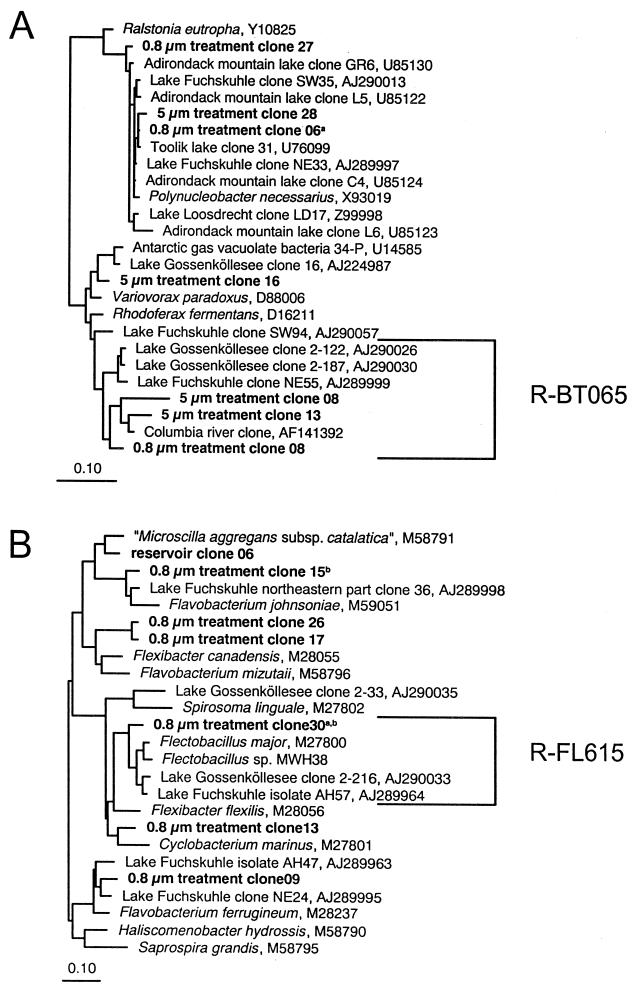
Plasmid DNA from 38 selected 16S rDNA clones were partially sequenced (approximately 700 bp) by Taq cycle sequencing with universal 16S rRNA specific primers using an AB1377 (Applied Biosystems, Inc.) sequencer. Sequence data were analyzed with the ARB software package (Lehrstuhl für Mikrobiologie, Technische Universität München, Munich, Germany [http://www.micro.biologie.tu-muenchen.de]). Almost full-length sequences of 15 clones which affiliate with the beta subclass of Proteobacteria and the Cytophaga-Flavobacterium-Bacteroides (CFB) phylum were determined. One sequence was rejected as chimeric (http://www.cme.msu.edu/RDP/html/analyses.html). A phylogenetic tree was reconstructed for these sequences by maximum parsimony of all sequences of >1,400 nucleotides in the ARB database, using neighbor joining and maximum-likelihood analyses on various subsets for the evaluation of topologies (Fig. 1). Alignment positions at which fewer than 50% of sequences of the entire data set had the same residues were excluded from the calculations. Using the PROBE-DESIGN tool of ARB, specific oligonucleotide probes were designed for defined phylogenetic clusters within the beta Proteobacteria and CFB, which also contained sequences from the clone libraries (Fig. 1). Two probes were found to detect significant populations in situ. The sequences targeted by these probes are depicted in Fig. 1. Hybridization conditions for probe R-BT065 (5′-GTTGCCCCCTCTACCGTT-3′, E. coli positions 65 to 83 [5]), were adjusted by formamide series applied to different filters from the experiment, and brightness was evaluated by eye. Probe R-FL615 (5′-CACTGCAATCGTTGAGCGA-3′, E. coli positions 615 to 634) was hybridized to pure cultures of Flectobacillus major at increasing levels of stringency, and fluorescence intensities were quantified (22). For evaluation of environmental samples, both probes were used with 35% formamide in the hybridization buffer and incubated for 2 h at 46°C.
FIG. 1.
Unrooted phylogenetic trees of 16S rDNA clones from the beta proteobacteria (A) and the CF cluster (B) retrieved from the various treatments and from the reservoir. Brackets indicate sequences targeted by FISH probes R-BT065 and R-FL615. Superscripts indicate that sequences with identical restriction patterns were also found in clone libraries from reservoir samples (a) and the 5-μm treatment (b). Scale bars indicate 10% estimated sequence divergence.
FISH with oligonucleotide probes.
BCC was analyzed by in situ hybridization with fluorescence oligonucleotide probes on membrane filters (1, 10). For details of the hybridization procedure and error estimates, see reference 26. Briefly, bacterial cells from 10 to 20-ml subsamples were concentrated on white 0.2-μm filters (47 mm in diameter; Poretics Corp.), fixed on membrane filters by overlaying the filters with 4% paraformaldehyde in phosphate-buffered saline (pH 7.2), and stored at −20°C (1). The seven different group-specific oligonucleotide probes (Interactiva, Ulm, Germany) were targeted to the domain Bacteria (Eubacteria, EUB338), the alpha, beta, and gamma subclasses of the class Proteobacteria (ALF968, BET42a, and GAM42a), the CF (CF319a) phylum (2), and two more narrow phylogenetic clusters (the probes R-BT065 and R-FL615). The probes were fluorescently labeled with the indocarbocyanine dye Cy3 (BDS, Pittsburgh, Pa.). After hybridization, filter sections were stained with DAPI, and the percentage of hybridized bacterial cells were enumerated by epifluorescence microscopy (Olympus, AX70 PROVIS).
Protozoan grazing and abundance.
Protozoan grazing on bacteria was estimated using fluorescently labeled bacterioplankton (FLB) (34) concentrated from the reservoir water (for details, see reference 38). FLB uptake experiments were run with water from each of the bags containing 5-μm-filtered water as well as with unfiltered reservoir water at each time point. Samples (180 ml) were dispensed into acid-soaked and rinsed 250-ml flasks and preincubated at in situ temperature for 15 min. HNF and ciliate FLB uptake rates were determined in short-term FLB direct-uptake experiments with FLB equal to 5 to 15% of the natural bacterial concentration. Subsamples (25 ml) for protozoan enumeration and tracer ingestion determinations were taken at 0, 5, 10, 15, 20, and 30 min after tracer addition and fixed by adding 0.5% of alkaline Lugol solution, immediately followed by 2% (vol/vol) (final concentration) formaldehyde and several drops of 3% (wt/vol) sodium thiosulfate to clear the Lugol color (34). We determined ciliate grazing rates in time series from 5- to 15-min subsamples and flagellate grazing rates in subsamples from 10 to 30 min. Subsamples (5 ml for flagellates or 20 ml for ciliates) were stained with DAPI, filtered through 1-μm black Poretics filters, and inspected via epifluorescence microscopy. Nonpigmented HNF and plastidic flagellates were differentiated. At least 40 ciliates and 50 flagellates were inspected for FLB ingestion in each sample. To estimate total protozoan grazing, we multiplied average uptake rates of ciliates and flagellates by their in situ abundances as previously described (37, 38).
Viral abundance and viral mortality of bacteria.
Viruses in formaldehyde (2% [vol/vol] final concentration)-preserved samples were stained with SYBR Green I (10,000× in dimethyl sulfoxide [Molecular Probes chemical S-7567]) and enumerated by epifluorescence microscopy (23). Viruses were also enumerated using transmission electron microscopy as described previously (44). Briefly, viruses were collected quantitatively onto Formvar-coated, 400-mesh electron microscope grids by centrifugation in a swinging-bucket rotor (Sorval TH-641; 100,000 × g for 2.5 h), stained for 30 s with 1% (wt/vol) uranyl acetate, and rinsed three times with deonized distilled water.
The frequency of visibly infected bacteria (FVIB) and the burst size were determined by transmission electron microscopy (50). Due to restrictions of sample volume, subsamples from each replicate were pooled, yielding a single sample for each treatment for each time point. Replicate grids were prepared and processed to estimate measurement error. The FVIB was related to virus-mediated mortality of bacteria using conversion factors relating the frequency of all infected cells to visibly infected cells and assuming that the latent period is equivalent to the generation time (30), as was done by Binder (3): mortality = (1/g ln 2) {FVIB/[(1 − e) − FVIB]}, where g is the ratio of latent period to generation time (g = 1) and e is the fraction of the latent period that elapsed before the appearance of intracellular virus particles (e = 0.816 [average from reference 30]). Mortality is a fraction per generation.
Nucleotide sequence accession numbers.
The 16S rDNA sequences generated in this study were deposited in GenBank under accession numbers AF361192 to AF361205.
RESULTS
Bacterial and protistan community dynamics and activity.
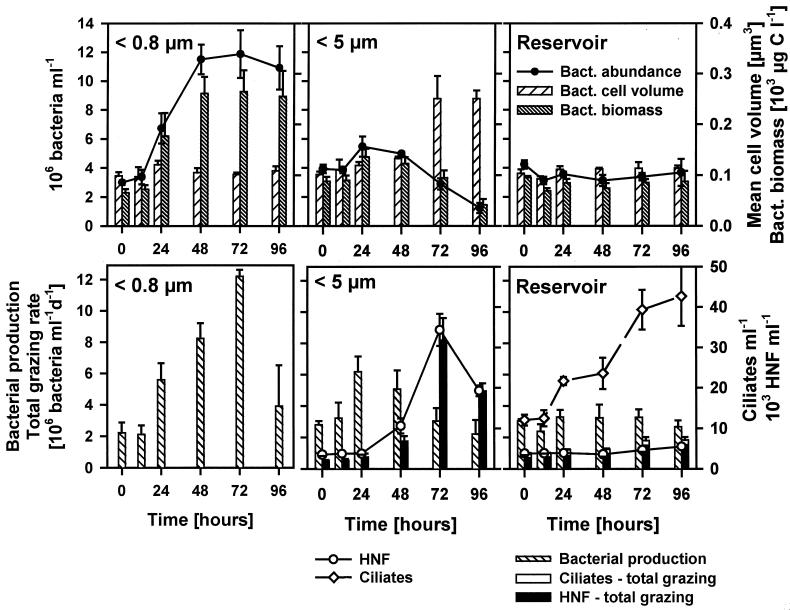
In the grazer-free treatment (<0.8 μm), bacterial numbers and total biomass covaried closely (Fig. 2); both increased between 12 and 48 h, with an apparent generation time of about 24 h (Table 1), and then remained relatively stationary at about 11 × 106 to 12 × 106 cells ml−1. Bacterial production (BP) data showed a similar trend but with a peak at 72 h followed by a decrease at the end of the study (Fig. 2). In contrast to bacterial numbers and total biomass, no marked changes in the mean cell volume of bacteria (MCV) were detected in the grazer-free treatments.
FIG. 2.
Changes in microbial parameters in different size fractionation treatments (<0.8 and <5 μm) exposed in dialysis bags compared to unfiltered reservoir water during the experiment. (Top) Bacterial abundances, biomasses, and mean cell volumes. (Bottom) Abundances of HNF, ciliates, bacterial production, and total protistan bacterivory subdivided into HNF and ciliate grazing. Values are means for three replicate treatments, and vertical bars show SDs. Statistical relationships are given in Tables 2 and 3.
TABLE 1.
Net doubling times of ungrazed microorganisms in the predator-free treatment or in the <5-μm treatment
| Organism | Doubling time (h)c | Interval (h) |
|---|---|---|
| Bacterioplankton | 24.8 ± 2.2 | 0–48a |
| CF319a bacteria | 19.7 ± 2.0 | 0–48a |
| BET42a bacteria | 16.6 ± 1.4 | 0–48a |
| R-BT065 bacteria | 8.8 ± 1.6 | 0–24a |
| R-FL615 bacteria | 12.7 ± 1.3 | 72–96b |
| HNF | 15.1 ± 2.0 | 24–72b |
Treatment at <0.8 μm (bacterioplankton and different bacterial groups).
Treatment at <5 μm (grazing-resistant R-FL615-positive filaments and HNF).
The values are mean ± SD of three triplicate treatments. Rates were calculated for intervals with exponential growth.
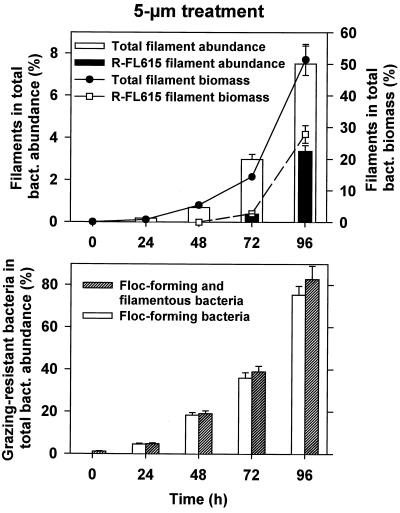
Microbial interactions in the <5-μm treatment yielded quite different trends. After a slight initial increase, bacterial abundance, biomass, and BP dropped between 24 and 96 h (e.g., bacterial numbers dropped from ∼5 × 106 to 1 × 106 cells ml−1 [Fig. 2]). A shift in bacterial cell morphology was evident as the bacterial MCV increased with bacterivore abundance, HNF (from ∼3 × 103 to 34 × 103 cells ml−1, -doubling time, 15.4 h [Table 1]), and bacterivory. HNF abundance and grazing were maximal at 72 h in the <5-μm treatment and then decreased toward the end of the experiment. While aggregate HNF grazing activity consumed 20 to 40% of BP during the first half of the study (Fig. 2), it represented about 200% of BP by the end of the incubation. The high HNF grazing pressure coincided with a marked, treatment-specific shift in bacterial size structure and morphology toward the dominance of grazing-resistant bacterial populations, i.e., filament- or floc-forming bacteria (Fig. 3). The clear increase in the MCV of bacteria reflected an exponentially increasing proportion of bacterial filaments (cells longer than 4 μm), accounting for >7% of the total bacteria and for >50% of the total bacterial biomass by the end of the experiment. Moreover, this process was also paralleled by a clear shift from the dominance of free-living single bacterial cells to that of floc-forming bacteria, which represented ∼75% of total bacteria by the end of the incubation.
FIG. 3.
Results for the <5-μm treatment. Shown are changes in proportions of the abundance and biomass of total filamentous bacterial cells and of only filaments targeted by the R-FL615 probe in total bacterial abundance and biomass (top) and abundance of floc-forming and of total grazing-resistant bacteria (a sum of floc- and filament-forming cells) during the experiment (bottom). Values are means for three replicate treatments, and vertical bars show SDs.
Compared to the <0.8- and <5-μm treatments, whole-water samples from the reservoir showed negligible changes in nearly all parameters throughout the experiment (Fig. 2). The only exception was a steady increase in ciliate abundance, probably reflecting decreasing grazer control of ciliates by large zooplankton during the late stage of the clear-water phase in the reservoir. However, it had a minor effect on the overall rates of protistan grazing on bacterioplankton in the reservoir.
Changes in bacterial community composition.
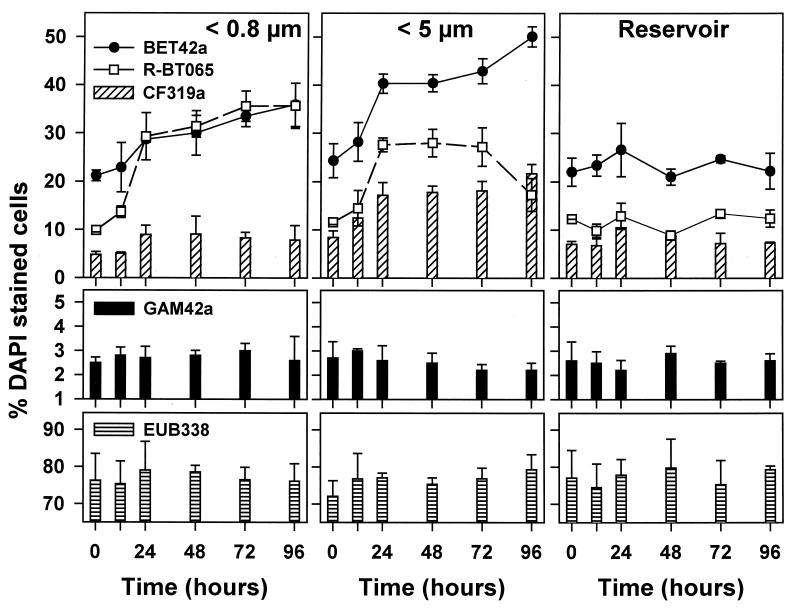
Of the 16S rDNA sequences obtained from the Římov Reservoir and from the different incubations (Fig. 1), 14 affiliated with the beta proteobacteria and the CF cluster, the majority with recently described cosmopolitan freshwater clades (beta I, beta II, cf I, cf II, and cf III) (12). Sequences were also affiliated with gamma proteobacteria, actinobacteria, and other gram-positive lineages (data not shown). An analysis of BCC based on FISH with group-specific probes did not show any significant differences among the treatments at time zero (analysis of variance and multiple Tukey honest significant distance [HSD] test, P < 0.5). Thus, at t0, the experimental manipulations yielded no marked bias apparent at our level of taxonomic resolution. During the course of the study, the manipulations resulted in treatment-specific changes in BCC indicated by means of both rRNA-targeted oligonucleotide probes (Fig. 4) and DGGE (data not shown). For the seven different rRNA-targeted oligonucleotide probes we used in this study, cells detected by ALF968, GAM42a, and EUB338 showed no clear trend during the course of the study in any treatment or sample group. The proportions of bacteria targeted by probe EUB338 ranged between 75 and 85% of total DAPI-stained bacterial cells (Fig. 4). The proportions of bacteria hybridizing with ALF968 (data not shown) and GAM42a were consistently low (<1% and 2 to 3%, respectively).
FIG. 4.
Time course changes in the phylogenetic composition of the bacterioplankton community in size fractionation treatments (<0.8 and <5 μm) exposed in dialysis bags compared to unfiltered reservoir water during the experiment. Shown are proportions of beta proteobacteria and cells detected by probe R-BT065 (top), gamma proteobacteria (middle), CF group (CF319a, top), and total FISH detection rates by the general bacterial probe EUB338 (bottom). Values are means for three replicate treatments, and vertical bars show SDs.
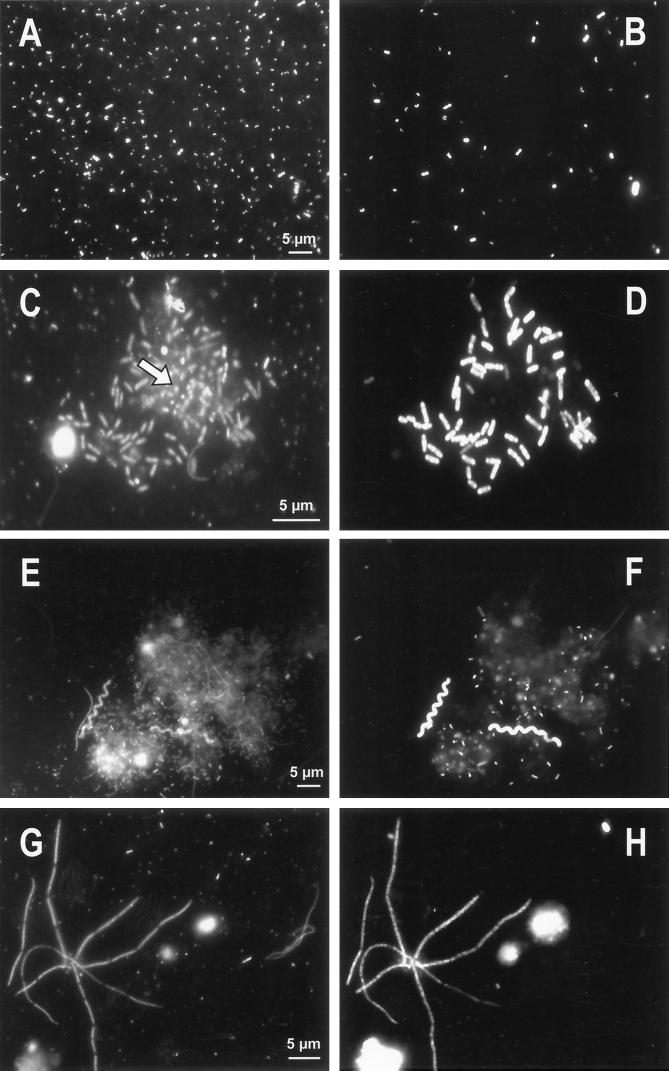
In the <5-μm treatment, significant changes in BCC occurred (Tukey HSD test, P < 0.05) compared with those of the <0.8-μm treatment and reservoir water. The shift consisted mainly of an increase in the proportions of members of the CF319a and BET42a lineages and an increase of a cluster of beta proteobacteria targeted by probe R-BT065 (Fig. 4), derived from the appearance of a treatment-specific DGGE band pattern (data not shown). Moreover, most of the very long (>5 μm), thin filaments (Fig. 5), which dominated the biomass of filamentous bacteria (Fig. 3), were detected by probe R-FL615 targeted to members of the Flectobacillus lineage (Fig. 1), accounting for 3.4% of total bacteria at t96. No cells targeted by the probe (more than 100 inspected per sample) were less than 5 μm long. Finally, the floc-forming bacteria, which by the end of the experiment numerically dominated the <5-μm treatment community (Fig. 3), showed a characteristic structure: the outer floc cells were large and hybridized with BET42a (Fig. 5), whereas the inner floc cells were detected by probe CF319a. Growth of these cells was clearly reflected in the appearance of treatment-specific bands in DGGE after a 48-h incubation (data not shown). It is noteworthy that neither R-FL615-positive cells nor flocs were found in the <0.8-μm treatment or the reservoir samples.
FIG. 5.
Micrographs of bacterial populations in experimental treatments. Total or DAPI-stained bacteria (left) and specifically Cy3-labeled bacteria (right) are shown. (A and B) Typical bacterioplankton at the beginning of the experiment in all treatments. (C to H) End of the experiment in the <5-μm treatment. Shown are the typical floc structure (C and D), with large beta proteobacteria in the outer part of flocs and small bacteria affiliated to the CF319a cluster usually located in the inner part of flocs (indicated by the position of the arrow at C); typical morphology of bacteria targeted by the CF319a probe (small cells bound in large flocs and large chain-forming cells) (F); and long filaments in DAPI preparations (G) targeted by the R-FL615 probe (H).
In the grazer-free treatment (<0.8 μm), no floc- or filament-forming bacteria appeared during the course of the study. A change in BCC occurred, compared to the untreated reservoir water, in the form of a significant increase in the proportions of BET42a-targeted bacteria, which was almost entirely due to a steep increase in the proportions of the cells affiliated to the R-BT065 cluster (Fig. 4). This treatment also yielded the shortest doubling time of R-BT065-positive bacteria compared to other phylogenetic lineages (Table 1). The DGGE banding pattern clearly indicated a qualitative shift in BCC after 72 h of incubation as well (data not shown).
In the reservoir population, neither group-specific probes nor DGGE indicated any changes in BCC during the study period, which corresponded well to the stability of other microbial parameters (compare Fig. 2 and 4).
Viral abundance and virus-mediated mortality of bacteria.
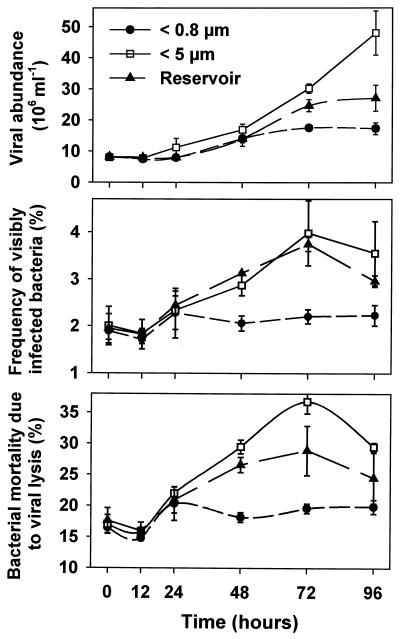
Marked differences in viral abundance, FVIC, and virus-induced mortality rates of bacteria were found among the <0.8-μm, <5-μm, and reservoir populations (Fig. 6). Viral parameters qualitatively appeared to vary inversely with bacterial abundance. For instance, in the heavily grazed <5-μm treatment, by the end of the experiment the largest numbers of viral particles (∼47 × 106 ml−1), FVIC, and calculated virus-induced mortality (30 to 37% day−1) of bacteria were detected, along with the lowest bacterial abundance (compare Fig. 2 and 6). In contrast, samples from the grazer-free treatment (<0.8 μm), in which bacteria became most abundant, yielded the lowest estimates of viral abundance (∼17 × 106 ml−1), FVIC, and virus-induced mortality (mean, 17% day−1).
FIG. 6.
Changes in total viral abundance, FVIC, and bacterial mortality due to virus-induced lysis in size fractionation treatments (<0.8 and <5 μm) exposed in dialysis bags compared to unfiltered reservoir water. Viral abundance values are the means of three replicates; error bars indicate SD. The FVIC and virus-induced mortality values are the means of duplicate subsamples from a single pooled sample representing all three replicates; the vertical bars show the range of the duplicate estimates. Statistical relationships are given in Tables 2 and 3.
As opposed to the markedly distinct trends between the grazer-free (<0.8-μm) and grazer-enhanced (<5-μm) treatments, samples from reservoir differed little from the <5.0-μm samples. In both reservoir and <5-μm treatment samples, viral abundance, FVIC, and estimates of mortality rates increased steadily from t24 to t72 and then declined slightly compared to those for the <0.8-μm treatment, in which viral parameters were relatively invariant.
Grazer versus virus-induced mortality.
In the grazer-enhanced (<5 μm) treatment, grazing mortality increased steadily during the experiment from about 25% to peak values of around 200% of bacterial production rates (Fig. 2) while virus-induced mortality increased in parallel from about 17 to 37% of bacterial stock per day (Fig. 6). In reservoir water samples, grazing by protists removed an average of about 50% and viral lysis removed about 25% of bacterial production. In comparison, bacterial mortality due to virus-induced lysis in the treatment without grazers (0.8 μm) was about 17% (Fig. 6).
Statistical relationships.
Pooling data across treatments (Table 2), bacterial abundance was negatively related to grazing rates as well as to MCV. This corresponded to the time course data from individual treatments (Fig. 2), showing the decrease in cell concentrations and increase in average bacterial cell size with increases in grazing. Considering only samples in which grazers were present (<5-μm and reservoir samples), MCV was positively related to grazing rates (r = 0.87, n = 35).
TABLE 2.
Correlation relationships of basic parameters
| Parametera | Correlation coefficient (r) | n | Significance (P) |
|---|---|---|---|
| [Bacteria] vs avg cell vol | −0.281 | 52 | 0.0434 |
| [Bacteria] vs Thy incorporation cell−1 h−1 | −0.039 | 51 | 0.7850 |
| [Bacteria] vs total grazing | −0.475 | 35 | 0.0035 |
| [Bacteria] vs HNF grazing | −0.453 | 35 | 0.0058 |
| [Bacteria] vs [Virus] | −0.133 | 52 | 0.3503 |
| Thy incorporation cell−1 h−1 vs avg cell volume | −0.041 | 51 | 0.7771 |
| Thy incorporation cell−1 h−1 vs total grazing | −0.110 | 35 | 0.5335 |
| Thy incorporation cell−1 h−1 vs HNF grazing | −0.095 | 35 | 0.5884 |
| Thy incorporation cell−1 h−1 vs [virus] | −0.073 | 51 | 0.6114 |
| Total grazing vs HNF grazing | 0.997 | 35 | <0.0001 |
| [Virus] vs avg cell vol | 0.694 | 52 | <0.0001 |
| [Virus] vs total grazing | 0.695 | 35 | <0.0001 |
| [Virus] vs HNF grazing | 0.683 | 35 | <0.0001 |
| Avg cell vol vs total grazing | 0.866 | 35 | <0.0001 |
| Avg cell vol vs HNF grazing | 0.876 | 35 | <0.0001 |
Bacterial concentration are indicated as [Bacteria], cell specific thymidine incorporation rates as Thy incorporation cell−1 h−1, average bacterial cell volume as avg cell vol, total protist grazing (ciliate and heterotrotrophic nanoflagellate) rate as total grazing, heterotrophic nanoflagellate grazing as HNF grazing, and concentration of viruses as [virus]. Significant relationships (P < 0.05) are shown in bold.
In contrast to grazing, the viral concentration was related neither to bacterial concentration nor to cell-specific incorporation rates of thymidine. Viral numbers were positively relatable to MCV (r = 0.70, n = 52) as well as to grazing rates (r = 0.70, n = 35). Correlations of FVIC (Table 3) largely followed those found for viral concentrations. Similar, close relationships between FVIC and MCV, as well as grazing rates (r = 0.70), were apparent. Infection frequencies were also positively correlated with ratios of virus to bacterial concentration and cell-specific thymidine incorporation rates.
TABLE 3.
Correlation of FVIC with bacterial and HNF parameters
| Parametera | Correlation coefficient (r) | Significance (P) |
|---|---|---|
| Bacterial abundance | −0.288 | 0.2507 |
| Avg cell volume | 0.699 | 0.0008 |
| [Virus]/[Bacteria] | 0.580 | 0.0103 |
| Production/cell | 0.546 | 0.0176 |
| Total grazing | 0.702 | 0.0090 |
| HNF grazing rate | 0.685 | 0.0120 |
For all correlations n = 18, except those with grazing (n = 12).
DISCUSSION
Overall, our results considerably extend the conclusions of previous studies concerning mortality-induced shifts in natural bacterioplankton populations. First, we demonstrated morphological and genotypic stability of an in situ community when production about equals moderate mortality (50 and 30% day−1 due to protists and virus, respectively). In the reservoir, both bacterial cell concentrations and MCV were quasi-constant within the study period (Fig. 2). The community composition showed no significant shifts with time in the proportions of cells detected by the CF319a, BET42a, R-BT065, and GAM42a probes (Fig. 4), and it is noteworthy that 75 to 85% of all bacterial cells were consistently detected by FISH. Further evidence of taxonomic stability was provided through DGGE since there were no obvious differences in the band patterns of reservoir populations when the band patterns of the t0, t48, and t72 populations were compared (data not shown). Earlier investigations have shown time course alterations in manipulated bacterioplankton over periods of a few days, but the in situ variability was not assessed (19, 20, 38, 45). Our data show that in situ populations can represent a stable background against which changes in manipulated populations may be judged. However, our data also show that a significant environmental change (e.g., a sudden relief from grazing or enhancement of grazing or a pulse of organic substrate), which disturbs the established balance between population-specific growth and mortality rates of bacteria, results in shifts in BCC (38).
Our findings with regard to the effects of grazers on bacterioplankton also extend the results of previous studies. Many changes in natural bacterioplankton communities have been associated with grazer activity. Major known phenomena are alterations in the size structure of the bacterial community, shifts in activity rates, and changes in taxonomic structure (see, e.g., reference 19, 20, 38, 40, and 45). Perhaps the best studied is the induction of filament or floc formation. This formation can be induced in a variety of taxa in natural populations (see, e.g., references 18, and 38) and may simply be growth rate associated (16). Interestingly, no filaments were found with mature phages. This phenomenon has been reported previously for the oxic layer of the Plußsee Lake (52), indicating that filaments can be not only resistant to grazing but also—for unknown reasons—resistant to viral infection. Such a life strategy may allow survival in the presence of competitive dominants in terms of organic matter acquisition.
A probe covering one specific lineage within the CF cluster (Fig. 1) detected a significant number of filamentous bacteria in the <5-μm treatment at t96 (Fig. 3). Bacteria targeted by R-FL615 formed >25% of total biomass (Fig. 3). This lineage so far contains one described species, Flectobacillus major. One 16S rDNA sequence from our clone libraries and several other sequences from bacteria from various lakes are also affiliated with this cluster (Fig. 1). A recently described isolate from this lineage, Flectobacillus sp. strain MWH38, forms filamentous cells in continuous culture when exposed to a mixotrophic flagellate (16). This perhaps explains our observation that the formation of filaments hybridized by R-FL615 was found only under conditions of increased HNF grazing. The filaments targeted by the R-FL615 probe also showed a very high growth rate (μ = 1.31 day−1), although this estimate was based only on the total number of R-FL615-positive filaments. The filaments were typically composed of a chain of cells, and they significantly lengthened between t72 and t96 of the experiment (from 12.9 ± 7.6 to 25.3 ± 12.2 μm, mean ± standard deviation [SD]). Therefore, our estimates of the growth rate of this bacterial group are conservative.
In our experiment, flocs eventually dominated the bacterial biomass in the grazing-enhanced <5-μm treatment. We found that flocs (usually 10 to 50 μm across) can have a distinct taxonomic structure, with cells targeted by the BET42a probe on the surface of the floc and cells targeted by the CF319a probe consistently in the interior (Fig. 5). While studies employing FISH have reported on the distinct taxonomic nature of particle-associated, as opposed to free-living, bacteria (see, e.g., references 27 and 54), a taxonomic structuring of flocs has not been revealed previously. However, we can only speculate about the mechanisms or factors controlling such taxonomic zonation. Along with the selective HNF grazing, there is the possibility of specific cometabolism of different phylotypes representing different metabolic types on such particles (35).
This study represents one of the very few attempts to directly estimate both grazer and virus-induced bacterial mortality and the effects of grazer and virus populations on natural bacterial communities. In studies focusing on the effects of grazing on BCC, viral activity is often ignored (19, 38, 45), while investigations of virus-induced bacterial mortality commonly estimate grazing mortality by simply multiplying grazer abundance by a conversion factor (see, e.g., references 32, 43, and 51). To our knowledge, there have been few attempts, using direct estimates of grazer and viral activity, to partition mortality between grazer- and virus-induced mortality in natural populations (9, 13). For instance, Fuhrman and Noble (9) using coastal seawater from southern California, estimated grazing mortality by using FLB disappearance rates and virus-induced mortality from (i) declines in radiolabeled DNA, (ii) production rates of virus-sized particles, and (iii) frequency of visibly infected bacterial cells. The three methods of estimating virus-induced mortality largely concurred and gave estimates similar to that for grazing, i.e., about 50% day−1 (9). Our estimates for the reservoir bacterioplankton mortality are in a comparable range; protist grazing was twice as high as virus-induced lysis, accounting for ∼50 and 25% of production, respectively.
We derived virus-induced mortality from infection frequencies. It should be noted that while such estimates appear robust (9), they are very sensitive to the conversion factors used, especially the factor relating the portion of the infection period during which viral particles are detectable (e) (3). When a virus dilution approach was used for estimating the frequency of infected cells (FIC) and the virus-induced mortality of bacteria (M. Weinbaues and M. G. Höfle, unpublished data) in samples from the surface water of the Baltic Sea and coastal and offshore waters of the Mediterranean Sea, the FIC was only similar to data obtained by TEM when the highest conversion factors were used for relating FVIC to FIC (Weinbauer and Höfle, unpublished). Since average conversion factors were used in the present study, the estimates of viral mortality of bacteria might be an underestimation. While our estimation of the absolute magnitudes of virus-induced mortality from infection frequencies perhaps should be treated with caution, the importance of virus-induced mortality relative to grazing did differ between treatments.
Cells in the “grazer-free” treatment, subject only to an apparently low mortality rate from virus-induced lysis, (∼17% of production [Fig. 6]) increased markedly in concentration (Fig. 2). While the average cell morphology, as indicated by the mean cell volume, appeared constant (Fig. 2), there was a shift in BCC. It consisted of an increase in the proportion of BET42a-positive bacteria, made up almost entirely of cells detected by probe R-BT065. This probe is targeted to a small cluster that contains three 16S rDNA sequences cloned from the <0.8-μm and <5-μm treatments and clone sequences from other aquatic habitats (Fig. 1). The cluster represents one branch of a recently described lineage of cosmopolitan freshwater beta proteobacteria (12). The DGGE band patterns of the t0 and t72 communities also differed, with the appearance of distinct bands at t72. Additionally, the significant shift in BCC was reflected by marked differences in doubling times of different bacterial groups (Table 1), with the highest values being detected for the R-BT065-positive cells in the “grazer-free” treatment.
Interestingly, viral concentrations were lowest in the grazer-free (<0.8-μm) treatment, intermediate in the reservoir population, and highest in the grazer-enhanced (<5-μm) treatment (Fig. 6). The same trend was found when TEM was used to count viruses in selected samples (data not shown). In marine mesocosms, a trend of virus concentrations increasing with flagellate grazer concentrations has been reported (32), while in another study, roughly parallel trends of bacterivory and virus-induced mortality were found (13). However, not previously reported is the close relationship we found between infection frequencies and grazing rates (Table 3).
We cannot reject the possibility that the differences in taxonomic composition influence “average” latent periods and hence the frequencies of visibly infected cells. However, not only infection frequencies but also viral abundances were highest in the <5-μm treatment. Furthermore, infection rates were also correlated with ratios of virus to bacterial abundance, which appears a reasonable relationship. The mechanism behind an apparent synergy between grazing and virus-induced mortality is unclear. We can only speculate that protist grazing may reduce bacterial diversity (20) and that there may be a reciprocal relationship between the diversity of a bacterial community and virus-induced mortality (46). Alternatively, infection rates may shift with changes in individual cells. Cell-specific production and activity are stimulated by grazing (29, 33, 39), e.g., due to the release of organic and inorganic nutrients. Higher growth rates might be associated with enhanced receptor formation on the cell surface, which may result in a greater chance of phage attachment and thus higher infection frequencies.
We observed large changes in the bacterioplankton communities in which grazing was greatly reduced or enhanced relative to the largely invariant natural reservoir community. The effects of both flagellate grazing and viruses appear to vary among different bacterial groups (compare Figs. 2 to 4 and 6 and Table 1). For example, in our experiment, filaments appeared resistant to grazing and viral infection. Given these differences, the invariant nature of the reservoir community suggests that among bacteria there are population-specific growth and removal rates. The changes in the bacterial communities which occurred in the manipulated treatments (<0.8- and <5-μm treatments) can thus be attributed to sudden alterations in the balance established in situ between population-specific growth and removal rates of bacteria.
ACKNOWLEDGMENTS
This study was supported by the Grant Agency of the Czech Republic (research grant 206/99/0028 awarded to K. Šimek); by CNRS international project “Regulation of Heterotrophic Planktonic Prokaryotes—PICS 1111”; by instrument grant AS CR “Microanalysis of microbial communities” program 1036, P 1011802; and by the Max-Planck Society.
We thank R. Psenner for valuable comments on an earlier version of the manuscript, F.-O. Glöckner for assistence with probe design and ARB, and R. Rossello-Mora for a tutorial on calculating phylogenetic trees.
REFERENCES
- 1.Alfreider A, Pernthaler J, Amann R, Sattler B, Glöckner F-O, Ville A, Psenner R. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake using in situ hybridization. Appl Environ Microbiol. 1996;62:2138–2144. doi: 10.1128/aem.62.6.2138-2144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binder B. Reconsidering the relationship between virally induced bacterial mortality and frequency of infected cells. Aquat Microb Ecol. 1999;18:207–215. [Google Scholar]
- 4.Bradbak G, Heldal M, Thingstad T F, Riemann B, Haslund O H. Incorporation of viruses into the budget of microbial C-transfer. A first approach. Mar Ecol Prog Ser. 1992;83:273–280. [Google Scholar]
- 5.Brosius J, Dull T J, Noller H F. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1980;77:201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchholz-Cleven B, Rattunde B, Straub K. Screening for genetic diversity of isolates of anaerobic Fe(II)-oxidizing bacteria using DGGE and whole-cell hybridization. Syst Appl Microbiol. 1997;20:301–309. [Google Scholar]
- 7.Fuchs B M, Zubkov M V, Sahm K, Burkill P H, Amann R. Changes in community composition during dilution cultures of marine bacterioplankton as assessed by flow cytometric and molecular biological techniques. Environ Microbiol. 2000;2:191–201. doi: 10.1046/j.1462-2920.2000.00092.x. [DOI] [PubMed] [Google Scholar]
- 8.Fuhrman J A. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 9.Fuhrman J A, Noble R T. Viruses and protists cause similar bacterail mortality in coastal seawater. Limnol Oceanogr. 1995;40:1236–1242. [Google Scholar]
- 10.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K-H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 11.Glöckner F O, Babenzien H-D, Wulff J, Amann R. Phylogeny and diversity of Achromatium oxaliferum. Syst Appl Microbiol. 1999;22:28–38. doi: 10.1016/S0723-2020(99)80025-3. [DOI] [PubMed] [Google Scholar]
- 12.Glöckner F-O, Zaichikov E, Belkova N, Denissova L, Pernthaler J, Pernthaler A, Amann R. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phyogenetic clusters including an abundant group of actinobacteria. Appl Environ Microbiol. 2000;66:5053–5065. doi: 10.1128/aem.66.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guixa-Boixereu N, Lysnes K, Pedros-Álio C. Viral lysis and bacteriovory during a phytoplankton bloom in a coastal water microcosm. Appl Environ Microbiol. 1999;65:1949–1958. doi: 10.1128/aem.65.5.1949-1958.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagström Ä, Pinhassi J, Zweifel U L. Biogeographical diversity among marine bacterioplankton. Aquat Microb Ecol. 2000;21:231–244. [Google Scholar]
- 15.Hahn M W, Höfle M G. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microb Ecol. 2001;35:113–121. doi: 10.1111/j.1574-6941.2001.tb00794.x. [DOI] [PubMed] [Google Scholar]
- 16.Hahn M W, Moore E R B, Höfle M G. Bacterial filament formation, a defense mechanism against flagellate grazing, is growth rate controlled in bacteria of different phyla. Appl Environ Microbiol. 1999;65:25–35. doi: 10.1128/aem.65.1.25-35.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollibaugh J T, Wong P S, Murrell M C. Similarity of particle-associated and free-living bacterial communities in northern San Francisco Bay, California. Aquat Microb Ecol. 2000;21:103–114. [Google Scholar]
- 18.Jürgens K, Güde H. The potential importance of grazing-resistant bacteria in planktonic systems. Mar Ecol Prog Ser. 1994;112:169–188. [Google Scholar]
- 19.Jürgens K, Pernthaler J, Schalla S, Amann R. Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl Environ Microbiol. 1999;65:1241–1250. doi: 10.1128/aem.65.3.1241-1250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lebaron P, Servais P, Troussellier M, Courties C, Vives-Rego J, Muyzer G, Bernard L, Guindulain T, Schäfer H, Stackebrandt E. Changes in bacterial community structure in seawater mesocosms differing in their nutrient status. Aquat Microb Ecol. 1999;19:255–267. [Google Scholar]
- 21.Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans A D L, Van Elsas J D, De Bruijn F J, editors. Molecular microbial ecology manual. 2nd ed. Vol. 3. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–27. [Google Scholar]
- 22.Neef A, Zaglauer A, Meler H, Amann R, Lemmer H, Schleifer K H. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl Environ Microbiol. 1996;62:4329–4339. doi: 10.1128/aem.62.12.4329-4339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noble R T, Fuhrman J A. Breakdown and microbial uptake of marine viruses and other lysis product. Aquat Microb Ecol. 1998;20:1–11. [Google Scholar]
- 24.Norland S. The relationship between biomass and volume of bacteria. In: Kemp P, Sherr B F, Sherr E B, Cole J, editors. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis; 1993. pp. 303–308. [Google Scholar]
- 25.Pernthaler J, Glöckner F-O, Unterholzner S, Alfreider A, Psenner R, Amann R. Seasonal community and population dynamics of pelagic Bacteria and Archaea in a high mountain lake. Appl Environ Microbiol. 1998;64:4299–4306. doi: 10.1128/aem.64.11.4299-4306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pernthaler J, Posch T, Šimek K, Vrba J, Amann R, Psenner R. Contrasting bacterial strategies to coexist with a flagellate predator in an experimental microbial assemblage. Appl Environ Microbiol. 1997;63:596–601. doi: 10.1128/aem.63.2.596-601.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ploug H, Grossart H-P, Azam F, Jorgensen B B. Photosynthesis, respiration, and carbon turnover in sinking marine snow from surface waters of Southern California Bight: implications for the carbon cycle in the ocean. Mar Ecol Prog Ser. 1999;179:1–11. [Google Scholar]
- 28.Posch T, Pernthaler J, Alfreider A, Psenner R. Cell-specific respiratory activity of aquatic bacteria studied with the tetrazolium reduction method, cyto-clear slides, and image analysis. Appl Environ Microbiol. 1997;63:867–873. doi: 10.1128/aem.63.3.867-873.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posch T, Simek K, Vrba J, Pernthaler S, Nedoma J, Sattler B, Sonntag B, Psenner R. Predator-induced changes of bacterial size-structure and productivity studied on an experimental microbial community. Aquat Microb Ecol. 1999;18:235–246. [Google Scholar]
- 30.Proctor L A, Okubo A, Fubrman J A. Calibrating estimates of phage-induced mortality in marine bacteria: ultrastructural studies of marine bacteriophage development from one-step growth experiments. Microb Ecol. 1993;25:161–182. doi: 10.1007/BF00177193. [DOI] [PubMed] [Google Scholar]
- 31.Riemann B, Søndergaard M. Carbon dynamics in eutrophic, temperate lakes. Amsterdam, The Netherlands: Elsevier Science Publishers; 1986. [Google Scholar]
- 32.Riemann L, Steward G F, Azam F. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl Environ Microbiol. 2000;66:578–587. doi: 10.1128/aem.66.2.578-587.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders R W, Caron D A, Berninger U G. Relationship between bacteria and heterotrophic nanoplankton in marine and freshwaters: an inter-ecosystem comparison. Mar Ecol Prog Ser. 1992;86:1–14. [Google Scholar]
- 34.Sherr E B, Sherr B F. Protistan grazing rates via uptake of fluorescently labelled prey. In: Kemp P, Sherr B F, Sherr E B, Cole J, editors. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis; 1993. pp. 695–701. [Google Scholar]
- 35.Sieburth J M. The trophic roles of bacteria in marine ecosystems are complicated by synergistic-consortia and mixotrophic cometabolism. Prog Oceanogr. 1998;21:117–128. [Google Scholar]
- 36.Šimek K, Bobková J, Macek M, Nedoma J, Psenner R. Ciliate grazing on picoplankton in a eutrophic reservoir during the summer phytoplankton maximum: a study at the species and community level. Limnol Oceanogr. 1995;40:1077–1090. [Google Scholar]
- 37.Šimek K, Jürgens K, Nedoma J, Comerma M, Armengol J. Ecological role and bacterial grazing of Halteria spp.: small oligotrichs as dominant pelagic ciliate bacteriovores. Aquat Microb Ecol. 2000;22:43–56. [Google Scholar]
- 38.Šimek K, Kojecka P, Nedoma J, Hartman P, Vrba J, Dolan D R. Shifts in bacterial community composition associated with different microzooplankton size fractions in a eutrophic reservoir. Limnol Oceanogr. 1999;44:1634–1644. [Google Scholar]
- 39.Šimek K, Macek M, Sed'a J, Vyhnálek V. Possible food chain relationships between bacterioplankton, protozoans, and cladocerans in a reservoir. Int Rev Gesanter Hydrobiol. 1990;75:583–596. [Google Scholar]
- 40.Šimek K, Vrba J, Pernthaler J, Posch T, Hartman P, Nedoma J, Psenner R. Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl Environ Microbiol. 1997;63:587–595. doi: 10.1128/aem.63.2.587-595.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sime-Ngando T, Bouredier G, Amblard C, Pinel-Alloul B. Short-term variations in specific biovolumes of different bacterial forms in aquatic ecosystems. Microb Ecol. 1991;21:211–226. doi: 10.1007/BF02539155. [DOI] [PubMed] [Google Scholar]
- 42.Simon M, Azam F. Protein content and protein synthesis rates of planktonic marine bacteria. Mar Ecol Prog Ser. 1989;51:201–213. [Google Scholar]
- 43.Steward G F, Smith D C, Azam F. Abundance and production of bacteria and viruses in the Bering and Chukchi Seas. Mar Ecol Prog Ser. 1996;131:287–300. [Google Scholar]
- 44.Suttle C A. Enumeration and isolation of viruses. In: Kemp P, Sherr B F, Sherr E B, Cole J, editors. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis; 1993. pp. 121–133. [Google Scholar]
- 45.Suzuki M T. Effect of protistan bacterivory on coastal bacterioplankton diversity. Aquat Microb Ecol. 1999;20:261–272. [Google Scholar]
- 46.Thingstad T F. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr. 2000;45:1320–1328. [Google Scholar]
- 47.Tuomi T, Kuuppo P. Viral lysis and grazing of bacteria in nutrient- and carbon-manipulated brackish water enclosures. J Plankton Res. 1997;21:923–937. [Google Scholar]
- 48.van Hannen E J, Veninga M, Bloem J, Grons H J, Laanbroek H J. Genetic changes in the bacterial community structure associated with protistan grazers. Arch Hydrobiol. 1999;145:25–38. [Google Scholar]
- 49.Verhagen F J M, Laanbroek H J. Effects of grazing by flagellates on competition for ammonium between nitrifying and heterotrophic bacteria in chemostats. Appl Environ Microbiol. 1992;58:1962–1969. doi: 10.1128/aem.58.6.1962-1969.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinbauer M, Fuks D, Peduzzi P. Distribution of viruses and dissolved DNA along a coastal trophic gradient in the northern Adriatic Sea. Appl Environ Microbiol. 1993;59:4074–4082. doi: 10.1128/aem.59.12.4074-4082.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinbauer M, Höfle M G. Significance of viral lysis and flagellate grazing as controlling factors of bacterioplankton production in an eutrophic lake. Appl Environ Microbiol. 1998;64:431–438. doi: 10.1128/aem.64.2.431-438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinbauer M, Höfle M G. Size-specific mortality of lake bacterioplankton by natural virus communities. Aquat Microb Ecol. 1998;15:103–113. [Google Scholar]
- 53.Weinbauer M, Peduzzi P. Significance of viruses versus heterotrophic nanoflagellates for controlling bacterial abundance in the Northern Adriatic Sea. J Plankton Res. 1995;17:1851–1856. [Google Scholar]
- 54.Weiss P, Schweitzer B, Aman R, Simon M. Identification in situ and dynamics of bacteria on limnetic organic aggregates (lake snow) Appl Environ Microbiol. 1996;62:1998–2005. doi: 10.1128/aem.62.6.1998-2005.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]