Abstract
To study the influence of phosphoglucomutase (PGM) activity on exopolysaccharide (EPS) synthesis in glucose- and lactose-growing Streptococcus thermophilus, a knockout PGM mutant and a strain with elevated PGM activity were constructed. The pgmA gene, encoding PGM in S. thermophilus LY03, was identified and cloned. The gene was functional in Escherichia coli and was shown to be expressed from its own promoter. The pgmA-deficient mutant was unable to grow on glucose, while the mutation did not affect growth on lactose. Overexpression of pgmA had no significant effect on EPS production in glucose-growing cells. Neither deletion nor overexpression of pgmA changed the growth or EPS production on lactose. Thus, the EPS precursors in lactose-utilizing S. thermophilus are most probably formed from the galactose moiety of lactose via the Leloir pathway, which circumvents the need for a functional PGM.
Exopolysaccharides (EPSs) produced by Streptococcus thermophilus have received a great deal of interest recently, since they are important for the rheological behaviour and texture of fermented milk (for a review, see reference 7). Furthermore, polysaccharides with defined properties produced by safe lactic acid bacteria have a potential as thickeners, stabilizers and texturizers. The biosynthetic pathways for EPSs from precursors in the form of nucleotide sugars have been the focus of recent studies (26, 27), and the genes coding for the enzymes necessary for EPS synthesis in S. thermophilus have been cloned (8, 25, 26). However, less is known about the regulation of the metabolic flux toward the precursor nucleotide sugars. UDP galactose and UDP glucose are formed from galactose 1-phosphate and glucose 1-phosphate (α-G1P), and the genes and encoded enzymes in the Leloir pathway which are involved have been characterized in S. thermophilus (20, 21).
Most strains of S. thermophilus are considered galactose negative, since they do not ferment galactose, and the galactose moiety of lactose is secreted when they are grown on lactose (28). However, the genes necessary for galactose catabolism are present, even though they are normally not expressed (6, 11). At the branching point between glycolysis and the Leloir pathway, α-phosphoglucomutase (PGM) is present, which interconverts α-G1P and glucose 6-phosphate. It is thus a key enzyme between glycolysis and the Leloir enzymes for EPS synthesis. When glucose is the carbon source, PGM is required for the synthesis of α-G1P, which is used by uridyltransferases for the production of nucleotide sugars. In S. pneumoniae, deletion of a gene coding for PGM decreased capsule biosynthesis to less than 10% (9). On the other hand, deletion of PGM in Escherichia coli leads to the accumulation of intracellular polysaccharide in galactose-growing cells since this prevents α-G1P from entering glycolysis (1, 14), showing that the role of PGM is dependent on the principal flux direction across the enzyme. Recent results showed a linear relationship between PGM activity and EPS production on different sugars in S. thermophilus (4). However, the direction of the metabolic flux across PGM has not been determined in S. thermophilus.
In this study, we describe the role of PGM in glucose and lactose metabolism of S. thermophilus and its relationship to EPS production. The results indicate that the Leloir pathway is responsible for the generation of EPS precursors in lactose-growing streptococci.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
The strains and plasmids used are listed in Table 1. S. thermophilus strains were cultivated at 37 or 42°C. E. coli strains were grown in Luria-Bertani medium or Terrific broth. S. thermophilus was maintained in Elliker broth (Difco) supplemented with 1% meat extract (Merck) or in M17 (Oxoid). In controlled batch experiments, a modified MRS medium (5) with 25 g of Bacteriological peptone (Oxoid) per liter and 5 g of yeast nitrogen base without amino acids (Difco) per liter was used. Lactose (75 g liter−1) or glucose (50 g liter−1) was used as the carbohydrate source. Spectinomycin and ampicillin were used at concentrations of 100 μg ml−1. Erythromycin was used at a concentration of 200 μg ml−1 for E. coli and 0.2 μg ml−1 for S. thermophilus. In batch experiments, only the precultures were grown under selective pressure and cell extracts from the late exponential growth phase were checked for PGM activity. The batch cultures were performed in duplicate in Bioflo III fermentors (New Brunswick Scientific Co., Edison, N.J.) at 42°C with an inital volume of 1.5 liters. Anaerobic conditions were maintained by nitrogen flushing in the headspace. The pH was adjusted to 6.2 by the automatic addition of 10 N NaOH.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | Cloning host | Life Technologies Inc. |
| W1485 pgmΔ::tet | pgm mutant | 14 |
| TMB2001 | W1485 pgmΔ::tet with pFL36 | This study |
| S. thermophilus strains | ||
| LY03 | Commercial yoghurt strain | IMDST01b |
| TMB6001 | LY03 with integrated pFL41; Emr | This study |
| TMB6002 | LY03 with pFL42; Specr | This study |
| Plasmids | ||
| pUC19 | Cloning vector; Ampr, lacZ′ | 17 |
| pFL36 | pgmA in pUC19 | This study |
| pLZ12spec | Shuttle vector; Specr | 10 |
| pG+host9 | Thermosensitive shuttle vector; Emr | 15 |
| pFL38 | pLZ12spec with pgmA from pFL36 in forward orientation, lacking promoter | This study |
| pFL39 | pLZ12spec with pgmA from pFL36 in reverse orientation, lacking promoter | This study |
| pFL41 | pG+host9 with internal pgmA fragment | This study |
| pFL42 | pLZ12spec with pgmA with native promoter, forward orientation | This study |
Emr, erythromycin resistant; Specr, spectinomycin resistant; Ampr, ampicillin resistant.
IMDST01 is the culture collection name at Vrije University, Brussels, Belgium.
DNA techniques and cloning procedure.
Plasmid DNA was isolated from E. coli using Quantum kits (Bio-Rad Laboratories AB). For S. thermophilus plasmid preparations, the Quantum miniprep kit was modified so that the cell pellets were dissolved in 140 μl of cell resuspension solution plus 60 μl of 100 mg of lysozyme solution ml−1 and incubated at 37°C for 15 min. Chromosomal DNA was isolated using Qiagen genomic tips. DNA digestion, dephosphorylation, agarose gel electrophoresis, and ligation were performed by standard methods (3). All DNA enzymes were obtained from Roche Diagnostics Scandinavia AB. Gel fragments were purified using Qiaquick kits (Qiagen Inc., Santa Clarita, Calif.). Ultracompetent E. coli strains were prepared and transformed as previously described (12). S. thermophilus was transformed as previously described (18). PCR was performed with Pwo DNA polymerase, and Inverse PCR was performed with an Expand High Fidelity system, using standard conditions as recommended by the manufacturer (Roche). Cycle sequencing was performed with the dRhodamine Terminator Ready kit (Perkin-Elmer, Boston, Mass.). Degenerate primers 5′-GGAATTCCACNGCNGGNATGMGNGG-3′ (restriction sites underlined) and 5′-GCTCTAGAGCRTCNGGRTCNGTNGC-3′ were used to amplify an internal fragment of pgmA. The product was cut with EcoRI and XbaI, inserted into pUC19, and sequenced. Chromosomal S. thermophilus DNA was cleaved in separate reactions with XbaI, EcoRI, and HindIII. The cleaved DNA was religated and used as a template for inverse PCR with primers 5′-GAAACTGCAGTTGGACGAAGGCTCTCGA-3′ and 5′-GAAACTGCAGATGCCGACGTATTGGTTG-3′. The products were cloned in pUC19 and used to sequence the remainder of pgmA and to map the surroundings. The whole gene was finally amplified from chromosomal DNA by PCR and sequenced. The gene was sequenced in both directions, using DNA templates from at least two independent PCR amplicons.
Overexpression of pgmA.
For overexpression of pgmA in E. coli, the gene was amplified from S. thermophilus LY03 with primers 5′-CGGGATCCTTTAGTTGTGATACAATGTAAG-3′ and 5′-TGCGAGCTCTTGGTGTAGCAGCGAAAG-3′, cleaved with BamHI and SacI, and inserted into pUC19, yielding pFL36. The resulting plasmid was transformed into W1485pgmΔ::tet, giving TMB2001. For promoter analysis in S. thermophilus, pFL36 was digested with BamHI and partially digested with EcoRI and the pgmA gene was inserted into pLZ12spec, resulting in pFL38. The pgmA gene was also cloned in the other direction in pFL39 by ligating the pgmA gene from pFL36, obtained by BamHI and partial Asp700I cleavage, into XbaI-blunt and BamHI-digested pLZ12spec. pFL42 was created by PCR-amplifying the pgmA gene with a promoter using primers CG GGATCCCGTAATTCTACTCAGCAGTGGA and 5′-TGCGAGCTCTTGGTGTAGCAGCGAAAG-3′. The product was cleaved with BamHI and inserted into pLZ12spec (BamHI-EcoRI blunt).
Inactivation of pgmA.
An internal 1,091-bp EcoRI-HindIII fragment of pgmA was cut from pFL36 and inserted into pG+host9, yielding pFL41. pFL41 was transformed into LY03, and the transformants were recovered at 30°C. Plasmid integration was selected for by growing the cells at 37°C in M17 containing erythromycin. Colonies on agar plates were checked by PCR for single integration. One integrant was chosen and checked for pgmA activity. The strain was named TMB 6001 and maintained at 42°C or −80°C to prevent loss of the integration.
Measurement of growth, substrate consumption, and product formation.
The optical density at 620 nm was used to monitor cell growth after appropriate dilution of samples. Samples for substrate and product determination were filtered through 0.2-μm-pore-size filters immediately after sampling and kept at 4°C until analysis. Lactose, galactose, and lactate were separated at 65°C on a cation-exchange column (Aminex HPX-87H; Bio-Rad) and quantified using a refractive index detector (RID 6A; Shimadzu Co.). The mobile phase was 5 mM H2SO4 at a flow rate of 0.6 ml min−1. At least three samples were taken for EPS analysis during the exponential growth phase. EPSs were isolated by spinning after removal of cells and proteins with trichloracetic acid and precipitation with acetone, as described by Degeest and De Vuyst (5). The anthrone method with a glucose standard was used for quantification of the isolated EPS (16). The EPS concentrations were converted to moles of carbon (cmol) by using 30 g cmol−1, as in the repeating units consisting of galactose and glucose (5).
PGM and PMM measurements.
Cell extracts were prepared from cells harvested in the mid-exponential phase that were washed twice in 50 mM potassium phosphate buffer (pH 7.0). E. coli was lysed by incubation of the cells in buffer containing lysozyme, DNase, RNase, and phenylmethylsulfonyl fluoride, followed by freezing and thawing. S. thermophilus cell extracts were prepared in an X-press (AB Biox). Cell debris was removed by centrifugation at 15,000 × g for 15 min at 4°C. PGM and phosphomannomutase (PMM) activities were measured in 50 mM triethanolamine buffer (pH 7.2) with 5 mM MgCl2 in coupled assays at 37°C by monitoring the formation of NADPH spectrophotometrically. The assay mixture for PGM contained 0.4 mM NADP+, 65 μM glucose 1,6-bisphosphate, and 2 U of glucose 6-phosphate dehydrogenase ml−1, and the assay was started by addition of α-G1P to 1 mM (23). The PMM assay mixture contained 1 mM NADP+, 25 mM glucose 1,6-bisphosphate, 0.5 U of phosphomannose isomerase ml−1, 0.5 U of phosphoglucose isomerase ml−1, and 0.5 U of glucose 6-phosphate dehydrogenase ml−1, and the assay was started with 1 mM mannose 1-phosphate (24). Protein concentrations were determined by the Micro BCA method (Pierce, Rockford, Ill.).
Bioinformatic methods.
PGM protein sequences were downloaded from Swissprot, Sptrembl, and GenBank. Putative sequences coding for PGMs and PMMs were obtained from Streptococcus pyogenes, Streptococcus pneumoniae, Streptococcus mutans, and Enterococcus faecalis sequencing projects by using BlastX (2) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Multiple alignment of protein sequences was performed with the ClustalX program (29). Trees were constructed with the ClustalX program and visualized with TreeView (19).
Nucleotide sequence accession number.
The gene sequence described in this section has been submitted to the EMBL database, accession number AJ243290.
RESULTS AND DISCUSSION
Functional analysis of the pgmA gene.
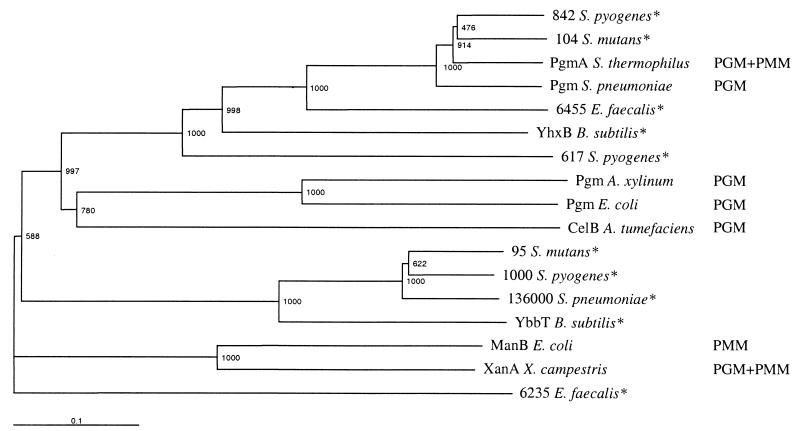
Enzymes with PGM and/or PMM activity can be phylogenetically grouped according to their substrate specificity (31). A search through current sequencing projects revealed that S. pyogenes, S. pneumoniae, and S. mutans all had at least two PGM and/or PMM homologues (Fig. 1). Based on alignments, these could be divided into two groups, and the group closest to E. coli PGM was sought for unique motifs since none of the sequences had been experimentally evaluated at the time of cloning. The motifs TAGMRG and ATDPDA were chosen for construction of degenerate primers for PCR. These primers were used for amplification of an internal fragment of a gene, pgmA, which was homologous to the expected group of putative PGMs. The remainder of the gene was cloned by inverse PCR and then sequenced. After the cloning of pgmA from S. thermophilus, the pgm gene of S. pneumoniae, which also belongs to the same group, was characterized and shown to have PGM activity (9). The 573-amino-acid protein product of pgmA showed 82% identity to PGM of S. pneumoniae, 46% identity to YHXB of Bacillus subtilis, and 24% identity to PGM of E. coli.
FIG. 1.
Phylogenetic tree with characterized PGMs and/or PMMs and putative proteins from sequencing projects. Bootstrap values from 1,000 bootstrap trials are marked in the branches. The number in front of the organism name is the contig number, or the location of the sequence in the case of the single contig of S. pneumoniae. Experimentally confirmed enzyme functions are indicated to the right and uncharacterized proteins are marked with an asterisk. Accesion numbers: XanA, P29955; ManB, P24175; YbbT, O87090; CelB, Q44417; Pgm A. xylinum, P38569; Pgm E. coli, P36938; YhxB, P18159; Pgm S. pneumoniae, Q9RP94.
To verify the function of pgmA, a 1,863-bp fragment containing the gene was expressed under the lac promoter of pUC19 in E. coli W1485 pgmΔ::tet, which lacks PGM. The resulting strain showed a PGM activity of 15 U mg of protein−1, while the host E. coli strain had an activity of <0.01 U mg of protein−1.
To establish whether pgmA is the only gene coding for PGM activity in S. thermophilus LY03, a knockout mutant, TMB 6001, was constructed by single-crossover recombination. No detectable PGM activity was found in TMB 6001 grown on lactose, while the parent strain had a PGM activity of 0.9 U mg of protein−1, showing that the gene is coding for a unique PGM in this strain.
Sequence analysis showed potential promoter regions upstream of the gene, and to confirm this, the pgmA gene was cloned onto a multicopy vector. When 50 bp upstream of the gene was included in the insert (pFL38), no elevated PGM activity was observed in S. thermophilus LY03. However, when 210 bp upstream of the gene was included (pFL42), the PGM activity in the resulting strain, TMB 6002, was 7 U mg of protein−1 in lactose-growing cells.
To investigate if pgmA also encodes PMM activity, cell extracts from LY03, TMB 6001, and TMB 6002 were checked for PMM activity. The specific PMM activities in lactose-grown LY03 and TMB 6002 were 0.2 and 2 U mg of protein−1, respectively, while no detectable PMM activity could be found in TMB 6001. Thus, there was a linear relationship between the PGM and PMM activities in the cell extracts, with the PMM activity being about fourfold lower than the PGM activity. This shows that the enzyme is bifunctional, like XanA from Xanthomonas campestris (13). The results also suggest that some presumed PGMs could have PMM activity, and they question the function of the other group of putative PGM homologues in the phylogenetic tree (Fig. 1).
Role of PGM in EPS production.
To investigate the role of PGM at the branching point between catabolism and anabolism in S. thermophilus LY03, strains with different levels of PGM activity were cultivated in batch cultures under conditions optimal for EPS production (5). With glucose as the carbon source, growth was rapid in LY03 and TMB 6002, and the yields of lactate and EPS were similar in these two strains (Table 2). The EPS yields decreased at the end of the exponential growth phase (reference 5 and data not shown), and for comparison a point was chosen in the late exponential phase. When the PGM-deficient TMB 6001 was transferred from M17-lactose to M17-glucose agar plates, no colonies appeared. Adaption to glucose was also done in liquid cultures that were transferred from MRS-lactose to MRS-glucose. After a long lag phase, this resulted in growth on glucose, but when the PGM activity was assayed in the cultures it had been restored to that of the host strain, even under antibiotic pressure, indicating that PGM activity was essential for growth on glucose. To verify that the growth deficiency on glucose was due to lack of PGM activity and not a result of polar effects from the insertion inactivation of pgmA, the terminal part of pgmA was integrated with another pG+host9 derivative, which kept pgmA intact but abolished downstream transcription (data not shown). In this insertion mutant, growth on glucose was unaffected. Furthermore, sequence analysis showed a putative gene downstream of pgmA in the opposite direction, encoding a protein with homology to methylentetrahydrofolate reductases.
TABLE 2.
Maximum specific growth rates and yields on glucose
| Strain | μmax (h−1)a | Yglucose, lactate (cmol/cmol)a | Yglucose, EPS (cmol/cmol)a |
|---|---|---|---|
| LY03 | 1.27 ± 0.06 | 0.87 ± 0.01 | 0.0083 ± 0.0014 |
| TMB6001 | —b | — | — |
| TMB6002 | 1.26 ± 0.02 | 0.87 ± 0.06 | 0.0072 ± 0.0019 |
Yields given are those after consumption of 28 g of glucose per liter. Values are from duplicate cultivations. μmax is the maximum specific growth rate; Y represents yield.
—, TMB6001 did not grow on glucose.
PGM mutants have been constructed from a number of gram-negative bacteria (1, 13, 14, 30, 32) and recently from the gram-positive bacterium S. pneumoniae (9). No growth defects have been reported in any of the gram-negative bacteria. However, in all cases important effects on polysaccharide production have been observed. These are probably due to changed levels of α-G1P that serves as a precursor for UDP sugars, but not to a total cessation in the supply, since the UDP sugars are needed in cell wall biosynthesis and for polysaccharide formation. For S. pneumoniae, the pgm mutants grew slowly and promotion of second-site suppressor mutations outside the pgm gene was observed, which restored growth to the normal level (9). It is not clear whether these mutations resulted in PGM activity or whether other pathways were activated, since these authors could not measure PGM activity in their cell extracts due to NADPH oxidases. Our results show that PGM activity is necessary for the growth of S. thermophilus on glucose, and PGM seems to be the only way to provide the cells with α-G1P on this substrate. Furthermore, this study demonstates that pgmA is the unique gene coding for PGM in S. thermophilus LY03.
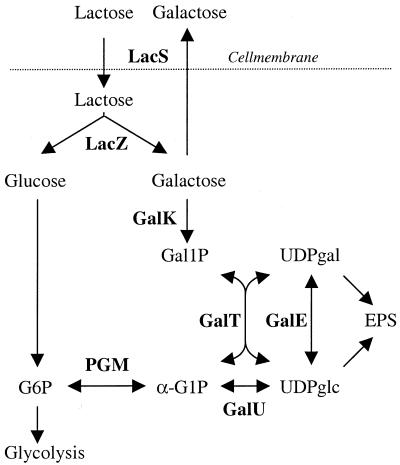
Lactose, the carbohydrate source of interest in milk fermentation, is transported into S. thermophilus by LacS, which can act either as a lactose-galactose antiporter or as a proton symporter (22). Lactose is split by β-galactosidase into glucose and galactose (Fig. 2). The glucose moiety enters the glycolysis, yielding lactate, while LacS normally secretes galactose in exchange for lactose. Even if LY03 does not grow on galactose, it assimilates some of the galactose derived from lactose fermentation (5). It was thus not evident how altered PGM levels would affect the EPS production on lactose. With lactose as the carbon source, there was no significant difference in maximum specific growth rate between the strains (Table 3). The EPS and galactose yields decreased during fermentation, whereas the lactate yield increased. The cells entered the stationary phase when about 50 g of lactose liter−1 had been consumed, and for comparison a point just before this was chosen (Table 3). No differences between the strains could be seen at this stage. Interestingly, neither deletion nor overexpression of pgmA had any significant effect on growth or EPS production in the lactose-grown strains. This implies that the precursors for EPS production originate from the galactose moiety of lactose and also that the net flux over PGM is close to zero in lactose-growing S. thermophilus LY03. Furthermore, sufficient galactose is taken up to provide precursors for the cell wall and EPS, but only small amounts enter the central metabolism. After prolonged fermentation into the stationary phase, more of the galactose was metabolized, resulting in lower galactose yields and higher lactate yields (Table 4). At this stage, the strain lacking PGM activity, TMB 6001, had secreted more galactose than the other strains, demonstrating that PGM activity is necessary to ferment galactose to lactate.
FIG. 2.
Lactose metabolism in S. thermophilus. Abbreviations for metabolites: Gal1P, galactose 1-phosphate; UDPgal, UDP galactose; UDPglc, UDP glucose; G6P, glucose 6-phosphate. Enzymes are in bold: LacS, lactose transporter; LacZ, β-galactosidase; GalK, galactokinase; GalT, galactose 1-phosphate uridyltransferase; GalE, UDP galactose 4-epimerase; GalU, UDP glucose pyrophosphorylase.
TABLE 3.
Maximum specific growth rates and yields on lactose
| Strain | μmax (h−1)a | Ylactose, galactose (cmol/cmol)a | Ylactose, lactate (cmol/cmol)a | Ylactose, EPS (cmol/cmol)a |
|---|---|---|---|---|
| LY03 | 1.01 ± 0.11 | 0.39 ± 0.02 | 0.47 ± 0.02 | 0.0021 ± 0.0003 |
| TMB6001 | 0.98 ± 0.08 | 0.42 ± 0.02 | 0.49 ± 0.01 | 0.0022 ± 0.0010 |
| TMB6002 | 0.96 ± 0.02 | 0.40 ± 0.00 | 0.48 ± 0.00 | 0.0023 ± 0.0000 |
Yields given are those after consumption of 45 g of lactose per liter. Values are from duplicate cultivations. μmax is the maximum specific growth rate; Y represents yield.
TABLE 4.
Yields 25 h after inoculation
| Strain | Ylactose, galactose (cmol/cmol)a | Ylactose, lactate (cmol/cmol)a |
|---|---|---|
| LY03 | 0.28 | 0.54 |
| TMB6001 | 0.34 | 0.51 |
| TMB6002 | 0.27 | 0.58 |
Y represents yield.
The flux-controlling function of PGM was investigated by deletion and overexpression of the pgmA gene. Recent studies of the same strain showed a linear relationship between the activity of PGM, UDP galactose 4-epimerase, and UDP glucose pyrophosphorylase and EPS production in S. thermophilus LY03 (4). Their data suggested that PGM might play a controlling role in the flux from glucose 6-phosphate to EPS production. However, our results after overexpressing pgmA indicated that the physiological amount of PGM was not limiting for EPS production (Table 2 and 3).
The major implication of these results is that it is possible to decouple lactose metabolism in S. thermophilus by deletion of pgmA. The glucose moiety is used for energy metabolism, while the galactose moiety can be used for anabolic reactions. This implies a great potential for metabolic engineering of EPS production in this organism, which could be fine-tuned by controlled expression of the enzymes in the Leloir pathway.
ACKNOWLEDGMENTS
We thank Emmanuelle Maguin for the kind gift of pG+host9.
This work was supported by the European Community FAIR programme, contract CT-98-4267.
REFERENCES
- 1.Adhya S, Schwartz M. Phosphoglucomutase mutants of Escherichia coli K-12. J Bacteriol. 1971;108:621–626. doi: 10.1128/jb.108.2.621-626.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1996. [Google Scholar]
- 4.Degeest B, De Vuyst L. Correlation of activities of the enzymes α-phosphoglucomutase, UDP-galactose 4-epimerase, and UDP-glucose pyrophosphorylase with exopolysaccharide biosynthesis by Streptococcus thermophilus LY03. Appl Environ Microbiol. 2000;66:3519–3527. doi: 10.1128/aem.66.8.3519-3527.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Degeest B, De Vuyst L. Indication that the nitrogen source influences both amount and size of exopolysaccharides produced by Streptococcus thermophilus LY03 and modelling of the bacterial growth and exopolysaccharide production in a complex medium. Appl Environ Microbiol. 1999;65:2863–2870. doi: 10.1128/aem.65.7.2863-2870.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Vos W M, Vaughan E E. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol Rev. 1994;15:217–237. doi: 10.1111/j.1574-6976.1994.tb00136.x. [DOI] [PubMed] [Google Scholar]
- 7.De Vuyst L, Degeest B. Heteropolysaccharides from lactic acid bacteria. FEMS Microbiol Rev. 1999;23:153–177. doi: 10.1111/j.1574-6976.1999.tb00395.x. [DOI] [PubMed] [Google Scholar]
- 8.Griffin A M, Morris V J, Gasson M J. The cpsABCDE genes involved in polysaccharide production in Streptococcus salivarius ssp. thermophilus strain NCBF 2393. Gene. 1996;183:23–27. doi: 10.1016/s0378-1119(96)00405-2. [DOI] [PubMed] [Google Scholar]
- 9.Hardy G G, Caimano M J, Yother J. Capsule biosynthesis and basic metabolism in Streptococcus pneumoniae are linked through the cellular phosphoglucomutase. J Bacteriol. 2000;182:1854–1863. doi: 10.1128/jb.182.7.1854-1863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Husmann L K, Scott J R, Lindahl G, Stenberg L. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect Immun. 1995;63:345–348. doi: 10.1128/iai.63.1.345-348.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutkins R, Morris H A, McKay L L. Galactokinase activity in Streptococcus thermophilus. Appl Environ Microbiol. 1985;50:777–780. doi: 10.1128/aem.50.4.777-780.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 13.Köplin R, Arnold W, Hötte B, Simon R, Wang G, Pühler A. Genetics of xanthan production in Xanthomonas campestris: the xanA and xanB genes are involved in UDP-glucose and GDP-mannose biosynthesis. J Bacteriol. 1992;174:191–199. doi: 10.1128/jb.174.1.191-199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu M, Kleckner N. Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli. J Bacteriol. 1994;176:5847–5851. doi: 10.1128/jb.176.18.5847-5851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maguin E, Prevost H, Ehrlich S D, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris D L. Quantitative determination of carbohydrates with Dreywood's anthrone reagent. Science. 1948;107:254–255. doi: 10.1126/science.107.2775.254. [DOI] [PubMed] [Google Scholar]
- 17.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 18.O'Sullivan T F, Fitzgerald G F. Electrotransformation of industrial strains of Streptococcus thermophilus. J Appl Microbiol. 1999;86:275–283. doi: 10.1046/j.1365-2672.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- 19.Page R D. Tree View: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 20.Poolman B. Energy transduction in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:125–147. doi: 10.1111/j.1574-6976.1993.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 21.Poolman B, Royer T J, Mainzer S E, Schmidt B F. Carbohydrate utilization in Streptococcus thermophilus: characterization of the genes for aldose 1-epimerase (mutarotase) and UDPglucose 4-epimerase. J Bacteriol. 1990;172:4037–4047. doi: 10.1128/jb.172.7.4037-4047.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poolman B, Royer T J, Mainzer S E, Schmidt B F. Lactose transport system of Streptococcus thermophilus: a hybrid protein with homology to the melibiose carrier and enzyme III of phosphoenolpyruvate-dependent phosphotransferase systems. J Bacteriol. 1989;171:244–253. doi: 10.1128/jb.171.1.244-253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian N, Stanley G A, Hahn-Hägerdal B, Rådström P. Purification and characterization of two phosphoglucomutases from Lactococcus lactis subsp. lactis and their regulation in maltose- and glucose-utilizing cells. J Bacteriol. 1994;176:5304–5311. doi: 10.1128/jb.176.17.5304-5311.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sa-Correia I, Darzins A, Wang S K, Berry A, Chakrabarty A M. Alginate biosynthetic enzymes in mucoid and nonmucoid Pseudomonas aeruginosa: overproduction of phosphomannose isomerase, phosphomannomutase, and GDP-mannose pyrophosphorylase by overexpression of the phosphomannose isomerase (pmi) gene. J Bacteriol. 1987;169:3224–3231. doi: 10.1128/jb.169.7.3224-3231.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stingele F, Mollet B. Disruption of the gene encoding penicillin-binding protein 2b (pbp2b) causes altered cell morphology and cease in exopolysaccharide production in Streptococcus thermophilus Sfi6. Mol Microbiol. 1996;22:357–366. doi: 10.1046/j.1365-2958.1996.00121.x. [DOI] [PubMed] [Google Scholar]
- 26.Stingele F, Neeser J R, Mollet B. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J Bacteriol. 1996;178:1680–1690. doi: 10.1128/jb.178.6.1680-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stingele F, Newell J W, Neeser J R. Unraveling the function of glycosyltransferases in Streptococcus thermophilus Sfi6. J Bacteriol. 1999;181:6354–6360. doi: 10.1128/jb.181.20.6354-6360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas T D, Crow V L. Selection of galactose fermenting Streptococcus thermophilus in lactose-limited chemostat cultures. Appl Environ Microbiol. 1984;48:186–191. doi: 10.1128/aem.48.1.186-191.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uttaro A D, Cangelosi G A, Geremia R A, Nester E W, Ugalde R A. Biochemical characterization of avirulent exoC mutants of Agrobacterium tumefaciens. J Bacteriol. 1990;172:1640–1646. doi: 10.1128/jb.172.3.1640-1646.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Videira P A, Cortes L L, Fialho A M, Sa-Correia I. Identification of the pgmG gene, encoding a bifunctional protein with phosphoglucomutase and phosphomannomutase activities, in the gellan gum-producing strain Sphingomonas paucimobilis ATCC 31461. Appl Environ Microbiol. 2000;66:2252–2258. doi: 10.1128/aem.66.5.2252-2258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou D, Stephens D S, Gibson B W, Engstrom J J, McAllister C F, Lee F K, Apicella M A. Lipooligosacharide biosyntesis in pathogenic Neisseria. Cloning, identification, and characterization of the phosphoglucomutase gene. J Biol Chem. 1994;269:11162–11169. [PubMed] [Google Scholar]