Abstract
Through the national surveillance program for Campylobacter spp., nine broiler chicken farms that were infected with Campylobacter jejuni in at least five rotations in 1998 were identified. One additional farm, located at the island of Bornholm where divided slaughter is used extensively, was also selected. Twelve broiler houses located on 10 farms were included in the study. The C. jejuni isolates collected from the selected houses during the surveillance were typed using fla typing and macrorestriction profiling (MRP), and a subset of the isolates, representing each of the identified clones, was serotyped according to the Penner scheme. Pulsed-field gel electrophoresis typing using SmaI and KpnI revealed that the majority of houses (11 of 12) carried identical isolates in two or more broiler flocks. Such persistent clones were found in 63% of all flocks (47 of 75). The majority of persistent clones (7 of 13) had fla type 1/1, but MRPs distinguished between isolates from different houses, and fla type 1/1 clones belonged to different serotypes. Seven houses carried persistent clones that covered an interval of at least four broiler flock rotations, or at least one half year. The dominant fla type (1/1) was represented by 44% of isolates, or by at least one isolate from 31 of 62 broiler flocks. This significantly exceeded the prevalence of fla type 1/1 C. jejuni isolates that we have estimated from other studies and suggests that isolates carrying this fla type are overrepresented in flocks with recurrent Campylobacter problems. The MRPs of clones belonging to fla type 1/1 serotype O:2 isolated from persistently infected flocks shared a high percentage of bands compared to the remaining isolates, indicating that some clones that have the ability to cause persistent infections in broiler farms are highly related to each other.
Campylobacter jejuni has become recognized as a major cause of human enteritis in many industrialized countries during the past 2 decades (4). In Denmark the incidence rate was 64 recorded cases per 100,000 inhabitants in 1998, and in 1999 it reached a peak, with 78 recorded cases per 100,000 inhabitants (1, 2). In developed countries the majority of human Campylobacter cases are sporadic and the consumption of undercooked poultry meat is considered an important source, whereas outbreaks are rare (4). In 1998 46% of Danish broiler flocks were infected with C. jejuni (26). Contamination control at the farm level would be one way of reducing consumer exposure; however, the epidemiology of C. jejuni in broiler flocks is still unclear.
It is well established that biosecurity measures such as improved hygiene barriers and staff education, together with pest and rodent control, reduce the frequency of infected flocks (3, 5, 26). However, full control of C. jejuni contamination has not been obtained yet. It has not been established whether common contamination reservoirs, for instance those that are attached to the broiler production chain (hatchery and abattoir, etc.), are of importance, although a British study (18) and a Danish study (26) point in that direction.
The diversity of C. jejuni is considerable, and investigations of contamination reservoirs therefore depend on the application of suitable typing tools. In recent years, various genotyping methods for C. jejuni have been described, with the most frequently used ones being PCR-restriction fragment length polymorphism (PCR-RFLP) analysis of the flaA gene (fla typing) and macrorestriction profiling (MRP) using pulsed-field gel electrophoresis (PFGE) (25). fla typing has the advantage of being fast and cheap. PFGE-MRP, on the other hand, is more laborious but has the advantage of being significantly more discriminatory and better able to describe the significant genetic variation of C. jejuni (16, 21). Penner serotyping has less discriminatory power than the genotyping tools, but on the other hand, it is standardized and has been used successfully in several studies worldwide (12, 14, 17).
The present study focused on farms where the proportion of positive flocks was above average in 1998. The purpose was to evaluate the potential epidemiological importance of farm-associated clones that occur in successive rotations versus clones that are common between different farms. An additional aim was to characterize such clones, when identified, using serotyping, fla typing, and MRP.
MATERIALS AND METHODS
Selection of isolates and culture conditions.
A total of 94 Campylobacter isolates were investigated. These isolates originated from a collection of fecal isolates sampled at the time of slaughter from healthy broiler chickens during 1998 as part of an ongoing surveillance program for Campylobacter spp. in Danish broilers (26).
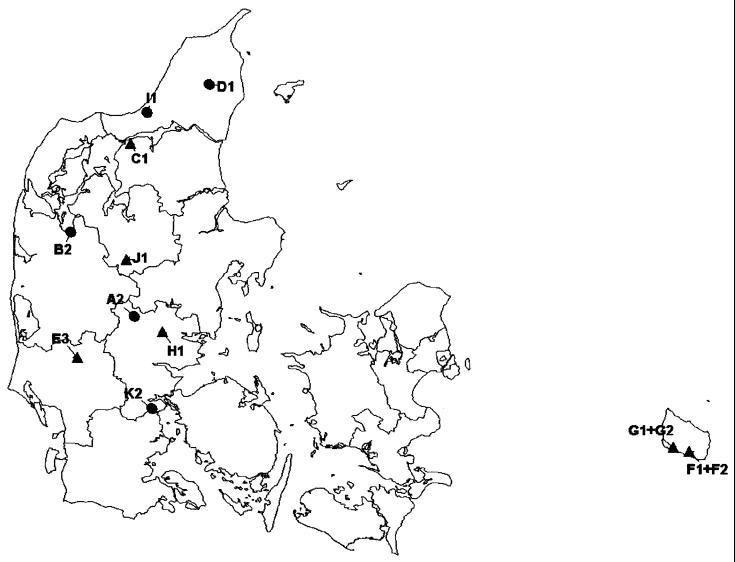
The 94 isolates originated from broiler farms where at least five flocks in most or all houses were positive for Campylobacter spp. in 1998. Nine farms in total (A, B, C, D, E, F, H, J, and K) met that criterion; eight were located on the peninsula of Jutland, and one was located on the island of Bornholm (Fig. 1). From each farm in Jutland one house that was infected by C. jejuni in at least five rotations was selected at random. If more than one house met this criterion, a house where flocks were slaughtered in a low number of batches was selected. Broiler house numbers are used in combination with farm designations below (e.g., A2 stands for farm A, house 2).
FIG. 1.
Geographical distribution of the broiler farms under study. ▴, farms contaminated by C. jejuni clones with fla type 1/1 or 1/1b; ●, other farms.
One additional farm (G) on the island of Bornholm was included. The Bornholm farms raised older chickens up to 49 days of age, produced five flocks each year, and slaughtered in several batches. Both farms consisted of two houses, and both houses were under study.
From all flocks (75 flocks in total, of which C. jejuni was not isolated from 10 flocks), one sample, consisting of a pool of 10 cloacal swabs taken at the abattoir, was collected from each batch at slaughter (Table 1). However, on farms F and G up to eight batches were slaughtered from each flock. In most cases one or two isolates were selected from the first batches that were slaughtered from each flock. From J1, one isolate from rotation 2 and one isolate from rotation 4 were lost during storage, and the same was the case with one isolate from H1 rotation 7.
TABLE 1.
Serotypes and genotypes of C. jejuni isolates from broiler flocks
| Farm and house no. | Rotation | Isolate no. | fla types (DdeI/AluI) | MRP type
|
Clonal groupa | Serotype | ||
|---|---|---|---|---|---|---|---|---|
| SmaI | KpnI | BamHI | ||||||
| A2 | 1 | P0181 | 48/48 | 22 | 27 | 1 | 12 | O:4 complex |
| 2 | P0182 | 1/1 | 10 | 9 | NDb | O:5 | ||
| P0183 | 1/1 | 10 | 10 | ND | O:5 | |||
| 3 | P0184 | 48/48 | 22 | 28 | 1 | (12) | ND | |
| 5 | P0185 | 48/48 | 22 | 27 | 1 | 12 | ND | |
| 6 | P0186 | 5/5 | 14 | 31 | ND | O:12 | ||
| B2 | 1 | P0173 | 11/11a | 12 | 14 | 21 | ND | |
| 2 | P0174 | 33/33 | 19 | 23 | 2 | 8 | O:4 complex | |
| 3 | P0175 | 11/11a | 12 | 15 | 21 | ND | ||
| 4 | P0176 | 33/33 | 19 | 23 | 2 | 8 | ND | |
| P0177 | 33/33 | 19 | 23 | 2 | 8 | ND | ||
| 5 | P0178 | 9/9 | 34 | 41 | ND | ND | ||
| 6 | P0179 | 66/66 | 19 | 23 | 2 | 8 | O:4 complex | |
| 7 | P0180 | 33/33 | 19 | 23 | 2 | 8 | ND | |
| C1 | 1 | P0187 | 7/7 | 30 | 38 | ND | ND | |
| 1 | P0188 | 51/51a | 25 | 32 | ND | ND | ||
| 2 | P0189 | 67/67 | 27 | 34 | ND | ND | ||
| 3 | P0190 | 1/1 | 8 | 8 | 6 | 5 | O:53 | |
| P0191 | 1/1 | 8 | 8 | 7 | 5 | ND | ||
| 4 | P0192 | 1/1 | 8 | 8 | 7 | 5 | ND | |
| 5 | P0193 | 1/1 | 8 | 8 | 8 | 5 | ND | |
| P0194 | 1/1 | 8 | 8 | 8 | 5 | ND | ||
| 6 | P0195 | 1/1 | 8 | 8 | 8 | 5 | ND | |
| 7 | P0196 | 1/1 | 8 | 8 | 8 | 5 | ND | |
| D1 | 1 | P0158 | 65/65 | 26 | 33 | ND | ND | |
| 2 | P0159 | 5/5 | 23 | 29 | ND | O:12 | ||
| P0160 | 5/5 | 23 | 29 | 22 | ND | |||
| 3 | P0161 | 2/2 | 14 | 17 | 9 | 13 | ND | |
| P0162 | 31/31b | 17 | 21 | ND | O:29(42) | |||
| 4 | P0163 | 1/1 | 9 | 13 | ND | O:4 complex | ||
| 6 | P0164 | 8/8 | 32 | 39 | ND | O:42 | ||
| E3 | 1 | P0204 | 1/1b | 5 | 4 | 10 | 1 | O:2 |
| P0205 | 1/1b | 5 | 4 | 10 | 1 | ND | ||
| 2 | P0206 | 1/1b | 5 | 4 | 10 | 1 | ND | |
| 3 | P0207 | 1/1b | 5 | 4 | 10 | 1 | ND | |
| 4 | P0208 | 1/1b | 5 | 4 | 10 | 1 | ND | |
| P0209 | 33/33 | 18 | 22 | 3 | 9 | ND | ||
| 5 | P0210 | 66/68 | 20 | 25 | 4 | (9) | O:4 complex | |
| 6 | P0211 | 66/66 | 18 | 22 | 5 | 9 | ND | |
| 7 | P0212 | 1/1b | 6 | 5 | 11 | (1) | O:2 | |
| P0213 | 1/1b | 6 | 5 | 12 | (1) | ND | ||
| F1 | 1 | P0103 | 1/1 | 4 | 7 | 13 | 6 | O:2 |
| 2 | P0102 | 1/1 | 4 | 7 | 13 | 6 | ND | |
| P0166 | 30/30 | 15 | 19 | 14 | 11 | O:11 | ||
| P0117 | 30/30 | 15 | 19 | 14 | 11 | ND | ||
| 3 | P0100 | 2/2 | 14 | 17 | 9 | 13 | O:1,44 | |
| P0118 | 2/2 | 14 | 17 | 9 | 13 | ND | ||
| P0120 | 30/30 | 15 | 19 | 14 | 11 | ND | ||
| P0121 | 30/30 | 15 | 19 | 14 | 11 | ND | ||
| 4 | P0105 | 1/1 | 7 | 6 | 15 | 7 | ND | |
| P0106 | 1/1 | 37 | 12 | 20 | O:1,44 | |||
| 5 | P0104 | 1/1 | 7 | 6 | 15 | 7 | O:2 | |
| F2 | 1 | P0112 | 1/1 | 4 | 7 | 13 | 6 | ND |
| 2 | P0110 | 1/1 | 4 | 7 | 13 | 6 | ND | |
| P0111 | 1/1 | 4 | 7 | 13 | 6 | O:2 | ||
| 3 | P0122 | 2/2 | 14 | 17 | 9 | 13 | O:1,44 | |
| P0123 | 2/2 | 14 | 17 | 9 | 13 | ND | ||
| P0124 | 30/30 | 15 | 19 | 14 | 11 | ND | ||
| P0125 | 30/30 | 15 | 19 | 14 | 11 | ND | ||
| P0108 | 1/1 | 4 | 7 | 13 | 6 | ND | ||
| 4 | P0115 | 39/14a | 13 | 16 | ND | ND | ||
| 5 | P0113 | 1/1 | 7 | 6 | 15 | 7 | ND | |
| P0114 | 1/1 | 7 | 6 | 15 | 7 | ND | ||
| G1 | 3 | P0141 | 2/2 | 14 | 17 | 9 | 13 | O:1,44 |
| 4 | P0132 | 8/8 | 31 | 40 | ND | O:42 | ||
| P0133 | 1/1 | 1 | 1 | 16 | 2 | ND | ||
| 5 | P0134 | 2/2 | 14 | 17 | 9 | 13 | ND | |
| P0135 | 2/2 | 14 | 17 | 9 | 13 | ND | ||
| G2 | 3 | P0140 | 1/1 | 4 | 7 | 13 | 6 | O:2 |
| 4 | P0136 | 2/2 | 14 | 17 | 9 | 13 | ND | |
| 5 | P0137 | 5/5 | 24 | 30 | ND | O:12 | ||
| P0138 | 1/1 | 11 | 11 | ND | O:4 complex | |||
| P0139 | 2/2 | 14 | 17 | 9 | 13 | ND | ||
| H1 | 1 | P0165 | 1/1 | 1 | 1 | 16 | 2 | 0:2 |
| 2 | P0166 | 1/1 | 2 | 1 | 16 | (2) | ND | |
| 3 | P0167 | 1/1 | 1 | 1 | 16 | 2 | ND | |
| P0168 | 9/9 | 35 | 42 | ND | O:31 | |||
| 4 | P0169 | 1/1 | 1 | 1 | 16 | 2 | ND | |
| 5 | P0170 | 1/1 | 1 | 1 | 16 | 2 | ND | |
| 6 | P0171 | 1/1 | 1 | 1 | 16 | 2 | ND | |
| 7 | P0172 | 69/69 | 28 | 36 | ND | O:21 | ||
| J1 | 1 | P0142 | 1/1 | 3 | 2 | 17 | 3 | O:2 |
| 4 | P0147 | 1/1 | 3 | 2 | 17 | 3 | ND | |
| 5 | P0149 | 31/31 | 16 | 20 | ND | NTc | ||
| 6 | P0153 | 44/44 | 21 | 26 | 23 | ND | ||
| P0154 | 1/1 | 4 | 3 | 18 | 4 | ND | ||
| 7 | P0156 | 1/1 | 4 | 3 | 18 | 4 | ND | |
| K2 | 1 | P0197 | 7/7 | 29 | 37 | 19 | 10 | O:4 complex |
| 2 | P0198 | 68/68 | 36 | 35 | ND | O:31 | ||
| 3 | P0199 | 35/35 | 38 | 18 | ND | O:1,44 | ||
| 5 | P0200 | 7/7 | 29 | 37 | 19 | 10 | ND | |
| 6 | P0201 | 7/7 | 29 | 37 | 19 | 10 | ND | |
| 7 | P0202 | 7/7 | 29 | 37 | 19 | 10 | ND | |
| P0203 | 7/7 | 29 | 37 | 19 | 10 | ND | ||
Parentheses indicate that the isolate showed a high degree of similarity to the clonal group. Clonal groups that were found on more than one farm are indicated in boldface.
ND, not done.
NT, nontypeable.
Bacteriological analysis was done as described previously (26). In brief, swabs were left for approximately 2 min in a tube containing 3 ml of sterile water, whereafter 10 μl was streaked onto modified CCDA plates (Oxoid CM 739/SR 155). The plates were incubated in a microaerobic atmosphere for 48 h. Isolates were identified to species level by standard procedures: oxidase (positive), catalase (positive), the ability to hydrolyze hippurate (positive), the production of indoxyl acetase (positive), and resistance to nalidixic acid (negative) and cephalothin (positive) (15).
MapInfo Professional (version 5.5; MapInfo Corporation, Troy, N.Y.) was used for topographic visualization of the farms and fla type results.
Typing methods. (i) Serotyping.
Serotyping was performed according to the Penner serotyping scheme as previously described (14) but with the use of all 66 antisera of the system (47 C. jejuni antisera and 19 Campylobacter coli antisera).
(ii) PCR-RFLP.
PCR-RFLP profiles of the flaA gene were produced as previously described using restriction endonucleases DdeI and AluI (19). Computer-assisted identification using GelCompar (Applied Maths, Kortrijk, Belgium) was used for identification of RFLP profiles, as previously described (13).
(iii) PFGE.
PFGE analysis was performed as previously described (19) using restriction endonucleases SmaI (Gibco), KpnI (Gibco), and BamHI (Gibco), with the following ramping parameters for BamHI digests: 2 to 5 s, 11 h; 6 to 12 s, 6 h; and 15 to 20 s, 5 h. MRPs were compared visually and assigned to arbitrarily defined profile types. Repeated gel runs were done to confirm profile identity or similarity. Isolates that could not be distinguished by MRPs using SmaI, KpnI, or BamHI were considered to belong to the same clone (16). Epidemiologically related isolates (isolated on the same farm) that could be distinguished by minor differences in BamHI MRPs but not by other MRPs were considered to belong to the same clone (22, 23). Epidemiologically related isolates (isolated on the same farm) that could be distinguished by not more than two band differences in each of the SmaI and KpnI MRPs and shared significant similarity in the BamHI MRPs were presumed to be related (22).
RESULTS
Twenty-three fla types, 37 SmaI MRP types, and 42 KpnI MRP types were found in this study (Table 1). Representative fla profiles are shown in Fig. 2, and MRPs are shown in Fig. 3. General features of fla types (13, 19) and of MRPs (16, 19) were as previously described.
FIG. 2.
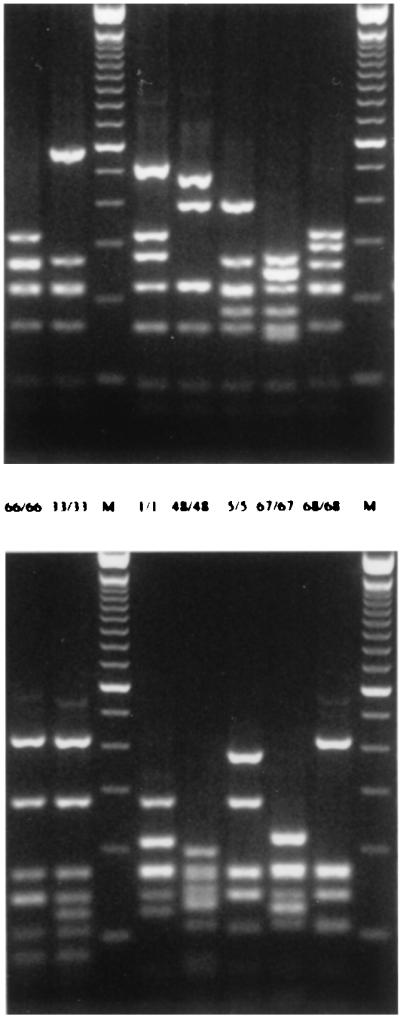
fla profiles of representative isolates using restriction endonucleases DdeI (upper panel) and AluI (lower panel). The numbers between the photographs indicate the DdeI or AluI profile type.
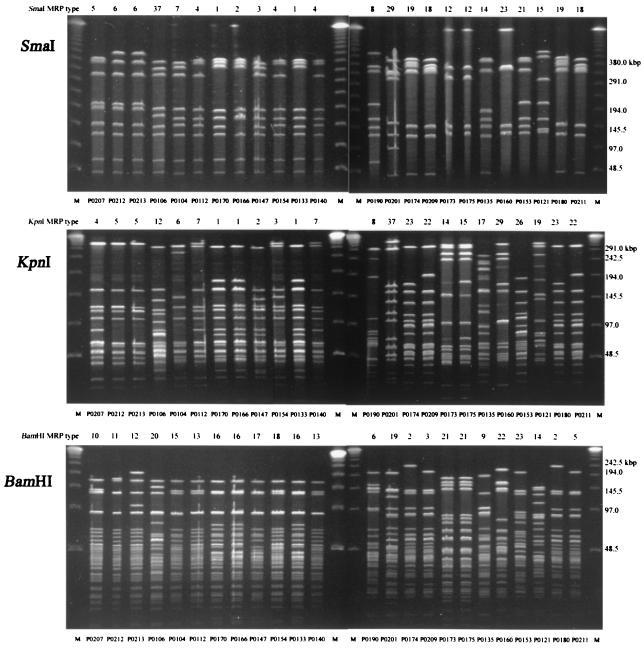
FIG. 3.
Macrorestriction profiles of representative isolates of C. jejuni using restriction endonucleases SmaI, KpnI, and BamHI. The numbers below each photograph indicate the isolate numbers as listed in Table 1. The numbers above the photographs indicate MRP types, as shown in Table 1.
Thirteen clones that occurred in more than one broiler flock were identified (Table 1). Minor differences in BamHI MRPs discriminated within two such clones; both were restricted to one broiler farm each. In clone 5, isolates P0193, P0194, P0195, and P0196 could be distinguished from isolates P0190, P0191, and P0192 by an upward band shift at around 155 kb, and in addition isolate P0190 could be distinguished by a difference in the intensity in one band and the presence of one extra band at around 48 kb. In clone 9, isolate P0209 could be distinguished from P0211 by a minor band shift at 130 kb (Fig. 3, lanes 19 and 28).
In addition, there were five isolates from four different flocks that had slightly different MRPs using SmaI and/or KpnI (not more than two band differences in each set of MRPs) than clones from the same farm (Table 1).
Clones 2 (and isolate P0166), 3, 4, 6, and 7 all had fla type 1/1, whereas clone 1 (and isolates P0212 and P0213) had the slightly different fla type 1/1b, which differed from 1/1 by a minor band shift in the AluI profile. Representative isolates from each of those clones all had serotype O:2. Furthermore, a high proportion of bands in the MRPs obtained using either restriction endonuclease were shared by all or several clones (Fig. 3).
Isolates from clones 8 and 9 were distinguished from each other by single band shifts in each set of MRPs. The positions of the upper bands in the BamHI MRPs (240 and 200 kb, respectively), the positions of two bands in the 350- to 400-kb range in the SmaI MRPs, and the positions of the second-largest band in the KpnI MRPs (165 and 190 kb, respectively) distinguished isolates from the two farms (Fig. 3, lanes 18, 19, 27, and 28). Among these isolates, fla types 33/33 and 66/66 were found on both farms, and one additional fla type, 66/68, was found on farm E3. fla profiles 33 and 66 differed by two bands using DdeI and by one band using AluI. AluI profile 68 resembled profile 66 (two band differences) more than it resembled profile 33 (Fig. 2).
Ten clones were restricted to one farm each, two clones (clones 2 and 6) were both found on two different farms, and one clone (clone 13) was found on three farms. One or more isolates from each clone were serotyped. Representatives from six clones were serotype O:2, three were serotype O:4 complex, and the remaining belonged to serotypes O:1,44; O:11, and O:53.
Isolates that were identical or highly similar to isolates from other rotations in the same farm were found in 47 flocks, or 72% of the positive flocks (63% of all flocks). A unique isolate was found in 21 flocks, or 32% of the positive flocks (28% of all flocks).
fla type 1/1 or 1/1b was represented by 41 isolates (44%), of which 33 were distributed on seven clones, including the six O:2 clones and O:53. The remaining clones had fla types 2/2 (O:1,44), 7/7 (O:4 complex), 30/30 (O:11), and 48/48 (O:4 complex), and two clones (O:4 complex) had fla type 33/33 or 66/66.
DISCUSSION
Farm-specific as well as unspecific clones were found in this study. The finding of farm-specific clones that occur in successive rotations strongly indicates that those clones persist on the individual farms, either inside the broiler house or in the near environment. In some cases, stretches of flocks where a clone was isolated were interrupted by flocks where a different isolate or no isolate was found. This can be due to shortcomings of the cultivation method or to the sampling size. Some flocks may be infected by two different strains, but only one isolate from each batch is obtained by the method used. Therefore, the importance of persistent clones, as well as of sporadic isolates, can be underestimated here. Evidently, farm-specific sources are of primary importance, as they can account for the majority of the positive flocks in this study, at least the 63% of all flocks where a persistent clone was actually isolated. In contrast, sporadic or unique isolates were found in 28% of flocks, strongly indicating that sporadic sources were of minor importance in the farms under study.
No obvious contamination source could explain the finding of clones 2 and 13 on farms located in different parts of the country. The Bornholm farms but not the remaining farms used a feed mill and abattoir located on the island of Bornholm and a Swedish hatchery. Therefore, feed, hatchery, or abattoir is not likely to be the source of common clones. This finding can be explained by a ubiquitous occurrence of the highly stable clones 2 and 13. The occurrence of two different clones on both of the Bornholm farms could be explained by the fact that the broiler flocks were divided into a high number of batches at slaughter, thereby increasing the risk of introducing contamination by new clones to the farms, in combination with the ability of those clones to persist on the farm after introduction.
Studies in several European countries (3, 5, 8) have identified risk factors such as the lack of appropriate hygiene barriers around broiler houses and infestation with insects or rodents. A possible explanation for the persistence or survival of Campylobacter clones on farms could be that local populations of rodents or beetles, etc., that are able to evacuate the house during cleaning and disinfection serve as reservoirs (3, 8, 9). It is also possible that this problem may be related to the handling of used litter. Used litter from broiler houses presumably contains large numbers of campylobacteria when removed from the house. Experiments have shown that C. jejuni has a good ability to survive during anaerobic digestion of animal waste (10, 11). An earlier study showed that houseflies can function as a vector for C. jejuni when a reservoir is located near the broiler house (20). If litter is not handled properly on the farm, it therefore has the potential to function as a continuous source of campylobacteria for broiler flocks housed on the farm. Further epidemiological investigations may clarify the role of litter handling in campylobacter colonization of broiler flocks.
Genotypic variation in C. jejuni and its impact on the applicability of genotyping methods for epidemiological studies have been extensively debated in recent years (6, 7, 24, 25). Minor differences in MRPs obtained using the frequently cutting enzyme BamHI were observed in epidemiologically related isolates, but according to generally accepted criteria for the interpretation of genotyping data, this is consistent with a clonal relationship (22, 23). Single isolates that were distinguished from clones 1, 2, 9, and 12 by minor differences in the MRPs obtained using SmaI and/or KpnI were found. The epidemiological information supports the argument that isolates P0166, P0184, P0210, P0212, and P0213 originated from clones 2, 12, 9, and 1, respectively. Assuming that those five isolates were genotypic variants of the clones, the occurrence of genotypic variants in this study was sporadic. We speculate that the identified clones may be representatives of C. jejuni strains that are well adapted for the colonization of broiler flocks and for survival around broiler houses. Successful as they are, their survival skills may not easily be improved by random genetic events, and genetically modified variants will remain outnumbered by the mother strains.
Clones 1 to 4 and clones 6 and 7 shared a high degree of similarity in MRPs. Furthermore, representative isolates from each clone all had serotype O:2, and all isolates had fla type1/1 or 1/1b. The results indicated that this group of clones may be representatives of a subgroup of C. jejuni that is largely clonal. Further investigations may clarify the importance of the identified clones in human infections.
The variable fla type identified in clones 8 and 9 showed an interesting pattern in that both clones, isolated from two different broiler farms (E3 and B2), expressed the same two fla types, which are highly similar. Harrington and colleagues (7) described intergenomic recombination events in the fla genes of some strains, and similar events may explain our finding. It is possible that fla type variation is a feature of some clonal lineages, and this should be kept in mind during epidemiological studies.
In conclusion, we have shown that local contamination reservoirs are of importance for the colonization of broiler farms with C. jejuni. Colonization by sporadic isolates was of lesser importance here but may be more significant in farms where persistent clones do not propagate. The possible existence of common contamination reservoirs is also indicated but in this study accounted for a minor proportion of infected flocks. Highly stable clones of C. jejuni were identified, of which a subgroup belonging to serotype O:2 appear to be interrelated. Further typing studies involving human and broiler isolates might help in elucidating the importance of the identified clones in relation to human infections.
ACKNOWLEDGMENTS
This study was partly supported by grants from the Danish Broiler Meat Association and from the Danish Ministry of Food, Agriculture and Fisheries.
We thank Karl Pedersen for critical reading of the manuscript, Stephen L.W. On for helpful discussions, and Connie Jenning Sørensen and Mette Hansen for excellent technical assistance.
REFERENCES
- 1.Anonymous. Campylobacter coli/jejuni. In: Hald T, Wegener H C, Jorgensen B B, editors. Annual report on zoonoses in Denmark 1998. Copenhagen, Denmark: Danish Zoonosis Centre; 1999. pp. 18–10. [Google Scholar]
- 2.Anonymous. Campylobacter coli/jejuni. In: Brondsted T, Hald T, Jorgensen B B, editors. Annual report on zoonoses in Denmark 1999. Copenhagen, Denmark: Danish Zoonosis Centre; 2000. pp. 20–23. [Google Scholar]
- 3.Berndtson E, Emanuelson U, Engvall A, Danielsson-Tham M-L. A 1-year epidemiological study of campylobacters in 18 Swedish chicken farms. Prev Vet Med. 1996;26:167–186. [Google Scholar]
- 4.Friedman C R, Neimann J, Wegener H C, Tauxe R V. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. In: Nachamkin I, Blaser M J, editors. Campylobacter. Washington D.C.: ASM Press; 2000. pp. 121–138. [Google Scholar]
- 5.Hald B, Wedderkopp A, Madsen M. Thermophilic Campylobacter spp. in Danish broiler production: a cross-sectional survey and a retrospective analysis of risk factors for occurrence in broiler flocks. Avian Pathol. 2000;29:123–131. doi: 10.1080/03079450094153. [DOI] [PubMed] [Google Scholar]
- 6.Hänninen M-L, Hakkinen M, Rautelin H. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1999;65:2272–2275. doi: 10.1128/aem.65.5.2272-2275.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrington C, Thomson-Carter F M, Carter P E. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implication for the flagellin gene typing scheme. J Clin Microbiol. 1997;35:2386–2392. doi: 10.1128/jcm.35.9.2386-2392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs-Reitsma W F, Van de Giessen A W, Bolder N M, Mulder R W A W. Epidemiology of Campylobacter spp. at two Dutch broiler farms. Epidemiol Infect. 1994;114:413–422. doi: 10.1017/s0950268800052122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapperud G, Skjerve E, Vik L, Hauge K, Lysaker A, Aalmen I, Ostroff M, Potter M. Epidemiological investigation of risk factors for Campylobacter colonization in Norwegian broiler flocks. Epidemiol Infect. 1993;111:245–255. doi: 10.1017/s0950268800056958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kearney T E, Larkin M J, Frost J P, Levett P N. Survival of pathogenic bacteria during mesophilic anaerobic digestion of animal waste. J Appl Bacteriol. 1993;75:215–219. doi: 10.1111/j.1365-2672.1993.tb02768.x. [DOI] [PubMed] [Google Scholar]
- 11.Kearney T E, Larkin M J, Levett P N. The effect of slurry storage and anaerobic digestion on survival of pathogenic bacteria. J Appl Bacteriol. 1993;74:86–93. doi: 10.1111/j.1365-2672.1993.tb03000.x. [DOI] [PubMed] [Google Scholar]
- 12.Nachamkin I, Ung H, Patton C M. Analysis of HL and O serotypes of Campylobacter strains by the flagellin gene typing system. J Clin Microbiol. 1996;34:277–281. doi: 10.1128/jcm.34.2.277-281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen E M, Engberg J, Fussing V, Petersen L, Brogren C-H, On S L W. Evaluation of phenotypic and genotypic methods for subtyping of Campylobacter jejuni isolates from humans, poultry, and cattle. J Clin Microbiol. 2000;38:3800–3810. doi: 10.1128/jcm.38.10.3800-3810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen E M, Engberg J, Madsen M. Distribution of serotypes of Campylobacter jejuni and C. coli from Danish patients, poultry, cattle and swine. FEMS Immunol Med Microbiol. 1997;19:47–56. doi: 10.1111/j.1574-695X.1997.tb01071.x. [DOI] [PubMed] [Google Scholar]
- 15.On S L W, Holmes B. Reproducibility of tolerance tests that are useful in the identification of campylobacteria. J Clin Microbiol. 1991;29:1785–1788. doi: 10.1128/jcm.29.9.1785-1788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.On S L W, Nielsen E M, Engberg J, Madsen M. Validity of SmaI-defined genotypes of Campylobacter jejuni examined by SalI, KpnI, and BamHI polymorphisms: evidence of identical clones infecting humans, poultry, and cattle. Epidemiol Infect. 1998;120:231–237. doi: 10.1017/s0950268898008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owen R J, Slater E, Telford D, Donovan T, Barnham M. Subtypes of Campylobacter jejuni from sporadic cases of diarrhoeal disease at different locations in England are highly diverse. Eur J Epidemiol. 1997;13:837–840. doi: 10.1023/a:1007497005152. [DOI] [PubMed] [Google Scholar]
- 18.Pearson A D, Greenwood M H, Feltham R K A, Healing T D, Donaldson J, Jones D M, Colwell R R. Microbial ecology of Campylobacter jejuni in a United Kingdom chicken supply chain: intermittent common source, vertical transmission, and amplification by flock propagation. Appl Environ Microbiol. 1996;62:4614–4620. doi: 10.1128/aem.62.12.4614-4620.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen L, On S L W. Efficacy of flagellin gene typing for epidemiological studies of Campylobacter jejuni in poultry estimated by comparison with macrorestriction profiling. Lett Appl Microbiol. 2000;31:14–19. doi: 10.1046/j.1472-765x.2000.00757.x. [DOI] [PubMed] [Google Scholar]
- 20.Shane S M, Montrose M S, Harrington K S. Transmission of Campylobacter jejuni by the housefly (Musca domestica) Avian Dis. 1984;29:384–391. [PubMed] [Google Scholar]
- 21.Steele M, McNab B, Fruhner L, DeGrandis S, Woodward D, Odumeru J A. Epidemiological typing of Campylobacter isolates from meat processing plants by pulsed-field gel electrophoresis, fatty acid profile typing, serotyping, and biotyping. Appl Environ Microbiol. 1998;64:2346–2349. doi: 10.1128/aem.64.7.2346-2349.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Struelens M J. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin Microbiol Infect. 1996;2:2–11. doi: 10.1111/j.1469-0691.1996.tb00193.x. [DOI] [PubMed] [Google Scholar]
- 23.Tenover F C, Arbeit R B, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wassenaar T M, Geilhausen B, Newell D G. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl Environ Microbiol. 1998;64:1816–1821. doi: 10.1128/aem.64.5.1816-1821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wassenaar T M, On S L W, Meinersmann R J. Genotyping and the consequences of genetic instability. In: Nachamkin I, Blaser M J, editors. Campylobacter. Washington D.C.: ASM Press; 2000. pp. 369–380. [Google Scholar]
- 26.Wedderkopp A, Rattenborg E, Madsen M. National surveillance of Campylobacter in broilers at slaughter in Denmark in 1998. Avian Dis. 2000;44:993–999. [PubMed] [Google Scholar]