Abstract
The Coronavirus disease 2019 (COVID-19) pandemic has affected more than 269 million worldwide, with more than five million deaths as of early December 2021. The main concerns in this pandemic include the asymptomatic nature of COVID-19, leading to the infection of many healthy people, the infectious nature of the pathogen, and its high spreading rate. The disease features have highlighted the importance of controlling this pandemic via vaccines. There has been a worldwide race to produce better, more protective, and efficacious vaccines. Simultaneously, different new variants of the virus are emerging. Therefore, there is a concern about the efficacy of the vaccines against new variants. The platform used for COVID-19 vaccine development needs to be flexible enough to enable the manufacturer to react suitably to new virus variants. We performed a comprehensive search in the online databases of PubMed, Scopus, Google Scholar, clinicaltrials.gov, WHO, ICTRP, and Cochrane until December 10th, 2021. There are 331 candidate vaccines in clinical development, with 194 in the preclinical stage and 137 in different clinical phases. Eleven platforms have been used for the development of COVID-19 vaccines, including inactivated/live attenuated virus, protein subunit, virus-like particle (VLP), non-replicating/replicating viral vectors (VVnr or VVr), VVr or VVnr plus antigen-presenting cell, bacterial antigen-spore expression vector, DNA, and RNA. The VLP-based vaccine platform is a safe, highly immunogenic, and flexible platform for developing vaccines. This review focuses on VLP-based vaccine platforms and explicitly discusses the six VLP-based COVID-19 vaccines in clinical trial phases.
Keywords: 2019-nCoV, COVID-19, Novel Coronavirus, SARS-CoV-2, Vaccines, Virus-Like Particle
1. Context
Emerging in December 2019 in China and rapidly spreading worldwide, the Coronavirus disease 2019 (COVID-19) outbreak became an international concern declared by the World Health Organization (WHO) (https://www.euro.who.int/en) in March 2020. This global pandemic has affected more than 269 million people globally, with more than five million deaths, as of December 12th, 2021 (https://www.worldometers.info/coronavirus/). Severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2), the cause of COVID-19, is a member of the Coronavirinae subfamily, with other members, including SARS-CoV and the Middle East respiratory syndrome Coronavirus (MERS-CoV). COVID-19 might be asymptomatic or have varying clinical symptoms, from minor flu-like symptoms to severe complications such as acute respiratory distress syndrome (ARDS), pneumonia, and death. As known, SARS-CoV is much more powerful, with higher lethality and aggressiveness than SARS-CoV-2. However, SARS-CoV-2 is much more infectious with a higher spreading rate. The other primary concern about SARS-CoV-2 is the asymptomatic nature of the disease (in some cases), leading to its spreading to many people via an asymptomatic patient.
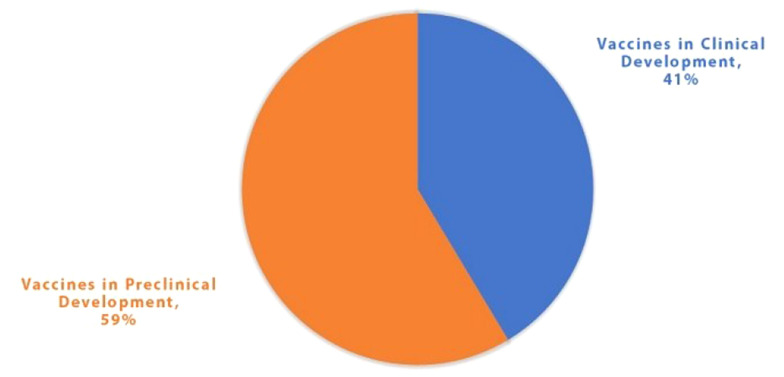
The considerable contagious nature of SARS-CoV-2 leading to the infection of millions of people globally has highlighted the importance of controlling this pandemic. This pandemic might be controlled through hygiene measures, masks, social distancing, a few antiviral drugs, and vaccines. Herd immunity is an option to acquire natural immunity through infection. However, the death rate and social implications would be catastrophic in this case (1-4). Therefore, the only practical approach to controlling the pandemic and obtaining immunity is to develop efficient vaccines. There is an attempt across the globe to create a competent vaccine against SARS-CoV-2. As of early December 2021, there were 137 vaccines in clinical development and 194 in the preclinical phase (https://www.who.int/publications/). A summary of the information on the vaccine products in clinical development (as of early December 2021) is illustrated in Figure 1.
Figure 1. Vaccine products in clinical development (as of early December 2021).
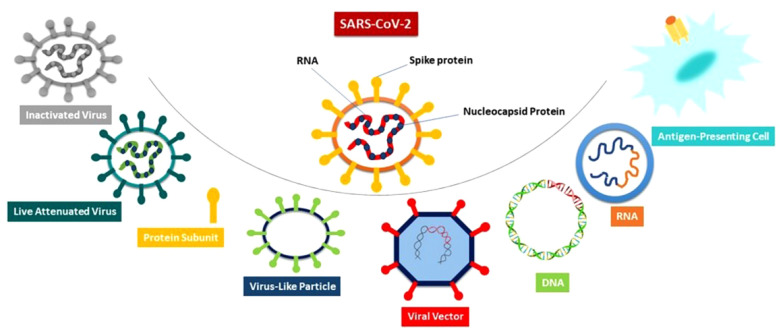
Many different platforms are used for developing vaccines against SARS-CoV-2, including the inactivated virus, Live-attenuated Virus (LAV), protein subunit, virus-like particle (VLP), viral vector (non-replicating) or VVnr, viral vector (replicating) or VVr, VVr plus antigen-presenting cell (APC), VVnr plus antigen-presenting cell, bacterial antigen-spore expression vector, DNA vaccine, and RNA vaccine (https://www.who.int/publications/). An overview of different SARS-CoV-2 vaccine platforms is done in Figure 2. This review focuses on VLP-based vaccine platforms for COVID-19, specifically six VLP-based vaccines in clinical trial phases.
Figure 2. An overview of different SARS-CoV-2 vaccine platforms.
2. Search Strategy
We performed a comprehensive literature search in the online databases of PubMed, Scopus, Google Scholar, clinicaltrials.gov, WHO, International Clinical Trials Registry Platform (ICTRP), Cochrane vaccine mapping tool, and the vaccine manufacturer's websites up to December 10th, 2021. The literature review was supposed to include SARS-CoV-2 structure, development processes and platforms used for COVID-19 vaccines, the rationale for selection of VLP platform for vaccine development, particular features of VLPs (advantages and disadvantages), the main findings in the development of VLP-based vaccine technology, WHO COVID-19 vaccine landscape for VLP-based COVID-19 vaccines in clinical trials, and the details of each VLP-based COVID-19 vaccine.
3. SARS-CoV-2 Structure
Coronaviruses have a single-stranded positive-sense RNA (+ssRNA) genome with a size of 27 - 32 kb, the most significant size among all RNA viruses (5). SARS-CoV-2 has a genome of about 29.9 kb (6) with 16 nonstructural proteins (nsp1-16) and four structural proteins (S, E, M, and N) (5, 7).
The main target for COVID-19 vaccine development is the S protein of SARS-CoV-2, an envelope glycoprotein expressed on the virus surface. This trimeric protein is composed of two major subunits: (1) S1, which is involved in receptor binding of the virus; and (2) S2, which regulates membrane fusion. Furthermore, the S protein has two major conformation states: (1) pre-, and (2) post-fusion states. There is significant diversity in coronaviruses' S proteins, which are mainly related to the S1 subunit. The S1 subunit consists of domains of receptor-binding (RBD) and N-terminal (NTD). The RBD has diversity among coronaviruses, hypothesized by the fact that different coronaviruses bind to different host-cell receptors upon entry. However, SARS-CoV and SARS-CoV-2 enter the host cell via the same receptor (angiotensin-converting enzyme 2, ACE2), which confirms the similarity in their RBD sequences (3, 7, 8).
Among the antigens mentioned above for SARS-CoV-2, it has been demonstrated that RBD serves as an important target for developing effective vaccines (9-11). In the next section, the development process of COVID-19 vaccines is summarized to show how it differs from the conventional vaccine development processes. Furthermore, a brief view of the platforms used to develop COVID-19 vaccines in clinical trials is provided.
4. Development Process and Platforms Used for COVID-19 Vaccines
Generally, new vaccine development is a long process taking about 10 to 15 years. However, the lowest period for the development of a vaccine was for MUMPS, with an approximate five-year period (for development and approval) (https://www.historyofvaccines.org/) (3). Therefore, it is understandable how developing a COVID-19 vaccine within a short period of 1-2 years might be challenging.
According to the World Health Organization report, as of early December 2021, there were 331 candidate vaccines in clinical development, with 194 vaccines in the preclinical stage and 137 vaccines in different clinical phases (42 vaccines in phase 1, 32 vaccines in phase 1/2, 11 vaccines in phase 2, 11 vaccines in phase 2/3, 29 vaccines in phase 3, and 10 vaccines in phase 4) (https://www.who.int/publications/). Eleven platforms have been used for the development of 137 vaccines that are in the clinical phase, including 18 (13%) vaccines with inactivated virus platform, two (1%) vaccines with a live attenuated virus platform, 47 (35%) vaccines with protein subunit platform, six (4%) vaccines with VLP platform, 20 (15%) vaccines with a viral vector (non-replicating) or VVnr platform, two (1%) vaccines with a viral vector (replicating) or VVr platform, two (1%) vaccines with VVr plus antigen-presenting cell platform, one (1%) vaccine with VVnr plus antigen-presenting cell platform, one (1%) vaccine with bacterial antigen-spore expression vector platform, 15 (11%) vaccines with DNA platform, and 23 (17%) vaccines with RNA platform (https://www.who.int/publications). Since there is no review on the VLP-based COVID-19 vaccines, this article focuses explicitly on this platform of COVID-19 vaccines. The following section discusses the pros and cons of each vaccine platform. In addition, the rationale for choosing VLPs as a vaccine development platform and their unique features are reviewed.
5. Virus-Like Particle-Based Vaccine Platforms
5.1. The Rationale for Selection of the VLP Platform for the Development of a Vaccine
The rationale for choosing the VLP platform for developing vaccines comes from excellent immune response stimulation associated with the traditional vaccines (attenuated and inactivated viruses). The excellent immune response stimulated by traditional vaccines is attributed to their noteworthy features: (1) attenuated viruses maintain their ability to replicate; (2) they have repetitive geometry on their surface; (3) they have a "particulate nature; and (4) they can induce non-specific (innate) and specific (adaptive) immune responses. These features are used as criteria for the fruitful development of VLP-based vaccines (12, 13).
The VLP-based vaccines represent the main characteristics of traditional vaccines, except for the replication ability, which is due to the lack of the viral genome in VLPs. According to the lack of the viral genome in VLPs, this platform is safe enough to be used as a vaccine platform. The VLP shell is an icosahedral or helical structure composed of many identical copies of proteins (12, 14, 15).
Different expression systems can produce VLPs, including bacteria, yeast, plant, insect, and mammalian cells. However, there are pros and cons to using each expression system for VLP production. Although used primarily for VLP expression, bacterial hosts are unsuitable for post-translational modifications such as glycosylation, which might be critical for some mammalian glycoproteins. Therefore, bacterial hosts are used for expression of the glycoproteins in which the carbohydrate chain is not essential for the biological function of the glycoprotein. In addition, bacterial hosts have been developed, introducing glycosylation at specific sites on proteins. The other drawback of bacterial hosts is the possibility of endotoxins as contaminants (12, 15-17). Yeasts have been used for VLP production in commercially available HPV (GARDSIL®) and HBV vaccines. Insect cells are also used for VLP production in another commercially available HPV vaccine (CERVARIX®) (12).
Significant advantages of VLPs, which make them suitable platforms for vaccine production, include the possibility of rapid and efficient production of VLPs in large quantities and their ability to be self-assembled. The self-assembly ability of VLPs helps the manufacturer spontaneously obtain the desired final folding, which resembles the structure of the original virus from which the VLP is derived. In general, the efficiency of VLP expression is lower in eukaryotic systems than in prokaryotic ones (12). By the points mentioned above about VLPs, the particular features of VLPs are discussed in the next section.
6. VLP-Based COVID-19 Vaccines in Clinical Trials
This section focuses on the details of the VLP-based COVID-19 vaccines in clinical trials. Up to early December 2021, there were six VLP-based COVID-19 vaccines in different phases of clinical studies (Table 1, key table). Based on the developer's name, these six VLP-based COVID-19 vaccines include: (1) Serum Institute of India with Accelagen Pty and SpyBiotech vaccine; (2) Medicago; (3) VBI Vaccines Inc.; (4) The Scientific and Technological Research Council of Turkey; (5) Radboud University; and (6) Yantai Patronus Biotech (https://www.who.int/publications/). Table 1 compares these six VLP-based COVID-19 vaccines in terms of the type of candidate vaccine, antigen, number of doses, schedule, route of administration, developers, and their status in the clinical trial studies. Some clinical studies in Table 1 are not registered on "clinicaltrials.gov." However, these studies are listed in the World Health Organization's International Clinical Trials Registry Platform (WHO ICTRP). The registration link for the clinical trials is provided in this table. Furthermore, the following sections of this article focus on the released information on the details of these vaccines based on published and available data on the internet.
Table 1. Virus-Like Particle-Based COVID-19 Vaccines in Clinical Trials a.
| No. | Type of Candidate Vaccine | Antigen | Number of Doses | Schedule | Route of Administration | Developers | Phase | Phase 1 | Phase 1/2 | Phase 2 | Phase 2/3 | Phase 3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RBD SARS-CoV-2 HBsAg VLP Vaccine | RBD | 2 | Day 0+28 | IM | Serum Institute of India+ Accelagen Pty + SpyBiotech | 1/2 | ACTRN12620000817943; ACTRN12620001308987 | ||||
| 2 | Coronavirus-like particle COVID-19 (CoVLP) | Stabilized prefusion form of SARS-CoV-2 spike protein fused with a transmembrane domain and cytoplasmic tail of influenza hemagglutinin | 2 | Day 0+21 | IM | Medicago | 3 | NCT04450004; Study Report | NCT04662697 | NCT04636697 | ||
| 3 | VBI-2902a. An enveloped virus-like particle (eVLP) of SARS-CoV-2 spike (S) glycoprotein and aluminum phosphate adjuvant | A modified and optimized prefusion form of SARS-CoV-2 spike protein | 2 | Day 0+28 | IM | VBI Vaccines Inc. | 1/2 | NCT04773665 | ||||
| 4 | SARS CoV-2 VLP vaccine | All four structural proteins of SARS-CoV-2 (S, E, M, and N) | 2 | Day 0 | SC | The Scientific and Technological Research Council of Turkey | 2 | NCT04818281 | NCT04962893 | |||
| 5 | ABNCoV2 capsid virus-like particle (cVLP) +/- adjuvant MF59 | RBD antigens were designed with boundaries aa319-591 of the SARS-CoV-2 sequence (Sequence; ID: QIA20044.1). | 2 | Day 0+28 | IM | Radboud University | 1 | NCT04839146 | ||||
| 6 | SARS-CoV-2 Vaccine LYB001, an RBD from SARS-CoV-2 and VLP vector, adjuvanted with Alum | RBD | 3 | Day 0, 28, 56 | IM | Yantai Patronus Biotech Co., Ltd | Phase 1 | NCT05125926 | NCT05137444 |
a Resource: https://www.who.int/publications/
6.1. SpyBiotech/Serum Institute of India (SpyBiotech/SIIPL)
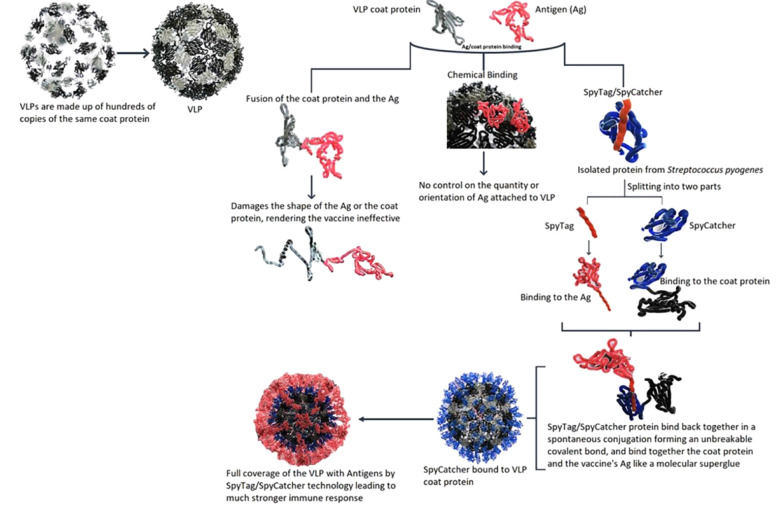
A VLP-based COVID-19 vaccine has been developed by a collaboration between SpyBiotech and Serum Institute of India Pvt. Co. Ltd. (SIIPL) and entered phase 1/2 clinical trials in Australia. In VLP-based vaccines, the antigen of interest is tightly packed on a virus-like particle, leading to a substantially increased immune response after vaccination. Hundreds of copies of the same coat protein make the composition of these VLPs. Attachment of antigens of interest to each of these coat proteins in VLPs leads to the presentation of many signals for the immune system, making the vaccine much more effective. However, the attachment of the antigens to the VLPs is associated with a few challenges. As illustrated in Figure 3, there have been two main methods for attaching the antigens to the VLPs. Binding the antigen to the coat protein by direct fusion may simply damage the folding of either the coat protein or the antigen, making the vaccine ineffective. An alternative approach is the application of chemical methods to make the antigen attached to the VLP. However, there is no control on the amount and orientation of the antigen-VLP binding. To solve these problems, Spybiotech developed a novel way of binding the antigen to the coat protein, called Spytag/Spycatcher technology which the Serum Institute of India employs to develop a VLP-based vaccine against COVID-19.
Figure 3. SpyTag/SpyCatcher technology for the development of VLP vaccines.
One of the main advantages of this vaccine technology is that the conjugation process is quite simple compared to most conjugated vaccines, in which the conjugation process is performed through complicated chemical approaches. The Spytag/Spycatcher technology is generated by splitting one of the immunoglobulin-like domains in Streptococcus pyogenes called CnaB2 from the fibronectin-binding protein FbaB. The domain in the native protein can spontaneously form an intramolecular bond between two residues (Lys-Asp) that helps the folding maintenance of the domain to be stable even in harsh conditions such as pH = 2 or temperatures up to 100°C. Howarth's group developed this technology at the University of Oxford. It was first used in malaria vaccine development, which is still ongoing at Jenner Institute (https://www.bioworld.com).
Figure 3 illustrates the SpyTag/SpyCatcher technology for the development of vaccines. The Spytag/Spycatcher system consists of two elements: (1) the catcher (Spycatcher), which is an engineered protein (138 amino acids) with a molecular weight of 10 kD; and (2) the tag (SpyTag), which is a short (13 amino acids) peptide. The DNA sequence of Spycatcher protein is accompanied by the gene sequence of the VLP part, and the DNA sequence of Spytag peptide is accompanied by the sequence considered for the antigen. When the products of these two genes are mixed, the SpyTag encounters the Spycatcher, and a covalent interaction called isopeptide bond formation spontaneously occurs. This isopeptide bond formation makes a reliable and straightforward means for the one-step conjugation of biomolecules. It is a molecular superglue-like binding method that allows VLPs to cover the target antigen fully. The full coverage of VLPs with the target antigen can lead to a substantially stronger immune response. In addition, this plug-and-display technology allows for fast and easy modifications in the development of new vaccine candidates against emerging viruses such as SARS-CoV-2. SARS-CoV-2 continues to have emerging variants; therefore, it seems necessary to have such a vaccine development platform allowing for a fast switch to the new mutated antigens. This technology can also effectively bind the antigens onto vaccine platforms other than VLPs. It has been applied to present many antigens, including HIV, Plasmodium spp., cancer cells, and the influenza A virus (https://www.bioworld.com) (18-20).
Yeast is the host used to produce the SpyBiotech/SIIPL VLP-based COVID-19 vaccine. The VLP part of the vaccine is composed of the surface antigen of HBV, which is quite commonly used in many approved vaccines. In the recombinant HBV vaccines, HbsAg is used as the antigen of interest against which the protective antibody is supposed to be produced in the body. However, in the VLP platforms used to develop vaccines against other diseases, HbsAg is used as a carrier for the heterologous antigen. One of the vaccines that use this VLP-based platform is a vaccine developed for malaria, named Mosquirix (RTS, S) (https://www.bioworld.com) (19).
6.2. Medicago
The next VLP-based COVID-19 vaccine is a plant-based CoVLP vaccine against COVID-19, developed by a collaboration between Canada-based Medicago and GlaxoSmithKline (GSK). GSK provides the adjuvant for the COVID-19 vaccine material developed by Medicago. The expression system which Medicago uses to produce VLPs is Nicotiana benthamiana. The unique ability of this tobacco plant species is its heterologous gene sequence expression, which makes it a commonly used plant expression system to produce biopharmaceuticals. The select antigen in this VLP-based COVID-19 vaccine was a stabilized prefusion form of SARS-CoV-2 spike protein fused with a transmembrane domain and cytoplasmic tail of influenza hemagglutinin transiently expressed in N. benthamiana (21-23).
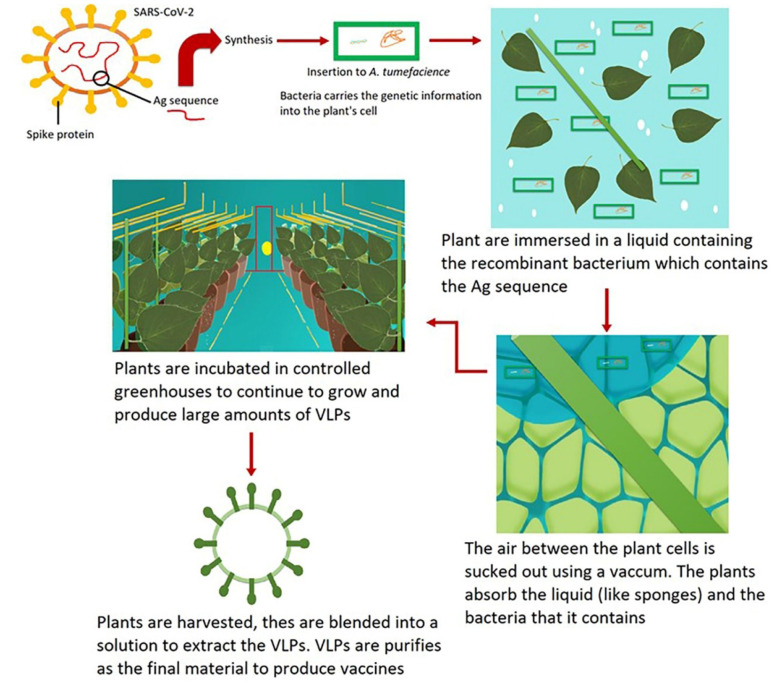
In 2019, Proficia®, Medicago's plant-based platform, was selected as the best new vaccine technology/platform at the world vaccine congress (https://www.medicago.com/en/media-room/). Medicago technology for developing plant-derived VLP vaccines is illustrated in Figure 4. This technology uses five main steps to produce the vaccine: Synthesis, infiltration, incubation, harvest, and purification. In the synthesis phase, the gene sequence of the antigen is synthesized, inserted into a plant-specific bacterial vector, and then multiplied. In the infiltration phase, the plants are immersed in a vector-containing solution, and the air is forced out of the cells by a vacuum. The pressure difference leads to the entrance of the vector to the plant cells. The third step is the incubation phase, in which the plant cells produce the VLPs in 4 - 6 days. In the harvest phase, the VLPs need to be extracted. Therefore, removing the leaves and blending them into a solution is performed, and the isolation and extraction of VLPs occur to provide the vaccine material. Finally, the purification of the VLPs is done by standard filtration and chromatographic steps, and the purified vaccine material is provided. This material is subject to sterilization and qualification analysis for formulation and finished product preparation. In this vaccine development method, a genetic modification of the plants does not occur, and the natural processes of plant cells are recruited for VLP manufacturing (https://www.medicago.com/en/technologies/) (24).
Figure 4. Medicago technology for the development of plant-based VLP vaccines.
The genetic modification is performed on Agrobacterium tumefaciens, which infects the plant by inserting its DNA into the plant cell to produce VLPs. Virus-like particles produced by this method resemble the viruses which induce immune responses. However, due to the absence of genetic material in these VLPs, they cannot replicate and cause virus-related diseases. Therefore, these may be safer vaccines than traditional ones. The utilization of Nicotiana benthamiana has the advantage of easy manipulation and infiltration procedure in which the infiltration of the bacteria into the leaves is performed. This assists a high expression level of the transgene, leading to a high VLP production level (25, 26).
Potential advantages of using plants as bioreactors for vaccine production include: (1) fast process rate, which provides the research doses of the vaccine in just 19 days and the clinical doses within 42 - 56 days; (2) accuracy, which provides a design ability to exactly match the target strains; (3) versatility and flexibility to be used for both antibody and vaccine production; and (4) simplicity of the scale-up process in which as many plants as needed could be grown in the same conditions (https://www.medicago.com/en/technologies/).
6.3. VBI COVID-19 Vaccine
VBI Vaccines Inc. develops the third VLP-based COVID-19 vaccine. VBI is a biopharmaceutical company with an R&D center in Canada that focuses on preventing and treating diseases. This company has an innovative eVLP technology used to develop potent vaccines that can stimulate the human immune system resembling the natural immune system stimulation by viruses. The commitment of VBI is to target and combat acute infectious diseases and aggressive cancers (https://www.biospace.com/). VBI starts a collaboration with the Coalition for Epidemic Preparedness Innovations (CEPI) to advance vaccine candidates against variants of SARS-CoV-2. CEPI gives donations from different organizations for financial support of vaccine development projects against emerging infectious diseases and cancers (https://cepi.net/).
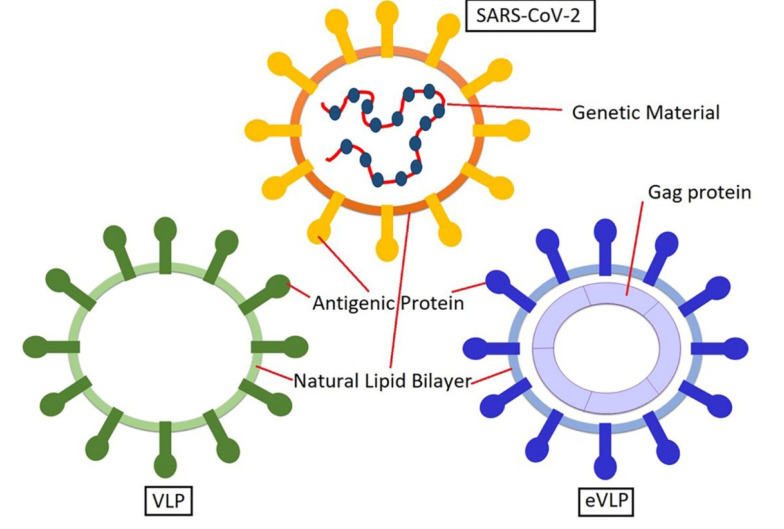
Based on virus structures, there are two types of VLPs (Figure 5): (1) non-enveloped VLPs; and (2) enveloped VLPs (eVLPs). eVLPs have more complex structures than non-enveloped VLPs. There are membranes in eVLPs derived from the host cell, which offers more opportunities for adjuvants and heterologous antigens incorporation. Furthermore, the composition and source of the envelope depend on three factors: (1) the family of the virus; (2) the process of virion assembly in its natural host; and (3) the specific host system used for production. Generally, there are two steps in the self-assembly process of eVLPs. At first, the inner structural proteins are formed, including core protein, capsid, or matrix. Then, for additional budding, the enclosure of the membrane occurs. The surface antigens of the virus, primarily glycoproteins, exist in the eVLP envelope. Therefore, it plays the role of an adjuvant and enhances immunogenicity. Two factors that affect the efficacy of an eVLP vaccine are the appropriate folding and proper glycosylation of antigens, which influence pathogenicity, stability, and immune recognition. Furthermore, the lipid membrane of an eVLP originates from the host cells, which occurs in a budding process. This process is similar to the developing process in the enveloped viruses (27).
Figure 5. Structure of VLP and eVLP. VBI developed its COVID-19 vaccine based on eVLP technology.
Coronavirus is an enveloped virus, and therefore, it is a suitable target for VBI's eVLP vaccine development platform. Collaborating with the National Research Council of Canada, VBI has started a program named VBI-2900 in which their eVLP technology is employed to develop vaccines against coronaviruses. This VBI technology uses eVLPs, comprising the murine leukemia virus (MLV) Gag protein sequence. This leads to the expression of proteins that make particles used as vaccine candidates. VBI Inc. developed two monovalent VLP-based COVID-19 vaccines, VBI-2902 and VBI-2905. It also created two multivalent eVLP-based vaccines for coronaviruses, VBI-2901 and the other undisclosed. VBI-2902 is an eVLP monovalent vaccine containing a modified and optimized prefusion form of SARS-CoV-2 spike protein with aluminum phosphate as the adjuvant. This vaccine is now under phase 1/2 clinical studies. These studies have a grant from the Strategic Innovation Fund of the Government of Canada. The studies were performed at nine different sites in Canada and were expected to reach the initial data by the end of June 2021. VBI-2905, the monovalent vaccine designed explicitly for the B.1.351 variant (the strain first identified in South Africa, which is also known as 501Y.V2), is now in the preclinical phase (https://www.vbivaccines.com/) (28). A multivalent pan-coronavirus vaccine candidate, VBI-2901, contains spike proteins of SARS-CoV-2, SARS-CoV, and MERS-CoV on the VLP's surface. This vaccine is in the preclinical phase. The other multivalent coronavirus vaccine, whose program has not been disclosed, is also in the preclinical phase (https://www.vbivaccines.com/) (27).
6.4. Scientific and Technological Research Council of Turkey
The fourth VLP-based COVID-19 vaccine has been developed by the Scientific and Technological Research Council of Turkey (TUBITAK), which is now under phase 2 clinical trial. This Turkish VLP-based COVID vaccine platform is a joint project between Middle East Technical University, led by Mayda Gürsel, and Bilkent University, led by Ihsan Gürsel. One of the significant differences between this VLP vaccine and other VLP-based COVID vaccines in the clinical phase is the inclusion of all four structural proteins of SARS-CoV-2 (S, E, M, and N) in the vectors as the antigenic part of the vaccine. The vectors are inserted into the host cell leading to the expression of these four antigens, which will be released as VLPs. After purification of the VLPs and formulation with adjuvants, they are subjected to clinical studies (https://covid19.tubitak.gov.tr/). This Turkish vaccine preparation has undergone phase 3 clinical studies (29). There is no further published data on this vaccine.
6.5. Radboud University
The fifth VLP-based COVID vaccine, ABNCoV2, has been developed based on capsid VLP (cVLP) technology by a partnership between ExpreS2ion and AdaptVac. The responsibility for the clinical studies of this vaccine has been given to Radboud University Medical Center in the Netherlands (NCT04839146) (https://news.cision.com/).
Capsid-like Particles (CLPs) are a subgroup of VLPs with specific characteristics, including rigidity and protein (non-lipid)-based nature of the particles. There are two methods used for antigen display on CLPs: (1) genetic fusion, and (2) modular antigen display, in which the antigens are attached to the surface of preassembled CLPs. In the latter method, there is a "split-protein (Tag/Catcher) conjugation" system in which vaccine antigens are covalently anchored onto CLPs (30).
The ABNCoV2 vaccine has been developed using Tag/Catcher-AP205 technology. In this technology, a bacterial protein is separated into a Tag (a reactive peptide) and a Catcher (a protein-binding part). Once the Tag and Catcher are mixed, they will rapidly form a spontaneous covalent bond called the isopeptide bond. The Tag/Catcher system is based on the genetic addition of the previously mentioned SpyTag/SpyCatcher to the capsid protein of Acinetobacter phage AP205. Therefore, the CLP backbone in this system is derived from a bacteriophage. Due to their uncomplicated structure, phage capsids are suitable for fast and economic recombinant bacterial expression. The attractive features of AP205 CLP as a vaccine platform include versatility, manufacturability, ease of use, immunogenicity, and structure. The general structure of the capsid is similar among all members of the RNA bacteriophage family. However, the exposed areas on the surface for genetic fusion are substantially different. The noteworthy feature of AP205 CLP is that the N- and C-termini are both surface-exposed and uniformly distributed on the assembled CLP. Furthermore, it is possible to make genetic fusion at both termini of the subunit protein without any change in the stability of the assembled CLP. Due to its cost-effectiveness and high production yield in E. coli, the Tag/Ctecher-AP205 system can be used for large-scale production and clinical development. In addition, AP205 CLP production in E. coli leads to bacterial RNA encapsulation, which plays a vital role as a Th1-type adjuvant via Toll-like Receptor (TLR) 7/8 (30-34).
Briefly, in the ABNCoV2 vaccine production system, the RBD is fused to the Catcher, and the CLP is added to the Tag. Genetic fusion of RBD antigens with either N- or C-terminus of the split-protein Catcher is done (called RBDn or RBDc, respectively). A secretion signal (BiP) at the N-terminus and a C-tag at the C-terminus for purification are added to the sequence of the antigen construct. The expression system used to produce the antigenic part (RBD-Catcher) is Drosophila melanogaster (Schneider-2 (ExpreS2) insect cell line), and the host used to make the Tag-CLP (split-protein Tag and coat protein of AP205) is E. coli in which the CLPs spontaneously represent the Tags on the surface (2).
6.6. Yantai Patronus Biotech Co., Ltd.
The last VLP-based COVID-19 vaccine recently added to the WHO COVID-19 vaccine landscape file is LYB001, a vaccine developed by Yantai Patronus Biotech company. This vaccine is now in phase 2/3 clinical trial. One of the differences between this vaccine and the other five VLP-based vaccines is its three-dose regimen. The schedule for the administration of this vaccine includes days 0, 28, and 56. No publications have been released for the LYB001 vaccine yet.
7. Conclusion
Virus-like Particle-based COVID-19 vaccine development is now under clinical studies; however, there are some unclear points: (1) Do VLP-based COVID-19 vaccines give sufficient protection against newly emerging variants of SARS-CoV-2? (2) Can VLP technology provide a platform as rapidly producing, safe, effective, and affordable as COVID-19 vaccine platforms in the market? Or can VLP technology be an acceptable substitution for other platforms? (3) How will current COVID-19 vaccine manufacturers react to the entry of VLP-based COVID-19 vaccines into the market? No VLP-based COVID-19 vaccines have gotten emergency approval yet. Therefore, it is too soon to judge the fate of these vaccines.
Acknowledgments
We would like to acknowledge the Tehran University of Medical Sciences and the Iran University of Medical Sciences for their support. We also thank Iman Tavassoly (MD, Ph.D.) for editing the paper.
Footnotes
Authors' Contribution: Study concept and design, N.M. D.; Acquisition of data, N. M. D., and M. S. R. R.; Drafting of the manuscript, N. M. D.; Critical revision of the manuscript for important intellectual content, M. S.; Study supervision, N. M. D.
Conflict of Interests: The authors declare no conflict of interest.
Funding/Support: This study received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Contributor Information
Mohammad Sharifzadeh, Email: msharifzadeh@tums.ac.ir.
Negar Mottaghi-Dastjerdi, Email: mottaghi.n@iums.ac.ir.
Mohammad Soltany Rezae Raad, Email: mohammad.soltany@gmail.com.
References
- 1.Randolph HE, Barreiro LB. Herd Immunity: Understanding COVID-19. Immunity. 2020;52(5):737–41. doi: 10.1016/j.immuni.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fougeroux C, Goksoyr L, Idorn M, Soroka V, Myeni SK, Dagil R, et al. Capsid-like particles decorated with the SARS-CoV-2 receptor-binding domain elicit strong virus neutralization activity. Nat Commun. 2021;12(1):324. doi: 10.1038/s41467-020-20251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma O, Sultan AA, Ding H, Triggle CR. A Review of the Progress and Challenges of Developing a Vaccine for COVID-19. Front Immunol. 2020;11:585354. doi: 10.3389/fimmu.2020.585354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tavassoly O, Safavi F, Tavassoly I. Heparin-binding Peptides as Novel Therapies to Stop SARS-CoV-2 Cellular Entry and Infection. Mol Pharmacol. 2020;98(5):612–9. doi: 10.1124/molpharm.120.000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brian DA, Baric RS. Coronavirus genome structure and replication. Curr Top Microbiol Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naqvi AAT, Fatima K, Mohammad T, Fatima U, Singh IK, Singh A, et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim Biophys Acta Mol Basis Dis. 2020;1866(10):165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorouri F, Emamgholipour Z, Keykhaee M, Najafi A, Firoozpour L, Sabzevari O, et al. The Situation of Small Molecules Targeting Key Proteins in combatting SARS-CoV-2: Synthesis, Metabolic Pathway, Mechanism of Action, and Potential Therapeutic Applications. Mini Rev Med Chem. 2022;22(2):273–311. doi: 10.2174/1389557521666210308144302. [DOI] [PubMed] [Google Scholar]
- 8.Saberi-Movahed F, Mohammadifard M, Mehrpooya A, Rezaei-Ravari M, Berahmand K, Rostami M, et al. Decoding Clinical Biomarker Space of COVID-19: Exploring Matrix Factorization-based Feature Selection Methods. medRxiv. 2021;Preprint doi: 10.1101/2021.07.07.21259699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang N, Shang J, Jiang S, Du L. Subunit Vaccines Against Emerging Pathogenic Human Coronaviruses. Front Microbiol. 2020;11:298. doi: 10.3389/fmicb.2020.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7(3):226–36. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38(1):1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 12.Mohsen MO, Zha L, Cabral-Miranda G, Bachmann MF. Major findings and recent advances in virus-like particle (VLP)-based vaccines. Semin Immunol. 2017;34:123–32. doi: 10.1016/j.smim.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Jennings GT, Bachmann MF. Designing recombinant vaccines with viral properties: a rational approach to more effective vaccines. Curr Mol Med. 2007;7(2):143–55. doi: 10.2174/156652407780059140. [DOI] [PubMed] [Google Scholar]
- 14.Adolph DW, Butler PJ. Studies on the assembly of a spherical plant virus. III. Reassembly of infectious virus under mold conditions. J Mol Biol. 1977;109(2):345–57. doi: 10.1016/s0022-2836(77)80038-7. [DOI] [PubMed] [Google Scholar]
- 15.Khudyakov Y, Pumpens P. Viral nanotechnology. Florida, USA: CRC Press; 2015. [Google Scholar]
- 16.Zhao Q, Allen MJ, Wang Y, Wang B, Wang N, Shi L, et al. Disassembly and reassembly improves morphology and thermal stability of human papillomavirus type 16 virus-like particles. Nanomedicine. 2012;8(7):1182–9. doi: 10.1016/j.nano.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Wacker M, Linton D, Hitchen PG, Nita-Lazar M, Haslam SM, North SJ, et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science. 2002;298(5599):1790–3. doi: 10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- 18.Reddington SC, Howarth M. Secrets of a covalent interaction for biomaterials and biotechnology: SpyTag and SpyCatcher. Curr Opin Chem Biol. 2015;29:94–9. doi: 10.1016/j.cbpa.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Tan TK, Rijal P, Rahikainen R, Keeble AH, Schimanski L, Hussain S, et al. A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nat Commun. 2021;12(1):542. doi: 10.1038/s41467-020-20654-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheridan C. Innovators target vaccines for variants and shortages in global South. Nat Biotechnol. 2021;39(4):393–6. doi: 10.1038/d41587-021-00001-x. [DOI] [PubMed] [Google Scholar]
- 21.Goulet MC, Gaudreau L, Gagne M, Maltais AM, Laliberte AC, Ethier G, et al. Production of Biopharmaceuticals in Nicotiana benthamiana-Axillary Stem Growth as a Key Determinant of Total Protein Yield. Front Plant Sci. 2019;10:735. doi: 10.3389/fpls.2019.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward BJ, Gobeil P, Séguin A, Atkins J, Boulay I, Charbonneau P, et al. Phase 1 trial of a Candidate Recombinant Virus-Like Particle Vaccine for Covid-19 Disease Produced in Plants. medRxiv . 2020;Preprint doi: 10.1101/2020.11.04.20226282. [DOI] [Google Scholar]
- 23.Klasse PJ, Nixon DF, Moore JP. Immunogenicity of clinically relevant SARS-CoV-2 vaccines in nonhuman primates and humans. Sci Adv. 2021;7(12) doi: 10.1126/sciadv.abe8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bally J, Nakasugi K, Jia F, Jung H, Ho SY, Wong M, et al. The extremophile Nicotiana benthamiana has traded viral defence for early vigour. Nat Plants. 2015;1:15165. doi: 10.1038/nplants.2015.165. [DOI] [PubMed] [Google Scholar]
- 25.Takeyama N, Kiyono H, Yuki Y. Plant-based vaccines for animals and humans: recent advances in technology and clinical trials. Ther Adv Vaccines. 2015;3(5-6):139–54. doi: 10.1177/2051013615613272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uthaya Kumar A, Kadiresen K, Gan WC, Ling APK. Current updates and research on plant-based vaccines for coronavirus disease 2019. Clin Exp Vaccine Res. 2021;10(1):13–23. doi: 10.7774/cevr.2021.10.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundstrom K. Viral Vectors for COVID-19 Vaccine Development. Viruses. 2021;13(2) doi: 10.3390/v13020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirchmeier M, Fluckiger AC, Soare C, Bozic J, Ontsouka B, Ahmed T, et al. Enveloped virus-like particle expression of human cytomegalovirus glycoprotein B antigen induces antibodies with potent and broad neutralizing activity. Clin Vaccine Immunol. 2014;21(2):174–80. doi: 10.1128/CVI.00662-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandal H. Achievements of the COVID-19 Turkey Platform in vaccine and drug development with an approach of "co-creation and succeeding together". Turk J Med Sci. 2021;51(SI-1):3139–49. doi: 10.3906/sag-2112-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aves KL, Goksoyr L, Sander AF. Advantages and Prospects of Tag/Catcher Mediated Antigen Display on Capsid-Like Particle-Based Vaccines. Viruses. 2020;12(2) doi: 10.3390/v12020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thrane S, Janitzek CM, Matondo S, Resende M, Gustavsson T, de Jongh WA, et al. Bacterial superglue enables easy development of efficient virus-like particle based vaccines. J Nanobiotechnology. 2016;14:30. doi: 10.1186/s12951-016-0181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brune KD, Leneghan DB, Brian IJ, Ishizuka AS, Bachmann MF, Draper SJ, et al. Plug-and-Display: decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci Rep. 2016;6:19234. doi: 10.1038/srep19234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci U S A. 2012;109(12):E690–7. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pumpens P, Renhofa R, Dishlers A, Kozlovska T, Ose V, Pushko P, et al. The True Story and Advantages of RNA Phage Capsids as Nanotools. Intervirology. 2016;59(2):74–110. doi: 10.1159/000449503. [DOI] [PubMed] [Google Scholar]