Abstract
Objective
To define the relationship of SARS-CoV-2 antigen, viral load determined by RT-qPCR, and viral culture detection. Presumptively, viral culture can provide a surrogate measure for infectivity of sampled individuals and thereby inform how and where to most appropriately deploy antigen and nucleic acid amplification-based diagnostic testing modalities.
Methods
We compared the antigen testing results from three lateral flow and one microfluidics assay to viral culture detection and viral load determination performed in parallel in up to 189 nasopharyngeal swab samples positive for SARS-CoV-2. Sample viral loads, determined by RT-qPCR, were distributed across the range of viral load values observed in our testing population.
Results
Antigen tests were predictive of viral culture positivity, with the LumiraDx microfluidics method showing enhanced sensitivity (90%; 95% CI 83–94%) compared with the BD Veritor (74%, 95% CI 65–81%), CareStart (74%, 95% CI 65–81%) and Oscar Corona (74%, 95% CI 65–82%) lateral flow antigen tests. Antigen and viral culture positivity were also highly correlated with sample viral load, with areas under the receiver operator characteristic curves of 0.94 to 0.97 and 0.92, respectively. A viral load threshold of 100 000 copies/mL was 95% sensitive (95% CI, 90–98%) and 72% specific (95% CI, 60–81%) for predicting viral culture positivity. Adjusting for sample dilution inherent in our study design, sensitivities of antigen tests were ≥95% for detection of viral culture positive samples with viral loads >106 genome copies/mL, although specificity of antigen testing was imperfect.
Discussion
Antigen testing results and viral culture were correlated. For culture positive samples, the sensitivity of antigen tests was high at high viral loads that are likely associated with significant infectivity. Therefore, our data provides support for use of antigen testing in ruling out infectivity at the time of sampling.
Keywords: Infectivity, Laboratory diagnostics, SARS-CoV-2 antigen test, SARS-CoV-2 viral culture, SARS-CoV-2 viral load
Introduction
SARS-CoV-2 nucleic acid amplification tests (e.g. PCR) are highly sensitive but expensive with a slow turn-around time. In contrast, antigen tests are fast, amenable to point-of-use testing, inexpensive, but less sensitive [1]. Nevertheless, a test with lower sensitivity may offer compelling benefit if able to reliably identify individuals who present an infectious risk to others [2].
An individual's infectivity may be approximated by the infectivity of a diagnostic sample. Antigen and PCR tests detect different components of the SARS-CoV-2 virus, neither of which necessarily reflect the presence of infectious virus. Tissue culture assays detect replicating virus, and, by inference, the infectivity of individuals from whom samples were obtained.
We compare antigen and quantitative PCR detection with viral culture results to further inform decisions on diagnostic test use.
Methods
Samples
Nasopharyngeal swabs were obtained in 3 mL of saline or viral transport medium at COVID-19 testing sites in Boston, MA from March through June 2021 for diagnostic purposes unrelated to this study and selected for analysis solely based on viral load distribution. Sample size was determined by available budget and desire to have 95% CIs of <30% when comparing antigen test sensitivity and viral culture results across log10 viral load bins. The testing population reflected both community testing sites and both symptomatic and asymptomatic individuals presenting for clinical care at our institution and affiliated practices. After PCR testing, de-identified samples were stored at 4ºC for a median of 3 days (IQR 1–3 days) from specimen collection until testing by viral culture. A minority of samples (n = 71) were frozen within one day of sample collection and then thawed prior to additional testing. Prior to use of frozen samples for culture analysis, we performed a validation with samples (n = 31) tested with and without freezing at –80ºC for ≤17 days, with minimal impact on isolation of viable virus. Our validation demonstrating lack of effect of freeze-thaw on viral viability will be reported in a separate study. Antigen testing was performed on the day of specimen retrieval above, or aliquots were frozen for later batch testing by all antigen testing methods in parallel.
Human subjects research with a waiver of informed consent was approved by the Institutional Review Boards at Beth Israel Deaconess Medical Center and the Harvard T.H. Chan School of Public Health.
A total of 206 samples positive for SARS-CoV-2 by RT-qPCR using the Abbott M2000 or Alinity M platforms (Abbott Laboratory, Abbott Park, IL) were analysed by LumiraDx (London, UK; n = 206), BD Veritor (Franklin Lakes, NJ; n = 204), CareStart (Access Bio, Inc., Somerset, NJ; n = 201), and Oscar Corona (Oscar Medicare Pvt., Ltd., New Deli, India; n = 193) antigen tests. For all but 13 samples, sufficient sample volume was available for testing with all four antigen methods. Of the 206 samples, 189 were tested by both LumiraDx and viral culture, 187 were tested by BD and viral culture, 184 were tested by CareStart and viral culture, and 176 were tested by Oscar Corona and viral culture.
Three hundred separate RT-qPCR antigen negative samples were selected sequentially and tested by all four antigen tests and not tested by viral culture.
RT-qPCR testing
SARS-CoV-2 RT-qPCR testing of samples and Vero cell culture supernatants was performed using the Abbott Molecular M2000 Real-Time or Alinity M SARS-CoV-2 assays. In addition to a qualitative result, both tests provide a fractional cycle number (FCN), a type of cycle threshold [3]. The assays were calibrated using standards ranging from 300 to 106 viral genome copies/mL (LGC Seracare, Milford, MA) consisting of replication-incompetent, enveloped, positive singled-stranded RNA Sindbis virus into which the whole genome of SARS-CoV-2 was cloned and titers determined using digital droplet PCR analysis by LGC SeraCare (Russell Garlick, LGC SeraCare, personal communication). The standards were run through all stages of sample preparation and extraction to allow appropriate comparison with identically processed patient samples. Coefficients of determination (R2) between log10 transformed viral load values and FCN values determined for standards were 0.997 for both assays. Slope and intercepts defined linear regression equations that were used to convert FCN to viral load values including extension above and below the level of calibrators tested. Analytical sensitivity of the Abbott M2000 and Alinity M SARS-CoV-2 PCR assays corresponded to a limit of detection of ∼100 genome copies/mL, as previously reported [4,5].
Antigen testing
The BD Veritor, LumiraDx, CareStart, and Oscar Corona antigen tests were performed according to the manufacturer's instructions with the exception that 250 μL of patient sample (nasopharyngeal swab sample eluted into 3 mL of saline or viral transport medium) was pipetted into each kit's extraction vial rather than direct insertion of the nasal swab into the extraction vial. We estimate a 17- to 18-fold dilution of sample based on this procedure (see Supplementary Methods). Indeterminates or invalid antigen test results, specifically due to lack of control line development on lateral flow assay (LFA), or control failure for the LumiraDx instrument were not observed in our study.
SARS-CoV-2 viral culture
Vero E6 (ATCC CRL-1586) cells were seeded on a 6-well flat bottom plate at 0.3 × 106 cells per well in Eagle's minimum essential media (EMEM) containing 1% antibiotic-antimycotic, 1% HEPES and 5% fetal calf serum (FCS, Gibco; Thermo Fischer Scientific, Inc., Pittsburgh, PA); grown to confluence at approximately 1 × 106 cells per well [[6], [7], [8]]; inoculated with 250 μl of patient sample; and then incubated at 37ºC for 24 hours for viral adsorption. Simultaneously, a negative control was also inoculated with 250 μL of viral growth media. Carryover of nonviable viral RNA present in samples was limited by washing cell cultures after the 24-hour viral adsorption and adding fresh EMEM composite media with reduced FCS to 2% for viral growth. Therefore, detectable virus should represent viable replicating virus. On days, 3, 6, and 13 to 14 days of culture, 800 μL of cell culture supernatant was removed and added to 800 μl of VXL buffer (QIAGEN, German, MD) (1:1 ratio) for subsequent nucleic acid extraction and SARS-CoV-2 real-time RT-qPCR. Cultures were re-fed with addition of 1 mL of EMEM with reduced FCS after sampling at each time point.
Supernatant viral loads below the LoD of the PCR assay were scored negative at that timepoint. Samples with at least two of three sequential supernatant viral loads exceeding the limit of detection (LoD) were considered positive, and, conversely, samples with either one or no viral loads exceeding the LoD were considered negative. The scoring system was developed to mitigate against the detection of RNA carryover from the original specimen leading to a false positive signal. Based on our procedure, carryover RNA would be diluted out during the fresh media exchanges and/or degraded during the viral culture procedure and was therefore not expected to remain above the viral load assay limit of detection for more than one culture sampling point. Conversely, detection of RNA viral load above the limit of detection in two or more samples was expected to be consistent with the presence of replicating virus. Research personnel performing viral culture and antigen testing were blinded to data from assays other than their own.
Statistics
Statistical comparisons were performed with Stata version 13.1 (Stata Corporation, College Station, TX) and/or Prism 9 for MacOS (GraphPad, San Diego, CA). The multi-comparison P values reported for comparison of geometric mean viral loads associated with antigen test results were calculated using the Holm-Sidak method. Receiver operator characteristic curve sensitivity and specificity were calculated using Microsoft Excel (Microsoft Corp., Redmon, WA).
Results
The comparison of each antigen test to viral culture resulted in two-by-two contingency tables and sensitivities and specificities shown in Table 1 . The overall sensitivity was 90% for LumiraDx and 74% for the other methods. Specificity was 70% for LumiraDx and 91% to 92% for the other antigen test methods. Comparison of antigen test detection versus viral culture positivity was also broken down by log10 viral load bins in Table 2 . Notably, sensitivity of antigen testing for detecting positive cultures was 100% for samples with a viral load >107 and dropped to essentially zero, albeit with wide confidence intervals for samples with a viral load <104. Taking into account the sample dilution factor associated with antigen testing, it is expected that the next lowest viral load bin may be more representative of antigen test performance in actual use (Table 2, Table 3 ; Tables S1 and S2) (i.e. that antigen tests perform well for samples with a viral load >106 genome copies/mL with some variance depending on the specific antigen test examined).
Table 1.
Performance of antigen detection tests compared with viral culture
| Culture POS | Culture NEG | Statistic | Value | 95% CI | |
|---|---|---|---|---|---|
| LumiraDx antigen results versus viral culture | |||||
| Lumira POS | 101 | 23 | Sensitivity | 0.90 | 0.83–0.94 |
| Lumira NEG | 11 | 54 | Specificity | 0.70 | 0.59–0.79 |
| BD Veritor antigen results versus viral culture | |||||
| BD POS | 82 | 6 | Sensitivity | 0.74 | 0.65–0.81 |
| BD NEG | 29 | 70 | Specificity | 0.92 | 0.84–0.96 |
| CareStart antigen results versus viral culture | |||||
| CareStart POS | 80 | 7 | Sensitivity | 0.74 | 0.65–0.81 |
| CareStart NEG | 28 | 69 | Specificity | 0.91 | 0.82–0.95 |
| Oscar Corona antigen results versus viral culture | |||||
| Oscar POS | 75 | 6 | Sensitivity | 0.74 | 0.65–0.82 |
| Oscar NEG | 26 | 69 | Specificity | 0.92 | 0.84–0.96 |
Table 2.
Antigen test results versus viral culture stratified by viral load bins
| Viral load bin (genome copies/mL) | n | Sensitivity | 95% CI | Specificity | 95% CI |
|---|---|---|---|---|---|
| LumiraDx | |||||
| 108–1010 | 23 | 1.00 | 0.85–1.00 | 0.00 | 0.00–0.95 |
| 107 to <108 | 43 | 1.00 | 0.91–1.00 | 0.00 | 0.00–0.56 |
| 106 to <107 | 34 | 0.93 | 0.77–0.99 | 0.00 | 0.00–0.39 |
| 105 to <106 | 29 | 0.72 | 0.49–0.88 | 0.55 | 0.28–0.79 |
| 104 to <105 | 23 | 0.00 | 0.00–0.56 | 0.70 | 0.48–0.85 |
| 103 to <104 | 18 | N.D. | 0.94 | 0.74–1.000 | |
| <103 | 19 | N.D. | 1.00 | 0.83–1.00 | |
| BD Veritor | |||||
| 108–1010 | 23 | 1.00 | 0.85–1.00 | 0.00 | 0.00–0.95 |
| 107 to <108 | 43 | 1.00 | 0.91–1.00 | 0.33 | 0.02–0.88 |
| 106 to <107 | 33 | 0.63 | 0.44–0.78 | 0.67 | 0.30–0.94 |
| 105 to <106 | 29 | 0.28 | 0.13–0.51 | 1.00 | 0.74–1.00 |
| 104 to <105 | 22 | 0.000 | 0.00–0.56 | 0.95 | 0.75–1.00 |
| 103 to <104 | 18 | N.D. | 1.00 | 0.82–1.00 | |
| <103 | 19 | N.D. | 1.00 | 0.83–1.00 | |
| CareStart | |||||
| 108–1010 | 22 | 1.00 | 0.85–1.00 | 0.00 | 0.00–0.95 |
| 107 to <108 | 43 | 1.00 | 0.91–1.00 | 0.00 | 0.00–0.56 |
| 106 to <107 | 33 | 0.63 | 0.44–0.78 | 0.50 | 0.19–0.81 |
| 105 to <106 | 27 | 0.13 | 0.02–0.36 | 1.00 | 0.74–1.00 |
| 104 to <105 | 22 | 0.00 | 0.00–0.56 | 1.00 | 0.83–1.00 |
| 103 to <104 | 18 | N.D. | 1.00 | 0.82–1.00 | |
| <103 | 19 | N.D. | 1.00 | 0.83–1.00 | |
| Oscar Corona | |||||
| 108–1010 | 21 | 1.00 | 0.84–1.00 | 0.00 | 0.00–0.95 |
| 107 to <108 | 40 | 1.00 | 0.91–1.00 | 0.00 | 0.00–0.56 |
| 106 to <107 | 32 | 0.65 | 0.46–0.81 | 0.67 | 0.30–0.94 |
| 105 to <106 | 25 | 0.07 | 0.00–0.31 | 1.00 | 0.74–1.00 |
| 104 to <105 | 22 | 0.00 | 0.00–0.56 | 1.00 | 0.83–1.00 |
| 103 to <104 | 17 | N.D. | 1.00 | 0.82–1.00 | |
| <103 | 19 | N.D. | 1.00 | 0.83–1.00 | |
N.D., not defined as there are no false negatives and true positives.
Table 3.
Viral culture positivity versus viral load bin
| Viral load bin | Percent positivity (95% CI) (%) |
|---|---|
| 108–1010 | 96% (79–100) |
| 107 to <108 | 93% (81–98) |
| 106 to <107 | 82% (66–92) |
| 105 to <106 | 62% (44–77) |
| 104 to <105 | 13% (5–32) |
| 103 to <104 | 0% (0–18) |
| <103 | 0% (0–17) |
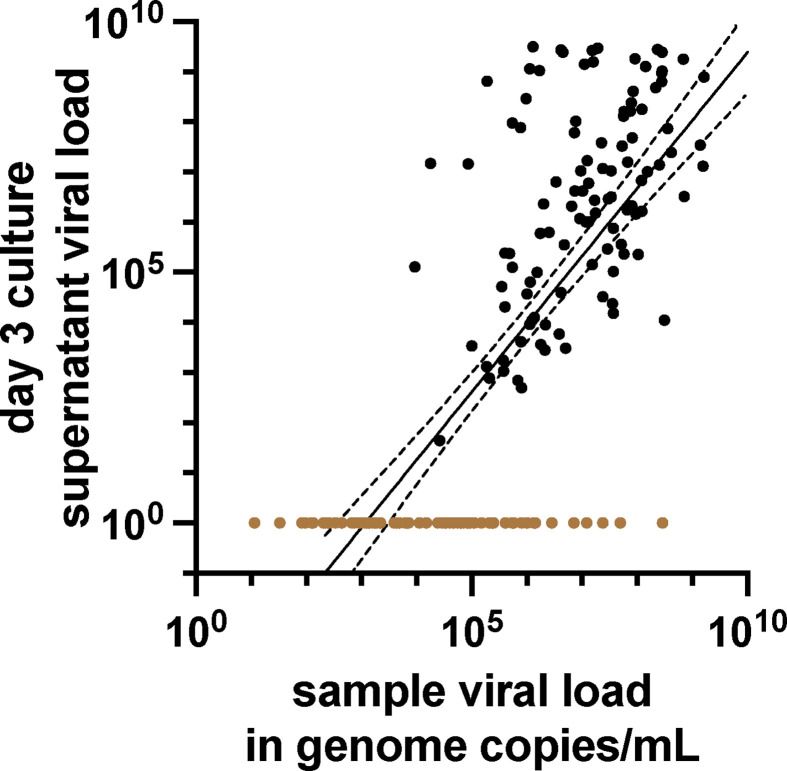
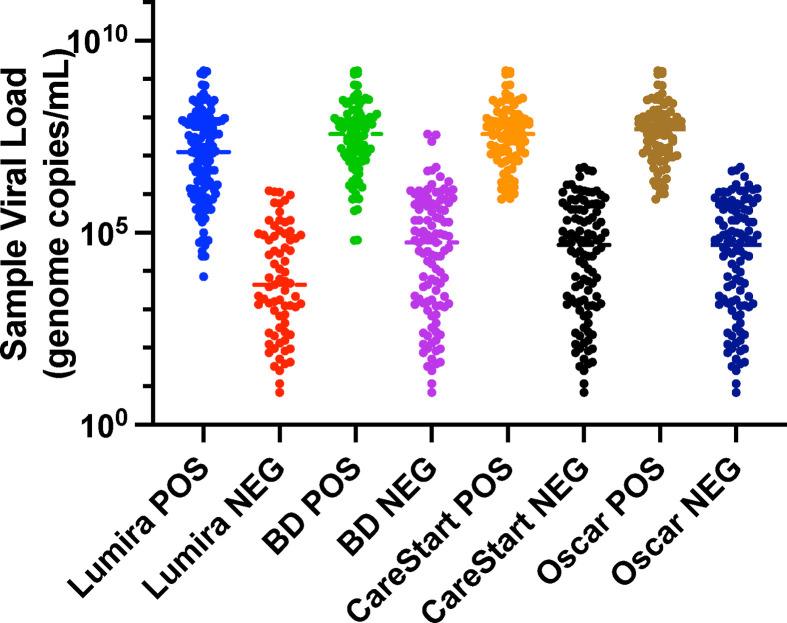
The original sample viral load determined by RT-qPCR and quantity of virus in day 3 culture supernatants was also reasonably correlated (R2 = 0.55) with a sharp loss of detection of viable virus in samples with viral loads below approximately 105 genome copies/mL (Fig. 1 ). Distributions of viral load results in samples, testing positive or negative, respectively, by respective antigen tests are shown in Fig. 2 . The LFA-based antigen tests detected samples with viral loads greater than approximately107 genome copies/mL, except for a small number of outliers. The microfluidic LumiraDx test detected samples with lower viral loads compared to the other antigen testing methods with a cutoff for consistent detection closer to 106 genome copies/mL. The differences between log10 transformed mean viral loads of samples positive by LumiraDx and positive by each of the LFA methods, respectively, were statistically significant for all pairwise comparisons (adjusted p < 0.05), as were differences in (geometric) mean viral loads of samples testing negative by LumiraDx and negative by each of the LFA methods, respectively. However, BD Veritor, CareStart, and Oscar Corona tests were indistinguishable in all pairwise comparisons with one another (adjusted p = 0.99).
Fig. 1.
Quantitative relationship between culturable virus and sample viral load. Day 3 viral culture supernatant from each cultured sample and its corresponding respiratory sample were each analysed by RT-qPCR to determine respective viral loads (n = 181). The viral load in genome copies/mL (logarithmic scale) of culture supernatant is plotted against the genome copies/mL (logarithmic scale) of the original patient sample. Linear regression (solid line) with 95% CIs (dashed lines) is shown. R2 = 0.55. Samples with negative viral cultures (see Materials and Methods) for representation are assigned a y-axis, value of 100, and are demarcated as coloured brown dots.
Fig. 2.
Antigen testing results compared with viral load. Viral load in genome copies/mL (logarithmic scale).
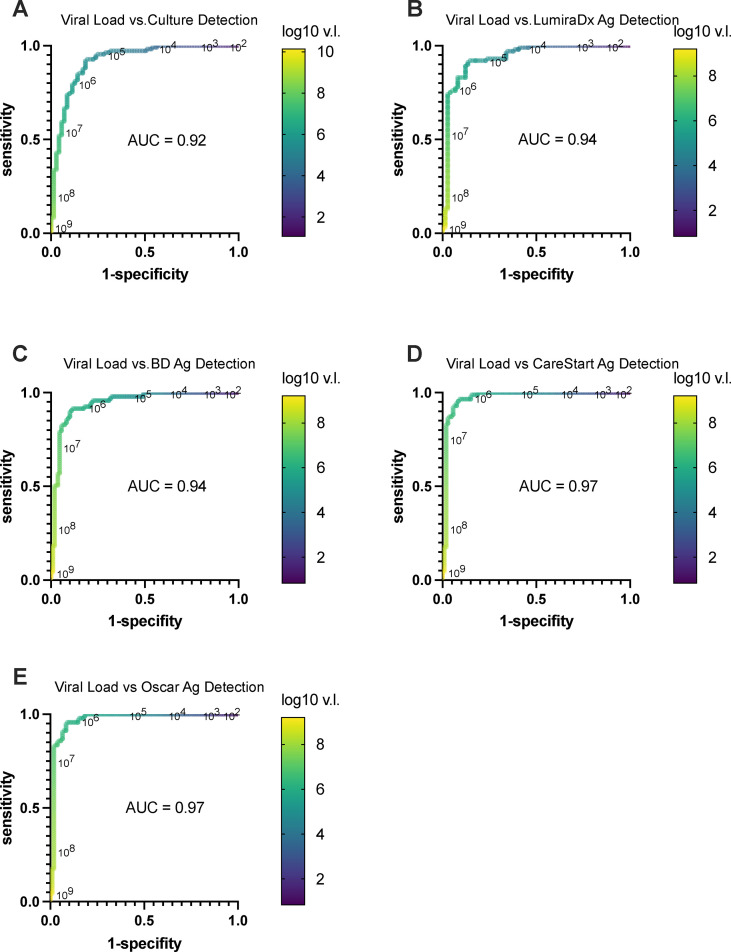
Receiver operator characteristic (ROC) curves were plotted to determine the viral load cutoffs that would reasonably predict detection by viral culture and antigen testing, respectively (Fig. 3 ). Notably, a viral load cutoff of ∼105 was highly sensitive for predicting a positive viral culture without undue loss of specificity. A viral load cutoff of ∼104 to 105 was reasonably sensitive for predicting a positive LumiraDx result and likewise a viral load cutoff of 105 to 106 was predictive of positive BD, Oscar, and CareStart test results. The ROC area under the curve was >0.94 for all comparisons (Figs. 3B–E), indicating that viral loads could serve as a reasonable surrogate for predicting the presence of culturable infectious virus and detectable antigen. Notably, viral load cutoffs for detecting positive viral cultures and antigen tests were similar, further supporting similar qualitative detection by viral culture and antigen tests.
Fig. 3.
Receiver operator characteristic curves (ROC) comparing SARS-CoV-2 sample viral load levels as a predictor of viral culture and antigen detection. For each plot, sensitivity versus 1-specificity was plotted for each viral load value (genome/copies/mL) determined by RT-qPCR for each sample in our study when used as a lower limit threshold for scoring positive and negative detection for all other viral load results with qualitative viral culture or antigen test determinations, respectively, as the comparators. (a) Viral load in genome copies/mL versus detection by viral culture. (b) Viral load versus LumiraDx antigen detection. (c) Viral load versus BD Veritor antigen detection. (d) Viral load versus Oscar Corona antigen detection. (e) Viral load versus CareStart antigen detection. Viral load values along the ROC curves are labeled in power of 10 logarithmic intervals and demarcated in colour as indicated in accompany heatmap legend bar. Area under the curve for each ROC curve is denoted on respective plots.
Three hundred SARS-CoV-2 RT-qPCR negative samples were also tested by all four antigen tests. All antigen tests were negative, supporting high specificity of the antigen assays.
Discussion
The COVID-19 pandemic response has required detection of both infected and infectious individuals. SARS-CoV-2 viral culture is considered a reasonable surrogate test for infectivity although it is impractical as a high throughput diagnostic test [9,10]. In the Syrian golden hamster model, transmissibility of SARS-CoV-2 correlated well with detection of infectious virus by culture, but not with positive qPCR results [11]. Previous studies of SARS-CoV-2 culture have reported culture of viable virus from samples with a viral load of 2.5 × 105 to 106 genome copies/mL [6,8] or expressed alternatively as cycle threshold value cutoffs of 24 to 35 [7,12] and/or during the presumptively high infectious period between 6 to 9 days after infection [8,10,13,14].
Importantly, our study showed that antigen tests predicted the ability to culture live virus and therefore infectivity. In addition, a viral load cutoff of 105 copies/mL was 95% sensitive (95% CI, 90–98%) and 72% specific (95% CI, 60–81%) for predicting viral culture positivity, similar to prior observations.
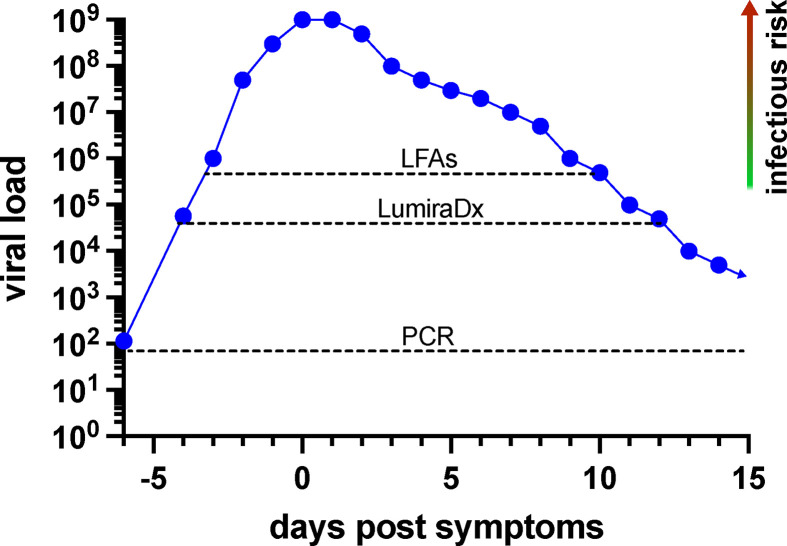
It should be stressed that viral loads for SARS-CoV-2 vary over nine orders of magnitude. The difference between the lowest viral loads, where virus is consistently detected by culture (105 copies/mL; Fig. 1, Fig. 3), and the highest observed viral loads (∼1 billion copies/mL) is at least four orders of magnitude. Furthermore, above the viral culture detection threshold, we found that the amount of viable virus in day 3 culture supernatants was roughly proportional to sample viral load determined by qPCR (Fig. 1), suggesting that the large range of viral loads determined by RT-qPCR corresponds to the range and degree of sample infectivity. Importantly, antigen test sensitivity is noted to be near 100% in the upper three orders of magnitude of viral loads observed (Fig. 2, Fig. 3, Fig. 4 ; Table 2; Table S1), suggesting that antigen tests are quite good in detecting individuals who shed larger amounts of virions and therefore would pose significant risk to others during casual contact.
Fig. 4.
Model of infectious risk versus SARS-CoV-2 detection by RT-qPCR and antigen tests. Both LumiraDx and lateral flow-based antigen tests (e.g. BD Veritor, CareStart, and Oscar Corona) were able to detect individuals with viable, culturable virus and who therefore pose an immediate infectious risk to others. Dotted lines indicate reliable detection threshold predicted for each method. Presumptively, infectious risk is proportional to the amount of culturable virus, which is roughly proportional to the viral load in samples. Antigen tests were excellent in detecting patients with the highest viral loads, which may be 104- to 105-fold greater than viral loads detected at the lowest levels where virus can be consistently cultured. The PCR and, to a lesser extent, the LumiraDx test can detect individuals before and after the expected infectious period and therefore may be more appropriate for screening programs where regular testing is performed at longer time intervals. The viral load curve shown is for representational purposes and may not reflect viral load kinetics in any specific individual.
It has been argued that cycle threshold values should not be made available to providers based on lack of clinical translatability or inadequate inter-assay comparability (reviewed in [15]). However, our data highlighted the meaningful association of viral load with infectivity and therefore supported conversion of assay-dependent cycle threshold values to assay-independent viral loads [16].
Our study had several strengths and some limitations. We sought to eliminate the source of inter-specimen variability by performing antigen, RT-qPCR, and viral culture testing on the same samples [16,17]. However, this led to the need to perform antigen testing outside of direct swab sampling testing recommendations, potentially leading to underestimation of antigen test sensitivity (see Supplemental Materials). A strength and weakness of the approach was the selection of real-world samples based on the distribution of viral loads observed during diagnostic testing [16]. Samples were not selected based on clinical symptoms and exposures. As we evaluated PCR positive and negative samples separately, it was possible that specificities determined for PCR positive samples may have been overestimated. Our study was conducted prior to Delta and Omicron emergence; verification of results with these and future variants is desirable.
The BD Veritor, CareStart, and Oscar Corona tests are chromatographic LFAs. The first two have received FDA EUA and CE Mark certification. In contrast, the LumiraDx, a microfluidic immunofluorescent detection test, appeared significantly more sensitive (see Fig. 1, Fig. 2, Fig. 3, Table 1), consistent with its high-level detection of infected patients during the first 12 days of symptom onset [18]. The overall observed lower sensitivity of antigen tests relative to PCR has been described previously [[19], [20], [21], [22]].
Taken together, our data supported the use of antigen detection tests immediately before communal events, same-day healthcare procedures, communal travel, and other settings with significant person-to-person contact, and where universal masking is neither feasible or desired. Similarly, we believe antigen tests will be useful in determining whether symptomatic individuals are infectious and need to self-isolate or can return to work or other activities. Importantly, for symptomatic individuals, it is prudent, based on variability in sampling, kinetics of viral replication, and varying sensitivity of different antigen tests (Fig. S1) to incorporate serial testing to increase negative predictive value. Notably, as a point-of-use testing modality, antigen test results can be available in minutes and inform timely mitigation of infectious risk to others and/or clinical management, and, when used at closely spaced time intervals, should also provide a lower cost alternative to nucleic acid amplification tests to address diagnostic goals of the pandemic.
Transparency declaration
This work was supported by an Accelerating Coronavirus Testing Solutions grant from the Massachusetts Life Sciences Cente. We thank LumiraDx for providing instrumentation and antigen test kits; Oscar Medicare Pvt. Ltd for providing Oscar Corona antigen test kits; and Ginkgo Bioworks (Boston, MA) for providing the CareStart antigen test kits. We thank LGC SeraCare for providing reagents used in calibrating SARS-CoV-2 viral load assays. Authors are solely responsible for study design, data analysis, and manuscript preparation.
We received support from Abbott Molecular unrelated to this study under a COVID-19 Diagnostics Evaluation Agreement. One co-author, RA, was also a recipient of grant support from Abbott Molecular under a clinical study agreement. LumiraDx, Oscar Medicare Pvt. Ltd, LGC SeraCare, Abbott Molecular, and Ginkgo Biosciences had no role in study design, manuscript preparation or decision to publish.
Author contributions
JEK, SR, and PJK conceived and designed the study. S Dutta, DH, AC, S Ditelberg, and PJK performed experiments and acquired data. JEK, SR, S Dutta, RA, CAC, AC, and PJK analysed and interpreted data. JEK, with input from SR, wrote the original manuscript draft. JEK, SR, RA, CAC, and PJK revised manuscript for intellectual content. All authors gave final approval for the submitted version. JEK and SR are co-first authors and equal contributors to manuscript.
Editor: M. Leeflang
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2022.07.010.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Koczula K.M., Gallotta A. Lateral flow assays. Essays Biochem. 2016;60:111–120. doi: 10.1042/EBC20150012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larremore D.B., Wilder B., Lester E., Shehata S., Burke J.M., Hay J.M., et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci Adv. 2021;7 doi: 10.1126/sciadv.abd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shain E.B., Clemens J.M. A new method for robust quantitative and qualitative analysis of real-time PCR. Nucleic Acids Res. 2008;36:e91. doi: 10.1093/nar/gkn408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith K.P., Cheng A., Chopelas A., DuBois-Coyne S., Mezghani I., Rodriguez S., et al. Large-scale, in-house production of viral transport media to support SARS-CoV-2 PCR testing in a multihospital health care network during the COVID-19 pandemic. J Clin Microbiol. 2020;58:e00913–e00920. doi: 10.1128/JCM.00913-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng A., Riedel S., Arnaout R., Kirby J.E. Verification of the Abbott Alinity m Resp-4-Plex assay for detection of SARS-CoV-2, influenza A/B, and respiratory syncytial virus. Diagn Microbiol Infect Dis. 2022;102 doi: 10.1016/j.diagmicrobio.2021.115575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C.G., Lee K.M., Hsiao M.J., Yang S.L., Huang P.N., Gong Y.N., et al. Culture-based virus isolation to evaluate potential infectivity of clinical specimens tested for COVID-19. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.01068-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 9.Rhee C., Kanjilal S., Baker M., Klompas M. Duration of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectivity: when is it safe to discontinue isolation? Clin Infect Dis. 2021;72:1467–1474. doi: 10.1093/cid/ciaa1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh K.A., Jordan K., Clyne B., Rohde D., Drummond L., Byrne P., et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81:357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singanayagam A., Patel M., Charlett A., Lopez Bernal J., Saliba V., Ellis J., et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R., et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020;382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W.D., Chang S.Y., Wang J.T., Tsai M.J., Hung C.C., Hsu C.L., et al. Prolonged virus shedding even after seroconversion in a patient with COVID-19. J Infect. 2020;81:318–356. doi: 10.1016/j.jinf.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhoads D.D., Pinsky B.A. The truth about SARS-CoV-2 cycle threshold values is rarely pure and never simple. Clin Chem. 2021;68:16–18. doi: 10.1093/clinchem/hvab146. [DOI] [PubMed] [Google Scholar]
- 16.Arnaout R., Lee R.A., Lee G.R., Callahan C., Cheng A., Yen C.F., et al. The limit of detection matters: the case for benchmarking severe acute respiratory syndrome coronavirus 2 testing. Clin Infect Dis. 2021;73:e3042–e3046. doi: 10.1093/cid/ciaa1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callahan C., Lee R.A., Lee G.R., Zulauf K., Kirby J.E., Arnaout R. Nasal swab performance by collection timing, procedure, and method of transport for patients with SARS-CoV-2. J Clin Microbiol. 2021;59 doi: 10.1128/JCM.00569-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drain P.K., Ampajwala M., Chappel C., Gvozden A.B., Hoppers M., Wang M., et al. A rapid, high-sensitivity SARS-CoV-2 nucleocapsid immunoassay to aid diagnosis of acute COVID-19 at the point of care: a clinical performance study. Infect Dis Ther. 2021;10:753–761. doi: 10.1007/s40121-021-00413-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford L., Lee C., Pray I.W., Cole D., Bigouette J.P., Abedi G.R., et al. Epidemiologic characteristics associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigen-based test results, real-time reverse transcription polymerase chain reaction (rRT-PCR) cycle threshold values, subgenomic RNA, and viral culture results from university testing. Clin Infect Dis. 2021;73:e1348–e1355. doi: 10.1093/cid/ciab303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karon B.S., Donato L., Bridgeman A.R., Blommel J.H., Kipp B., Maus A., et al. Analytical sensitivity and specificity of four point of care rapid antigen diagnostic tests for SARS-CoV-2 using real-time quantitative PCR, quantitative droplet digital PCR, and a mass spectrometric antigen assay as comparator methods. Clin Chem. 2021;67:1545–1553. doi: 10.1093/clinchem/hvab138. [DOI] [PubMed] [Google Scholar]
- 21.Krüger L.J., Klein J.A.F., Tobian F., Gaeddert M., Lainati F., Klemm S., et al. Evaluation of accuracy, exclusivity, limit-of-detection and ease-of-use of LumiraDx: an antigen-detecting point-of-care device for SARS-CoV-2. Infection. 2022;50:395–406. doi: 10.1007/s15010-021-01681-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollock N.R., Savage T.J., Wardell H., Lee R.A., Mathew A., Stengelin M., et al. Correlation of SARS-CoV-2 nucleocapsid antigen and RNA concentrations in nasopharyngeal samples from children and adults using an ultrasensitive and quantitative antigen assay. J Clin Microbiol. 2021;59 doi: 10.1128/JCM.03077-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.