Abstract
A new extracellular protease (PoSl; Pleurotus ostreatus subtilisin-like protease) from P. ostreatus culture broth has been purified and characterized. PoSl is a monomeric glycoprotein with a molecular mass of 75 kDa, a pI of 4.5, and an optimum pH in the alkaline range. The inhibitory profile indicates that PoSl is a serine protease. The N-terminal and three tryptic peptide sequences of PoSl have been determined. The homology of one internal peptide with conserved sequence around the Asp residue of the catalytic triad in the subtilase family suggests that PoSl is a subtilisin-like protease. This hypothesis is further supported by the finding that PoSl hydrolysis sites of the insulin B chain match those of subtilisin. PoSl activity is positively affected by calcium. A 10-fold decrease in the Km value in the presence of calcium ions can reflect an induced structural change in the substrate recognition site region. Furthermore, Ca2+ binding slows PoSl autolysis, triggering the protein to form a more compact structure. These effects have already been observed for subtilisin and other serine proteases. Moreover, PoSl protease seems to play a key role in the regulation of P. ostreatus laccase activity by degrading and/or activating different isoenzymes.
White rot basidiomycetes have received extensive attention because of their lignin-degrading activity. The biochemistry of lignin degradation is a complex process involving a series of enzymatic and nonenzymatic reactions. Extracellular enzymes which catalyze oxidative reactions are lignin peroxidases, laccases, manganese peroxidases, and hydrogen peroxide-producing enzymes (3, 13). Even though their catalyzed reactions have been studied in detail, their in vivo coordination and, possibly, synergistic action are not clearly understood.
Pleurotus ostreatus is a white rot basidiomycete which belongs to the subclass of ligninolytic microorganisms that produce laccases, manganese peroxidases, and veratryl alcohol oxidases but no lignin peroxidase. Among these enzymes, laccases have been the most widely studied and characterized (11, 12, 17). In a recent study (16), it has been demonstrated that a laccase isoenzyme (POXA1b; phenol oxidase A1b) is specifically degraded in the early phase of fungal growth by proteases present in P. ostreatus culture broth; hence, the disappearance of POXA1b seems to be correlated with the appearance of extracellular protease activity. A similar relationship was observed for lignin peroxidases in Phanerochaete chrysosporium, and, in this case, the extracellular proteases caused an almost complete disappearance of lignin peroxidase activity due to degradation of all lignin peroxidase isoenzymes (8). Furthermore, a recent report (21) suggests that both intracellular and extracellular proteases are involved in the regulation of ligninolytic activities in cultures of Trametes versicolor under nutrient limitation. In contrast, it has been reported (2) that proteases are not responsible for the decrease in peroxidase activity in Pleurotus pulmonarius cultures. Moreover, a purified protease from solid substrate cultures of Phanerochaete chrysosporium did not affect lignin peroxidase (5). Hence, it is not clear if there is a relationship between ligninolytic activity and protease secretion in white rot fungi.
Despite the fact that the production of extracellular proteolytic enzymes is a common feature among fungi, relatively few proteases secreted from lignin-degrading fungi have been characterized on a molecular level (5, 9, 15). This paper reports the purification and characterization of a novel protease (named PoSl) present in liquid culture of P. ostreatus which appears to belong to the serine protease family. Its structural and kinetic properties are significantly different from those of other proteases purified from P. ostreatus fruiting bodies (7). The purified enzyme is involved in POXA1b degradation and in activation of another recently characterized laccase isoenzyme (unpublished data). On the basis of these results, we hypothesize that extracellular proteases could play a regulatory role in laccase activity in P. ostreatus.
MATERIALS AND METHODS
Organism and culture conditions.
The white rot fungus P. ostreatus (Jacq.:Fr.) Kummer (type:Florida) was maintained through periodic transfer at 4°C on potato dextrose agar plates (Difco) in the presence of 0.5% yeast extract (Difco).
Incubation was carried out as previously described (17). The mycelium was grown in liquid basal medium (24 g of potato dextrose broth/liter, 5 g of yeast extract/liter) with one of the following additions: 150 μM CuSO4, 100 μM FeCl3, 150 μM CuSO4 plus 100 μM FeCl3, or 100 μM ZnSO4.
Fungal culture in the presence of phenylmethylsulfonyl fluoride (PMSF) was performed by adding 0.1 mM PMSF after 2 days of growth. The broth was filtered 24 h after the addition of PMSF.
Enzyme purification.
Proteins were precipitated from 3 liters of filtered medium supplemented with CuSO4 plus FeCl3 by the addition of (NH4)2SO4 up to 80% saturation at 4°C and centrifuged at 10,000 × g for 30 min. The precipitate was resuspended in 50 mM sodium phosphate buffer, pH 7.0, and extensively dialyzed against the same buffer. The sample was again centrifuged, and the supernatant, concentrated on an Amicon PM-10 membrane, was loaded on a DEAE-Sepharose Fast Flow (Pharmacia Biotech Inc.) column (1.5 by 40 cm) equilibrated with the phosphate buffer. The column was washed at a flow rate of 30 ml/h with 150 ml of buffer, and a 0 to 0.5 M NaCl linear gradient (200 ml) was applied. Fractions containing protease activity were pooled and concentrated on an Amicon PM-10 membrane.
The major active peak was then subjected to a size exclusion chromatography on a Superdex 75 PC 3.2/30 gel filtration column (Pharmacia). The column was eluted with 50 mM sodium phosphate buffer, pH 7.0, containing 150 mM NaCl (flow rate, 0.05 ml/min). Active fractions were finally equilibrated in 50 mM sodium phosphate buffer, pH 7.0 (buffer A), with an Amicon PM-10 membrane and then loaded onto an anion-exchange Mono-Q HR 5/5 column in a fast protein liquid chromatography system equilibrated with the same buffer. The enzyme (PoSl) was eluted with a linear gradient (buffer B = 50 mM sodium phosphate buffer [pH 7.0] plus 0.5 M NaCl; gradient: at t = 0, buffer B = 0%; at t = 10 min, buffer B = 0%; at t = 60 min, buffer B = 80%). The active fractions were pooled and desalted.
Enzyme assays.
Protease activity was assayed using N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide (SucAAPFpNA), N-succinyl-Ala-Ala-Pro-Leu-p-nitroanilide (SucAAPLpNA), azo dye-impregnated collagen (Azocoll) , Nα-p-tosyl-l-arginine methyl ester (TAME), and N-benzoyl-l-tyrosine ethyl ester (BTEE), all from Sigma, as substrates as follows.
(i) SucAAPFpNA as substrate.
The SucAAPFpNA assay mixture contained 5 mM SucAAPFpNA, 10 mM CaCl2, and 50 mM Tris-HCl buffer, pH 8.0, in a final volume of 1 ml. Hydrolysis of the substrate was monitored by noting the increase in absorbance at 405 nm (ɛ405 = 8,800 M−1 cm−1).
(ii) SucAAPLpNA as substrate.
The SucAAPLpNA assay mixture contained 5 mM SucAAPLpNA, 10 mM CaCl2, and 50 mM Tris-HCl buffer, pH 7.0, in a final volume of 1 ml. Hydrolysis of the substrate was followed by an increase in absorbance at 405 nm (ɛ405 = 8,800 M−1 cm−1).
(iii) Azocoll as substrate.
The protease activity assay using Azocoll as the substrate was performed as described by Datta (5). One unit of activity was defined as the amount of enzyme that catalyzed the release of enough azo dye to give a change in absorbance at 520 nm of 0.001 in 30 min.
(iv) TAME as substrate.
The TAME assay mixture contained 10 mM TAME, 10 mM CaCl2, and 50 mM Tris-HCl buffer, pH 8.0, in a final volume of 1 ml. Hydrolysis of TAME was monitored at room temperature by noting the increase in absorbance at 247 nm (ɛ247 = 540 M−1 cm−1).
(v) BTEE as substrate.
The BTEE assay mixture contained 0.5 mM BTEE, 10 mM CaCl2, and 50 mM Tris-HCl buffer, pH 8.0, in a final volume of 1 ml. Hydrolysis of BTEE was monitored at room temperature by noting the increase in absorbance at 256 nm (ɛ256 = 964 M−1 cm−1).
Unless otherwise reported, SucAAPFpNA was the substrate used to measure the protease activity.
The influence of pH values upon protease activity was determined at room temperature using McIlvaine citrate-phosphate buffer for the pH range of 6.0 to 8.0 and in 50 mM Tris-HCl buffer for pH 8.0 and 9.0.
The effect of different inhibitors on PoSl activity was tested by preincubating the enzyme with the different putative inhibitors (1:105, enzyme/inhibitor molar ratio) in 50 mM Tris-HCl, pH 7.0, for 30 min at room temperature before addition to the substrate.
Protein determination.
Protein concentration was determined using the Bio-Rad protein assay with bovine serum albumin as the standard.
Nondenaturing PAGE.
Polyacrylamide gel electrophoresis (PAGE) was performed at an alkaline pH under nondenaturing conditions. The separating and stacking gels contained 9 and 4% concentrations of acrylamide, respectively. The buffer solutions were 50 mM Tris-HCl (pH 9.5) for the separating gel and 18 mM Tris-HCl (pH 7.5) for the stacking gel; the electrode reservoir solution was 25 mM Tris–190 mM glycine (pH 8.4). Gels were stained for laccase activity using ABTS [2,2′-azinobis-(3-ethylbenzthiazolinesulfonic acid)] as the substrate as previously described (16).
Determination of molecular mass.
The molecular mass of native protease was determined using a Superdex 200 PC 3.2/30 gel filtration column (Pharmacia). The column was eluted with 50 mM sodium phosphate buffer, pH 7.0, containing 150 mM NaCl (flow rate, 1 ml/min). The calibration of the column was performed with alcohol dehydrogenase (150 kDa), hemoglobin (64 kDa), ovalbumin (45 kDa), and carbonic anhydrase (29 kDa) as standards.
Electrophoresis and isoelectrofocusing.
Polyacrylamide (9%) gel slab electrophoresis in 0.1% sodium dodecyl sulfate (SDS) was carried out as described by Laemmli (14). For molecular mass determinations, the gel was calibrated with rabbit muscle myosin (205 kDa), β-galactosidase (116 kDa), rabbit muscle phosphorylase b (97.4 kDa), bovine serum albumin (66 kDa), egg albumin (45 kDa), and carbonic anhydrase (29 kDa). Proteins were visualized by Coomassie blue staining.
Analytical isoeletric focusing in the pH range of 2.5 to 7.0 was performed on a 5.0% acrylamide gel slab with a Multiphor II electrophoresis system (Pharmacia) by following the manufacturer's instructions. Proteins were stained using the silver staining method.
Sequence analysis.
Automated N-terminal degradation of the electroblotted protein was performed with a Perkin-Elmer Applied Biosystems 477A pulsed-liquid protein sequencer equipped with a model 120A phenylthiohydantoin analyzer for the online identification and quantification of phenylthiohydantoin amino acids.
Lectin assay.
PoSl and control standard glycoproteins (supplied with the Boehringer Mannheim glycan differentiation kit; 1 μg) were directly spotted onto an Immobilon membrane and detected immunologically after binding to lectins conjugated with digoxigenin by following the manufacturer's instructions (Boehringer Mannheim). The lectins used were the following: Galanthus nivalis agglutinin, specific for terminal mannose; Sambucus nigra agglutinin, specific for sialic acid α(2-6)-galactose; Maackia amurenais agglutinin, specific for sialic acid α(2-3)-galactose; peanut agglutinin, specific for galactose β(1-3)-N-acetylglucosamine; and Datura stramonium agglutinin, specific for galactose β(1-4)-N-acetylglucosamine. The protein linked to the lectin was detected by a colorimetric reaction. The immunological detection was performed according to the manufacturer's instructions. This experiment was performed in duplicate on two different enzymatic preparations.
MALDI-MS analysis.
Matrix-assisted laser desorption ionization–mass spectrometry (MALDI-MS) analyses were carried out with a Voyager DE MALDI-time of flight MS (PerSeptive Biosystem, Boston, Mass.).
Molecular mass determination of protease was performed by loading a mixture of protease solution and 3,5-dimethoxy-4-hydroxycinnamic acid (sinapinic acid) on a sample slide and drying the slide in vacuo. Mass range was calibrated using apomyoglobin from horse heart (average molecular mass, 16,952.5 Da) and human serum albumin (average molecular mass, 66,431.0 Da).
Enzymatic hydrolysis.
Protease samples (1 nmol) were reduced by incubation with 200 nmol of dithiothreitol at 37°C for 2 h under a nitrogen atmosphere in 0.25 M Tris-HCl (pH 8.5)–1.25 mM EDTA containing 6 M guanidinium chloride. Free SH groups were alkylated by using an excess of iodoacetamide at room temperature in the dark under a nitrogen atmosphere. Protein samples were freed from salt and an excess of reagent by loading the reaction mixture onto a PD-10 prepacked column (Pharmacia) equilibrated and eluted in 0.4% ammonium bicarbonate, pH 8.5.
Enzymatic digestion with trypsin was carried out in 0.4% ammonium bicarbonate, pH 8.5, at 37°C overnight using an enzyme/substrate molar ratio of 1:50. The peptide mixture was deglycosylated by incubation with 0.15 U of peptide N-glycosidase F (Boehringer Mannheim) in 0.4% ammonium bicarbonate, pH 8.5, at 37°C for 16 h. The sample was directly analyzed by reverse-phase high-performance liquid chromatography (HPLC); sugar and peptide fractions were collected manually.
Oligosaccharide derivatization.
Oligosaccharides were permethylated as described by Dell (6) and directly analyzed by MALDI-MS by mixing permethylated oligosaccharides with 2,5-dihydroxybenzoic acid on a sample plate and drying the plate in vacuo. Mass range was calibrated with bovine insulin (average molecular mass, 5,734.6 Da).
Specificity assay with oxidized B chain of insulin.
Oxidized B chain from bovine insulin (Sigma) was hydrolyzed with PoSl, trypsin (bovine pancreas), subtilisin bacterial protease nagarse, chymotrypsin (bovine pancreas), and elastase (pig pancreas), all from Sigma. Incubations were performed in 0.4% ammonium bicarbonate, pH 8.5, at 37°C for 5 min, using an enzyme/substrate ratio of 1:1,000 (wt/wt). Proteolytic mixtures were analyzed at each time by MALDI-MS analysis.
Autocatalytic assay.
PoSl autocatalytic activity was checked using 2 μM solutions of PoSl incubated in 0.4% ammonium bicarbonate, pH 8.5, at 37°C. Aliquots were withdrawn at different incubation times (0, 1 h, 3 h, and 18 h), and catalytic activity was stopped by adding 20% trifluoroacetic acid (TFA).
The samples were analyzed by reverse-phase HPLC (Beckman Gold HPLC system) on a reverse-phase column (Vydac C4 narrow-bore column; 250 by 2.1 mm, 5-μm particle size) and eluted using a system consisting of solvent A (0.1% TFA in water) and solvent B (0.07% TFA in 95% acetonitrile). Proteins were eluted with a linear 20 to 85% gradient of solvent B in 30 min at a flow rate of 0.2 ml/min. The autodigestion of PoSl was calculated by comparing peak area at time zero with peak areas at subsequent times.
Incubation of PoSl with laccase isoenzymes.
A sample of P. ostreatus culture broth supplemented with copper and iron (pH ∼6) was collected after 2 days of growth, filtered, and incubated overnight at 25°C with PMSF (1 mM final concentration). After this treatment, no protease activity, using SucAAPFpNA as the substrate, was detected. Fifty microliters of this sample (0.15 U of total laccase activity) was incubated with 0.01 nmol of PoSl in the presence of 10 mM CaCl2. Aliquots of this sample (12 μl) after 1, 3, and 18 h were analyzed by nondenaturing PAGE.
Incubations of PoSl with POXA1b were performed at room temperature in McIlvaine buffer, adjusted to different pHs (5.0, 6.0, and 7.0), containing 10 mM CaCl2. Laccase activity was monitored over 2 days of incubation. Incubations of PoSl with POXA3 (purified from standard fungal culture broth or supplied with 0.1 mM PMSF) were performed at room temperature in McIlvaine buffer, at pH 7.0, containing 10 mM CaCl2, using a PoSl/POXA3 molar ratio of 1:10. POXA3 was also incubated, under the same conditions, in the presence of an equivalent number of enzymatic units of subtilisin, determined using SucAAPFpNA as the substrate.
RESULTS
Protease assay.
In order to identify the best substrate for detection of protease activity in P. ostreatus culture broth, four different protease substrates were tested. Activities of 10 and 0.03 U/ml were determined in the basal medium after 3 days of growth using Azocoll and SucAAPFpNA as substrates, respectively (the activity towards Azocoll is expressed in arbitrary units), while no activity was detected with TAME or BTEE as the substrate. Because the SucAAPFpNA assay allows the expression of protease activity in international units, SucAAPFpNA was used as the substrate to establish the catalytic parameters.
Protease purification.
Culture conditions which induce protease production were analyzed; two- and threefold-higher protease activities were obtained in the presence of 0.15 mM CuSO4 and 0.15 mM CuSO4 plus 0.1 mM FeCl3, respectively, after 3 days of incubation, than in the basal medium. No increase of protease activity was observed when ZnSO4 or FeCl3 was used. Culture broth supplemented with CuSO4 plus FeCl3 was fractionated after 3 days of growth by ammonium sulfate precipitation followed by anion-exchange chromatography at pH 7.0. Three different protease fractions were separated. A major peak (PoSl) was eluted with a saline gradient at approximately 0.4 M NaCl. Fractions corresponding to PoSl were collected and further purified by gel filtration chromatography (Superdex 75) and anion-exchange chromatography on a Mono Q column at pH 7.0. The purified protein appeared to be homogeneous when analyzed by SDS-PAGE, isoelectric focusing, and gel filtration chromatography. A summary of the purification procedure is shown in Table 1; about 70-fold purification was achieved, with a final yield of 13%.
TABLE 1.
Purification of PoS1 protease
| Purification step | Vol (ml) | Total activity (U) | Total proteins (mg) | Sp act (U/mg) | Recovery (%) |
|---|---|---|---|---|---|
| Broth | 2,220 | 279 | 111 | 2.5 | 100 |
| NH4(SO4)2 precipitate | 14 | 265 | 40 | 6.6 | 95 |
| DEAE Sepharose | 2 | 226 | 2.7 | 84 | 81 |
| Superdex 75 | 1 | 84 | 0.6 | 140 | 30 |
| Mono Q | 0.5 | 36 | 0.2 | 180 | 13 |
Physical and chemical properties.
The molecular mass of PoSl is 75 kDa under denaturing conditions (SDS-PAGE) and 74 kDa under native conditions (gel filtration chromatography), thus indicating the monomeric nature of this protein. A more accurate determination of the molecular mass of PoSl was performed by MALDI-MS: a broad peak centered at 75,163 Da was obtained. The isoelectric point of the purified PoSl is 4.5.
Sequences of the N terminus and of three tryptic peptides were also determined. The sequences of two internal peptides (Ser-Leu-Gly-Gly-Tyr-Ala-Asp-Gly-Trp-Thr-Glu-Ser-Val-Ser- Ala-Val-Val-Ala-Ser-Arg and Thr-Leu-Phe-Glu-Thr-Thr-Ala-Gln-Arg) and the N terminus (Gly-Pro-Asp-Asp-Pro-Ala-Leu-Pro-Pro-Asp-Ser-Glu-Xaa-Thr-His) did not show any significant homology to proteases, based on a search of different protein data banks. On the other hand, the sequence of the third internal peptide was homologous to the conserved sequence around the Asp residue of the catalytic triad in the subtilase protease family (S8) (18) (Fig. 1).
FIG. 1.
Sequence of a tryptic peptide from P. ostreatus PoSl protease. The sequence has been aligned with those of other proteases belonging to the subtilase family (S8). An asterisk indicates the Asp residue present in the catalytic triad.
PoSl samples were analyzed for the presence of specific oligosaccharides by lectin binding assays. The protein was specifically recognized by Galanthus nivalis agglutinin lectin, which binds to terminal mannose residues, and Datura stramonium agglutinin, specific for galactose β(1-4)-N-acetylglucosamine. On the basis of lectin specificity, the presence of both high-mannose and hybrid or complex-type N-linked glycans can be suggested. The intact N-linked oligosaccharides were released from the peptide backbone by peptide N-glycosidase F digestion of the tryptic peptide mixture. MALDI-MS analysis of permethylated glycans showed a major peak at m/z 1580.0 that was assigned to a glycosidic chain (hexose)5(N-acetylhexosamine)2. This structure was identified as a high-mannose type N-linked chain with two mannose residues linked to the pentasaccharide core. The other molecular ions at m/z 1784.0, 1987.5, 2191.5, and 2394.6 were identified as homologous structures having between three and six mannose residues linked to the pentasaccharide core.
Catalytic properties.
The activity of PoSl increased in the presence of CaCl2 in the assay mixture at concentrations up to 2 mM and then was nearly constant at concentrations up to 10 mM, giving rise to a twofold increase in activity. Like calcium, Mn2+ similarly influenced proteolytic activity, while Zn2+and Mg2 affected the activity to a lesser extent.
The specificity of purified extracellular protease PoSl was tested using the synthetic substrates TAME, BTEE, SucAAPFpNA, SucAAPLpNA, and Azocoll. All substrates except TAME were hydrolyzed by PoSl. The specific activities towards BTEE, SucAAPFpNA, SucAAPLpNA, and Azocoll were 129 U/mg, 180 U/mg, 34 U/mg, and 5.3 × 104 U/mg, respectively. Since PoSl is able to release azo dye from Azocoll and to hydrolyze BTEE, it was possible to conclude that PoSl can act as an endoprotease and esterase.
The optimum pH of the purified protease was determined over a pH range of 6.0 to 9.0 using SucAAPFpNA and SucAAPLpNA as substrates; the enzyme was found to be more active in the alkaline pH range, with maximums at pH 8.0 and 7.0 for SucAAPFpNA and SucAAPLpNA, respectively.
The catalytic parameters of PoSl with respect to SucAAPFpNA, in the presence or absence of CaCl2, were determined on the basis of typical Michaelis-Menten behavior (Table 2). The occurrence of excess substrate inhibition in the case of SucAAPLpNA, either in the presence or absence of Ca2+, led us to calculate the apparent kinetic parameters by extrapolation. PoSl protease shows a greater than 10-fold increase in catalytic efficiency (kcat/Km) in the presence of CaCl2 due to a significant increase in affinity (about 15-fold-lower Km) for both substrates in the presence of Ca2+. Moreover, the catalytic efficiencies determined for both substrates are very similar, indicating that PoSl does not distinguish between Phe and Leu in the P1 position of peptide amides.
TABLE 2.
Kinetic constants of PoS1 protease
| Substrate | Optimum pH | Km (mM) | kcat (min−1) | kcat/Km (min−1 mM−1) |
|---|---|---|---|---|
| SucAAPFpNA | 8 | 6.7 ± 0.9 | (1.3 ± 0.1) × 104 | 1.9 × 103 |
| SucAAPFpNA plus 10 mM CaCl2 | 8 | 0.38 ± 0.05 | (9.35 ± 0.03) × 103 | 2.46 × 104 |
| SucAAPLpNA | 7 | 1.7 ± 0.3 | (1.9 ± 0.1) × 103 | 1.1 × 103 |
| SucAAPLpNA plus 10 mM CaCl2 | 7 | 0.12 ± 0.01 | (4.4 ± 0.2) × 103 | 3.7 × 104 |
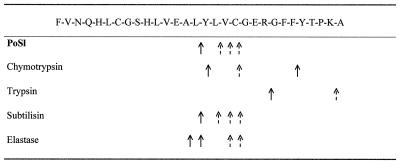
In order to better clarify the specificity of PoSl, proteolysis experiments were carried out to compare four different proteases (trypsin, chymotrypsin, elastase, and subtilisin). Enzymatic hydrolysis was performed on oxidized B chain of bovine insulin, and each peptide mixture was analyzed by MALDI-MS, thus allowing identification of preferential cleavage sites for each enzyme in the experimental condition used (Fig. 2). The spectrum of B chain hydrolyzed by PoSl showed two main signals at m/z 1715.40 and 1799.4, which were assigned to peptides 1 to15 and 16 to 30, respectively. At the same time, other minor signals which corresponded to subdigestion of the main peptides were present in the spectrum. The preferential cleavage site was at Leu15, even if other secondary hydrolysis sites could be identified at Leu17, Val18, or Cys19 (Fig. 2). Analysis of the proteolytic pattern also indicates that PoSl recognizes and hydrolyzes peptide bonds with a subtilisin-like specificity.
FIG. 2.
Comparison of cleavage sites of PoSl and other serine proteases in cleavage sites on oxidized bovine insulin B chain. Solid and broken arrows indicate preferential and secondary cleavage sites, respectively.
Autocatalytic assay.
In order to evaluate the autocatalytic activity of PoSl, the enzyme was incubated at pH 8.5 at 37°C. The residual activities after 1, 3, and 18 h were 86, 25, and 0%, respectively. The recoveries of undigested PoSl, determined by HPLC analysis after 1, 3, and 18 h of incubation, were 72, 40, and 0%, respectively. These values are similar to those measured in an analogous reference experiment performed with chymotrypsin. Furthermore, calcium protects PoSl against autolysis; in fact, when incubation was performed in the presence of 10 mM CaCl2, residual activities were 100 and 52% after 1 and 3 h, respectively.
Effect of inhibitors.
The effect of various compounds on the ability of PoSl to hydrolyze SucAAPFpNA is shown in Table 3. PoSl activity was almost completely inhibited by antipain, chymostatin, PMSF, 3,4-dichloroisocoumarin, and 4-amidinophenyl-methanesulfonyl fluoride at the indicated concentrations (Table 3). Partial inhibition was observed using aprotinin, another serine protease inhibitor. Among the heavy metal ions tested, only Cu2+ caused a slight inhibition. None of the thiol-blocking reagents or metal-chelating agents affected the protease activity. These results, together with those described above, strongly suggest that PoSl belongs to the serine protease family.
TABLE 3.
Effect of protease inhibitors on PoS1 activity
| Inhibitor (concn) | Residual activity (%) |
|---|---|
| HgCl2 (1 mM) | 93 |
| FeCl3 (1 mM) | 100 |
| CuSO4 (1 mM) | 77 |
| EDTA (1 mM) | 100 |
| o-Phenanthroline (1 mM) | 100 |
| p-Hydroxymercuribenzoate (1 mM) | 100 |
| p-Chloromercuribenzoate (1 mM) | 100 |
| E-64 (0.1 mM) | 93 |
| Chimostatin (0.1 mM) | 17 |
| Antipain (1 mM) | 2 |
| Pepstatin A (1 mM) | 100 |
| DCIa (0.5 mM) | 6 |
| PMSF (1 mM) | 1 |
| APMSFb (1 mM) | 6 |
| Aprotinin (1 mM) | 54 |
DCI, 3,4-dichloroisocoumarin.
APMSF, 4-amidinophenyl-methanesulfonyl fluoride.
Effect of PoSl on P. ostreatus laccase isoenzymes.
The effect of PoSl on some P. ostreatus laccase isoenzymes, namely, POXC (17), POXA1b (12), and POXA3 (data to be published), was analyzed.
Culture broth, after 2 days of growth, was filtered and incubated overnight with 1 mM PMSF in order to irreversibly inhibit serine proteases. After 12 h, PoSl was added. Samples were withdrawn at different incubation times (1, 3, and 18 h) and loaded on native PAGE gels (Fig. 3). An aliquot of PMSF-treated broth, kept at room temperature for 18 h, was used as the control.
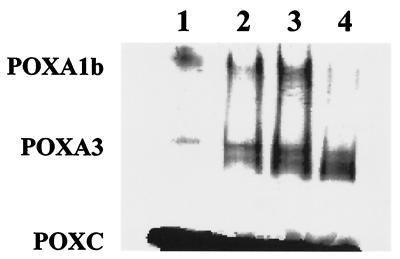
FIG. 3.
Zymogram of laccase isoenzymes incubated with purified PoSl protease. Fifty microliters of 2-day-old filtered culture broth that had been treated with PMSF (1 mM) was incubated with PoSl (0.01 nmol), and aliquots of equal volumes, withdrawn at different times, were loaded onto a native PAGE gel. Lane 1, control (sample incubated for 18 h in the absence of PoSl); lanes 2 to 4, samples incubated for 1, 3 and 18 h, respectively.
As shown in Fig. 3, POXC seems to be insensitive to PoSl; on the other hand, POXA1b activity is completely lost after a prolonged incubation. In order to establish whether this effect was due solely to the action of PoSl, purified POXA1b was incubated with PoSl. After 48 h, no effect on POXA1b (specific activity or intensity and mobility of the related SDS-PAGE band) was observed even when a PoSl/POXA1b molar ratio of 1:10 was used. Therefore, the observed POXA1b inactivation can be achieved proteolytically only in the case of PoSl activation of some, not-PMSF-inhibited, proteases.
POXA3 displays a peculiar behavior (Fig. 3); in fact, the intensity of the active band strongly increases during incubation, and a new active band, with higher electrophoretic mobility, appears. The results apparently suggest a sort of activation following a maturation process induced by PoSl. When different preparations of purified POXA3 were directly incubated with PoSl, the results were erratic. In order to exclude heterogeneity among different POXA3 preparations, possibly due to in vivo proteolytic aging, PMSF was added to the fungal culture and POXA3, purified from this broth, was incubated with PoSl. Under this condition, a reproducible increase of POXA3 activity was obtained (about 30%). The results are strongly suggestive of the generation of a more active POXA3 isoform(s) due to PoSl-induced proteolysis.
Taking into consideration the fact that PoSl is a subtilisin-like protease, we incubated culture broth with subtilisin under the same conditions used for PoSl. However, no effect on the POX isoenzymes was detectable in these experiments or on purified isoenzymes incubated with subtilisin.
DISCUSSION
Proteolytic enzymes can play different roles in the physiology of ligninolytic fungi, such as activation of zymogenic enzymes or release of enzymes from the fungal cell walls. It has been previously reported (4, 8) that, in Phanerochaete chrysosporium under ligninolytic conditions, the turnover of lignin peroxidases is due in part to enzyme proteolysis, and specific proteases associated with conditions which promote lignin degradation have been identified. To the best of our knowledge, no relationship between manganese peroxidase and protease activity has been evidenced.
P. ostreatus produces several laccase isoenzymes putatively involved in lignin degradation (11, 12, 17). Recently, we demonstrated that the amounts of these isoenzymes increase substantially in copper-supplemented cultures and the presence of proteases secreted into the copper-containing culture medium specifically affects at least one of the known laccase isoenzymes (16). In order to better clarify the relationship between laccases and proteases in P. ostreatus and the role of these extracellular proteases, we have purified and characterized a protease (PoSl) from the fungal culture broth. PoSl is a monomeric glycoprotein with a molecular mass of 75 kDa, which is larger than that determined for most fungal proteases (5, 7, 9, 15, 19, 22).
This protease displays an optimum pH in the alkaline range, and its ability to release azo dye from Azocoll suggests that it is an endoprotease; moreover, the inhibitory profile indicates that PoSl is a serine protease. The sequences of the N terminus and of three tryptic peptides have been determined. The homology of one internal peptide with the conserved sequence around the Asp residue of the catalytic triad in the subtilase family (S8) suggests that PoSl is a subtilisin-like protein. This hypothesis is further supported by the finding that PoSl hydrolysis sites of the insulin B chain match those of subtilisin.
PoSl activity is positively affected by calcium; calcium ions, in fact, have a significant effect upon Km values for two of the substrates tested (kcat is almost unaffected), which can reflect an induced structural change in the substrate recognition site region. Furthermore, Ca2+ binding slows PoSl autolysis, triggering the protein to form a more compact structure. Both of these effects have been observed previously for subtilisin-like and other serine proteases (1, 10, 20).
The kinetic and structural characteristics of PoSl define it as a novel subtilisin-like protease that is different from the already known proteases from white rot fungi.
Recent reports on laccase isoenzymes secreted by P. ostreatus indicated an involvement of extracellular proteases in the degradation of POXA1b isoenzyme (16). In the present study, the effect of PoSl on laccase isoenzymes has been investigated. Results show that PoSl is actually involved in degrading POXA1b. In fact, POXA1b was unaffected in the PMSF-treated control (Fig. 3), while it was degraded in the presence of PoSl. On the other hand, no effect of PoSl on purified POXA1b was observed. These results suggest that PoSl plays a role in POXA1b degradation through either (i) a cascade activation mechanism of other proteases present in the culture broth or (ii) proteolysis of POXA1b leading to a polypeptide moiety, which is a better substrate for other proteases. A more significant effect of PoSl on laccase isoenzymes is the activation of POXA3. This effect can be due to PoSl-dependent POXA3 proteolysis generation of, in this case, a more active isoform(s). On the other hand, the possibility cannot be excluded that the observed activation could also be due to the PoSl-mediated hydrolysis of a polypeptide type of inhibitor. Further investigations are needed to discriminate between these two hypotheses.
No effect on POX isoenzymes was detectable when subtilisin was used instead of the fungal protease, although its specificity is actually very similar to that of PoSl. However, the possibility cannot be excluded that some differences in the specificities of these two proteases could be observable under different experimental conditions or when specific target proteins are used.
Reported results allow the conclusion that P. ostreatus extracellular proteases, besides their nutritional role, may be involved in the regulation of laccase activity by degrading or activating different isoenzymes.
ACKNOWLEDGMENTS
This work was supported by grants from the Ministero dell'Università e della Ricerca Scientifica (Progetti di Rilevante Interesse Nazionale grant PRIN 98 and PRIN 2000) and from the Consiglio Nazionale delle Ricerche (Progetto Finalizzato Biotecnologie).
REFERENCES
- 1.Bajorath J, Hinrichs W, Saenger W. The enzymatic activity of proteinase K is controlled by calcium. Eur J Biochem. 1988;176:441–447. doi: 10.1111/j.1432-1033.1988.tb14301.x. [DOI] [PubMed] [Google Scholar]
- 2.Bockle B, Martinez M J, Guillen F, Martinez A T. Mechanism of peroxidase inactivation in liquid cultures of the ligninolytic fungus Pleurotus pulmonarius. Appl Environ Microbiol. 1999;65:923–928. doi: 10.1128/aem.65.3.923-928.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breen A, Singleton F L. Fungi in lignocellulose breakdown and biopulping. Curr Opin Biotechnol. 1999;10:252–258. doi: 10.1016/S0958-1669(99)80044-5. [DOI] [PubMed] [Google Scholar]
- 4.Dass S B, Dosoretz C G, Reddy C A, Grethlein H E. Extracellular proteases produced by the wood-degrading fungus Phanerochaete chrysosporium under ligninolytic and nonligninolytic conditions. Arch Microbiol. 1995;163:254–258. doi: 10.1007/BF00393377. [DOI] [PubMed] [Google Scholar]
- 5.Datta A. Purification and characterization of a novel protease from solid substrate cultures of Phanerochaete chrysosporium. J Biol Chem. 1992;267:728–736. [PubMed] [Google Scholar]
- 6.Dell A. Preparation and desorption mass spectrometry of permethyl and peracetyl derivatives of oligosaccharides. Methods Enzymol. 1990;193:647–660. doi: 10.1016/0076-6879(90)93443-o. [DOI] [PubMed] [Google Scholar]
- 7.Dohmae N, Hayashi K, Miki K, Tsumuraya Y, Hashimoto Y. Purification and characterization of intracellular proteinases in Pleurotus ostreatus fruiting bodies. Biosci Biotechnol Biochem. 1995;59:2074–2080. doi: 10.1271/bbb.59.2074. [DOI] [PubMed] [Google Scholar]
- 8.Dosoretz C G, Dass B, Reddy A, Grethlein H E. Protease-mediated degradation of lignin peroxidase in liquid cultures of Phanerochaete chrysosporium. Appl Environ Microbiol. 1990;56:395–400. doi: 10.1128/aem.56.11.3429-3434.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eriksson K E, Pettersson E. Purification and partial characterization of two acidic proteases from the white rot fungus Sporotrichum pulverulentum. Eur J Biochem. 1982;124:635–642. doi: 10.1111/j.1432-1033.1982.tb06641.x. [DOI] [PubMed] [Google Scholar]
- 10.Exterkate F A, Alting A C. Role of calcium in activity and stability of the Lactococcus lactis cell envelope proteinase. Appl Environ Microbiol. 1999;65:1390–1396. doi: 10.1128/aem.65.4.1390-1396.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giardina P, Aurilia V, Cannio R, Marzullo L, Amoresano A, Siciliano R, Pucci P, Sannia G. The gene, protein, and glycan structures of laccase from Pleurotus ostreatus. Eur J Biochem. 1996;235:508–515. doi: 10.1111/j.1432-1033.1996.00508.x. [DOI] [PubMed] [Google Scholar]
- 12.Giardina P, Palmieri G, Scaloni A, Fontanella B, Faraco V, Cennamo G, Sannia G. Protein and gene structure of a blue laccase from Pleurotus ostreatus. Biochem J. 1999;34:655–663. [PMC free article] [PubMed] [Google Scholar]
- 13.Hatakka A. Lignin-modifying enzymes from selected white-rot fungi: production and role in lignin degradation. FEMS Microbiol Rev. 1994;13:125–135. [Google Scholar]
- 14.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Mellon J E, Cotty P J. Purification and partial characterization of an elastinolytic proteinase from Aspergillus flavus culture filtrates. Appl Microbiol Biotechnol. 1996;46:138–142. [Google Scholar]
- 16.Palmieri G, Giardina P, Bianco C, Fontanella B, Sannia G. Copper induction of laccase isoenzymes in the ligninolytic fungus Pleurotus ostreatus. Appl Environ Microbiol. 2000;66:920–924. doi: 10.1128/aem.66.3.920-924.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmieri G, Giardina P, Bianco C, Scaloni A, Capasso A, Sannia G. A novel white laccase from Pleurotus ostreatus. J Biol Chem. 1997;272:31301–31307. doi: 10.1074/jbc.272.50.31301. [DOI] [PubMed] [Google Scholar]
- 18.Rawlings N D, Barret A J. Families of serine peptidases. Methods Enzymol. 1994;244:19–67. doi: 10.1016/0076-6879(94)44004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhodes W G, Lindberg R A, Drucker H. Purification and characterization of an extracellular protease from Neurospora crassa. Arch Biochem Biophys. 1983;223:514–520. doi: 10.1016/0003-9861(83)90616-1. [DOI] [PubMed] [Google Scholar]
- 20.Sipos T, Merkel J R. An effect of calcium ions on the activity heat stability and structure of trypsin. Biochemistry. 1970;9:2766–2775. doi: 10.1021/bi00816a003. [DOI] [PubMed] [Google Scholar]
- 21.Staszczak M, Zdunek E, Leonowicz A. Studies on the role of proteases in the white-rot fungus Trametes versicolor: effect of PMSF and chloroquine on ligninolytic enzyme activity. J Basic Microbiol. 2000;40:51–36. [PubMed] [Google Scholar]
- 22.Tapia G, Curotto E, O'Reilly S, Gonzales G. Isolation and partial characterization of an extracellular protease from Sporotrichum dimorphosporum. FEBS Lett. 1981;130:205–207. [Google Scholar]