Abstract
Integrin CD103 mediates the adhesion and tissue retention of T cells by binding to E-cadherin which is abundant on epithelial cells. Notably, CD103 is highly expressed on CD8 T cells but conspicuously absent on most CD4 T cells. The mechanism controlling such lineage-specific expression of CD103 remains unclear. Using a series of genetically engineered mouse models, here, we demonstrate that the regulatory mechanism of CD103 expression is distinct between CD4 and CD8 T cells, and that the transcription factor Runx3 plays an important but not an essential role in this process. We further found that the availability of integrin β7 which heterodimerizes with CD103 was necessary but also constrained the surface expression of CD103. Notably, the forced surface expression of CD103 did not significantly alter the thymic development of conventional T cells but severely impaired the generation of MHC-II-restricted TCR transgenic T cells, revealing previously unappreciated aspects of CD103 in the selection and maturation of CD4 T cells. Unlike its effect on CD4 T cell development, however, CD103 overexpression did not significantly affect CD4 T cells in peripheral tissues. Moreover, the frequency and number of CD4 T cells in the small intestine epithelium did not increase even though E-cadherin is highly expressed in this tissue. Collectively, these results suggest that most mature CD4 T cells are refractory to the effects of CD103 expression, and that they presumably utilize CD103-independent pathways to control their tissue retention and residency.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-03877-9.
Keywords: Integrin, Intraepithelial lymphocytes, Runx3, CD103, TGFβ
Introduction
Both CD4 helper and CD8 cytotoxic T cells are generated from a common precursor in the thymus [1]. The molecular mechanism that drives the bifurcation of immature thymocytes into CD4 versus CD8 lineages has been mostly deciphered and was found to involve the mutually exclusive expression and antagonistic functions of the two transcription factors ThPOK and Runx3 [2]. CD8 T cell identity is established by the runt-related transcription factor 3, Runx3, which suppresses the CD4-specifying effects of ThPOK [3]. Notably, Runx3 is absent in pre-selection thymocytes but induced upon positive selection by intrathymic cytokines that impose CD8 lineage fate [4–7]. Consequently, Runx3-deficient mice are impaired in CD8 T cell differentiation, and these mice display a substantial paucity of CD8 T cells in peripheral tissues [4, 8]. Runx3 was further found to be necessary for CD8 lineage commitment, as it silences CD4 coreceptor expression and upregulates CD8 gene expression [4]. Additionally, Runx3 is also required for CD8 effector T cell function because it establishes cytotoxic features, such as the expression of granzyme B, perforin, IFNγ and eomesodermin, and guards against their deviation into follicular helper T lineage cells [9, 10]. Thus, Runx3 plays a central role in specifying the phenotypic and functional features of CD8 lineage T cells.
The expression of the integrin CD103, which is also referred to as αE, is induced during CD8 lineage differentiation in the thymus. CD103 is selectively found on CD8 T cells but not on CD4 T cells. As such, CD103 has been considered to be a CD8 lineage-specific molecule whose expression is induced downstream of Runx3 [11–13]. CD103 mediates cell adhesion to epithelial cells by binding to its ligand, E-cadherin. Because E-cadherin is highly expressed on gut epithelial cells, the selective expression of CD103 on CD8 T cells confers them an advantage for the migration and tissue residency in the gut [14, 15]. Indeed, among all peripheral tissues, the small intestinal mucosa is unique, as it contains more CD8 T cells than CD4 T cells. In most other tissues, CD4 T cells predominate over CD8 T cells [16–18]. It remains unclear whether such preferential accumulation of CD8 T cells in the gut is solely mediated by the selective CD103 expression on CD8 T cells or whether forced expression of CD103 on CD4 T cells would also cause them to be recruited and retained in the gut mucosa. In this regard, the biological implications of CD8 lineage-specific CD103 expression remain unexplored. It is also unclear whether CD4 T cells lack CD103 simply because they are absent for Runx3. Moreover, CD103 is highly expressed on tissue-resident memory T cells (TRM), where it mediates their tissue residency and prevents their entry into the circulation [15]. Thus, it is conceivable that CD103 expression is suppressed on CD4 T cells to prevent their tissue residency and to facilitate their tissue trafficking and migration. However, these possibilities have not yet been examined. Thus, the biological significance of the CD8 T cell-specific expression of CD103 or the suppression thereof on CD4 T cells remains to be clarified.
Here, we addressed these questions using experimental mice in which CD103 was engineered to be ectopically expressed on CD4 T cells (CD103Tg). To maximize surface CD103 expression, we further introduced an integrin β7 transgene (β7Tg) into CD103Tg mice to generate CD103, β7 double transgenic mice (CD103Tgβ7Tg). Surface staining for CD103 confirmed that the CD4 T cells of these mice expressed copious amounts of CD103 protein. However, CD103 expression on CD4 T cells did not alter their distribution and accumulation in peripheral tissues, indicating that CD103 is unlikely to be involved in tissue tropism of CD4 T cells under steady-state conditions. Thus, surprisingly, CD4 T cells are refractory to the tissue retention-inducing and gut-tropic effects of CD103, and these findings necessitate the re-evaluation of the biological significance of CD103 on CD4 T cells.
Results
Runx3 promotes but is not required for integrin CD103 expression
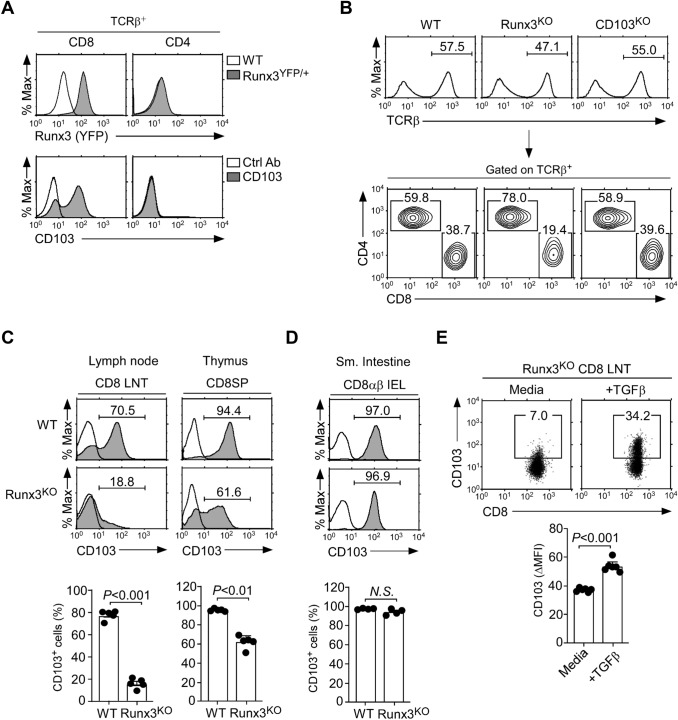
We embarked on this study to understand why CD103 would be only expressed on CD8 T cells and not CD4 T cells, and whether such selective CD103 expression would be required for normal T cell immunity. Earlier studies suggested a requirement for Runx3 to express CD103 on CD8 T cells [11–13]. In agreement, we confirmed that Runx3 gene reporter expression, i.e., Runx3-YFP, highly correlated with surface CD103 expression, as demonstrated in lymph node (LN) T cells and thymocytes of Runx3YFP/+ reporter mice (Fig. 1A and Supplemental Fig. 1A) [13]. We further affirmed that Runx3 is critical for CD103 upregulation by utilizing Runx3YFP/YFP homozygous mice where both alleles of Runx3 are disrupted by insertion of YFP reporter constructs [13]. Runx3YFP/YFP homozygous mice are unable to produce Runx3 (i.e., Runx3KO), and they contained significantly lower frequencies of CD8 LN T cells compared to WT mice (Fig. 1B middle). Notably, the decrease in peripheral CD8 T cell frequency was due to the lack of Runx3 and not because of the lack of CD103 expression because CD103 deficiency (CD103KO) [19] did not affect the percentages or numbers of CD8 LN T cells (Fig. 1B right and Supplemental Fig. 1B). In fact, we also did not find any major changes in T cell development in the thymus of CD103KO mice (Supplemental Fig. 1C). An important function of Runx3 is the upregulation of cytotoxic lineage molecules, such as granzyme B and perforin [9]. The lack of CD103, however, did not affect their expression which are induced downstream of Runx3 (Supplemental Fig. 1D). Therefore, CD103 expression is dispensable for the development and differentiation of CD8 T cells, and the requirement for Runx3 in CD8 T cell development is not due to a requirement for CD103 expression.
Fig. 1.
Runx3 is associated with but not required for CD103 expression on CD8 T cells. A Runx3 gene reporter (top) and surface CD103 expression (bottom) in CD8 and CD4 LN T cells of Runx3YFP/+ and WT mice, respectively. The results are representative of at least 3 independent experiments with a total of 5 WT and 3 Runx3YFP/+ mice. B CD8 versus CD4 profiles in LN T cells in WT, Runx3KO, and CD103KO mice. The results are representative of at least 5 independent experiments with a total of 7 WT, 5 Runx3KO, and 7 CD103KO mice. C Surface CD103 expression on CD8 LN T cells (LNT) and mature CD8SP thymocytes of WT and Runx3KO mice. Histograms show staining of anti-CD103 (shaded) versus isotype control antibody (open). Data are representative (top), and the bar graphs show a summary (bottom) of 5 independent experiments with a total of 5 WT and 5 Runx3KO mice. D Surface CD103 expression on CD8αβ TCRβ+ IELs of WT and Runx3KO mice. Histograms show the staining of anti-CD103 (shaded) versus isotype control antibody (open). Data are representative (top), and the bar graph shows a summary (bottom) of 4 independent experiments (bottom) with a total of 4 WT and 4 Runx3KO mice. E Surface CD103 expression on Runx3KO CD8 LN T cells that were FACS-sorted for CD103-negative cells and then cultured in vitro for 4 days with 5 ng/mL recombinant TGFβ. Dot plots are representative of 2 independent experiments
To further understand the biological implication of CD103 expression being linked with Runx3 expression, we next aimed to affirm whether Runx3 is a prerequisite for CD103 expression. To this end, we assessed CD103 abundance on CD8 T cells from WT and Runx3KO mice. In agreement with previous reports [4, 11], we confirmed that Runx3 deficiency resulted in significantly reduced CD8 T cell numbers in LN, spleen, and small intestine intraepithelial lymphocytes (IELs) (Supplemental Fig. 2A). The protein abundance of CD103 was also dramatically decreased on Runx3-deficient CD8 LN T cells (Fig. 1C left and Supplemental Fig. 2B left). While these results indicated that Runx3 is necessary to induce CD103 expression, we were surprised to find that the majority of CD8 single-positive (SP) thymocytes in Runx3KO mice still expressed CD103 even in the absence of Runx3 (Fig. 1C right). Moreover, we found that CD8 IELs showed abundant CD103 expression at levels indistinguishable from those of WT CD8 IELs (Fig. 1D and Supplemental Fig. 2B right). These results suggested that Runx3 is not essential to upregulate the expression of CD103 on CD8 T cells. In fact, the cytokine TGFβ, which is an inducer of CD103 expression (Supplemental Fig. 2C) [20], still upregulated CD103 expression on Runx3KO CD8 T cells, revealing a Runx3-independent pathway of CD103 expression (Fig. 1E). Nonetheless, Runx3 was required to maximize CD103 upregulation by TGFβ, which we demonstrated by the diminished de novo expression of CD103 in Runx3KO CD8 T cells upon in vitro TGFβ stimulation of FACS-sorted CD103-negative CD8 T cells (Supplemental Fig. 2D). Thus, Runx3 clearly promotes but is not an essential requirement for CD103 expression in CD8 T cells. These findings indicated that the lack of CD103 on CD4 T cells is not simply a consequence of lacking Runx3, but a developmentally controlled event with potential biological significance.
Forced Runx3 expression is not sufficient for robust CD103 expression on CD4 T cells
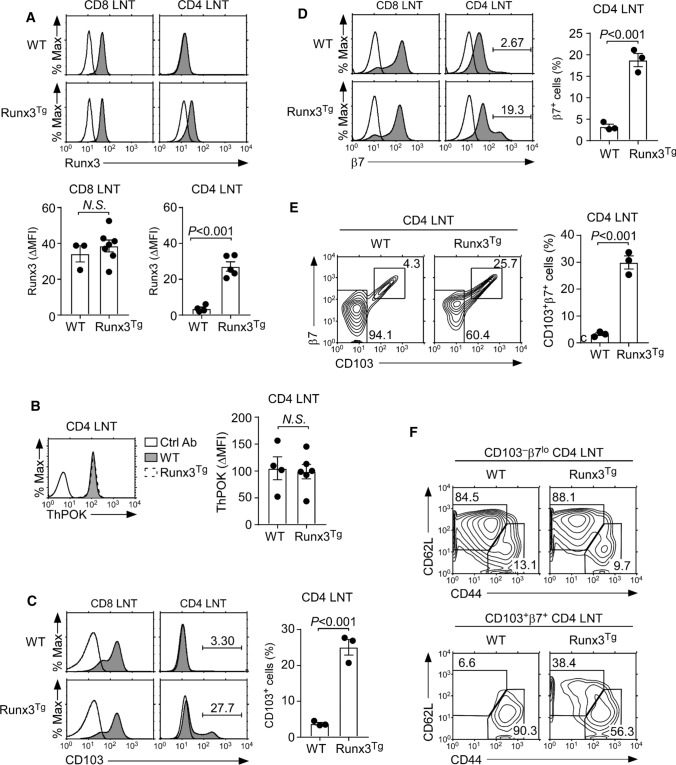
Because Runx3 was not required to induce CD103 expression on CD8 T cells (Fig. 1C, D), we further wished to know if Runx3 would be sufficient to induce CD103 expression on CD4 T cells. Contrary to CD8 T cells, CD4 T cells normally lack the expression of both Runx3 and CD103 [11, 21]. Thus, we generated transgenic mice that overexpressed mouse Runx3 cDNA under the control of the human CD2 promoter/enhancer (Runx3Tg) to assess potential upregulation of CD103. Intracellular staining of Runx3 proteins in Runx3Tg T cells demonstrated the successful expression of transgenic Runx3 proteins in CD4 T cells (Fig. 2A). While Runx3 imposes CD8 lineage fate in the thymus, Runx3Tg CD4 T cells remained committed to the CD4 lineage. ThPOK, a zinc-finger transcription factor that specifies CD4 lineage fate [2], was also expressed in similar amounts in Runx3Tg CD4 T cells as in WT CD4 T cells (Fig. 2B). The ectopic expression of Runx3, however, failed to induce CD103 on most CD4 T cells, except on a small subset (Fig. 2C). The upregulation of CD103 on Runx3Tg CD4 T cells was associated with the increased expression of integrin β7 which pairs with CD103 to form the αEβ7 heterodimer (Fig. 2D) [22]. Thus, Runx3Tg CD4 LN T cells contained a significantly increased population of CD103, β7 coexpressing cells (Fig. 2E), that was also present in CD4 T cells from the spleen and small intestine IELs of Runx3Tg mice (Supplemental Fig. 3A and B). The cellular identity of these CD4 T cells that upregulated CD103 expression is not yet clear to us. However, we found them highly enriched for CD44hiCD62Llo memory phenotype cells (Fig. 2F bottom). On the other hand, such an increase in memory phenotype cells did not affect the overall CD4 T cell numbers in peripheral lymphoid organs (Supplemental Fig. 3C). Collectively, these results demonstrated that both CD103 and β7 can be forced to be expressed, at least in a subset of CD4 T cells, but that the transgenic expression of Runx3 alone was clearly insufficient to fully upregulate CD103 and β7 expression on all CD4 T cells.
Fig. 2.
Forced expression of Runx3 induces CD103 expression on CD4 T cells. A Intracellular Runx3 expression in CD4 and CD8 LN T cells of WT and Runx3Tg mice. Histograms show staining of anti-Runx3 (shaded) versus isotype control antibody (open). Data are representative (top), and bar graphs show a summary (bottom) of 3 independent experiments with a total of 3 WT and 7 Runx3Tg mice (CD8) and 4 WT and 5 Runx3Tg mice (CD4). B Intracellular ThPOK expression in CD4 LN T cells of WT and Runx3Tg mice. Histogram is representative (left) and the bar graph shows the summary (right) of 3 independent experiments with a total of 4 WT and 6 Runx3Tg mice. C, D Surface CD103 (C) and β7 (D) expression was assessed on CD4 and CD8 LN T cells of WT and Runx3Tg mice. Histograms show staining of anti-CD103 or anti-β7 (shaded) versus isotype control antibody (open). Data are representative (left), and bar graphs show a summary of 3 independent experiments (right) with a total of 3 WT and 3 Runx3Tg mice. E Coexpression of CD103 and β7 in CD4 LN T cells of WT and Runx3Tg mice. CD4 LN T cells were assessed for CD103 and β7, and the frequency of CD103, β7 coexpressing cells were determined. Data are representative (left), and bar graphs (right) show a summary of 2 independent experiments with a total of 3 WT and 3 Runx3Tg mice. F Phenotype of CD103+β7+ coexpressing CD4 LN T cells. Naïve and memory phenotype was assessed by CD44 versus CD62L staining on CD103+β7+ (bottom) and CD103-negative β7lo (top) CD4 LN T cells. Data are representative of 2 independent experiments with a total of 3 WT and 3 Runx3Tg mice
Forced expression of the integrin β7 does not increase Runx3-induced CD103 expression
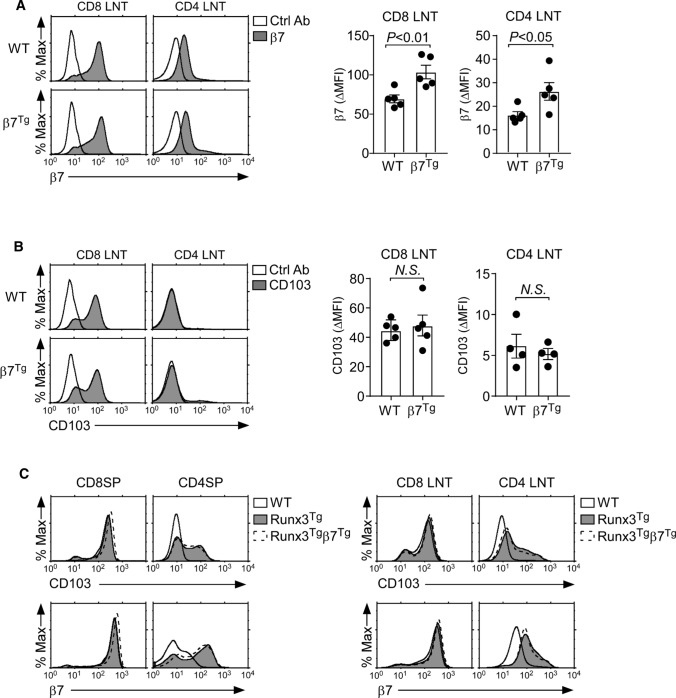
CD103 is expressed as a heterodimer with the integrin β7 [22]. It is unclear whether CD103 can be transported to the cell surface by itself or whether β7 is required for the cell surface expression of CD103. Thus, we wished to rule out the possibility that the availability of integrin β7 is limiting and constrains the CD103 expression in Runx3Tg CD4 T cells. To this end, we aimed to overexpress integrin β7 in CD4 T cells. We generated transgenic mice that overexpressed the cDNA of mouse Itgb7, which encodes integrin β7, under the control of the human CD2 promoter/enhancer (β7Tg). We confirmed a modest but statistically significant increase in surface β7 expression on both CD8 and CD4 LN T cells of β7Tg mice (Fig. 3A). However, the increased β7 availability did not increase the amount of CD103 on β7Tg CD8 LN T cells or CD4 LN T cells (Fig. 3B). These results suggested that, at least in mature T cells, the abundance of surface CD103 was not limited by the availability of β7 (Fig. 3B). We also found that increasing the amount of β7 failed to further upregulate CD103 expression on Runx3Tg thymocytes and LN T cells (Fig. 3C). Thus, the CD103 abundance on β7-transgenic Runx3Tg (Runx3Tgβ7Tg) T cells remained similar to CD103 expression on T cells expressing Runx3Tg alone (Fig. 3C). Specifically, the amounts of CD103 and β7 on Runx3Tgβ7Tg thymocytes and LN T cells were similar to those on Runx3Tg cells (Fig. 3C). Collectively, these results indicated that CD103 expression is controlled by distinct mechanisms between CD4 and CD8 T cells, and they further suggest that factors other than Runx3 and β7 would be necessary to fully induce the expression of CD103 on CD4 T cells.
Fig. 3.
Increased availability of integrin β7 is insufficient to upregulate CD103 expression on Runx3Tg CD4 T cells. A Surface β7 expression on CD4 and CD8 LN T cells of WT and β7Tg mice. Data are representative (left), and bar graphs show the summary of 5 independent experiments (right) with a total of 5 WT and 5 β7Tg mice. B Surface CD103 expression on CD8 and CD4 LN T cells of WT and β7Tg mice. Data are representative, and bar graphs show the summary of 4 independent experiments with a total of 4 WT and 5 β7Tg mice (CD8) and 4 WT and 4 β7Tg mice (CD4). C Surface CD103 and β7 expression on CD4SP and CD8SP thymocytes (left) and CD4 and CD8 LN T cells (right) of Runx3Tgβ7Tg mice. The results are representative of 2 independent experiments with a total of 2 WT, 2 Runx3Tg and 2 Runx3Tgβ7Tg mice
Forced expression of integrin CD103 and β7 on thymocytes
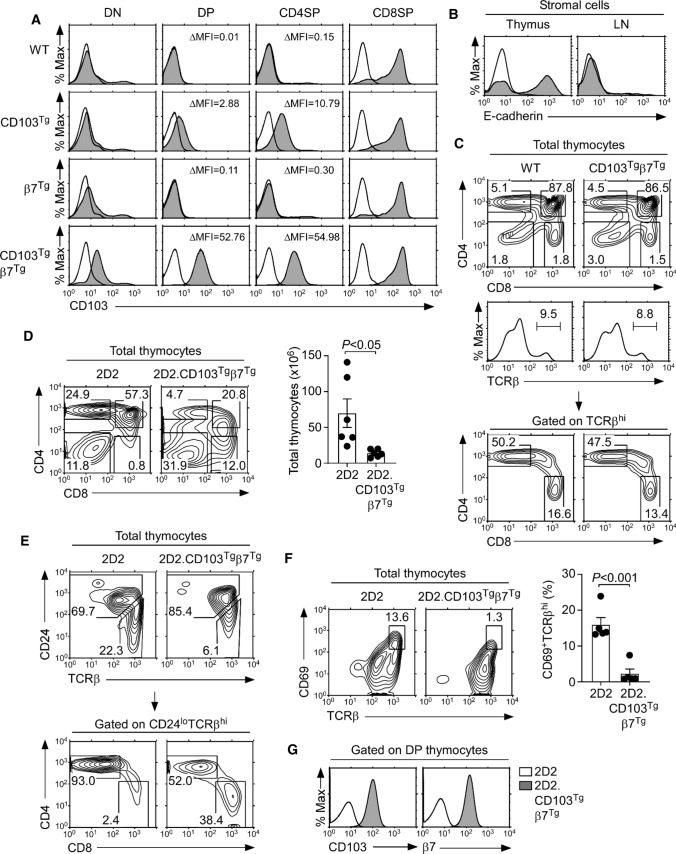
As an alternative and direct approach to express CD103 on CD4 T cells, we generated transgenic mice that overexpressed CD103 under the control of the human CD2 promoter/enhancer elements (CD103Tg). While the forced expression of CD103 did not increase the abundance of CD103 in immature double-negative thymocytes, the CD103Tg was sufficient to upregulate surface CD103 expression on immature double-positive (DP) and CD4SP thymocytes that normally do not express CD103 (Fig. 4A, second row). Such an effect of CD103Tg on CD103 expression was distinct to the effect of the β7Tg, which failed to increase CD103 abundance on DP and CD4SP thymocytes (Fig. 4A, third row). The co-expression of β7Tg with CD103Tg (CD103Tgβ7Tg), however, dramatically increased the amount of surface CD103 proteins on all thymocytes (Fig. 4A, last row). As such, the abundance of CD103 proteins on CD103Tgβ7Tg CD4SP cells increased more than fivefold compared to CD103Tg CD4SP cells that do not coexpress the β7Tg (Fig. 4A, last row). This was also the case for CD103Tgβ7Tg DP thymocytes which expressed more than 15-fold greater amounts of surface CD103 than CD103Tg DP cells without the β7Tg. These results revealed that the surface expression of CD103 is constrained by the availability of β7, and that CD103 requires heterodimerization with β7 to maximize cell surface expression. Conversely, our results also revealed that the cell surface abundance of β7 is controlled by the availability of CD103, as demonstrated by the β7 expression in thymocytes of β7Tg and CD103Tgβ7Tg mice (Supplemental Fig. 4). While the surface β7 expression on DP and CD4SP thymocytes of β7Tg mice was significantly increased compared to WT control mice (Supplemental Fig. 4, first versus third row), it required the co-expression of the CD103Tg to dramatically and fully upregulate β7 expression in these cells (Supplemental Fig. 4, last row). Collectively, these findings establish CD103, β7 heterodimerization as a new mechanism to control the surface abundance of CD103 and β7 proteins. Thus, maximizing the expression of CD103 in CD103Tg mice requires an accompanying increase in β7 expression.
Fig. 4.
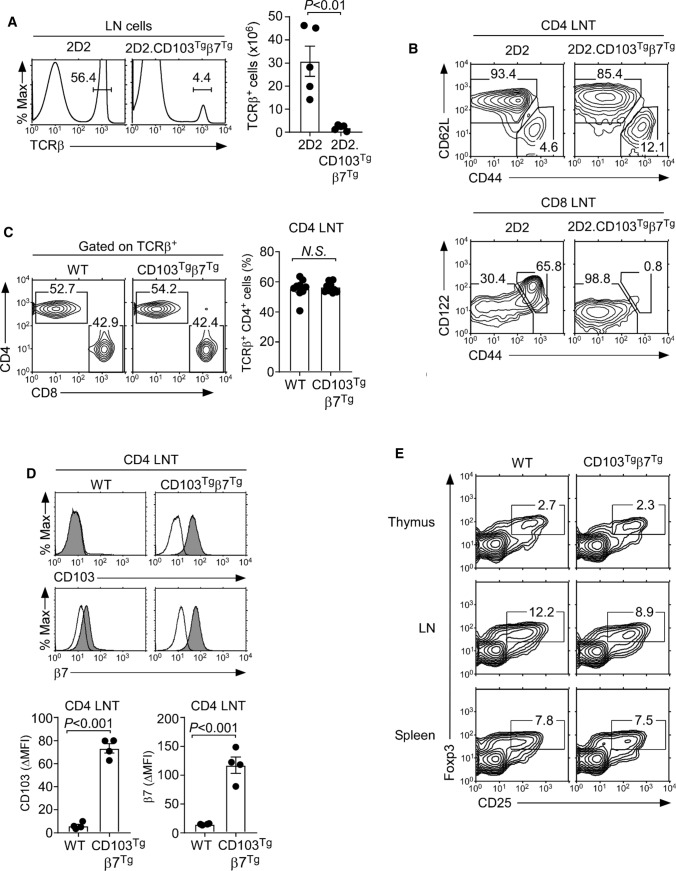
Generation of CD103+ CD4 T cells by transgenic expression of CD103 and β7. A Surface CD103 expression on DN, DP, and mature CD4SP and CD8SP thymocytes of WT, CD103Tg, β7Tg, and CD103Tgβ7Tg mice. Shaded histograms show anti-CD103 staining; open histograms indicate control antibody staining. ∆Mean Fluorescence Intensity (∆MFI) of CD103 expression was determined by subtracting the control antibody MFI from CD103 MFI. The results are representative of 7 independent experiments with a total of 7 WT, 7 CD103Tg, 7 β7Tg, and 7 CD103Tgβ7Tg mice. B E-cadherin expression on thymus and LN stromal cells. Shaded histograms show anti-E-cadherin staining; open histograms indicate control antibody staining. Results are representative of at least 2 independent experiments. C CD4 versus CD8 profile of total (top) or TCRβ+-gated (middle) mature thymocytes of WT and CD103Tgβ7Tg mice. The results are representative of 8 independent experiments with a total of 10 WT and 10 CD103Tgβ7Tg mice. D CD4 versus CD8 thymocyte profiles and cell numbers of 2D2 and 2D2.CD103Tgβ7Tg mice. The contour plot is representative (left) and the graph (right) is the summary of 5 independent experiments with 6 2D2 and 6 2D2.CD103Tgβ7Tg mice. E CD24loTCRβhi mature CD4 and CD8 T cells in 2D2 and 2D2.CD103Tgβ7Tg mice. Mature thymocytes were identified by CD24 versus TCRβ staining (top), and CD4 versus CD8 expression were determined among CD24loTCRβhi thymocytes (bottom). Results are representative of 4 independent experiments with 5 2D2 and 5 2D2.CD103Tgβ7Tg mice. F CD69+TCRβhi thymocytes in 2D2 and 2D2.CD103Tgβ7Tg mice. Thymocytes undergoing positive selection were identified by the co-expression of CD69 and TCRβ. The contour plot is representative (left) and the graph (right) is the summary of 4 independent experiments with 5 2D2 and 5 2D2.CD103Tgβ7Tg mice. G Surface CD103 and β7 expression on DP thymocytes of 2D2 and 2D2.CD103Tgβ7Tg mice. Results are representative of 4 independent experiments with 5 2D2 and 5 2D2.CD103Tgβ7Tg mice
T cell development in CD103Tgβ7Tg mice
Having achieved our goal to force CD103 expression on T cells, we next aimed to examine the effect of forced CD103 expression on the tissue distribution of CD103Tgβ7Tg T cells. CD103 binds to E-cadherin, and the thymic medulla is enriched in E-cadherin-expressing stromal cells [23]. In fact, we found that thymic stromal cells but not LN stromal cells expressed substantial amounts of E-cadherin (Fig. 4B). Thus, we first assessed whether newly generated CD103Tgβ7Tg CD4 T cells would accumulate in the thymus because of potential thymic retention mediated by binding to E-cadherin. Interestingly, the forced CD103 expression on thymocytes did not affect thymocyte development or positive selection as documented by CD69 and TCRβ analysis in WT and CD103Tgβ7Tg mice which did not differ from each other (Supplemental Fig. 5A). Moreover, the frequency of mature TCRβhi thymocytes was unaffected (Supplemental Fig. 5B), and we did not find any substantial changes in the frequency of CD8SP cells among total thymocytes and no significant increase in CD4SP cells when gating on TCRβhi mature thymocytes (Fig. 4C and Supplemental Fig. 5C). In fact, the frequency of mature CD4SP cells was slightly reduced, whereas the frequency of CD8SP cells remained unaltered (Supplemental Fig. 5C). Therefore, the forced expression of CD103 did not result in the accumulation of CD4 T cells in the thymus.
Immature thymocytes undergo TCR rearrangement that permits the acquisition of new TCR specificities which could circumvent potential detrimental effects of premature or ectopic CD103 expression. Thus, we next examined the T cell development in CD103Tgβ7Tg mice that express a fixed T cell receptor. The 2D2 is an MHC-II-restricted TCR which imposes CD4 lineage fate onto developing thymocytes, permitting us to track the generation of a monoclonal CD4 T cell population [24]. In agreement with previous observations [24], 2D2 TCR transgenic mice generated a large population of CD4SP cells but virtually no CD8SP thymocytes (Fig. 4D, left). Surprisingly, 2D2 TCR transgenic mice that were forced to express CD103 and β7 (2D2.CD103Tgβ7Tg) were severely impaired in their thymopoiesis, with dramatically reduced thymocyte numbers and inefficient CD4SP cell generation (Fig. 4D, right). To pinpoint the developmental block in 2D2.CD103Tgβ7Tg mice, we assessed the TCRβ versus CD24 expression to visualize pre-selection (CD24hiTCRβlo) and post-selection (CD24loTCRβhi) thymocytes (Fig. 4E). The 2D2 TCR transgene permitted effective positive selection in the thymus, thus resulting in the generation of a large number of mature CD4 T cells (Fig. 4E, left). In marked contrast, the frequency of CD24loTCRβhi post-selection thymocytes in 2D2.CD103Tgβ7Tg mice was dramatically reduced (Fig. 4E, right). CD4 lineage differentiation was also substantially impaired, resulting in the appearance of lineage-mismatched CD8 T cells expressing the 2D2 TCR transgene (Fig. 4E and Supplemental Fig. 5D). To directly assess the effect of forced CD103 and β7 expression on the positive selection of 2D2 thymocytes, we next stained for CD69, a classical marker of thymic positive selection [25]. Compared to 2D2 mice, the frequency of TCRβhiCD69+ cells which correspond to thymocytes undergoing positive selection was significantly reduced in 2D2.CD103Tgβ7Tg mice (Fig. 4F). Thus, the forced expression of CD103 and β7 interferes with and impairs the positive selection of 2D2 thymocytes. Because immature DP thymocytes of 2D2.CD103Tgβ7Tg mice express large amounts of CD103 and β7 (Fig. 4G), these results suggest that the premature expression of CD103 is detrimental for thymocyte development, presumably by binding to thymic stromal E-cadherin and by interfering with their intrathymic migration.
Tissue distribution of CD4 T cells that are forced to express CD103
Once thymocytes emigrate from the thymus, they move into and repopulate peripheral lymphoid tissues. To assess the effect of CD103, β7 overexpression on peripheral T cells, we next examined the frequencies and numbers of LN T cells in 2D2 and 2D2.CD103Tgβ7Tg mice. Compared to 2D2 mice, 2D2.CD103Tgβ7Tg mice were severely lymphopenic in peripheral tissues as shown for LN T cells (Fig. 5A). These results indicated that the forced expression of CD103 and β7 interfered with and suppressed the homeostasis of 2D2 T cells. Moreover, the frequency of LN CD4 T cells was significantly reduced, while the frequency of LN CD8 T cells was substantially increased in 2D2.CD103Tgβ7Tg mice (Supplemental Fig. 6A). CD8 T cells normally express large amounts of CD103, so that the forced expression of CD103 would have less detrimental effects on CD8 T cells compared to CD4 T cells. The CD8 T cells in 2D2 mice are TCR/coreceptor mismatched because they express the MHC-II-restricted 2D2 TCR in association with the CD8 coreceptor which binds to MHC-I. Whether such TCR/coreceptor mismatch would alleviate any detrimental effects of CD103 overexpression in 2D2.CD103Tgβ7Tg CD8 T cells is currently unclear to us. It is also unclear why CD8 T cells in 2D2.CD103Tgβ7Tg mice retained a naïve phenotype, whereas CD8 T cells in 2D2 mice contain a large population of memory phenotype cells (Fig. 5B, bottom). We aim to address the molecular basis of such distinct phenotypes in follow-up studies.
Fig. 5.
Phenotypic characterization of CD103Tg CD4 T cells. A LN T cell frequency and numbers in 2D2 and 2D2.CD103Tgβ7Tg mice. The histogram is representative (left) and the graph (right) is the summary of 4 independent experiments with 5 2D2 and 5 2D2.CD103Tgβ7Tg mice. B Phenotype of 2D2 and 2D2.CD103Tgβ7Tg LN T cells. Naïve versus memory phenotype was assessed by CD44 versus CD62L staining in CD4 LN T cells (top) and by CD44 versus CD122 staining in CD8 LN T cells (bottom). Results are representative of 3 independent experiments with 4 2D2 and 4 2D2.CD103Tgβ7Tg mice. C CD4 versus CD8 profiles of TCRβ+-gated LN T cells from WT and CD103Tgβ7Tg mice (left). The bar graph shows CD4 T cell frequencies from LNs of WT and CD103Tgβ7Tg mice (right). The results show the summary of 8 independent experiments with a total of 9 WT and 9 CD103Tgβ7Tg mice. D CD103 and β7 expression were determined on CD4 LN T cells of the indicated mice (top). Histograms show staining of anti-CD103 or anti-β7 (shaded) versus isotype control antibody (open). Bar graphs show the ΔMFI of CD103 and β7 on CD4 LN T cells of the indicated mice (bottom). The results are a summary of 4 independent experiments with a total of 4 WT and 4 CD103Tgβ7Tg mice. E Frequency of Foxp3+ Treg cells in the thymus, LN, and spleen of WT and CD103Tgβ7Tg mice. The results are representative of 6 independent experiments with a total of 6 WT and 6 CD103Tgβ7Tg mice
In contrast to CD8 T cells, the CD4 T cells in 2D2 and 2D2.CD103Tgβ7Tg mice were both enriched for CD44hiCD62Llo memory phenotype cells (Fig. 5B, top). Thus, the CD4 T cells in these mice are phenotypically comparable, and it would have been interesting to further analyze them for the effects of forced CD103, β7 expression. However, we found the 2D2 TCR transgenic mouse model being inadequate to assess the effect of CD103 expression in CD4 T cells, because CD4 T cell numbers were dramatically reduced in 2D2.CD103Tgβ7Tg mice (Supplemental Fig. 6A). Under these circumstances, it would be difficult to discriminate the role of CD103 in the survival versus tissue distribution of peripheral CD4 T cells.
Thus, to examine whether forced CD103 expression on CD4 T cells affects their tissue distribution, we assessed the frequency and phenotype of CD4 T cells in the LN and spleen of WT and CD103Tgβ7Tg mice (Fig. 5C and Supplemental Fig. 6B). In both organs, we found that the CD4 T cell frequencies were unaffected by forced CD103 expression. Notably, peripheral CD4 T cells of CD103Tgβ7Tg mice expressed copious amounts of both CD103 and β7 (Fig. 5D), whereas CD4 T cells of WT mice were devoid of CD103 and only expressed low levels of β7 (Fig. 5D). Additionally, the ectopic expression of CD103 on CD4 T cells did not alter their distribution in nonlymphoid tissues, as we did not find increased accumulation of CD4 T cells among CD45+-gated T cells in the liver and in the lung of CD103Tgβ7Tg mice (Supplemental Fig. 6C). Altogether, these results indicate that CD4 T cells under steady-state condition are refractory to the cell retention effect of CD103 in peripheral tissues.
Finally, to examine if such was the case for all CD4 T cell subsets, we also assessed the frequency of CD25+Foxp3+ regulatory T cells (Tregs) among CD4 T cells of CD103Tgβ7Tg mice. We did not find any significant changes in the CD25+Foxp3+ Treg cell population because the frequencies of Treg cells were similar in the thymus, LN and spleen of WT and CD103Tgβ7Tg mice (Fig. 5E and Supplemental Fig. 6D). Collectively, and contrary to our expectation that forced CD103 expression would alter the tissue distribution of CD4 T cells, these results demonstrated that CD103 expression in CD4 T cells did not affect their frequencies and did not alter their presence in peripheral tissues when compared to normal CD4 T cells that lack CD103.
Characterization of CD103-expressing CD4 IEL T cells
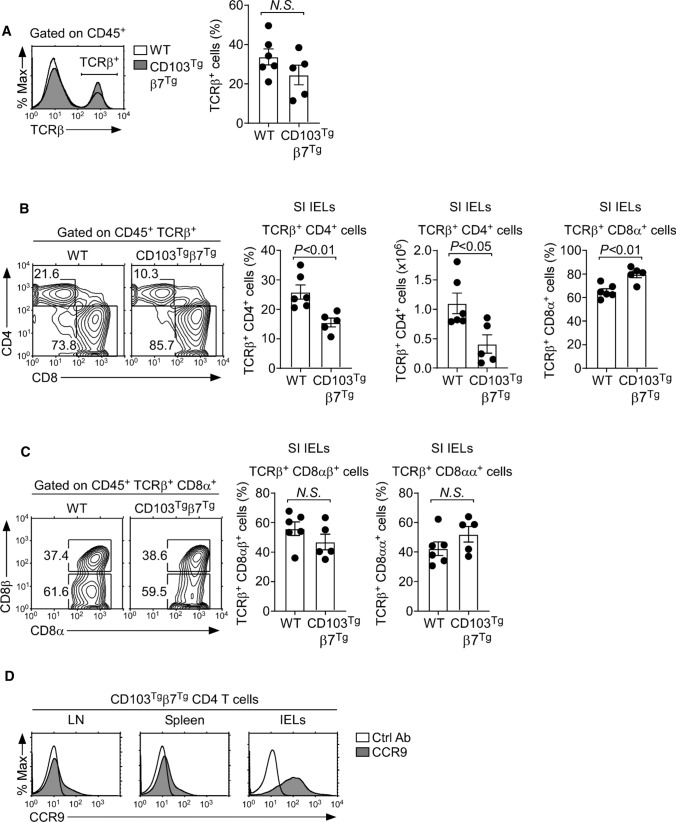
The forced CD103 expression on CD4 T cells did not cause the enrichment of CD4 T cells in lymphoid tissues, which might be explained by the insufficient numbers of E-cadherin+ cells in these sites. Thus, we wished to examine whether mature CD4 T cells would accumulate in E-cadherin-rich tissues, specifically in the small intestinal (SI) epithelium. The SI epithelium is a critical barrier tissue that harbors the largest number of lymphocytes in the body, which are collectively referred to as IELs [26]. CD103 plays a critical role in the tissue retention of SI IELs, and CD8 T cells express large amounts of CD103 [15]. Notably, the SI IEL αβ T cell population is uniquely enriched in CD8 T cells, so that the SI is the only tissue where CD8 T cells outnumber CD4 T cells [16]. The selective expression of CD103 on CD8 T cells could have facilitated the retention of CD8 T cells in this E-cadherin-rich tissue. Thus, we aimed to examine whether the forced expression of CD103 would promote the accumulation of CD4 T cells among SI IELs.
Surprisingly, the overall frequency of αβ T cells did not differ between WT and CD103Tgβ7Tg SI IELs (Fig. 6A), and the frequency of SI IEL CD4 T cells also did not increase by the forced expression of CD103 and β7 (Fig. 6B). Instead, both the frequency and number of CD4 T cells were reproducibly reduced among CD103Tgβ7Tg IELs (Fig. 6B). In contrast, the overexpression of CD103, β7 increased the frequency of CD8 T cells within the SI IEL population (Fig. 6B). Importantly, such an increase was not due to the accumulation of CD8αα T cells which are prevalent in IELs (Fig. 6C) [27]. These results document a dichotomy in the effect of CD103 on SI IEL tissue residency. CD103 promotes the tissue retention of CD8 T cells in the SI epithelium, but CD4 T cells are refractory to the increased abundance of CD103. It remains unclear why CD103 expression only promotes the SI IEL tissue residency of CD8 T cells and not that of CD4 T cells. Here, we wish to point out that peripheral CD4 T cells, unlike CD8 T cells, only express small amounts of the CC chemokine receptor 9 (CCR9) which is considered an important gut-tropic chemokine receptor (Supplemental Fig. 6E) [28]. In fact, the CCR9 abundance remained very low on CD103Tgβ7Tg CD4 T cells from the LN and spleen, compared to the abundance on SI IELs (Fig. 6D). Therefore, the inefficient homing of CD103Tgβ7Tg CD4 T cells to SI epithelial tissues could provide a potential explanation why CD103 expression on CD4 T cells did not result in increased accumulation in the gut. We are currently addressing these possibilities, and we aim to obtain mechanistic insights in future studies.
Fig. 6.
Small intestine CD4+ IELs in CD103Tgβ7Tg mice. A Identification of CD45+-gated αβ T cells in WT and CD103Tgβ7Tg mice (left). Bar graphs show frequencies of αβ T cells among CD45+-gated SI IELs (right). Data are representative of 5 independent experiments with a total of 6 WT and 5 CD103Tgβ7Tg mice. B Contour plots show CD4 versus CD8α profiles of αβ T cells among SI IELs (left). Bar graphs show frequency and number of CD4+ and the frequency of CD8α+ T cells among CD45+ TCRβ+-gated SI IELs in WT and CD103Tgβ7Tg mice (right). Results are the summary of 5 independent experiments with a total of 6 WT and 5 CD103Tgβ7Tg mice. C Frequency of CD8αβ and CD8αα αβ T cells among IELs of WT or CD103Tgβ7Tg mice. Contour plots are representative (left), and bar graphs are a summary of 5 independent experiments with a total of 6 WT and 5 CD103Tgβ7Tg mice. D CCR9 expression on CD4 T cells from spleen, LN, and IELs of CD103Tgβ7Tg mice. The results are representative of 3 independent experiments
Discussion
The integrin CD103 is selectively expressed on CD8 lineage T cells and conspicuously absent on most CD4 T cells [11, 12]. Whether CD103 is detrimental for CD4 T cells, and thus, needs to be excluded from CD4 T cells is unknown. It also remains unclear whether CD103 impedes T cell recirculation by imposing tissue residency if expressed on CD4 T cells. Here, we addressed these questions by forcing CD103 expression on CD4 T cells using transgenic approaches. We report that ectopic expression of CD103 did not alter the tissue distribution or cause the accumulation of CD4 T cells in peripheral tissues. CD103-expressing CD4 T cells also did not preferentially migrate to and accumulate in the gut, which is a site with high abundance of E-cadherin—the only known ligand of CD103 [29]. Thus, CD4 T cells differ from CD8 T cells, as they are refractory to the tissue retention effect of CD103. Moreover, these results indicated that CD103 is absent on CD4 T cells, not because CD103 expression would cause adverse effects that need to be avoided. Instead, we consider it more likely that the molecular machinery that controls CD103 expression is absent in CD4 T cells but is triggered upon CD8 lineage differentiation, resulting in the distinct CD103 expression on CD4 and CD8 T cells.
The molecular basis of CD8 lineage-specific CD103 expression has been mostly attributed to the effect of the CD8 lineage-specifying factor Runx3, which is specifically expressed by CD8 T cells and not by CD4 T cells [2]. Consequently, the initial studies which determined that CD103 expression occurs downstream of Runx3 led to the assumption that CD103 would be associated with T cell functions that are specific to CD8 lineage cells. Because Runx3 induces the cytolytic transcriptional program in CD8 effector T cells [8, 9], it has also been suggested that CD103 might be a part of the CD8 T cell cytotoxic program. In support of this idea, CD103 engagement by E-cadherin was found to trigger polarization of lytic granules in tumor-infiltrating cytotoxic T lymphocytes (CTLs) and to promote degranulation and cytotoxicity. Furthermore, CD103 facilitated CTL conjugate formation with E-cadherin-expressing target cells and increased CTL cytotoxicity during allogenic T cell response [30]. Thus, CD103 can contribute to the cytotoxic function of CD8 T cells.
On the other hand, there were no major defects in CD8 T cell immune responses when tested in CD103-deficient mice, and CD103-deficiency did not impact CD8 memory responses after viral infections either [31]. These results indicated the potential redundancy of CD103 with other cell adhesion molecules whose identities are unknown. Nonetheless, CD8 T cell immunity in CD103KO mice is not entirely without impairment. For example, CD103KO CD8 T cells failed to induce destruction of the intestinal epithelium in graft-versus-host disease [32], and they also failed to infiltrate and destroy pancreatic islet allografts in an autoimmune mouse model [33]. Because CD103KO CD8 T cells were fully competent in mounting CTL responses in vitro but defective in graft rejection, these results indicated that the major function of CD103 is the tissue migration and retention of CD8 T cells rather than the direct regulation of cytolytic activity.
In this regard, we initially hypothesized that CD103 would be absent on CD4 T cells because it could otherwise interfere with their normal tissue migration and distribution. CD103 mediates tissue retention by binding to E-cadherin, which is highly expressed on epithelial cells, including thymic medullary epithelial cells and the SI epithelial cells [34]. Thus, we expected that CD103+ CD4 T cells would preferentially accumulate in these sites. In fact, some tissue-resident CD4 memory T cells were previously found to express CD103, but its expression appeared to be less stringently associated with tissue residency compared to that of CD8 tissue-resident memory cells [35]. Detailed analysis of CD4 T cells from CD103Tgβ7Tg mice further supported such a distinct role for CD103 in CD4 versus CD8 T cells. Along these lines, we found no significant differences in the CD4 T cell frequencies between CD103Tgβ7Tg and WT mice when assessing their thymocytes and SI epithelial lymphocytes. We also did not find any changes in the CD4 T cell frequencies in other lymphoid organs, including the LN and spleen. Collectively, these results indicated that CD4 T cells are refractory to the tissue retention effect of CD103 expression. These findings also suggested that CD103 expression alone is possibly insufficient to drive CD4 T cell accumulation in the gut or in other tissues where E-cadherin is abundant. As a potential explanation, we noted that CD4 T cells also differ from CD8 T cells in the expression of other gut homing receptors and specifically considered that CD4 T cells lack the chemokine receptor CCR9 [28]. CCR9 is the receptor for the chemokine CCL25, which is specifically expressed in the thymus and the SI [36]. Curiously, CCR9 is only expressed on CD8 T cells and is not present in CD4 T cells [28]. Thus, CD4 T cells lack the ability to undergo chemotaxis in response to the CCL25 gradient. On the other hand, it is proposed that the expression of CCR9 on CD8 T cells would license their migration into gut tissues, resulting in the preferential accumulation of CD8 T cells in the SI epithelium [37]. In this regard, CD103Tgβ7Tg CD4 T cells might fail to take tissue residency in the SI epithelium because they lack the chemotactic signal that guides them to the gut in the first place. Forced expression of CCR9 in CD103Tgβ7Tg mice could determine whether it is indeed the lack of gut-tropic signals that impedes the accumulation of CD4 T cells in the SI epithelium. Transgenic CCR9 mice have been previously described [28], and we aim to make use of these mice to generate CD4 T cells that express both CD103 and CCR9 and examine their migration and tissue residency compared to those of WT CD8 T cells that naturally express both molecules.
While most CD4 T cells do not express either CCR9 or CD103, nonetheless, there is a substantial fraction of CD4 T cells that do express these molecules and that reside in the SI epithelium. Therefore, CD4 T cells evidently can express CD103 under steady-state conditions. However, the developmental signals that induce CD103 expression remain mostly unknown. Because TGFβ can induce CD103 expression, it is not surprising that Foxp3+ CD4+ Treg cells, which require TGFβ for their generation, represent a prominent population among CD4 T cells that express CD103 [38]. In this regard, CD103+ CD4 T cells in humans mostly correspond to Foxp3+ Treg cells [21], whereas in mice, CD103 expression in Treg cells is more heterogeneous, and the CD103+ subset has been referred to as “effector/memory”-like Treg cells with potent anti-inflammatory function [39]. However, it has been uncertain whether CD103 expression is induced to promote the generation of Foxp3+ Treg cells or whether CD103 is induced as a consequence of Foxp3+ Treg cell differentiation. Our current data suggest the latter to be the case because forced CD103 expression did not alter the frequency of Foxp3+ cells among CD4 T cells in CD103Tgβ7Tg and WT mice.
The literature also documents cases where CD103 is expressed on CD4 T cells that are not Foxp3+ Treg cells. For example, in autoimmune Scurfy mice that lack Foxp3, some CD4 effector T cells express CD103, and these CD103+ CD4 T cells contribute to the disease phenotype, because CD103 deficiency ameliorated autoimmunity in scurfy mice [40]. The acquisition of CD103 expression in these autoimmune CD4 T cells was mediated by IL-2 because IL-2-deficiency reduced the CD103 expression on these CD4 effector T cells.
Additionally, it is noted that a fraction of tissue-resident memory CD4 T cells express CD103 akin to their CD8 T cell counterparts [41]. Thus, it is likely that memory CD4 T cells utilize CD103 for their tissue retention in sites that are abundant in E-cadherin. Because antigen activation and cytokine-driven differentiation drive the generation of memory T cells, it is evident that cellular signals distinct from TGFβ can also induce CD103 expression. The identification of these cues will clarify the molecular pathways of CD103 expression on CD4 T cells.
Along these lines, we wish to emphasize that the current study also revealed a new regulatory mechanism of CD103 expression that depends on the integrin β7. While CD103 is expressed as a heterodimer with β7, it was not clear whether the surface expression of CD103 depends on β7. Detailed analysis of thymocyte subpopulations in CD103Tg and CD103Tgβ7Tg mice demonstrated this to be the case. CD103 was only minimally expressed on immature DP thymocytes in CD103Tg mice. However, the additional transgenic expression of β7 dramatically increased the surface expression of CD103, and we hypothesize that it presumably does so because CD103 must heterodimerize with β7 to be transported or to be stably expressed on the cell surface. Assessing either of these possibilities will need further investigation. Notably, we also found that β7 expression is limited by the availability of CD103, so that the overexpression of β7 was insufficient to fully upregulate surface expression of β7 in DP and CD4SP thymocytes. Instead, the co-expression of CD103Tg was required to fully upregulate β7. Altogether, here, we presented evidence that the availability of β7 limits the surface expression of CD103 and vice versa, thus introducing a new regulatory element that determines the abundance of CD103 and β7. The physiological significance of this complex regulatory mechanism of CD103 expression is showcased in the 2D2.CD103Tgβ7Tg mice where the thymic development of MHC-II-restricted CD4 T cells was severely impaired, presumably by the ectopic expression of CD103 in pre-selection thymocytes. While the molecular basis for the developmental arrest of 2D2 thymocytes remains unclear, we are considering a scenario where the CD103 binding to E-cadherin in 2D2.CD103Tgβ7Tg immature DP thymocytes would make them adhere to the thymic cortex and impair their access to positively selecting peptide/MHC-II complexes. Consistent with this hypothesis, we found that the frequency of CD69+ thymocytes in 2D2.CD103Tgβ7Tg mice was dramatically decreased. These results indicate that pre-selection thymocytes require effective intrathymic trafficking to engage ligands and to undergo positive selection. Thymocytes with fixed TCR specificities, as is the case for 2D2 TCR transgenic thymocytes, would have greater difficulties in engaging the cognate ligand when their trafficking is constrained, resulting in the severely impaired positive selection and thymocyte maturation as described for 2D2.CD103Tgβ7Tg CD4 T cells.
Finally, our study also re-examined the role of Runx3 as a master regulator of CD103 expression, which made us question the use of CD103 as a marker for CD8 lineage T cells. Using mouse models of loss- or gain-of-Runx3 function, we showed that Runx3 is neither necessary for CD103 expression on CD8 T cells nor sufficient to fully induce CD103 expression on all CD4 T cells. Moreover, this study indicates that the lack of CD103 on CD4 T cells is not a simple consequence of the lack of Runx3 expression and that CD103 expression alone is insufficient to significantly alter CD4 T cell tissue distribution and residency.
Materials and methods
Mice
C57BL/6 (B6 or WT) mice of both sexes were obtained from the Charles River Laboratories (Frederick, MD). CD103KO mice [19] were purchased from the Jackson Laboratory. Runx3YFP/+ and Runx3YFP/YFP mice were previously reported [13] and acquired through the Jackson Laboratory. 2D2 TCR transgenic mice have been described [24], and they were kindly provided by Dr. V. Kuchroo (Harvard University). Runx3Tg mice were generated by transgenesis in which a mouse Runx3 cDNA was placed under the control of the human CD2 promoter/enhancer. In the same way, β7Tg mice were also generated by transgenesis by placing a mouse β7 cDNA under the control of the human CD2 promoter/enhancer. Runx3Tgβ7Tg mice were generated by crossing Runx3Tg mice with β7Tg mice. CD103Tg mice were generated in this study by placing a mouse Itgae cDNA under the control of the human CD2 promoter/enhancer. CD103Tgβ7Tg animals were generated by crossing CD103Tg with β7Tg mice. Animal experiments were performed with 6- to 12-week-old mice of both sexes. Approval of animal experiments was granted by the NCI Animal Care and Use Committee. All mice were cared for in accordance with NIH guidelines.
Flow cytometry
Mice were sacrificed, and the lymphoid organs were harvested and processed into single cell suspensions in 10% fetal calf serum (FCS) in RPMI media. Cells were stained with the following antibodies: CD44 (IM7), TCRβ (H57-597), Foxp3 (FJK-16), CD103 (2E7), β7 (FIB504), and CD8β (eBioH35-17.2), CD24 (M1/69) from ThermoFisher eBioscience; CD4 (GK1.5 and RM4.5), CD8α (53-6-7), CD62L (MEL-14), CD69 (H1.2F3), CD25 (PC61.5), CD122 (TM-β1), Runx3 (R3-5G4), and ThPOK (T43-94) from BD Biosciences (San Jose, CA); and CD45 (30-F11), E-cadherin (DECMA-1), CCR9 (CW-1.2), Vα3.2 (RR3-16), Vβ11 (KT11) from BioLegend (San Diego, CA). For Foxp3 staining, the Foxp3 intracellular staining buffer set was used according to the manufacturer’s instructions (Thermo Fisher eBioscience). For intracellular Runx3 and ThPOK staining, intracellular IC fixation and permeabilization buffers from Thermo Fisher eBioscience were used according to the manufacturer’s instructions. Flow cytometry data were acquired on LSRFortessa, X-20, or LSRII flow cytometers from BD Biosciences.
Lymphocyte isolation from nonlymphoid tissues
Lymphocytes were enriched from the lung and liver using Percoll gradients as previously described [42, 43]. For IEL isolation, the small intestines of mice were harvested and flushed with ice-cold 2% FCS in HBSS twice to remove all fecal material. The lumen of the small intestine was cut open longitudinally, diced transversely into 1 cm pieces and washed with fresh 2% FCS in HBSS twice. The pieces were then placed into 25 mL of solution A media (10% FCS, 0.0154 g DL-dithiothreitol, and 1% 0.5 M EDTA in HBSS, prewarmed to 37 °C) and shaken at 245 rpm for 45 min at 37 °C. The SI pieces were passed through a 70 µm filter, and the suspension was pelleted at 1,500 rpm for 7 min, washed in ice-cold PBS, and resuspended in 10% FCS in HBSS at 20 × 106 cells/mL. SI epithelial cells were removed from the cell suspension by negative selection upon incubation with 10 µL/mL of anti-EpCAM antibody (eBiosciences, clone G8.8) for 30 min on ice and washed in 10% FCS in HBSS, followed by incubation with BioMag beads (Qiagen) and magnetic separation. IELs were then analyzed by flow cytometry.
In vitro stimulation with TGFβ
T cells were enriched from the lymph nodes (LNs) of WT and Runx3KO mice by negative selection using anti-mouse IgG BioMag beads (Qiagen). The purity of the isolated cells was confirmed by flow cytometry. The LN T cells were resuspended in IMDM media at 1–2 × 106 cells/mL and stimulated with 5 ng/mL TGFβ (R&D Systems) for 4 days at 37 °C. On day 4, stimulated cells were analyzed by flow cytometry.
Isolation of stromal cells
For stromal cell isolation, thymus and pooled LNs (inguinal, brachial, and axillary) were teased into small pieces with sharp tweezers, resuspended in HBSS with 2% FCS, and then treated for enzymatic digestion with collagenase IV (1 mg/ml) and DNAse I (40 μg/ml) at 37 °C for 30 min under continuous agitation. Undigested tissues from this step were then treated for additional 15 min after adding collagenase D (3.5 mg/ml) and DNAse I (40 μg/ml), followed by passing through a 70 μm cell strainer (Falcon, Corning) to remove undigested debris. Cells were then resuspended in cell culture media before further analysis.
Quantitative real-time PCR
TCRβ+ CD8 LN T cells were purified from WT and CD103KO mice. Total RNA was isolated using the RNeasy kit (Qiagen) and then reverse-transcribed into cDNA with the QuantiTect Reverse Transcription kit (Qiagen) per manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was then performed with the QuantiTect SYBR Green detection system (Qiagen) on the QuantiStudio 6 RT-PCR instrument (Life Technologies) by following the manufacturer’s directions. The primer sequences were as follows: β-actin (forward: 5’-GAGAGGGAAATCGTGCGTGA-3’; reverse: 5’-ACATCTGCTGGAAGGTGG-3’), granzyme B (forward: 5’-CCTCCAGGACAAAGGCAGGGGA-3’; reverse: 5’-CCCACATATCGCCTCAGGCTGC-3’), and perforin (forward: 5’- ATGTGGCCGCAGCCAAGGTC-3’; reverse: 5’- AGGCGCGCATGTTTGCCTCT-3’).
Statistical analysis
Statistical analysis was conducted using an unpaired two-tailed Student’s t test using GraphPad Prism 7. Data represent the mean ± standard error of the mean (SEM). P values less than 0.05 were considered significant. P values greater than or equal to 0.05 were considered nonsignificant (N.S.).
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the General Surgery Department and Residency Program, Guthrie Robert Packer Hospital, for support in allowing Hilary R. Keller an interruption in her residency training to join the National Institutes of Health Cancer Research Training Award fellowship program.
Author contributions
HRK, DLL, and CL designed and performed the experiments, analyzed the data, and contributed to the writing of the manuscript. SH, MAL, PP, NL, AC, and VL performed the experiments, analyzed the data, and commented on the manuscript. YKP and JHP conceived the project, analyzed the data, and wrote the manuscript.
Funding
This study has been supported by the Intramural Research Program of the US National Institutes of Health, National Cancer Institute, Center for Cancer Research, and by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI18C0479).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
All authors consent to the publication of this study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Davinna L. Ligons and Can Li have equally contributed to this work.
Contributor Information
Yoo Kyoung Park, Email: ypark@khu.ac.kr.
Jung-Hyun Park, Email: parkhy@mail.nih.gov.
References
- 1.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol. 2008;8(10):788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taniuchi I. CD4 helper and CD8 cytotoxic T cell differentiation. Annu Rev Immunol. 2018;36:579–601. doi: 10.1146/annurev-immunol-042617-053411. [DOI] [PubMed] [Google Scholar]
- 3.Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, Okuda T, Taniuchi I. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319(5864):822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 4.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204(8):1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehlers M, Laule-Kilian K, Petter M, Aldrian CJ, Grueter B, Wurch A, Yoshida N, Watanabe T, Satake M, Steimle V. Morpholino antisense oligonucleotide-mediated gene knockdown during thymocyte development reveals role for Runx3 transcription factor in CD4 silencing during development of CD4-/CD8+ thymocytes. J Immunol. 2003;171(7):3594–3604. doi: 10.4049/jimmunol.171.7.3594. [DOI] [PubMed] [Google Scholar]
- 6.Park J-H, Adoro S, Guinter T, Erman B, Alag AS, Catalfamo M, Kimura MY, Cui Y, Lucas PJ, Gress RE. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat Immunol. 2010;11(3):257. doi: 10.1038/ni.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Etzensperger R, Kadakia T, Tai XG, Alag A, Guinter TI, Egawa T, Erman B, Singer A. Identification of lineage-specifying cytokines that signal all CD8(+)-cytotoxic-lineage-fate 'decisions' in the thymus. Nat Immunol. 2017;18(11):1218–1227. doi: 10.1038/ni.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, Bernstein Y, Goldenberg D, Brenner O, Berke G, Levanon D, Groner Y. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A. 2003;100(13):7731–7736. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, Groner Y, Rao A. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med. 2009;206(1):51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan Q, Zeng Z, Xing S, Li F, Hartwig SM, Gullicksrud JA, Kurup SP, Van Braeckel-Budimir N, Su Y, Martin MD, Varga SM, Taniuchi I, Harty JT, Peng W, Badovinac VP, Xue HH. The transcription factor Runx3 guards cytotoxic CD8(+) effector T cells against deviation towards follicular helper T cell lineage. Nat Immunol. 2017;18(8):931–939. doi: 10.1038/ni.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grueter B, Petter M, Egawa T, Laule-Kilian K, Aldrian CJ, Wuerch A, Ludwig Y, Fukuyama H, Wardemann H, Waldschuetz R. Runx3 regulates integrin αE/CD103 and CD4 expression during development of CD4−/CD8+ T cells. J Immunol. 2005;175(3):1694–1705. doi: 10.4049/jimmunol.175.3.1694. [DOI] [PubMed] [Google Scholar]
- 12.Yarmus M, Woolf E, Bernstein Y, Fainaru O, Negreanu V, Levanon D, Groner Y. Groucho/transducin-like Enhancer-of-split (TLE)-dependent and -independent transcriptional regulation by Runx3. Proc Natl Acad Sci U S A. 2006;103(19):7384–7389. doi: 10.1073/pnas.0602470103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9(10):1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermiston ML, Gordon JI. In vivo analysis of cadherin function in the mouse intestinal epithelium: essential roles in adhesion, maintenance of differentiation, and regulation of programmed cell death. J Cell Biol. 1995;129(2):489–506. doi: 10.1083/jcb.129.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity. 2014;41(6):886–897. doi: 10.1016/j.immuni.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14(10):667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 17.Camerini V, Panwala C, Kronenberg M. Regional specialization of the mucosal immune-system - intraepithelial lymphocytes of the large-intestine have a different phenotype and function than those of the small-intestine. J Immunol. 1993;151(4):1765–1776. [PubMed] [Google Scholar]
- 18.Parrott DM, Tait C, MacKenzie S, Mowat AM, Davies MD, Micklem HS. Analysis of the effector functions of different populations of mucosal lymphocytes. Ann N Y Acad Sci. 1983;409(1):307–320. doi: 10.1111/j.1749-6632.1983.tb26879.x. [DOI] [PubMed] [Google Scholar]
- 19.Schön MP, Arya A, Murphy EA, Adams CM, Strauch UG, Agace WW, Marsal J, Donohue JP, Her H, Beier DR. Mucosal T lymphocyte numbers are selectively reduced in integrin αE (CD103)-deficient mice. J Immunol. 1999;162(11):6641–6649. [PubMed] [Google Scholar]
- 20.Robinson PW, Green SJ, Carter C, Coadwell J, Kilshaw PJ. Studies on transcriptional regulation of the mucosal T-cell integrin alphaEbeta7 (CD103) Immunology. 2001;103(2):146–154. doi: 10.1046/j.1365-2567.2001.01232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allakhverdi Z, Fitzpatrick D, Boisvert A, Baba N, Bouguermouh S, Sarfati M, Delespesse G. Expression of CD103 identifies human regulatory T-cell subsets. J Allergy Clin Immunol. 2006;118(6):1342–1349. doi: 10.1016/j.jaci.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 22.Bertoni A, Alabiso O, Galetto AS, Baldanzi G. Integrins in T cell physiology. Int J Mol Sci. 2018;19(2):485. doi: 10.3390/ijms19020485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutlesa S, Wessels JT, Speiser A, Steiert I, Muller CA, Klein G. E-cadherin-mediated interactions of thymic epithelial cells with CD103+ thymocytes lead to enhanced thymocyte cell proliferation. J Cell Sci. 2002;115(Pt 23):4505–4515. doi: 10.1242/jcs.00142. [DOI] [PubMed] [Google Scholar]
- 24.Bettelli E, Pagany M, Weiner HL, Linington C, Sobel RA, Kuchroo VK. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J Exp Med. 2003;197(9):1073–1081. doi: 10.1084/jem.20021603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashita I, Nagata T, Tada T, Nakayama T. Cd69 cell-surface expression identifies developing thymocytes which audition for T-cell antigen receptor-mediated positive selection. Int Immunol. 1993;5(9):1139–1150. doi: 10.1093/intimm/5.9.1139. [DOI] [PubMed] [Google Scholar]
- 26.Pabst R, Russell MW, Brandtzaeg P. Tissue distribution of lymphocytes and plasma cells and the role of the gut. Trends Immunol. 2008;29(5):206–208. doi: 10.1016/j.it.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Zhou C, Qiu Y, Yang H. CD4CD8alphaalpha IELs: they have something to say. Front Immunol. 2019;10:2269. doi: 10.3389/fimmu.2019.02269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uehara S, Grinberg A, Farber JM, Love PE. A role for CCR9 in T lymphocyte development and migration. J Immunol. 2002;168(6):2811–2819. doi: 10.4049/jimmunol.168.6.2811. [DOI] [PubMed] [Google Scholar]
- 29.Higgins JM, Mandlebrot DA, Shaw SK, Russell GJ, Murphy EA, Chen YT, Nelson WJ, Parker CM, Brenner MB. Direct and regulated interaction of integrin alphaEbeta7 with E-cadherin. J Cell Biol. 1998;140(1):197–210. doi: 10.1083/jcb.140.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smyth L, Kirby J, Cunningham A. Role of the mucosal integrin αE (CD103) β7 in tissue-restricted cytotoxicity. Clin Exp Immunol. 2007;149(1):162–170. doi: 10.1111/j.1365-2249.2007.03385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fousteri G, Dave A, Juntti T, von Herrath M. CD103 is dispensable for anti-viral immunity and autoimmunity in a mouse model of virally-induced autoimmune diabetes. J Autoimmun. 2009;32(1):70–77. doi: 10.1016/j.jaut.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Asady R, Yuan R, Liu K, Wang D, Gress RE, Lucas PJ, Drachenberg CB, Hadley GA. TGF-{beta}-dependent CD103 expression by CD8(+) T cells promotes selective destruction of the host intestinal epithelium during graft-versus-host disease. J Exp Med. 2005;201(10):1647–1657. doi: 10.1084/jem.20041044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng Y, Wang DH, Yuan RW, Parker CM, Farber DL, Hadley GA. CD103 expression is required for destruction of pancreatic islet allografts by CD8(+) T cells. J Exp Med. 2002;196(7):877–886. doi: 10.1084/jem.20020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boller K, Vestweber D, Kemler R. Cell-adhesion molecule uvomorulin is localized in the intermediate junctions of adult intestinal epithelial cells. J Cell Biol. 1985;100(1):327–332. doi: 10.1083/jcb.100.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen QP, Deng TZ, Witherden DA, Goldrath AW. Origins of CD4(+) circulating and tissue-resident memory T-cells. Immunology. 2019;157(1):3–12. doi: 10.1111/imm.13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wurbel MA, Philippe JM, Nguyen C, Victorero G, Freeman T, Wooding P, Miazek A, Mattei MG, Malissen M, Jordan BR, Malissen B, Carrier A, Naquet P. The chemokine TECK is expressed by thymic and intestinal epithelial cells and attracts double- and single-positive thymocytes expressing the TECK receptor CCR9. Eur J Immunol. 2000;30(1):262–271. doi: 10.1002/1521-4141(200001)30:1<262::AID-IMMU262>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 37.Staton TL, Habtezion A, Winslow MM, Sato T, Love PE, Butcher EC. CD8+ recent thymic emigrants home to and efficiently repopulate the small intestine epithelium. Nat Immunol. 2006;7(5):482–488. doi: 10.1038/ni1319. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Zhang P, Li J, Kulkarni AB, Perruche S, Chen W. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat Immunol. 2008;9(6):632–640. doi: 10.1038/ni.1607. [DOI] [PubMed] [Google Scholar]
- 39.Huehn J, Siegmund K, Lehmann JCU, Siewert C, Haubold U, Feuerer M, Debes GF, Lauber J, Frey O, Przybylski GK, Niesner U, de la Rosa M, Schmidt CA, Bauer R, Buer J, Scheffold A, Hamann A. Developmental stage, phenotype, and migration distinguish naive- and effector/memory-like CD4(+) regulatory T cells. J Exp Med. 2004;199(3):303–313. doi: 10.1084/jem.20031562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma R, Sung SSJ, Abaya CE, Ju ACY, Fu SM, Ju ST. IL-2 Regulates CD103 expression on CD4(+) T cells in scurfy mice that display both CD103-dependent and independent inflammation. J Immunol. 2009;183(2):1065–1073. doi: 10.4049/jimmunol.0804354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szabo PA, Miron M, Farber DL. Location, location, location: tissue resident memory T cells in mice and humans. Sci Immunol. 2019 doi: 10.1126/sciimmunol.aas9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park JY, DiPalma DT, Kwon J, Fink J, Park JH. Quantitative difference in PLZF protein expression determines iNKT lineage fate and controls innate CD8 T cell generation. Cell Rep. 2019;27(9):2548–2557. doi: 10.1016/j.celrep.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang JH, Dong ZJ, Zhou RB, Luo DM, Wei HM, Tian ZG. Isolation of lymphocytes and their innate immune characterizations from liver, intestine. Lung Uterus Cell Mol Immunol. 2005;2(4):271–280. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.