Abstract
There is considerable interest in regenerating the injured heart by reprogramming resident fibroblasts into new functional cardiomyocytes. Cardiac reprogramming has been achieved via transcription factors or miRNAs. Transcription factor combinations appear to be species-specific as evidenced by the fact that combinations of transcription factors which are effective for the reprogramming of mouse fibroblasts are ineffective in pigs and humans. Whether miRNA based cardiac reprogramming suffers from the same limitation is unknown. We have previously demonstrated that mouse cardiac fibroblasts can be directly converted into cardiomyocytes both in vitro and in vivo via a combination of four microRNAs (miR-1, miR-133a, miR-208a and miR-499) termed “miR combo.” To assess species-specificity, miR combo was transfected into cardiac fibroblasts isolated from the left ventricle of dogs, pigs and humans. QPCR analysis indicated that miR combo effectively reprogrammed fibroblasts from all of the tested mammalian species. Significant upregulation of cardiac developmental, sarcomere, and cardiac ion channel genes was observed. Through Actn2+ staining, we also found that miR combo transfection induced dog, pig and human cardiac fibroblasts to develop into cardiomyocyte-like cells. In conclusion, we have demonstrated that in contrast to transcription factor based approaches, miR combo effectively reprograms mammalian cardiac fibroblasts into cardiomyocyte-like cells.
Keywords: Reprogramming, Fibroblasts, Cardiomyocytes, miRNAs
Highlights
-
•
Fibroblasts are reprogrammed into cardiomyocytes via transcription factors/miRNAs.
-
•
Transcription factor combinations differ between mammals.
-
•
miR combo reprogrammed fibroblasts from mice, pigs, dogs and humans.
1. Introduction
Myocardial infarction (MI) is the permanent death of heart muscle. As such, there is considerable interest in regenerating the injured heart by reprogramming resident fibroblasts into new functional cardiomyocytes. Direct cardiac reprogramming has been achieved via transcription factors or miRNAs. Transcription factor combinations appear to be species-specific. In the mouse, the transcription factors Gata4, Mef2C and Tbx5 are effective in converting fibroblasts into cardiomyocytes [1,2]. In contrast, Gata4, Mef2C and Tbx5 have proven to be ineffective in converting human [3,4] and pig [5] cardiac fibroblasts to cardiomyocytes. This has spurred researchers to identify additional transcription factors. For reprogramming human fibroblasts, various additional transcription factors have been proposed. These additional transcription factors differ between studies with one proposing Esrrg and Mesp1 [3] while an alternative study proposes the transcription factor Myocd [4]. The inability of a distinct combination of transcription factors to universally reprogram all mammalian fibroblasts has implications for clinical applications as well as for our understanding of cardiac development. For clinical applications, it suggests that transcription factor based research in the mouse may not be applicable to human disease. Similarly, the studies also suggest that transcription factors may not play a uniquely causal role in cardiomyogenesis.
The alternative approach to reprogramming fibroblasts to cardiomyocytes is via miRNAs. We have previously identified a combination of four miRs (miR-1, miR-133a, miR-208a and miR-499) termed “miR combo” which directly reprograms mouse cardiac fibroblasts into cardiomyocyte-like cells both in vitro and in vivo [[6], [7], [8], [9], [10]]. When administered into the infarcted myocardium, miR combo promoted reprogramming; two months post-MI, 10% of the cardiomyocytes in the infarct border zone were of a fibroblast origin [11]. Additionally, an improvement in cardiac function was observed post-MI as well as a significant decrease in fibrosis [11]. A recent report suggests that miR combo reprograms human fibroblasts into cardiomyocytes [12], indicating that miR combo may function as a universal reprogrammer.
In this report, we sought to the test the species specificity of miR combo. To this end, cardiac fibroblasts isolated from the left ventricle of dogs, pigs and humans were transfected with miR combo. In all species, we observed marked upregulation of cardiac developmental, sarcomere, and cardiac ion channel genes. Moreover, miR combo transfection induced dog, pig, and human cardiac fibroblasts to develop into cardiomyocyte-like cells. These findings validate the potential of miR combo as a therapeutic modality for myocardial regeneration following myocardial injury in humans.
2. Materials & methods
Cardiac fibroblast isolation and culture: Left ventricle from dog (6-week old beagle) and pig (6-week old Yorkshire mini-pigs) was shipped overnight in CoStorSol® by North Western Bio. Cardiac fibroblasts were immediately isolated according to method employed for mouse cardiac fibroblasts [6] and cultured in growth media containing DMEM (ATCC, Catalogue number 30–2002) supplemented with 15%v/v FBS (Gemini Bio BenchMark™ Fetal Bovine Serum, Catalogue number 100–106, Lot number A16H74K) and 1%v/v penicillin/streptomycin (Gibco, Catalogue number 15140-122, 100units Penicillin, 100ug/ml Streptomycin). Fibroblasts were passaged once the cells had reached 70–80% confluence using 0.05% w/v trypsin (Gibco, Catalogue number 25300-054). Freshly isolated fibroblasts were labelled as Passage 0. Experiments were conducted with cells at passage 2.
Fetal (24-weeks gestation) left ventricular primary human cardiac fibroblasts were commercially obtained from Cell Applications (Catalogue number 306K-05f). Cells were cultured in Fibroblast Growth Media (Catalogue number 316–500). They were passaged once they had reached 80% confluence using Trypsin/EDTA (Catalogue number 070–100). Experiments were conducted with cells at passage 2.
Animal Experiment Care Statement: All animal experiments complied with the National Research Council's Guide for the Care and Use of Laboratory Animals.
Cardiac reprogramming with miR combo: Cells were seeded at 5000 cells/cm2 in growth media in a 12-well plate. After 24 h, the cells were transfected with transfection complexes containing the transfection reagent (Dharmafect-I, ThermoScientific) and either a non-targeting miR (negmiR) or miR combo at 50 nmol/L. One day later, transfection complexes were removed and replaced with fresh growth media. Media was replaced every two to three days for the duration of the experiment
qPCR: Total RNA was extracted using Quick-RNA MiniPrep Kit according to the manufacturer's instructions (Zymo Research). Total RNA (50 ng–100ng) was converted to cDNA using a high capacity cDNA reverse transcription kit according to the manufacturer's instructions (Applied Biosystems). cDNA was used in a standard qPCR reaction involving FAM conjugated gene specific primers and TaqMan Gene Expression Master Mix according to the manufacturer's instructions (Applied Biosystems). The following qPCR primers were obtained commercially (ThermoFisher):
Human: Gapdh Hs99999905; Gata4 Hs00171403_m1; Mef2c Hs00231149_m1; Tbx5 Hs00361155_m1; Hand2 Hs00232769_m1; Actc1 Hs01109515; Actn2 Hs00153809_m1; Myh6 Hs01101425_m1; Nebl Hs01067284_m1; Tnnc1 Hs00268524_m1; Tnni3 Hs00165957_m1; Tnnt2 Hs00943911_m1; Ttn Hs00399225_m1.
Pig: Gapdh (Ss03375435_u1); Actn2 (Ss00473654_m1); Cacna1c (Ss04394001_m1); Nebl (Ss04306506_m1).
Dog: Gapdh Cf04419463; Actn2 (Cf01552484_m1); Cacna1c (Cf00930467_m1); Nebl (Cf02656135_m1).
Please note that the sequences are not provided by the manufacturer.
RNA-Seq: Fetal human cardiac fibroblasts were transfected with the cardiac reprogramming cocktail miR combo as described above. Fourteen days post-transfection, total RNA was extracted using the Quick-RNA MiniPrep Kit (Zymo Research, 11–328). High-throughput sequencing was performed by the Duke Genomic Core. Libraries were generated with a HiSeq 4000 kit (Illumina). Libraries were pooled and run in duplicate (50bp paired-end) with an Illumina HiSeq 4000. Sequencing depth was >25 × 106 individual reads per sample. Individual bioinformatics programs within the Galaxy suite were used to analyze gene expression as described in Ref. [13].
Immunofluorescence & staining for sarcomeres: Cells were fixed with 2%v/v paraformaldehyde (EMS) as described previously [14]. Fixed cells were blocked in antibody buffer (1%w/v BSA, 0.3%v/v Triton X-100, in PBS) for 1 h at room temperature and then incubated with an Actn2 antibody (Sigma Aldrich, A7811, 1:100) overnight at 4 °C in antibody buffer. The next day, Alexa-Fluor conjugated secondary antibodies (Invitrogen, Goat-Anti-mouse 594 nm) was used at 1:500 dilution in antibody buffer for 1hr at room temperature. Nuclei were stained by DAPI at 1 μg/ml for 30 min at room temperature in antibody blocking buffer.
Statistics: Graphs display individual data points along with summary statistics (mean, SEM). Significance was determined by Student T-tests.
Images: Images were processed with Zeiss software ZenBlue and Axiovision.
3. Results & discussion
3.1. miR combo reprograms human cardiac fibroblasts into cardiomyocyte-like cells
We have previously shown that a combination of miRNAs called miR combo directly reprograms mouse cardiac fibroblasts into cardiomyocytes [[6], [7], [8], [9], [10]]. To assess the conservation of miR direct cardiac reprogramming across mammalian species, we first examined reprogramming in human cardiac fibroblasts.
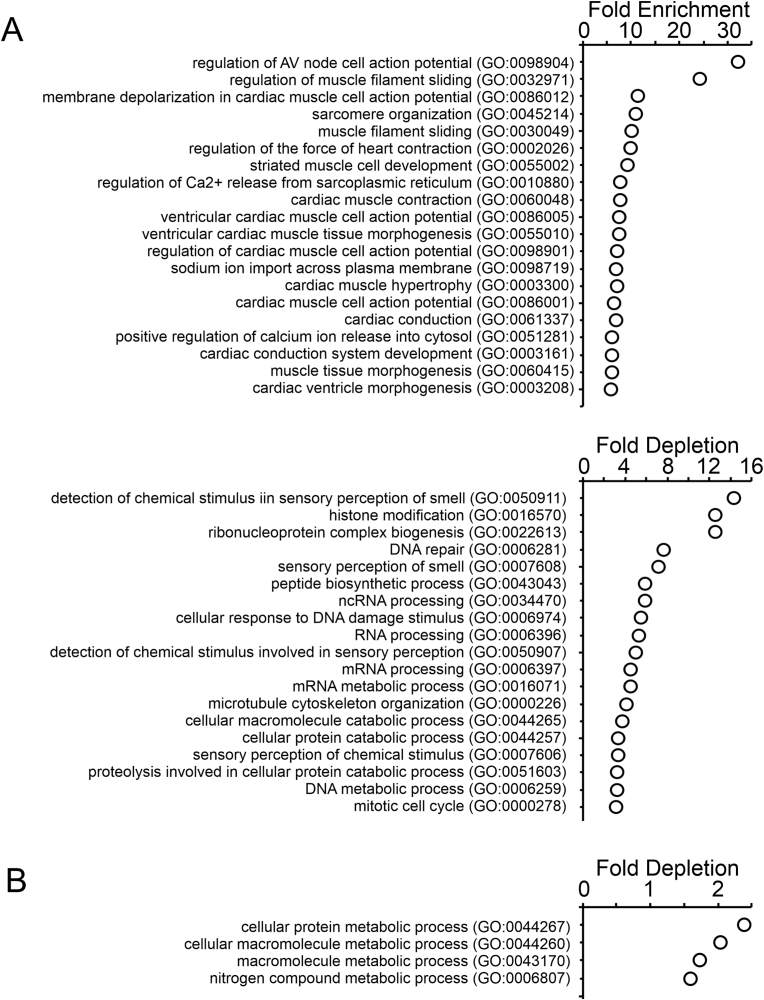
To test this hypothesis we first performed RNA-seq. Human cardiac fibroblasts were transfected with either a control non-targeting miRNA (negmiR) or miR combo. After 14 days, RNA was isolated and submitted for high-throughput sequencing. Analysis of the RNA-seq data indicated that miR combo upregulated 746 genes (Supplementary Table 1) and downregulated 470 genes (Supplementary Table 2) by more than 3-fold when compared to control cells. To determine the biological relevance gene ontology was performed. With respect to the genes up-regulated by miR combo, there was significant enrichment in gene ontology terms associated with cardiomyocyte formation and function (Fig. 1A. Full list provided in Supplementary Table 3). There were also a number of gene ontology terms that were significantly depleted in the data set. These gene ontology terms were mainly associated with DNA and RNA processing, suggesting that miR combo may impact such pathways for reprogramming (Fig. 1B. Full list provided in Supplementary Table 3). With regards to genes down-regulated by miR combo, the data set was enriched for general cellular processes (Fig. 1B, Full list provided in Supplementary Table 4).
Fig. 1.
miR combo induces cardiomyocyte gene expression in human cardiac fibroblasts.
Fetal human cardiac fibroblasts isolated from the left ventricle were transfected with miR combo or the non-targeting control miRNA negmiR. After 14 days, RNA was extracted and pooled from three separate transfections. Pooled RNA was analyzed by high-throughput sequencing. Sequences were matched to the human genome and genes significantly up- and down-regulated by miR combo were determined. Up- and down-regulated genes were analyzed via gene ontology to determine their function.
(A) Enrichment analysis displaying Gene Ontology (GO) terms which are over-represented in the set of genes up-regulated by miR combo.
(B) Enrichment analysis displaying Gene Ontology (GO) terms which are under-represented in the set of genes up-regulated by miR combo.
(C) Enrichment analysis displaying Gene Ontology (GO) terms which are over-represented in the set of genes down-regulated by miR combo.
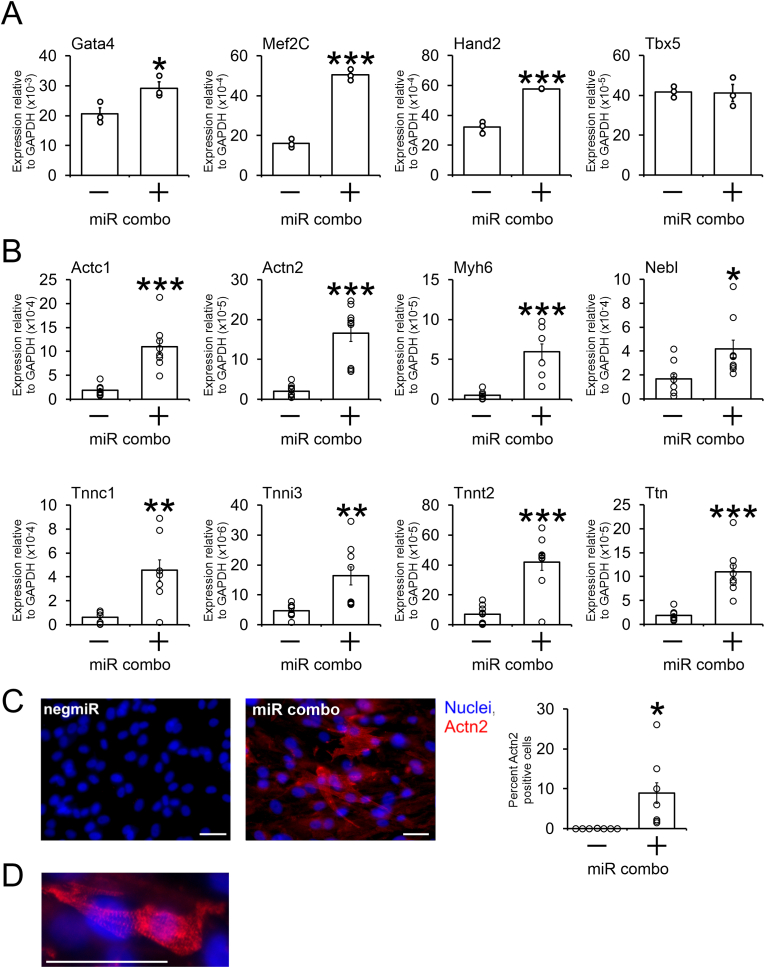
To verify the RNA-seq data, fresh human cardiac fibroblasts were obtained and transfected with miR combo. In support of the RNA-seq data, we found that miR combo strongly induced the expression of genes believed to be important for cardiac commitment such as Gata4, Hand2 and Mef2C (Fig. 2A). There was no effect on Tbx5 (Fig. 2A). Cardiac commitment does not necessarily mean the cells go on to become cardiomyocytes. Therefore we focused on a defining feature of the cardiomyocyte, the sarcomere. Expression of various sarcomere components was readily and significantly observed in the miR combo transfected cells. Expression of Actc1, Actn2, Myh6, Nebl, Tnnc1, Tnni3, Tnnt2 and Ttn mRNAs were all significantly increased by miR combo (Fig. 2B). Similarly, robust staining for the Actn2 protein indicated that miR combo was inducing the formation of cardiomyocyte-like cells (Fig. 2C). Moreover, the cardiomyocyte-like cells showed evidence of sarcomere formation (Fig. 2D). Spontaneous beating was not observed within the time-frame of the experiment. Based on our previous work, beating may require modulation of cell density or the addition of RNA-sensing receptor agonists [7]. This is a focus for future studies.
Fig. 2.
miR combo reprograms human cardiac fibroblasts into cardiomyocyte-like cells.
Fetal human cardiac fibroblasts were transfected with the cardiac reprogramming cocktail miR combo. A non-targeting control miRNA (negmiR) were used as a control.
(A) Four days post-transfection, RNA was extracted and analyzed for the expression of the cardiac commitment markers Gata4, Mef2C, Hand2, and Tbx5 by qPCR. Expression values are shown relative to the house keeping gene Gapdh. N = 3. *P < 0.05, **P < 0.01, ***P < 0.001.
(B) Fourteen days post-transfection, RNA was extracted and analyzed for the expression of the indicated sarcomere genes by qPCR. Expression values are shown relative to the house keeping gene Gapdh. N = 4–7. *P < 0.05, **P < 0.01, ***P < 0.001.
(C) Cells were fixed fourteen days post-transfection and incubated with Actn2 binding antibodies to determine the number of cardiomyocyte-like cells. Representative images are shown (scale bar 100 μm) with quantification provided. N = 7. ***P < 0.001.
(D) A representative image showing sarcomere formation in the cardiomyocyte-like cells. Scale bar 50 μm.
3.1.1. miR combo is a universal reprogrammer of fibroblasts to cardiomyocytes
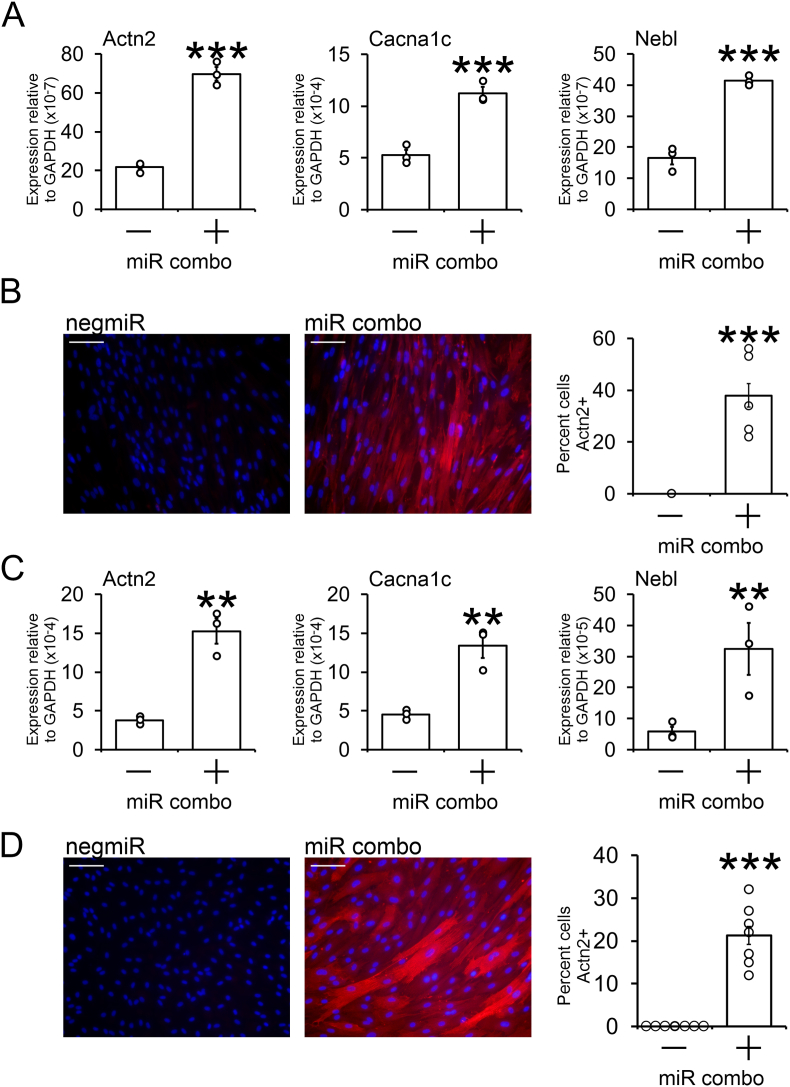
Following on from the studies with human cardiac fibroblasts, we wanted to determine if miR combo would also reprogram fibroblasts from other large mammals. To that end, we obtained cardiac fibroblasts from the left ventricle of pigs and dogs. Again cardiac fibroblasts from these animals were transfected with miR combo and reprogramming assessed 14 days after transfection. Effective fibroblast to cardiomyocyte reprogramming was observed in both pig and dog cardiac fibroblasts.
Pig cardiac fibroblasts transfected with miR combo expressed sarcomere genes Actn2 and Nebl (Fig. 3A). In addition, the miR combo transfected cells also expressed the cardiomyocyte specific ion channel Cacna1c (Fig. 3A). Following analysis of cardiomyocyte-specific mRNAs, the cells were stained with the Actn2 antibody to determine the number of cardiomyocyte-like cells. As shown in Fig. 3B, miR combo transfection strongly induced the formation of cardiomyocyte-like cells.
Fig. 3.
miR combo reprograms pig and dog cardiac fibroblasts into cardiomyocyte-like cells.
(A) Pig cardiac fibroblasts from the left ventricle were transfected with miR combo or the non-targeting miR negmiR. Fourteen days post-transfection, RNA was extracted and analyzed for the expression of the indicated cardiomyocyte-specific genes by qPCR. Expression values are shown relative to the house keeping gene Gapdh. N = 3. *P < 0.05, **P < 0.01, ***P < 0.001.
(B) Pig cells were fixed fourteen days post-transfection and incubated with Actn2 binding antibodies to determine the number of cardiomyocyte-like cells. Representative images are shown (scale bar 100 μm) with quantification provided. Cardiomyocyte-like cell counts were determined from individual wells (N = 5). ***P < 0.001.
(C) Dog cardiac fibroblasts from the left ventricle were transfected with miR combo or the non-targeting miR negmiR. Fourteen days post-transfection, RNA was extracted and analyzed for the expression of the indicated cardiomyocyte-specific genes by qPCR. Expression values are shown relative to the house keeping gene Gapdh. N = 3. *P < 0.05, **P < 0.01, ***P < 0.001.
(D) Dog cells were fixed fourteen days post-transfection and incubated with Actn2 binding antibodies to determine the number of cardiomyocyte-like cells. Representative images are shown (scale bar 100 μm) with quantification provided. There were two populations of Actn2+ cells: low Actn2 expression and high Actn2 expression. Cell counts were determined for the high Actn2 expression population (N = 7). ***P < 0.001.
Similar findings were observed with dog cardiac fibroblasts. Again, transfection of dog cardiac fibroblasts with miR combo strongly induced the expression of sarcomere and cardiac ion channels (Fig. 3C). In addition, ∼60% of the cells were cardiomyocyte-like (Fig. 3D).
In summary, we demonstrate direct reprogramming of pig, dog, and human cardiac fibroblasts into cardiomyocyte-like cells via miR combo. This finding is significant because it validates miR combo as a potential therapeutic modality for myocardial regeneration following myocardial injury in humans. Mechanistic studies have revealed that both miR combo and GMT rely on chromatin remodeling; opening up inaccessible genes by forcing changes in histone architecture particularly mediated by a shift from inhibitory Histone-H3 lysine-27 tri-methylation (H3K27me3) to stimulatory Histone-H3 lysine-4 tri-methylation (H3K4me3) [10,15]. MiRNAs and transcription factors work differently from each other. Transcription factors bind genes and activate them. In contrast, miRNAs are inhibitors which influence cell behavior by binding and destroying mRNAs. The fact that miR combo can induce reprogramming implies that fibroblasts actively repress cardiomyocyte genes. Considering that miR combo appears to be a universal reprogrammer it also suggests that these repressive mechanisms are conserved. It is possible that H3K27me3 is one such repressive mechanism. However, when contrasting the opposing fortunes of GMT and miR combo in reprogramming human fibroblasts, it is possible that other mechanisms are more important. One possible alternative mechanism is via transcription repressors and this is an avenue which we are actively investigating.
4. Conclusions
Transcription factor combinations for fibroblast to cardiomyocyte reprogramming are markedly different for each mammalian species. In contrast, the constituent miRNAs of miR combo will effectively reprogram fibroblasts of any mammalian species into cardiomyocytes.
Funding and additional information
This work was funded by the National Institutes of Health (R01 HL131814-01A1) and the Fred & Edna Mandel Foundation (no grant ID). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Declaration of competing interest
Conrad Hodgkinson is a co-founder of Recardia Therapeutics. This company is focused on developing miRNAs that reprogram fibroblasts into cardiomyocytes.
Acknowledgements
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2022.101310.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Song K., Nam Y.J., Luo X., Qi X., Tan W., Huang G.N., Acharya A., Smith C.L., Tallquist M.D., Neilson E.G., Hill J.A., Bassel-Duby R., Olson E.N. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qian L., Huang Y., Spencer C.I., Foley A., Vedantham V., Liu L., Conway S.J., Fu J.D., Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu J.D., Stone N.R., Liu L., Spencer C.I., Qian L., Hayashi Y., Delgado-Olguin P., Ding S., Bruneau B.G., Srivastava D. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. 2013;1:235–247. doi: 10.1016/j.stemcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nam Y.J., Song K., Luo X., Daniel E., Lambeth K., West K., Hill J.A., DiMaio J.M., Baker L.A., Bassel-Duby R., Olson E.N. Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vp Singh M.M., Xu X., Patel V.K., Belaguli N.S., Gibson B.W., Cooney A.J., Rosengart T.K. Reprogramming of pig cardiac fibroblasts to cardiomyocyte fate: implications for gene therapy to treat myocardial infarction. Circ. Res. 2014;115 [Google Scholar]
- 6.Jayawardena T.M., Egemnazarov B., Finch E.A., Zhang L., Payne J.A., Pandya K., Zhang Z., Rosenberg P., Mirotsou M., Dzau V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodgkinson C.P., Pratt R.E., Kirste I., Dal-Pra S., Cooke J.P., Dzau V.J. Cardiomyocyte maturation requires TLR3 activated nuclear factor kappa B. Stem Cell. 2018;36:1198–1209. doi: 10.1002/stem.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang M.H., Hu J., Pratt R.E., Hodgkinson C.P., Asokan A., Dzau V.J. Optimizing delivery for efficient cardiac reprogramming. Biochem. Biophys. Res. Commun. 2020;533:9–16. doi: 10.1016/j.bbrc.2020.08.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J., Hodgkinson C.P., Pratt R.E., Lee J., Sullenger B.A., Dzau V.J. Enhancing cardiac reprogramming via synthetic RNA oligonucleotides. Mol. Ther. Nucleic Acids. 2021;23:55–62. doi: 10.1016/j.omtn.2020.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dal-Pra S., Hodgkinson C.P., Mirotsou M., Kirste I., Dzau V.J. Demethylation of H3K27 is essential for the induction of direct cardiac reprogramming by miR combo. Circ. Res. 2017;120:1403–1413. doi: 10.1161/CIRCRESAHA.116.308741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jayawardena T.M., Finch E.A., Zhang L., Zhang H., Hodgkinson C.P., Pratt R.E., Rosenberg P.B., Mirotsou M., Dzau V.J. MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circ. Res. 2015;116:418–424. doi: 10.1161/CIRCRESAHA.116.304510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paoletti C., Divieto C., Tarricone G., Di Meglio F., Nurzynska D., Chiono V. MicroRNA-mediated direct reprogramming of human adult fibroblasts toward cardiac phenotype. Front. Bioeng. Biotechnol. 2020;8:529. doi: 10.3389/fbioe.2020.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodgkinson C.P., Gomez J.A., Baksh S.S., Payne A., Schmeckpeper J., Pratt R.E., Dzau V.J. Insights from molecular signature of in vivo cardiac c-Kit(+) cells following cardiac injury and beta-catenin inhibition. J. Mol. Cell. Cardiol. 2018;123:64–74. doi: 10.1016/j.yjmcc.2018.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodgkinson C.P., Naidoo V., Patti K.G., Gomez J.A., Schmeckpeper J., Zhang Z., Davis B., Pratt R.E., Mirotsou M., Dzau V.J. Abi3bp is a multifunctional autocrine/paracrine factor that regulates mesenchymal stem cell biology. Stem Cell. 2013;31:1669–1682. doi: 10.1002/stem.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z., Chen O., Zheng M., Wang L., Zhou Y., Yin C., Liu J., Qian L. Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem Cell Res. 2016;16:507–518. doi: 10.1016/j.scr.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.