Abstract
The species Bifidobacterium lactis, with its main representative strain Bb12 (DSM 10140), is a yoghurt isolate used as a probiotic strain and is commercially applied in different types of yoghurts and infant formulas. In order to ensure the genetic identity and safety of this bacterial isolate, species- and strain-specific molecular tools for genetic fingerprinting must be available to identify isolated bifidobacteria or lactic acid bacteria from, e.g., various clinical environments of relevance in medical microbiology. Two opposing rRNA gene-targeted primers have been developed for specific detection of this microorganism by PCR. The specificity of this approach was evaluated and verified with DNA samples isolated from single and mixed cultures of bifidobacteria and lactobacilli (48 isolates, including the type strains of 29 Bifidobacterium and 9 Lactobacillus species). Furthermore, we performed a Multiplex-PCR using oligonucleotide primers targeting a specific region of the 16S rRNA gene for the genus Bifidobacterium and a conserved eubacterial 16S rDNA sequence. The specificity and sensitivity of this detection with a pure culture of B. lactis were, respectively, 100 bacteria/ml after 25 cycles of PCR and 1 to 10 bacteria/ml after a 50-cycle nested-PCR approach.
The genera Bifidobacterium and Lactobacillus constitute a significant proportion of the probiotic cultures used in the food industry (15, 24, 25). The utilization of strains belonging to Bifidobacterium animalis, Bifidobacterium longum, Bifidobacterium bifidum, and Bifidobacterium infantis as probiotic starter cultures is due to their important role played in the large intestine, namely, control of the pH value and reduction of growth of many potential pathogens and putrefactive bacteria (4, 8, 19). An accurate species and strain identification is mandatory in clinical studies involved in monitoring the probiotic value of cells during passage through the human gastrointestinal tract or potentially any public health-related monitoring. Furthermore, such monitoring of Bifidobacterium cells directly from yoghurt, cheese, infant formula, and other supposedly Bifidobacterium-containing products is an important quality control tool of this probiotic strain in culture.
An identification of bifidobacteria at the genus and species level is possible by use of several probes targeting different genes, e.g., 16S rRNA (11, 16, 17), 23S rRNA, and recA (13). Sequence comparison of 16S and 23S rRNAs has demonstrated its suitability as a genetic tool for identification of many bacterial species (6, 28). The advantages of using these genes as targets in hybridization experiments or in amplification reactions depend highly on the fact that they exist in large copy numbers within one single bacterial cell, exceeding by far the number of chromosomally encoded genes.
Several studies have been carried out on Bifidobacterium ribosomal DNA (rDNA) sequences in order to develop species-specific primers. Furthermore, Kaufmann et al. (11) described PCR oligonucleotide primers based on specific 16S rRNA sequences for the identification of isolates belonging to the genus Bifidobacterium. Recently, interest has been focused on the generation of 16S rRNA gene probes, allowing the exclusive detection of bifidobacteria in fecal samples through an in situ hybridization approach (14). Another way to utilize rDNA sequences in Bifidobacterium identification is to generate rDNA by a PCR approach and then separate the achievable amplicons in a sequence-specific manner in a temperature gradient gel electrophoresis or denaturing gradient gel electrophoresis (21, 27). Specific Multiplex-PCR assays based on the gene amplification of, e.g., parts of the 16S rRNA or surface layers by two pairs of primers in a single reaction were already demonstrated to be useful for a species-specific identification in the genus Lactobacillus (20, 29).
In the present study, a new set of PCR primers for a specific species identification within a mixture of bifidobacteria or as a pure culture of B. lactis was developed. This probiotic species is used in various infant formulas and yoghurts and is technologically favored by its high tolerance of increased oxygen levels and its unusual acid tolerance (18). The aim of our work was the development of a rapid and reliable method for the highly specific detection, identification, and safety assurance (due to genetic identity) of B. lactis isolates (by means of PCR amplification) frequently detectable in infant feces and from different commercially available products. This was undertaken by a Multiplex-PCR approach using simultaneously strain- and species-specific (for B. lactis), genus-specific (for bifidobacteria), and eubacterium-specific conserved oligonucleotide primers.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in the present study are listed in Table 1. Strains were grown anaerobically in MRS broth (Difco, Detroit, Mich.) generally supplemented with 0.5 g of cysteine hydrochloride per liter and incubated for 16 h at 37°C under anaerobic conditions (Gas-pack, AnaeroGen; Oxoid, Basingstoke, United Kingdom).
TABLE 1.
Bacterial type strains (29 bifidobacteria and 9 lactobacilli) and nine additional Bifidobacterium and Lactobacillus isolates utilized to evaluate the specificity of the Multiplex-PCR
| Species | Straina | PCR results
|
Originb | ||
|---|---|---|---|---|---|
| Bflact2- Bflact5 | P0- Lm3 | P0- 338F | |||
| B. lactis | DSM 10140T | + | + | + | Yoghurt |
| B. lactis | NCC 311 | + | + | + | Human feces |
| B. lactis | NCC 363 | + | + | + | Human feces |
| B. adolescentis | ATCC 15703T | − | + | + | Intestine of adult |
| B. adolescentis | ATCC 15704 | − | + | + | Intestine of adult |
| B. animalis | ATCC 25527T | − | + | + | Rat feces |
| B. animalis | ATCC 27672 | − | + | + | Rat feces |
| B. animalis | ATCC 27673 | − | + | + | Sewage |
| B. bifidum | ATCC 29521T | − | + | + | Infant feces |
| B. bifidum | ATCC 15696 | − | + | + | Intestine of infant |
| B. breve | ATCC 15700T | − | + | + | Intestine of infant |
| B. breve | ATCC 15701 | − | + | + | Intestine of infant |
| B. catenulatum | ATCC 27539T | − | + | + | Intestine of adult |
| B. coryneforme | DSM 20216T | − | + | + | Honeybee hindgut |
| B. cuniculi | ATCC 27916T | − | + | + | Feces of rabbit |
| B. dentium | ATCC 27534T | − | + | + | Dental caries |
| B. infantis | ATCC 15697T | − | + | + | Intestine of infant |
| B. infantis | ATCC 25962 | − | + | + | Intestine of infant |
| B. angulatum | DSM 20098T | − | + | + | Human feces |
| B. longum | LMG 13197T | − | + | + | Intestine of adult |
| B. magnum | ATCC 27540T | − | + | + | Rabbit feces |
| B. pseudocatenulatum | DSM 20438T | − | + | + | Feces of infant |
| B. pseudolongum | DSM 20099T | − | + | + | Swine feces |
| B. pullorum | DSM 20433T | − | + | + | Feces of chicken |
| B. merycicum | DSM 6492T | − | + | + | Bovine rumen |
| B. minimum | DSM 20102T | − | + | + | Sewage |
| B. ruminantium | DSM 6489T | − | + | + | Bovine rumen |
| B. saeculare | DSM 6531T | − | + | + | Rabbit feces |
| B. subtile | DSM 20096T | − | + | + | Sewage |
| B. thermophilum | DSM 20210T | − | + | + | Swine feces |
| B. asteroides | DSM 20089T | − | + | + | Honeybee hindgut |
| B. boum | DSM 20432T | − | + | + | Rumen of cattle |
| B. gallicum | DSM 20093T | − | + | + | Human feces |
| B. gallinarum | DSM 20670T | − | + | + | Chicken cecum |
| B. inopinatum | DSM 10107T | − | + | + | Dental caries |
| B. choerinum | ATCC 27686T | − | + | + | Swine feces |
| B. suis | ATCC 27533T | − | + | + | Swine feces |
| L. acidophilus | ATCC 4356T | − | − | + | Human |
| L. crispatus | DSM 20584T | − | − | + | ND |
| L. casei | ATCC 393T | − | − | + | Cheese |
| L. fermentum | ATCC 14931T | − | − | + | ND |
| L. gasseri | DSM 20243T | − | − | + | ND |
| L. johnsonii | ATCC 33200T | − | − | + | Human blood |
| L. johnsonii | NCC 533 (La1) | − | − | + | Human isolate |
| L. paracasei | ATCC 334T | − | − | + | Emmental cheese |
| L. reuteri | ATCC 23272T | − | − | + | Human feces |
| L. rhamnosus | ATCC 7469T | − | − | + | ND |
ATCC, American Type Culture Collection; DSM, Deutsche Sammlung von Mikroorganismen; LMG, Bacteria Collection Universiteit Gent; NCC, Nestlé Culture Collection.
ND, not determined.
Preparation of template DNA.
DNA was extracted following a new rapid method of cell lysis: 2 ml of an overnight culture was collected by centrifugation at 12,000 × g (10,000 rpm) for 10 min at 4°C, and the pellet was washed twice with 2 ml of TE buffer (10 mM Tris [pH 8], 10 mM EDTA). One gram of glass beads (106 μm; Sigma, St. Louis, Mo.) was added to the bacterial cell suspension. Maximal occurring cell lysis was performed with the Mini-Beadbeater (Biospec product) for 3 min at maximum speed. Subsequently, the suspension was centrifuged at 12,000 × g (10,000 rpm) for 2 min at 4°C, and 5 μl of the supernatant was applied directly into the PCR. Chromosomal DNA was further purified by phenol-chloroform extraction (26) only for those experiments designed to detect the limit of sensitivity of the applied PCR amplifications. Chromosomal DNA was quantified on the basis of the absorbance (λ) at 260 and 280 nm (26).
Development of specific strain primers.
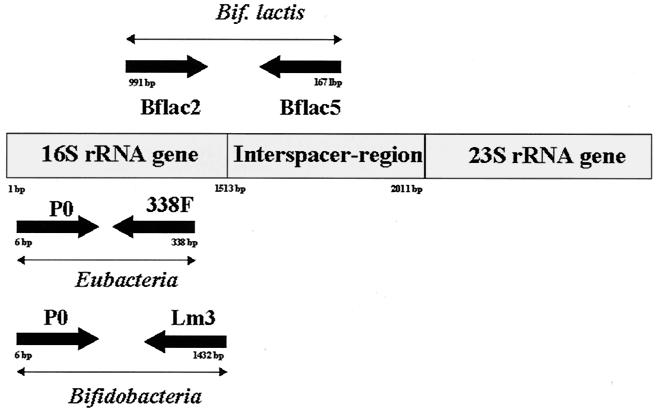
All available bifidobacterial 16S rDNA and 16S-23S rDNA spacer region sequences were retrieved from two databases, EMBL and GenBank, and aligned by using the Clustal program. Two potentially strain- and species-specific primers for B. lactis could be identified. These primers were synthesized (MWG Oligo Synthesis) and used with the other listed primers in this study as outlined in Fig. 1 and summarized in Table 2.
FIG. 1.
Schematic location of the primers used and the overall PCR approach. We utilized one pair of primers, P0 and 338F, resulting in an amplicon for all eubacteria. The utilization of oligonucleotides Lm3 and P0 resulted in a PCR amplicon of about 1,400 bp specific for the genus Bifidobacterium. An amplification product with the primer pair Bflact2-Bflact5 could be achieved only for the species B. lactis.
TABLE 2.
Primer sequences used
| Probe | Locationa | Sequence 5′→3′ | Source or reference |
|---|---|---|---|
| Lm3 | 1412–1432 (16S rRNA) | CGGGTGCTNCCCACTTTCATG | 11 |
| P0 | 6–28 (16S rRNA) | GAGAGTTTGATCCTGGCTCAG | 5 |
| 338F | 321–338 (16S rRNA) | GCTGCCTCCCGTAGGAGT | 1 |
| Bflact2 | 991–1009 (16S rRNA) | GTGGAGACACGGTTTCCC | This study |
| Bflact5 | 1651–1671 (16S-23S spacer region) | CACACCACACAATCCAATAC | This study |
PCR amplification.
Amplification reactions were performed in a total volume of 50 μl of a solution containing 1.5 mM MgCl2; 10 mM Tris HCl, pH 8.3; 50 mM KCl; each deoxynucleoside triphosphate at 200 μM; 10 pmol of P0 primer and Lm3 primer; 5 pmol of 338F primer; 50 pmol of each of the two B. lactis-specific primers, Bflact2 and Bflact5; 5 μl of the respective template DNA (from the supernatants after rapid DNA extraction, which equals about 25 ng of DNA); and 2.5 U of Taq polymerase (Gibco BRL). Amplifications were performed with a DNA thermocycler (Cetus 9700; Perkin Elmer, Norwalk, Conn.) with the following temperature profiles: 1 cycle of 95°C for 5 min; 30 cycles of 95°C for 30 s, 58°C for 1 min, and 72°C for 4 min; and finally 1 cycle of 72°C for 7 min.
Gel electrophoresis.
Aliquots of each amplification reaction (15 μl each) were analyzed by 1.5% (wt/vol) agarose gel electrophoresis in Tris-acetate-EDTA buffer (26) at a constant voltage of 7 V cm−1 and visualized with ethidium bromide (0.5 μg/ml) and photographed under UV light (wavelength, 260 nm).
Detection of B. lactis in food systems.
Different commercially available dairy products and infant formula containing bifidobacteria and lactobacilli were investigated for the presence of the species B. lactis. In summary, 1 g of product was dissolved in 1 ml of sterilized water. An equivalent amount of glass beads (Sigma) was added to the suspension, and the mixture was treated with the Mini-Beadbeater (Biospec product) for 3 min at maximum speed in order to achieve a maximal cell lysis. After cell lysis, the suspension was centrifuged 12.000 × g (10,000 rpm) for 2 min at 4°C to remove the majority of cell fractions and debris. The DNA in the supernatant was purified by using DNAzol (Gibco BRL) according to the supplier's instructions. About 25 ng of the chromosomal DNA was loaded into the PCR amplification under the conditions described above in order to apply this Multiplex-PCR approach directly on various final, commercially available products.
RESULTS
Selection of a specific primer pair for PCR.
The analysis of the 16S rDNA sequence of the B. lactis type strain (DSM 10140) indicated that the strain is very closely related (98.6% similarity) to B. animalis ATCC 25527 (18). Based on the comparison of both nucleotide sequences, one PCR primer pair (Bflact2 and Bflact5) was designed for the specific detection of only B. lactis. The oligonucleotide Bflact2 targets a region of the 16S rRNA gene, whereas the Bflact5 primer was designed to target the 16S-23S rRNA spacer region. The specificity of this primer pair could be demonstrated in a convincing manner with all DNA samples coming from strains (type strains and various isolates) summarized in Table 1.
Species-specific and genus-specific detection with the Multiplex-PCR.
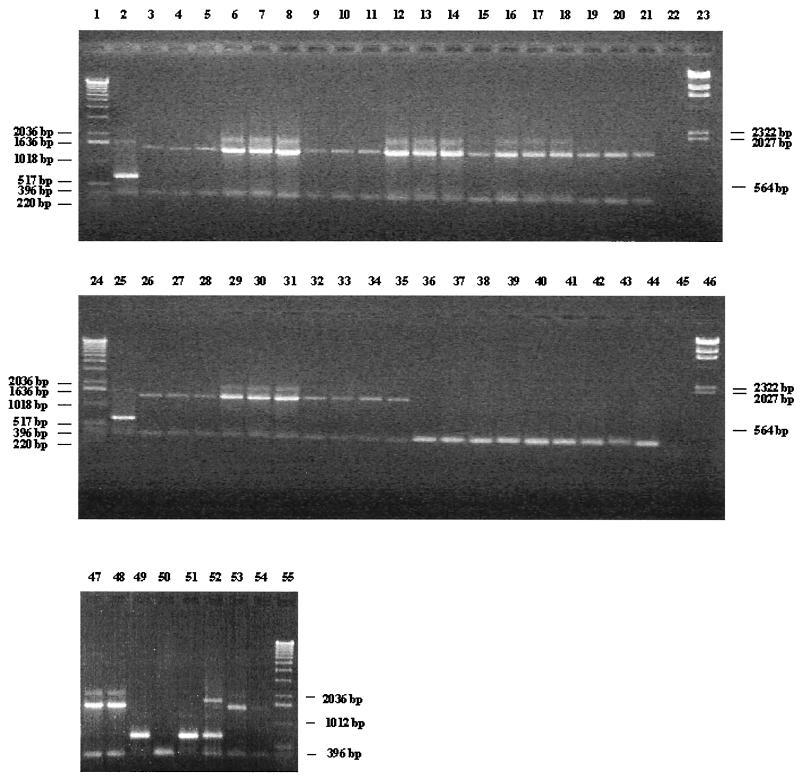
The application of the oligonucleotide pair Bflact2-Bflact5 resulted in a PCR amplicon of 680 bp only with DNA derived from B. lactis DSM 10140, whereas absolutely no PCR product could be detected with those primers for any other Bifidobacterium and Lactobacillus isolates, including 38 type strains (Fig. 2).
FIG. 2.
Multiplex-PCR products of several Bifidobacterium and Lactobacillus species. Lanes 2 and 25, B. lactis DSM 10140; lane 3, B. animalis ATCC 25527; lane 4, B. bifidum ATCC 29521; lane 5, B. breve ATCC 15700; lane 6, B. catenulatum ATCC 27539; lane 7, B. adolescentis ATCC 15703; lane 8, B. coryneforme DSM 20216; lane 9, B. cuniculi ATCC 27916; lane 10, B. dentium ATCC 27534; lane 11, B. infantis ATCC 15697; lane 12, B. angulatum DSM 20098; lane 13, B. magnum ATCC 27540; lane 14, B. longum LMG 13197; lane 15, B. pseudocatenulatum DSM 20438; lane 16, B. pseudolongum DSM 20099; lane 17, B. pullorum DSM 20433; lane 18, B. merycicum DSM 6492; lane 19, B. minimum DSM 20102; lane 20, B. ruminantium DSM 6489; lane 21, B. saeculare DSM 6531; lane 26, B. subtile DSM 20096; lane 27, B. thermophilum DSM 20210; lane 28, B. asteroides DSM 20089; lane 29, B. boum DSM 20432; lane 30, B. gallicum DSM 20093; lane 31, B. gallinarum DSM 20670; lane 32, B. inopinatum DSM 10107; lane 33, B. choerinum ATCC 27686; lane 34, B. suis ATCC 27533; lane 35, B. bifidum ATCC 15696; lane 36, L. casei 393; lane 37, L. paracasei ATCC 334; lane 38, L. rhamnosus ATCC 7469; lane 39, L. fermentum ATCC 14931; lane 40, L. johnsonii ATCC 33200; lane 41, L. johnsonii NCC 533; lane 42, L. gasseri DSM 20243; lane 43, L. acidophilus ATCC 4356; lane 44, L. reuteri ATCC 23272; lane 47, B. adolescentis ATCC 15704; lane 48, B. breve ATCC 15701; lane 49, B. lactis NCC 311; lane 50, L. crispatus DSM 20584; lane 51, B. lactis NCC 363; lane 52, B. animalis ATCC 27536; lane 53, B. animalis ATCC 27673; lane 54, B. infantis ATCC 25962; lanes 22 and 45, negative control (complete PCR mixture without DNA), lanes 1, 24, and 55, 1-kb DNA ladder (Gibco BRL); lanes 23 and 46, DNA molecular weight marker II (Boehringer Mannheim).
Furthermore, due to the expected variability in PCR conditions in processing complete bacterial cells (in the very rapid cell lysis procedure described here), the absence of any PCR product should be attributed not only to the general absence of any DNA target but also to an overall failure of the amplification reaction. To distinguish between these two events, we utilized a pair of primers, P0 (5) and 338F (1), targeting a conserved region of 332 bp within the 16S rRNA gene (the sizes of all achievable PCR amplicons were calculated from databases and estimated by gel electrophoresis).
Use of this primer pair is a valid and necessary positive PCR control step, but unfortunately it is unable to yield a specific taxonomic allocation and identification of the genus Bifidobacterium. In order to confirm this genus, a second primer was used in parallel in the same PCR preparation. The utilization of oligonucleotide Lm3 (11) resulted with P0 primer in a PCR using any DNA template extracted from bifidobacterium cells in an amplicon of about 1,400 bp specific for the genus Bifidobacterium. A PCR band of 2,000 bp achieved for the genus Bifidobacterium is currently under investigation. An amplification product due to Bflact2-Lm3 can be demonstrated only in B. lactis; no PCR fragment could be found with the primer pair Lm3-P0, due to a competitive reaction between the primer pairs (P0-Lm3 and Bflact2-Lm3) and their respective target sites on the chromosomal DNA. When a Multiplex-PCR was performed with this mixture of five primers in the same reaction, three major products of all expected sizes were detected only in the presence of DNA originating from B. lactis. The amplification of other Bifidobacterium DNA species resulted in the typical pattern of only two amplicons, and amplification with DNA of lactobacilli yielded a single 332-bp product. Furthermore, for B. lactis it was also possible to yield an additional PCR amplicon of 1,665 bp with the primer pair P0-Bflact5 (Fig. 2).
Sensitivity of amplification.
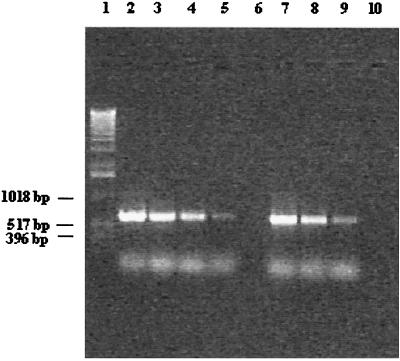
Generally, a PCR amplification allows a very sensitive detection of specific DNA sequences. The sensitivity of amplifications was evaluated by using whole cells or extracted pure chromosomal DNA. Lysed bacterial cells were serially diluted, and aliquots were assayed for any occurring and detectable sensitivity with Bflact2 and Bflact5. After 25 PCR cycles, this set of primers yielded, with a quantitative PCR approach, a product of the expected size with a culture dilution of as few as 100 bacterial cells per ml. Sensitivity can be improved further by the application of a nested-PCR approach (Fig. 3). In such a nested-PCR approach, 2 μl of the reaction mixture was transferred after the first 25 PCR cycles to a second (not yet inoculated) reaction mixture and amplified for an additional 25 PCR cycles. In such a way, we have been able to reduce the detection limit to 1 to 10 bacterial cells per ml. In order to establish the sensitivity of the method described here, different amounts of chromosomal DNA from B. lactis DSM 10140 were used in various PCRs. Visible amplicons were still obtained from the amplification of 3.4 pg of DNA template, even in a mixture with 60 or 200 ng of DNA derived from other bifidobacterial species (data not shown).
FIG. 3.
Quantitative PCR results and nested-PCR approach with regard to the detection limit for B. lactis. Lane 1, 1-kb molecular marker ladder (Gibco BRL); lanes 2 through 6, direct detection of B. lactis from culture concentrates (no direct DNA isolation) and subsequent 10-fold dilutions containing 105 to 101 cells per reaction tube; lanes 7 to 9, nested-PCR approach for detection of B. lactis from culture concentrates (no direct DNA isolation) and subsequent dilutions containing 1.5 × 103 to 1.5 × 101 cells per reaction tube (lane 7, 1.5 × 103 CFU/ml; lane 8, 1.5 × 102 CFU/ml; lane 9, 1.5 × 101 CFU/ml); lane 10, negative control.
Efficacy of primers for the application of B. lactis from various food systems.
To evaluate the ability of Bflact2 and Bflact5 primers to monitor B. lactis in various food systems such as yoghurts and infant formula, 18 commercially available products claiming to contain B. lactis were analyzed. In 10 samples we were able to detect B. lactis, whereas a further 7 samples resulted in PCR amplicons specific for only the genus Bifidobacterium and only one sample contained bacterial species not belonging to the genus Bifidobacterium at all (Table 3). All Multiplex-PCR results for the various products analyzed for the presence of B. lactis were in agreement with the labeling of bacterial strains as stated by the producers. Furthermore, our Multiplex-PCR was also performed with an additional 56 bifidobacterial strains previously isolated from infant feces and identified with species-specific primers (16, 17). B. lactis-specific primers resulted in a PCR product of the expected size with DNA of only two additional isolates, which are currently under investigation for their taxonomic affiliation (NCC 311 and NCC 363).
TABLE 3.
Direct Multiplex-PCR analysis of 18 commercially available product samples with declared content of bifidobacteria (samples 1 to 4 and 6 to 18), lactobacilli (sample 5), or other LABb
| Sample no. | PCR results
|
||
|---|---|---|---|
| Bflact2-Bflact5 | Lm3-P0a | P0-338F | |
| 1 | + | ± | + |
| 2 | + | ± | + |
| 3 | − | + | + |
| 4 | − | + | + |
| 5 | − | − | + |
| 6 | + | ± | + |
| 7 | + | ± | + |
| 8 | − | + | + |
| 9 | − | + | + |
| 10 | + | ± | + |
| 11 | + | ± | + |
| 12 | + | ± | + |
| 13 | + | ± | + |
| 14 | + | ± | + |
| 15 | + | ± | + |
| 16 | − | + | + |
| 17 | − | + | + |
| 18 | − | + | + |
±, PCR results after a run of 25 cycles. For B. lactis, a competitive reaction between the resulting PCR amplicons for the species B. lactis and the genus Bifidobacterium might occur (primer pair Bflact2-Lm3).
LAB, lactic acid bacteria.
DISCUSSION
In modern bacterial taxonomy, analysis of 16S and 23S rDNA sequences is considered to be an overall powerful tool to investigate phylogenetic relationships. These sequences are used to design species-specific and genus-specific oligonucleotide probes for a rapid identification of, e.g., lactic acid bacteria and bifidobacteria.
Several reports have already described the use of PCR-specific primers targeting rDNA sequences to identify various Bifidobacterium species. Cai et al. (3) reported that the relative taxonomic position of B. lactis is still under discussion and that B. lactis could be considered a junior subjective synonym for B. animalis. A decision on this issue by the International Committee on Systematic Bacteriology is still outstanding (9). However, our study demonstrated clear differences in rDNA sequences between B. lactis DSM 10140 and the type strain of B. animalis, ATCC 25527, overcoming the so far rather difficult selective and reliable species distinction between these two species. Only a few nucleotide mismatches were found when the rDNA sequences of these two Bifidobacterium species were compared. To design primers that would specifically identify B. lactis species, we had to screen at least four regions in the 16S-23S interspacer region showing the highest variability within these two closely related species. Subsequently, for each species-specific PCR amplification each of the putative species-specific primers was paired with the primer Bflact2, originating from the 16S rRNA gene. The four primers designed were subjected to further specificity testing using DNA from all strains listed in Table 1. Only the primer pair Bflact2-Bflact5 allowed a very specific amplification for the species B. lactis and not for any other Bifidobacterium or Lactobacillus species (Fig. 2). No rapid discrimination tool for these closely related species is available so far (18).
Unfortunately, at present the species B. lactis comprises basically only one strain, the type strain DSM 10140, isolated from a dairy product (yoghurt), as described by Meile et al. (18). Prasad et al. (22) and Kok et al. (12) described two other supposed B. lactis isolates, potentially members of the same species, confirming their allocation to the species B. lactis after sequencing of their 16S-23S interspacer region and comparison to the deposited B. lactis DSM 10140 sequence. Complete nucleotide sequence analysis of 16S rRNA and 16S-23S spacer region genes provides the most accurate basis for an identification of B. lactis; however, such a complete gene sequencing cannot currently be considered to be a reasonable approach for any routine identification of multiple isolates from clinical and environmental studies or from a product or contamination tracing. While the utilization of B. lactis species-specific primers described here could allow the rapid identification of further isolates, they should be also considered as a valuable tool for use in identifying new strains belonging to these rather limited bifidobacterial species.
The investigation of infant fecal samples for B. lactis by using this very specific set of primers allowed us to identify two additional strains as being potential members of the species B. lactis, but the strains expressed clearly different and distinguishable randomly amplified polymorphic DNA patterns (data not shown). Further investigation will be necessary to redefine with various oligonucleotides an overall strain allocation or relocation from and to the species B. animalis. As described by Roy et al. (23), B. animalis ATCC 27536 has unique pulsed-field gel electrophoresis patterns, therefore being more similar to the strain B. lactis DSM 10140 than to the type strain of its own species, B. animalis. This raises questions with regard to the taxonomic borderlines between these two species (B. lactis and B. animalis) specifically and among other bifidobacterial species in general (7). DNA isolated from B. animalis ATCC 27536 yielded an amplicon of 680 bp by use of the B. lactis-specific primers. Further analysis for this specific strain seems to be required to finally confirm its taxonomic assignment (23).
The application of this Multiplex-PCR in the analysis of commercially available products can be a very useful tool for any rapid monitoring of the species B. lactis (product claims, bacterial counts, strain and species identification) and might simultaneously give indications of other occurring bifidobacterial cells. This Multiplex-PCR can be considered, in terms of reduction of tedious labor time, easy application, reliable and repeatable PCR results and intrinsic controls, and low costs, a uniquely powerful tool for studying any bifidobacterial ecology (e.g., that of the gastrointestinal tract) and furthermore the fecal environment of newborns, babies, toddlers, and adults fed with products containing members of the genus Bifidobacterium in general and B. lactis specifically. Unfortunately, such a PCR amplification that uses chromosomal DNA as a target cannot distinguish between viable and nonviable bacterial cells (10). Therefore, in the determination of the shelf life of a product containing B. lactis with regard to bacterial stability and viability, it remains essential that all detectable and viable bacterial cells are additionally plate counted. Due to the extremely short half-life of the majority of bacterial mRNAs, it seems to be possible and maybe more appropriate to select mRNA as a PCR-based target to detect bacteria by PCR amplification. In particular, total RNA extraction followed by a reverse transcriptase PCR using primers Bflact2 and Bflact5 will represent a reliable tool for detection of only the viable fraction of B. lactis cells present in various food systems.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 3.Cai Y, Matsumoto M, Benno Y. Bifidobacterium lactis Meile et al. 1997 is a subjective synonym of Bifidobacterium animalis (Mitsuoka 1969) Scardovi and Trovatelli 1974. Microbiol Immunol. 2000;44:815–820. doi: 10.1111/j.1348-0421.2000.tb02568.x. [DOI] [PubMed] [Google Scholar]
- 4.Charteris W P, Kelly P M, Morelli L, Collins J K. Selective detection, enumeration and identification of potentially probiotic Lactobacillus and Bifidobacterium species in mixed bacterial populations. Int J Food Microbiol. 1997;35:1–27. doi: 10.1016/s0168-1605(96)01222-6. [DOI] [PubMed] [Google Scholar]
- 5.Di Cello F P, Fani R. A molecular strategy for the study of natural bacterial communities by PCR-based techniques. Minerva Biotec. 1996;8:126–134. [Google Scholar]
- 6.Drake M, Small C L, Spence K D, Swanson B G. Rapid detection and identification of Lactobacillus ssp. in dairy products by using the polymerase chain reaction. J Food Prot. 1996;59:1031–1036. doi: 10.4315/0362-028X-59.10.1031. [DOI] [PubMed] [Google Scholar]
- 7.Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 8.Gibson G R, Roberfroid M B. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. 1995;125:1410–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 9.International Committee on Systematic Bacteriology. Minutes of the meetings, 22 and 23 September 1999, Veldhoven, The Netherlands. Int J Syst Evol Microbiol. 2001;51:259–261. [Google Scholar]
- 10.Josephon K L, Gerba C P, Pepper I L. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl Environ Microbiol. 1993;59:3513–3515. doi: 10.1128/aem.59.10.3513-3515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaufmann P, Pfefferkorn A, Teuber M, Meile L. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl Environ Microbiol. 1997;63:1268–1273. doi: 10.1128/aem.63.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kok R, De Waal A, Schut F, Welling G W, Weenk G, Hellingwerf K J. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl Environ Microbiol. 1996;62:3668–3672. doi: 10.1128/aem.62.10.3668-3672.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kullen M J, Brady L J, O'Sullivan D J. Evaluation of using a short region of the recA gene for rapid and sensitive speciation of dominant bifidobacteria in the human large intestine. FEMS Microbiol Lett. 1997;154:377–383. doi: 10.1111/j.1574-6968.1997.tb12670.x. [DOI] [PubMed] [Google Scholar]
- 14.Langendijk P S, Schut F, Jansen G J, Raangs G C, Kamphuis G R, Wilkinson M H H, Welling G W. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995;61:3069–3075. doi: 10.1128/aem.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langhendries J P, Detry J, van Hees J, Lamboray J M. Effect of a fermented infant formula containing viable bifidobacteria on the fecal flora composition and pH of healthy full-term infants. J Pediatr Gastroenterol Nutr. 1995;21:177–181. doi: 10.1097/00005176-199508000-00009. [DOI] [PubMed] [Google Scholar]
- 16.Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol. 1999;65:4506–4512. doi: 10.1128/aem.65.10.4506-4512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuki T, Watanabe K, Tanaka R, Oyaizu H. Rapid identification of human intestinal bifidobacteria by 16S-rRNA targeted species- and group-specific primers. FEMS Microbiol Lett. 1998;167:113–121. doi: 10.1111/j.1574-6968.1998.tb13216.x. [DOI] [PubMed] [Google Scholar]
- 18.Meile L, Ludwig W, Rueger U, Gut C, Kaufmann P, Dasen G, Wenger S, Teuber M. Bifidobacterium lactis sp. nov., a moderately oxygen tolerant species isolated from fermented milk. Syst Appl Microbiol. 1997;20:57–64. [Google Scholar]
- 19.Modler H W, McKellar R C, Yaguchi M. Bifidobacteria and bifidogenic factors. Can Inst Food Sci Technol J. 1990;23:29–41. [Google Scholar]
- 20.Muller M R A, Ehrmann M A, Vogel R F. Multiplex PCR for detection of Lactobacillus pontis and two related species in a sourdough fermentation. Appl Environ Microbiol. 2000;66:2113–2116. doi: 10.1128/aem.66.5.2113-2116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prasad J, Harsharanjit G, Smart J, Pramod G. Selection and characterisation of Lactobacillus and Bifidobacterium strains for use as probiotics. Int Dairy J. 1998;8:993–1002. [Google Scholar]
- 23.Roy D, Ward P, Champagne G. Differentiation of bifidobacteria by use of pulsed-field gel electrophoresis and polymerase chain reaction. Int J Food Microbiol. 1996;29:11–29. doi: 10.1016/0168-1605(95)00013-5. [DOI] [PubMed] [Google Scholar]
- 24.Saavedra J M, Baumann N A, Oung I. Feeding of Bifidobacterium lactis and Streptococcus thermophilus to infant in hospital for prevention of diarrhoea and shedding of rotavirus. Lancet. 1994;344:1046–1049. doi: 10.1016/s0140-6736(94)91708-6. [DOI] [PubMed] [Google Scholar]
- 25.Saavedra J M, Abi-Hanna A, Moore N, Yolken R. Effect of long term consumption of infant formulas with bifidobacteria and S. thermophilus on stool pattern and diaper rash in infants. J Pediatr Gastroenterol Nutr. 1998;27:483–486. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Satokari R M, Vaughan E E, Akkermans A D, Saarela M, de Vos W M. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2001;67:504–513. doi: 10.1128/AEM.67.2.504-513.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tilsala-Timisjarvi A, Alatossava T. Development of oligonucleotide primers from the 16S–23S intergenic sequences for identifying different dairy and probiotic lactic acid bacteria by PCR. Int J Food Microbiol. 1997;35:49–56. doi: 10.1016/s0168-1605(97)88066-x. [DOI] [PubMed] [Google Scholar]
- 29.Ventura M, Callegari M L, Morelli L. S-layer as a molecular marker for identification Lactobacillus helveticus. FEMS Microbiol Lett. 2000;189:275–279. doi: 10.1111/j.1574-6968.2000.tb09243.x. [DOI] [PubMed] [Google Scholar]