Abstract
Low oxygen levels and extremely cold weather in high-altitude environments requires more energy intake to maintain body temperature in animals. However, little is known about the characteristics of cecal and ileac microbiota in Tibetan chicken and how the high and low altitude environments affect the gut microbiota communities in Tibetan chicken. In the present study, In the present study, Tibetan chickens (Group HA, 3572 m, 578.5 Pa) and their introduced flatland counterparts (Group LA, 580 m, 894.6 Pa) in the cecum and ileum to identify the possible bacterial species that are helpful for their host in environmental adaption. High-throughput sequencing was used to sequence the V3 to V4 hypervariable regions of the bacterial 16S rRNA gene. By comparing the gut microbial diversity of HA chicken with that of LA, the results indicated that the microbial diversity of the cecum and ileum in group HA was significantly lower (P < 0.05) than those in group LA. The cecum microbiome maintained higher population diversity and richness than the ileum (P < 0.05). Four phyla Firmicutes, Bacterioidetes, Actinobacteria, and Proteobacteria were dominant in two groups. Interestingly, there were significant differences in abundance ratio among the four groups (P < 0.05). The predominant bacteria in HA and LA ileum belong to Proteobacteria and Firmicutes, whereas in cecum, Bacterioidetes and Actinobacteria were predominant in both groups (P < 0.05). Correlation analysis showed that Sporosarcina, Enterococcus, and Lactococcus were strongly related to air pressure, and Peptoclostridium and Ruminococcaceae_UCG-014 are related to altitude and gut microbiota of LA group was influenced by altitude, while HA group affected by air pressure. Meanwhile, the Ruminococcus-torques-group was negatively correlated with the relative abundance of Paenibacillus, and positive correlated with those of other microorganisms. Furthermore, HA has higher abundance of microbiota involved in energy and glycan biosynthesis metabolism pathway, while LA has higher abundance of microbiota involved in membrane transport, signal transduction, and xenobiotics biodegradation and metabolism. Generally, our results suggested that the composition and diversity of gut microbes changed after Tibetan chickens were introduced to the plain. Tibetan chicken may adapt to new environment via reshaping the gut microbiota. Gut microbes may contribute to the host adaption to high altitude environments by increasing host energy and glycan biosynthesis.
Key words: Tibetan chicken, high-altitude adaption, gut microbiota, 16S rRNA, environment
INTRODUCTION
The Tibetan plateau is regarded as the third pole on earth for its extreme environments, such as low oxygen levels and high radiation, for the survival of humans and other mammalian species (Zhou et al., 2016; Guo et al., 2016). The native Tibetans and animals living in the Tibet plateau (more than 3,000 m above sea level) have adapted to the special plateau environment and have developed unique genetic predispositions, lifestyle, and dietary habits (Li et al., 2016, Wang et al., 2015). Tibetan chickens (Gallus gallus) have been well adapted to live in such harsh high-altitude environments (2,200–4,100 m) for thousands of years (Zhang et al., 2021, Zhang et al., 2021; Zhang et al., 2007). In most pastoral areas, Tibetan chickens primarily provide meat and eggs in conjunction with a high sodium intake as a Tibetan food source. In recent years, Tibetan chickens have been introduced to the plains area because of domestication demands to satisfy the inland market (Zhang et al., 2016; Ling et al., 2017).
Previous studies have indicated that gut microbes play an essential role in maintaining the stability of their internal environment, promoting growth performance and immunity of chickens (Yang et al., 2012; Bai et al., 2013). Many factors affect the composition of the gut microbiome in animals, such as age (Zhu et al., 2002), diet (Kohl, 2012), antibiotic feeding (Bueno et al., 2017), and altitude (Adak et al., 2013) and so on. It has been reported that cecal microflora diversity of Tibetan chickens from five high-altitude areas is different and the degree of similarity of the intestinal microbiota was correlated with geographic distance (Zhou et al., 2002). In addition, different gastrointestinal tracts of chickens play different roles in feed digestion (Wen et al., 2021), nutrient absorption (Yuan et al., 2020), immunity (Abaidullah et al., 2019), metabolism (Kong et al., 2020; Zhang et al., 2021), energy harvest (Yin et al., 2021), and intestinal health (Diaz et al., 2019). The Ileum is the main site of nutrient absorption. Cecum is not only helpful for absorbing nutrients (Partridge, 1978), but also helps to detoxify harmful substances and prevent pathogen colonization (Clench and Mathias, 1996; Gong et al., 2007; Yan et al., 2017), and the cecum is the main place for bacterial fermentation (McLelland, 1989).
The structural differences and related adaptive mechanisms of human gut microbiota at different geographical locations have been clarified gradually (Xu et al., 2015). However, little is known about the cecal and ileal microbiomes of Tibetan chickens. Previous studies indicated that the growth performance of chickens was improved when birds were introduced to the low altitude area (Yılmazdikmen et al., 2014). However, how the gut microbiota changes when Tibetan chickens are introduced into the plain is not clear. We hypothesized that changes in the gut microbiota in chickens may contribute to environmental adaption according to the establishment of numerous intestinal microflora. Thus, we compared the composition and diversity of gut microbiota in Tibetan chickens both at high altitudes and in plains to investigate the altitude-adaption of gut microbiota in Tibetan chickens.
MATERIALS AND METHODS
Ethics Statement
Animal management in this study was approved by the Institutional Animal Care and Use Committee of Sichuan Agricultural University with the permit number DKY-S20163638. All experiments were performed in strict accordance with the relevant guidelines of the Sichuan Agricultural University (SAU) Laboratory Animal Welfare and Ethics.
Experimental Design and Sampling
A total of twelve 180-day-old Tibetan chickens were used to collect ileal and cecal contents including 6 birds (3 males and 3 females) from high altitude (HA, Shannan, southern of Tibetan Plateau, 3,572 m) and other 6 (3 males and 3 females) from plain (LA, Sichuan Ya,an, western margin of Sichuan basin, 580 m), and the average air pressure data of the 2 places were obtained through China Meteorological Website (http://www.nmc.cn/publish/forecast/AXZ/linzhi.html, and http://haiba.qhdi.com). All chickens originated from Shannan, Tibet. The LA chickens were hatched on the farm of Sichuan Agricultural University in Ya'an by their parents transported from Shannan as fertilized eggs, and the HA chickens were hatched at the same time.
Chickens were raised on the indoor ground and covered with wood chips and rice husks with outdoor playgrounds. During the first week after hatching, the house temperature was maintained at 35°C to 36°C by a coal-fired boiler, and it was gradually reduced to 24°C from the second week. The chickens raised in high and low altitude farms were fed the same diets ad libitum. Diets (Table 1) were formulated based on the feeding standards for yellow-feather chickens in China (NY/T 33,2004). Birds can freely access water freely. The chicks were vaccinated with Marek's disease (d 1), H9 avian influenza (d 8), Newcastle disease (d 10), and H5 avian influenza (d 24) accordingly. Antibiotics were not administered during the feeding process. The body weights of the chickens were measured at 0, 2, 3, 4, and 6 wk.
Table 1.
Dietary formulation and composition.
| Ingredients, % | 1-28 d | 29-42 d | 43-180 d |
|---|---|---|---|
| Corn | 61.77 | 69.95 | 68.4 |
| Soybean | 29.4 | 22.10 | 23.80 |
| Soybean oil | 2.70 | 2.70 | 2.50 |
| Calcium hydrogen phosphate | 1.90 | 1.70 | 1.64 |
| Shell powder | 2.00 | 1.93 | 2.02 |
| Salt | 0.35 | 0.35 | 0.36 |
| Methionine | 0.18 | 0.10 | 0.08 |
| Lysine | 0.00 | 0.05 | 0.08 |
| Zeolite powder | 1.38 | 0.80 | 0.80 |
| Vitamin premix1 | 0.16 | 0.16 | 0.16 |
| Mineral premix2 | 0.16 | 0.16 | 0.16 |
| Total | 100.0 | 100.0 | 100.0 |
| Caculated analysis, % | |||
| Metabolizable energy, Kcal/kg | 2897.0 | 2998.0 | 3819.0 |
| Crude protein | 20 | 19 | 17 |
| Lysine | 0.89 | 0.87 | 0.58 |
| Methionine | 0.36 | 0.32 | 0.21 |
| Calcium, % | 1.18 | 0.85 | 0.72 |
| Total phosphorus, % | 0.65 | 0.62 | 0.5 |
| Non-phytate phosphorous, % | 0.41 | 0.38 | 0.26 |
Provided per kilogram of diet: 12,8000IU vitamin A, 1,600IU vitamin D3, 60IU vitamin E, 1.6 mg vitamin K3, 0.12 mg biotin, 50 mg choline, 1.2 mg folic acid, 32 mg nicotinic acid, 16 mg pantothenic acid,4.8 mg riboflavin, 2.4 mg thiamine (VB1), 3.2 mg vitamin B6, and 0.03 mg vitamin B12.
Provided per kilogram of diet: Mg, 79 mg as manganese oxide; Zn, 60 mg as zinc oxide; Cu, 100 mg as copper sulfate; Fe, 120 mg as iron sulfate; I, 0.96 mg as potassium iodine; and Se, 0.24 mg as sodium selenite.
We collected the contents from the whole ileum and cecum in each bird (HI = ileal content of HA chicken; HC = cecal content of HA chicken; LI = ileal content of LA chicken; LC = cecal content of LA chicken). Samples were wrapped in foil and immediately frozen in liquid nitrogen before they were transferred to the laboratory where the samples were stored at −80°C until DNA extraction.
DNA Extraction and Sequencing
Sixty milligrams of sample for each bird were used to extract the total bacterial DNA using the E.Z.N.A. stool DNA kit (Omega) according to the manufacturer's protocol. Extracted DNA was quantified using a NanoDrop ND-2000 spectrophotometer (Thermo Scientific) to assess the concentration and quality of the DNA. The integrity of purified the DNA was determined using 2% agarose gels in buffer (Tris/Boric acid/EDTA) and was observed using Universal Hood's Gel Imaging System. The qualified samples were stored at −80°C for further processing. Sequencing was performed using an Illumina HiSeq 2500 platform. Briefly, the V3 to V4 hypervariable regions of the 16S rRNA gene were amplified from each bacterial DNA sample using the primer pairs (338F: 5’- ACTCCTACGGGAGGCAGCAG -3’ and 533R 5’- GGACTACHVGGGTWTCTAAT -3’) combined with adapter sequences and barcode sequences (Yin et al., 2010). PCR amplification was performed in a total volume of 50 μL (including 10 μL Buffer, 0.2 μL Q5 High-Fidelity DNA Polymerase, 10 μL High GC Enhancer, 1 μL dNTP, 10 μM of each primer and 60 ng genomic DNA). The reactions were performed on Healforce T960 PCR (Likang, China) with the following amplification conditions: initial denaturation at 95°C for 5 min; followed by 25 cycles of denaturation at 95°C for 60 s, annealing at 50°C for 60 s, elongation at 72°C for 60 s; and final extension at 72°C for 7 min. The amplification products were gel purified, quantified, pooled, and sequenced according to the manufacturer's protocol.
Data Analysis and Statistics
Raw data were preprocessed and assigned to samples according to specific bar-coded primers. Paired-end sequencing reads were merged using FLASH (version v1.2.7) (Magoč and Salzberg, 2011). Read preprocessing included filtering of low-quality reads, removing contaminated adapters, undetermined nucleotides and low complexity using QIIME 1.8 (Caporaso et al., 2010). The resulting sequences were clustered into operational taxonomic units (OTUs) based on 97% sequence identity. Chimeric OTUs were detected and removed using UCHIME v4.2 (Edgar, 2017) and gained effective tags. Then representative sequences for each OTU were picked and aligned to different taxonomic level based on the Ribosomal Database Project classifier v2.2. For alpha and beta diversity analysis, the number of reads per sample was randomly subsampled to 54,698 reads. The alpha diversity index of Shannon and Inver-Simpson, and the beta diversity index of Jaccard and Bray-Curtis were calculated using Mothur software (Kozich et al., 2013). The beta diversity was visualized using principal coordinate analysis (PCoA) (Sakaki et al., 1994).
Bioinformatic and Statistical Analysis
The alpha and beta diversities were statistically tested with Wilcoxon-rank-sum analysis (Wilcoxon test), P < 0.05 was considered statistically significant. As no differences between the genders within the group (same altitude and intestinal segment) were significant (P > 0.05), hence, the gender effect was not considered in further analysis. In addition, T-test and Wilcox-test were used to investigate the differences in the microbial community abundance between the high and low altitude group using Metastats and R software.
Linear discriminant analysis effect size (LEfSe) was performed to analyze the statistical significance and biological relevance (Segata et al., 2011). PICRUSt analysis was used to predict the functional differences in the microbiota between the high and low altitude groups (Parks et al., 2014). The functional differences in the different groups were observed by KEGG metabolic pathways. Moreover, Canonical Correspondence analysis (CCA) was conducted to reflect the relationship between gut microbiota or sample and the environmental factors (Lindström et al., 2005).
RESULTS
Sequences summary
Totally, 1,908,162 raw reads were obtained from the 24 samples. After removing low quality reads and chimeras, a total of 1,423,855 high-quality sequences with an average of 59,327 reads per sample (ranging from 54,095 to 61,791) were used for downstream analysis. These sequences were assigned to 897 operational taxonomic units (OTUs) based on 97% sequence similarity. The sequencing depth index coverage estimates are summarized in Table 2. Sample evaluation showed a mean coverage of 99.87 ± 0.0004 % at 97% similarity level, indicating that the sequences generated from the sample were sufficient to define and compare the diversity within the sample.
Table 2.
Alpha-diversity indices of the bacterial communities.
| 97% similarity |
||||||
|---|---|---|---|---|---|---|
| Sample | No. of reads | OTUs | Chao1 | Inver simpson | Shannon | Coverage |
| HC-1 | 59,302 | 449 | 483.871 | 42.2074 | 4.3438 | 0.9991 |
| HC-2 | 58,247 | 408 | 485.1429 | 15.9477 | 3.5733 | 0.9985 |
| HC-3 | 61,497 | 524 | 576.5 | 63.8118 | 4.6674 | 0.9988 |
| HC-4 | 54,698 | 403 | 460.973 | 34.1970 | 4.0749 | 0.9985 |
| HC-5 | 57,004 | 454 | 503.7632 | 43.8651 | 4.3763 | 0.9988 |
| HC-6 | 58,612 | 476 | 515.90 | 21.2400 | 4.0809 | 0.9989 |
| HI-1 | 58,111 | 357 | 510.2703 | 5.9546 | 2.3252 | 0.9978 |
| HI-2 | 59,060 | 205 | 276.0789 | 3.5145 | 1.7428 | 0.9985 |
| HI-3 | 61,633 | 254 | 367.1351 | 3.2535 | 1.9709 | 0.9984 |
| HI-4 | 61,047 | 168 | 233.5556 | 1.8394 | 1.0154 | 0.9988 |
| HI-5 | 57,953 | 464 | 511.6341 | 5.3513 | 2.8719 | 0.9988 |
| HI-6 | 58,697 | 368 | 440.4043 | 2.5360 | 1.8012 | 0.9983 |
| LC-1 | 62,430 | 550 | 576.8966 | 41.4205 | 4.7924 | 0.9993 |
| LC-2 | 61,334 | 548 | 598.6471 | 37.6940 | 4.7299 | 0.9992 |
| LC-3 | 59,572 | 529 | 584.7143 | 45.7384 | 4.8045 | 0.9992 |
| LC-4 | 60,591 | 522 | 544.6579 | 35.3496 | 4.545 | 0.9992 |
| LC-5 | 61,875 | 546 | 566.027 | 59.1678 | 4.7398 | 0.9993 |
| LC-6 | 61,995 | 499 | 548.1707 | 24.3588 | 4.1391 | 0.9988 |
| LI-1 | 60,686 | 362 | 435.1 | 2.4048 | 1.8981 | 0.9984 |
| LI-2 | 62,298 | 575 | 620.0238 | 10.2628 | 3.6187 | 0.9989 |
| LI-3 | 62,488 | 621 | 652.3182 | 8.5493 | 3.5361 | 0.999 |
| LI-4 | 61,532 | 615 | 653.7692 | 33.3607 | 4.6514 | 0.9989 |
| LI-5 | 59,623 | 317 | 380.4054 | 1.5019 | 0.9418 | 0.9988 |
| LI-6 | 59,484 | 484 | 555.5 | 37.8631 | 4.0108 | 0.9978 |
The number of reads, OTUs, richness estimator Chao1, diversity estimator Shannon, and Inver simpson were calculated at the 97% similarity level. HC and HI = the cecum and ileum of birds living in high altitude; LC and LI = the cecum and ileum of birds living in low altitude.
Gut Microbiota Diversity of Tibetan chickens Between High and Low Altitude Groups
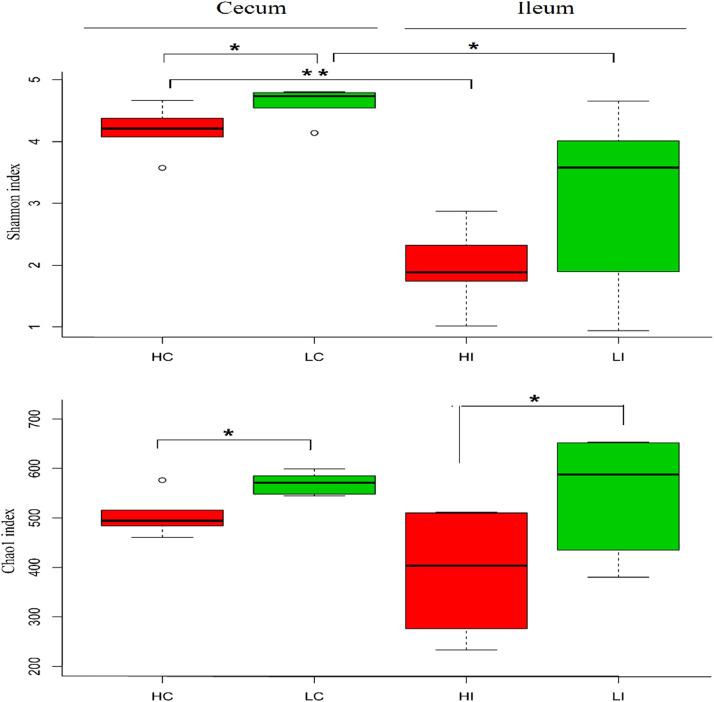
The gut microbial diversity was measured by Shannon, Chao1 (estimated OTUs) and the Inver-Simpson index (Table 2). The Chao1 index indicated that the LA group had significantly higher bacterial diversity than the HA group in both segments (P < 0.05, Figure 1). The Shannon index showed that the chicken cecum microbiota had higher community diversity than that of the ileum in both high and low altitude groups (P < 0.05, Figure 1), while no significant difference in community richness was observed.
Figure 1.
Microbial diversity and composition of cecum and ileum of chickens in HA and LA groups. HC and HI = the cecum and ileum of birds living in high altitude; LC and LI = the cecum and ileum of birds living in low altitude. *P < 0.05 and **P < 0.01.
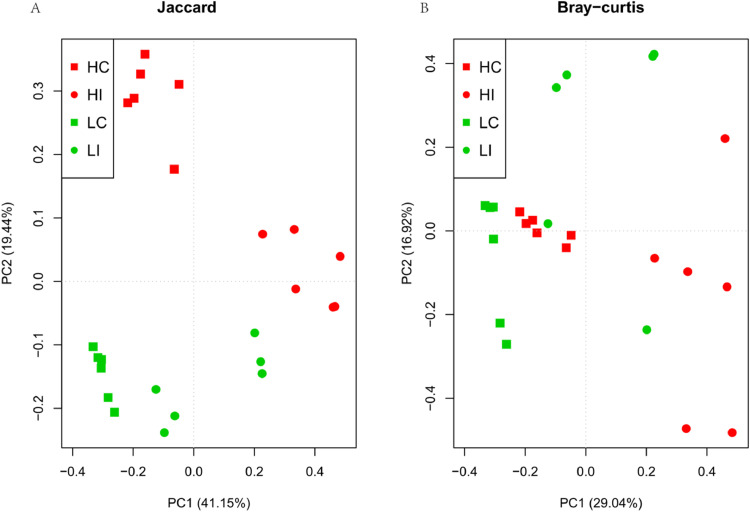
We further explored the dissimilarity between community structure and community membership by calculating the distance of Bray-Curtis and Binary-Jaccard indices, respectively. From the PCoA plot, clear separation of community membership was observed between the HA and LA groups (Figure 2A). In addition, community structure analysis showed that the cecum bacterial communities clustered tightly and were separated from the ileum communities along PC1 (Figure 2B), indicating that LC was highly similar to HC. Notably, the variation within the ileum communities was significantly higher than that within the cecum communities (P < 0.05, Wilcoxon-test).
Figure 2.
Principal Coordinate Analysis (PCoA) of the community membership (A) and structure (B) using Jaccard and Bray Curtis distances, respectively. Red squares and circles represent HA Tibetan chickens’ cecum and ileum bacterial communities, green squares and circles represent LA Tibetan chickens’ cecum and ileum bacterial communities, respectively. Distances between symbols on the ordination plot reflect relative dissimilarities in community memberships or structures.
The Gut Microbiota Composition is Associated with Altitude
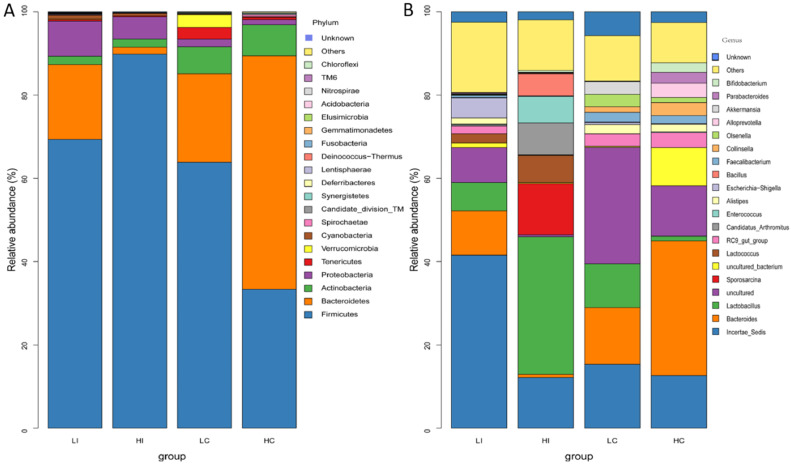
Analysis of Metastats indicated that the abundance of microbial communities between the LA and HA groups differed significantly (P < 0.05) at different microbial classification levels. The dominant phyla in the LA and HA groups were Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria (Figure 3A). The relative abundance of each taxon differed significantly between the HA and LA groups (P < 0.05). The diversities of Proteobacteria and Firmicutes in the ileum were higher than those in the cecum in both groups, while Bacteroidetes and Actinobacteria in the ileum were lower than those in the cecum in both groups (P < 0.05). The ratio of Firmicutes to Bacteroidetes (F/B) differed between the LI (3.86) and HI (53.19) groups, and between the LC (3.01) and HC (0.59) groups. At the genus level, Incertae Sedis was the most abundant genus followed by Bacteroides and Lactobacillus (Figure 3B).
Figure 3.
Relative abundance of top 20 OTUs at the phylum (A) and genus (B) level in ileal and cecal microbiota from HA and LA chickens. HC and HI = the cecum and ileum of birds living in high altitude; LC and LI = the cecum and ileum of birds living in low altitude.
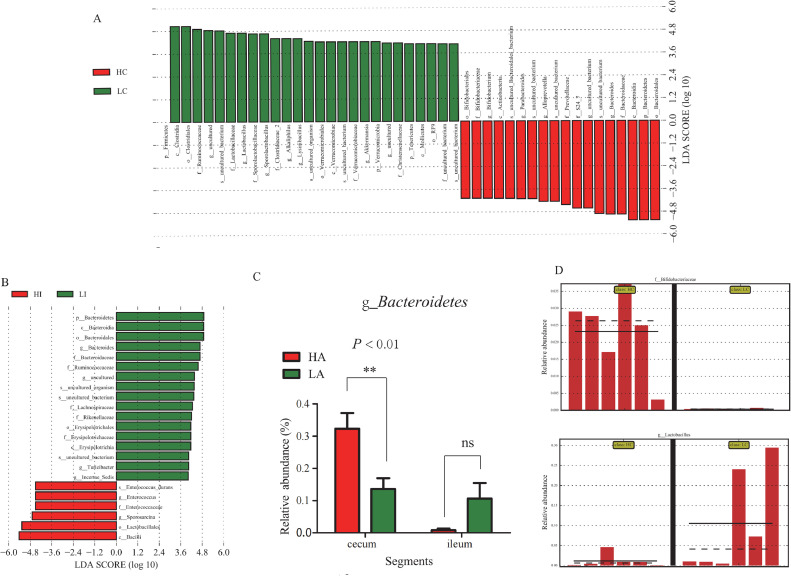
To identify the specific bacterial species that were distributed among the LC, LI, HC, and HI groups, we performed LEfSe analysis. The results showed that 45 bacterial species were significantly different between LC and HC (Figure 4A), and 23 differential taxa existed in LI and HI (Figure 4B). HC had higher abundances of Bacteroidaceae, Prevotellaceae, and Bifidobacteriaceae; LC was richer in Ruminococcaceae, Lactobacillaceae, Verrucomicrobiaceae, and Chirstensenellaceae; HI had a higher abundance of Enterococcaceae, and LI had a higher abundance of Bacteroidaceae, Ruminococcaceae, and Lachonspiraceae. The distribution of representative Bacteroidetes microbes is illustrated in Figure 4C. In cecum, relative abundance of Bifidobacteriaceae in HA was higher than LA group (P < 0.05, Figure 4D), but relative abundance of Lactobacillus was higher in LA group (P < 0.05, Figure 4D).
Figure 4.
Taxon's abundances between cecum and ileum of Tibetan chickens identified by linear discriminant analysis (LDA) coupled with effect size (LDA > 4) were shown in Part A and B (P < 0.05). The relative abundance of Bacteroidetes at genus level (g_Bacteroidetes) was compared among the four groups, shown in Part C. Microbiota abundances between different altitude birds was shown in Part D; some differentially distributed bacteria: e. g., f_Bifidobacterium and g_Lactobacillus were illustrated in Part D, respectively.
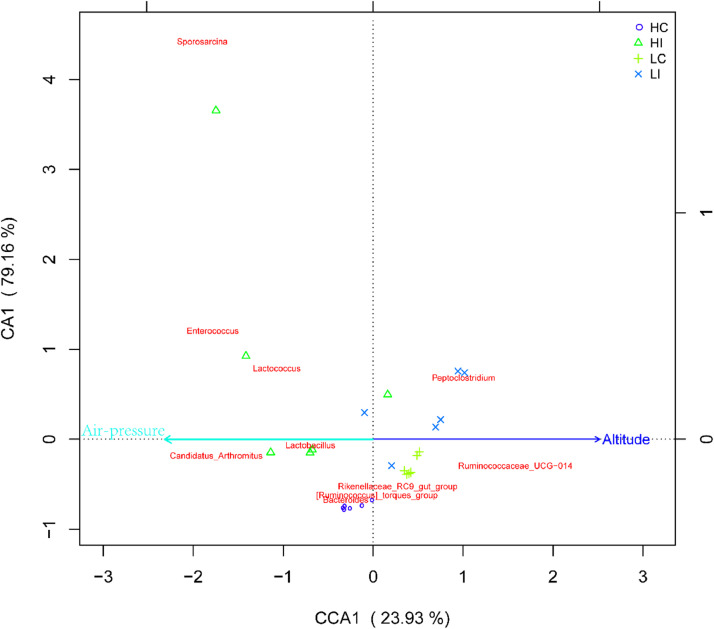
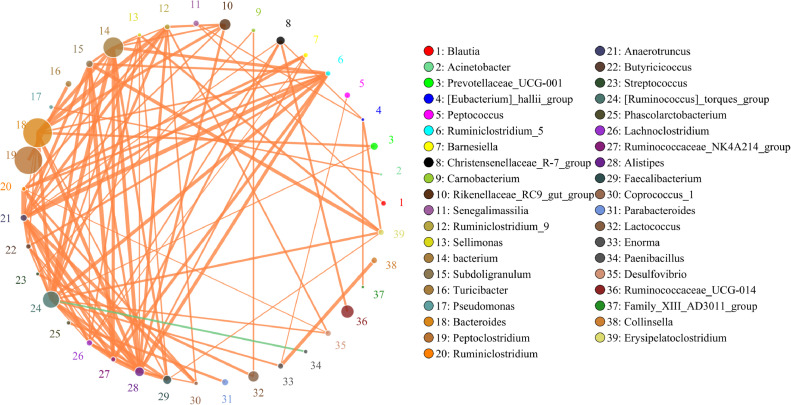
Additionally, Canonical Correspondence analysis (CCA) was used to reflect the relationship between the gut microbiota and environmental factors (Figure 5). The top ten microbiota species at the genus level were Sporosarcina, Enterococcus, Lactococcus, Peptoclostridium, Lactobacillus, Candidatus_Arthromitus, Ruminococcaceae_UCG-014, Rikenellaceae_RC9_gut_group, Ruminococcus_torques_group, Bacteroides from HC, HI, LC, and LI were considered here. The results indicated that Sporosarcina, Enterococcus, and Lactococcus were strongly related to air pressure, and Peptoclostridium and Ruminococcaceae_UCG-014 are related to altitude. Meanwhile the microbiota from HC and HI were mainly related to air pressure, whereas microbiota from LC and LI were mainly related to altitude. Based on the abundance and variation of each species, correlation analysis was calculated and shown in Figure 6, including positive correlation and negative correlation. At the genus level, Bacteroides, Peptoclostridium and Ruminococcusou had high expression abundance and were more correlated with other microbiota species.
Figure 5.
Canonical correspondence analysis of intestinal microbiota from Tibetan chickens responding to environmental factors. The scales on the horizontal and vertical coordinates were the values produced by sample or species when performing regression analysis with environmental factors; the dots represented samples; the red font indicated the top ten microbiota species at genus level; the blue and green arrows represent environmental factors, altitude and air pressure, respectively.
Figure 6.
Network diagram is a form of correlation analysis at the genus level (r > 0.1). Circle represents the species, circle size represents the expression abundance of species; lines represent the correlation between the two species, line thickness represents the strength of the correlation, line color: orange represents positive correlation, green represents negative correlation.
Predicted Metagenome Variation and Body Weight Between the LA and HA Groups
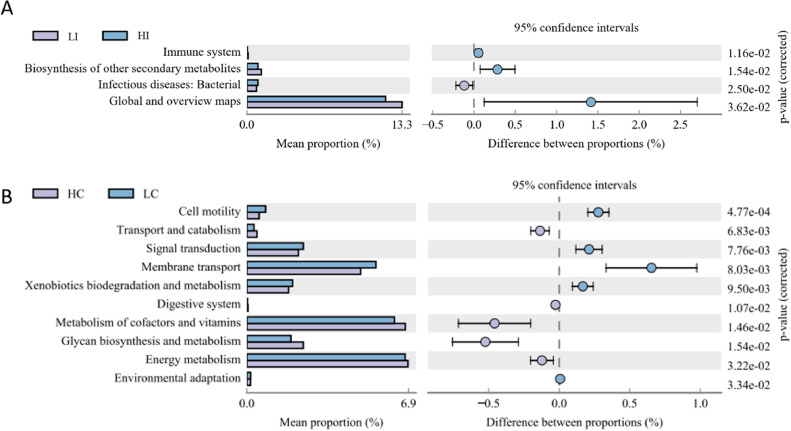
We used PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States), a predictive tool to predict the functional profiles of Tibetan gut microbiota from (KEGG) pathways. Comparison among groups indicated there were At KEGG level 2 and 4 KEGG pathways in the ileum and 10 pathways in the cecum in the high and low altitude groups, respectively (Figure 7). The ileal microbiota plays a key role in secondary metabolism, biosynthesis, and immunity, while the cecal microbiota contributes to membrane transport, signal transduction, energy metabolism, environmental adaptation, signaling transduction, and metabolism of cofactors and vitamins. In the ileum and cecum, the most significantly different functions, such as the function related to the immune system, transport and catabolism, metabolism of cofactors and vitamins, glycan biosynthesis and metabolism and energy metabolism, were enriched in the HA group compared with the LA group (P < 0.05).
Figure 7.
Analysis of the differences in KEGG metabolic pathways between groups at class level. The left side of the graph indicated the ratios of abundance of samples LI to HI (Upper), and of samples HC to LC (Lower) in different functions. The right side of the graph shows the proportions of functional abundance in 95% confidence interval, respectively.
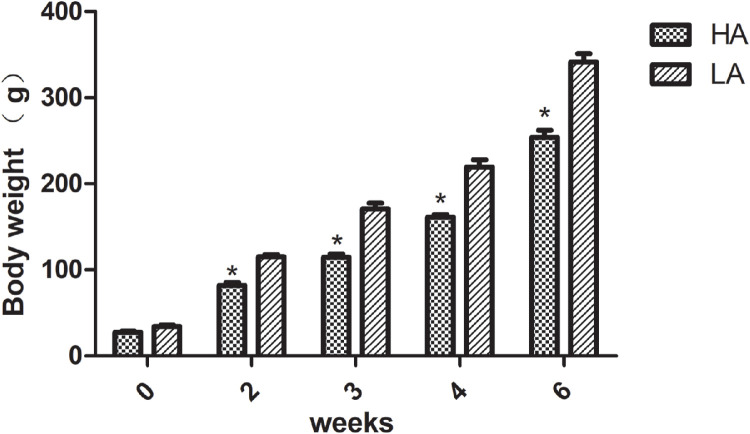
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) analyses revealed significant differences in microbiota metabolite pathways due to the genera of the antibiotic-responsive microbes (P < 0.01), especially the pathways related to cell growth and death, immune system diseases, carbohydrate metabolism, and nucleotide metabolism. Body weight is shown in Figure 8. There were no obvious differences between two groups in 0 wk. However, the body weight in the HA group were lower (P < 0.05) than those in the LA group in 2, 3, 4, and 6 wk.
Figure 8.
The body weight of the chickens in HA and LA groups were measured at 0, 2, 3, 4, and 6 weeks.*Difference in two groups (P<0.05).
DISCUSSION
It is well-known that the compositional diversity of the gut microbiota plays an important role in host environmental adaptation (Zhou et al., 2016; Zneg et al., 2017; Zhao et al., 2018). Much attention has been paid to the gut microbial communities in high altitude animals because of their unique living surroundings (Zhang et al., 2018; Zhao et al., 2018). Li et al. (2016) found that the Tibetan herders living at 4,800 m were enriched in butyrate-producing bacteria to adapt to high-altitude environments, compared with rural Tibetans living at an altitude of 3,600 m altitude. Zhang et al. (2018) reported that the community composition and structure of the lizard intestinal microbiota varied with altitude, and the proportion of Bacteroides increased significantly with altitude. In our results, we found that the Tibetan chicken gut microbiota diversity in the LA group was significantly higher than those in the HA group. Reports indicated that people living in high and low areas have dominant bacterial community at the phylum level (Li and Zhao, 2015). Similarly, our results showed that the microbial diversity in Tibetan chickens was significantly lower at high altitudes than at low altitudes similar to observations in humans and pigs (Zeng et al., 2017). We deduced that this may be due to the better climate with the change from high to low altitude. Similar observations have also been reported in lake water, which is the average taxon richness of bacteria in Tibetan lakes is relatively low compared with that in reference lakes at low altitudes (Xing et al., 2009). Moreover, to our knowledge, this is the first study to explore the Tibetan chicken gut microbiota patterns when it is introduced to low altitude environments from high altitudes.
We also observed that both gut segments (cecum and ileum) of chickens possess distinct microbiota composition along with altitude. The relative abundance of the phylum Firmicutes in the ileum of Tibetan chickens tended to increase with altitude. However, this phenomenon was not observed in the cecum. More detail indicated that the abundance of Bacteroides in the cecum was higher than that in the ileum at the genus level, and group HC had higher Bacteroides abundance than that of the LC group. This indicates that the cecum plays a more important role in fermentation than the ileum, which is related to the anatomical structure of the cecum. The fermentative ability of HA chicken was stronger than that of LA chicken. A previous study has shown that a higher ratio of Firmicutes to Bacteroidetes in a microbial community is correlated with higher absorption of food energy (Turnbaugh et al., 2006; Murphy et al., 2010). Many studies have indicated that Bacteroidetes as a probiotic improves host nutrient utilization and immunity by increasing polysaccharide decomposition (Hooper, 2004; Bai et al., 2013; Lee and Hase, 2014). In the present study, we further found that the HC group had a higher phylum Bacteroidetes abundance than the Firmicutes group. This may be helpful for Tibetan chickens to adapt to harsh environments. The Tibet rhesus macaques had higher abundances of Firmicutes and lower abundances of Bacteroide (Zhao et al., 2018). Nevertheless, compared with LI, the higher F/B ratio in the HI chicken group was probably due to environmental factors, since both groups had the same diets (corn - bean meal). However, a study also indicated that a high ratio of Firmicutes to Bacteroidetes in response to a high-fat diet did not affect energy harvesting (Murphy et al., 2010). Additionally, the dominant phyla Firmicutes and Bacteroidetes in the gut of Tibetan chickens accounted for over 85% of the 16S rRNA gene sequences, which may help to facilitate diet fermentation and degrade carbohydrates and proteins efficiently, according to previous reports (Ley et al., 2006; Jami et al., 2014; Waite and Taylor, 2014).
Previous studies reported that some Paenibacillus, such as P. macerans, P. polymyxa, and P. azoctoria, were rhizosphere dominant microorganism (Guemouriathmani et al., 2000; Von et al., 2000). They affect the promotion and inhibition of plant diseases (Bae et al., 2010). Paenibacillus spp have significant antimicrobial activity and can improve skin hydration (Reuter, 2001). Thus, we deduced that paenibacillus is likely to be a candidate microbial label indicating intestinal homeostasis.
Considering the altitude difference in chicken living areas, we conducted a LEfSe analysis and found a few distinct flora at the genus level, such as Bacteroides, Bifidobacterium, Sporelactobacillus, and Lactobacillus in the cecum, and Bacteroidetes and Enterococcus in the ileum. Studies have shown that Bifidobacterium, Sporelactobacillus, and Lactobacillus regulate gastrointestinal probiotics (Mountzouris et al., 2007; Sanders et al., 2003), and Sporelactobacillus are used as probiotics in commercial production. Bacteroides are associated with the degradation of cellulose and organic matter, while Enterococcus produces bacteriostatic substances and improves the intestinal microenvironment (Rojobezares et al., 2006; Kaakoush, 2015). This indicates that when environmental factors change, such as altitude variation, the intestinal micro-ecology adjusts the microenvironment by regulating microorganisms (flora and abundance), so that the organism can adapt to the environment better. This may indicate that birds living in high altitude areas have specific microbial species that make them adapt to harsh environments.
Furthermore, hypobaric pressure and high altitude, which are typical features of high-altitude regions have been proven to be important factors that strongly modulate the composition of gut microflora in mammals (Maity et al., 2012). Our results showed that hypoxia changed intestinal microbial composition in Tibetan chickens. An increase in the fraction of Lactobacillus and a decrease in the proportion of Peptoclostridium were accompanied by altitude variation. This phenomenon is similar to a previous study in which Lactobacillus was slightly affected by altered air pressures (Li et al., 2013). Bacteroides was correlated with air pressure, which was in accordance with a previous report that air pressure mainly affects the intestinal microbiota in animals at relatively high altitudes, and the gut microbes were mainly influenced by increased altitude (Zhang et al., 2018).
We also found that the microbial function of the cecum in high-altitude group chickens was related to glycan biosynthesis metabolism, environmental adaptation, transport and catabolism, and energy metabolism. Specifically, the functional enrichment of energy and glycan biosynthesis metabolism in the high-altitude group was higher than that of the low altitude groups, suggesting that the gut microbiota may associated with the altitude adaptation of the host. Energy metabolism-related genes that help the host adapt to the high-altitude environment in ruminants have been studied (Qiu et al., 2012; Li et al., 2013). Li et al. (2013) reported that genes related to energy metabolism and oxygen transmission can associated with the high-altitude adaptions of Tibetan antelope and Tibetan wild boars. Thus, we may deduce that the body regulates metabolism through intestinal microbes to adapt to environmental changes. Higher glycan biosynthesis metabolism was beneficial for high-altitude Tibetan chickens to survive in extreme environments (e.g., against cold, hypoxia etc.) by increasing feedstuff energy conversion rates (Xu et al., 2016).
CONCLUSION
In general, the diversity and composition of gut microbiota changed after Tibetan chickens were introduced to the flatlands. The distribution of bacterial genera was correlated with environmental factors. At the genus level, Bacteroides, Peptoclostridium and Ruminococcusou had high expression abundance and were more correlated with other microbiota species. Tibetan chickens gained weight faster on the flatlands than on the plateau. Gut microbes may contribute to the adaptation to high altitude environments in Tibetan chickens by increasing host energy and glycan biosynthesis. The findings of this study provide valuable information for understanding the adaptability of Tibetan chicken to the environment.
Acknowledgments
Acknowledgments
We would like to thank the financial support provided by the Modern Agricultural Technology System of Ministry of Agriculture and Rural Areas (Grant No. CARS-41), National Natural Science Foundation of China (Grant No. 31872347), the Key Research and Development Plan of the Department of Science and Technology of Tibet Autonomous Region (XZ202101ZY0002N), the Local Projects Guided by the Central Government from Razi County, Tibet Autonomous Region, the Projects Funded by the Central Government to Guide Local Scientific and Technological Development from Guizhou province [QIANKEZHONGYINDI (2021)4003], and Bazhong Municipal Government and University Cooperation Project Breeding, Development and Healthy Raising of Meihua chicken in Bazhong for funding this work.
Disclosures
All authors have no conflict of interest to declare.
REFERENCES
- Abaidullah M., Peng S., Kamran M., Song X., Yin Z. Current findings on gut microbiota mediated immune modulation against viral diseases in chicken. Viruses. 2019;25:681. doi: 10.3390/v11080681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adak A., Maity C., Ghosh K., Pati B.R., Mondal K.C. Dynamics of predominant microbiota in the human gastrointestinal tract and change in luminal enzymes and immunoglobulin profile during high-altitude adaptation. Folia. Microbiol. 2013;58:523–528. doi: 10.1007/s12223-013-0241-y. [DOI] [PubMed] [Google Scholar]
- Bae J.Y., Kim K.Y., Kim J.H., Lee K., Cho J.C., Cha C.J. Paenibacillus aestuarii sp. nov., isolated from an estuarine wetland. Int. J. Syst. Evol. Microbiol. 2010;60:644–647. doi: 10.1099/ijs.0.011544-0. [DOI] [PubMed] [Google Scholar]
- Bai S.P., Wu A.M., Ding X.M., Lei Y., Bai J., Zhang K.Y., Chio J.S. Effects of probiotic-supplemented diets on growth performance and intestinal immune characteristics of broiler chickens. Poult. Sci. 2013;92:663–670. doi: 10.3382/ps.2012-02813. [DOI] [PubMed] [Google Scholar]
- Bueno I., Williams J.N., Hwang H., Sargeant J.M., Nault A.J., Singer R.S. Impact of point sources on antibiotic resistance genes in the natural environment: a systematic review of the evidence. Anim. Health. Res. Rev. 2017;18:112–127. doi: 10.1017/S146625231700007X. [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clench M.H., Mathias J.R. Myoelectric activity of the cecum in fed and fasted domestic fowl (Gallus sp.) Comp. Biochem. Physiol. A. Physiol. 1996;115:253–257. doi: 10.1016/0300-9629(96)00056-4. [DOI] [PubMed] [Google Scholar]
- Diaz C.J.M., Casanova N.A., Fernández M.M.E. Microbiota, gut health and chicken productivity: what is the connection? Microorganisms. 2019;7:374. doi: 10.3390/microorganisms7100374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. SEARCH_16S: A new algorithm for identifying 16S ribosomal RNA genes in contigs and chromosomes. BioRxiv. 2017 [Google Scholar]
- Gong J., Si W., Forster R.J., Huang R., Yu H., Yin Y., Yang C., Han Y. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS. Microbiol. Ecol. 2007;59:147–157. doi: 10.1111/j.1574-6941.2006.00193.x. [DOI] [PubMed] [Google Scholar]
- Guemouri-Athmani S., Berge O., Bourrain M., Mavingui P., Thiéry J.M., Bhatnagar T., Heulin T. Diversity of paenibacillus polymyxa populations in the rhizosphereof wheat (triticum durum) in algerian soils. Euro. J. Soil Biol. 2000;36:149–159. [Google Scholar]
- Guo Y., Shen C., Meng H., Dong Q., Kong T., Yang C., Wang H., Jin R., Zhu B. Population Differentiations and Phylogenetic Analysis of Tibet and Qinghai Tibetan Groups Based on 30 InDel Loci. DNA Cell Biol. 2016;35:787–794. doi: 10.1089/dna.2016.3395. [DOI] [PubMed] [Google Scholar]
- Hooper L.V. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004;12:129–134. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Jami E., White B.A., Mizrahi I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS One. 2014;9:e85423. doi: 10.1371/journal.pone.0085423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush N.O. Insights into the Role of Erysipelotrichaceae in the Human Host. Front. Cell. Infect. Microbiol. 2015;5:84. doi: 10.3389/fcimb.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl K.D. Diversity and function of the avian gut microbiota. J. Comp. Physiol. B. 2012;182:591–602. doi: 10.1007/s00360-012-0645-z. [DOI] [PubMed] [Google Scholar]
- Kong A., Zhang C., Cao Y., Cao Q., Liu F., Yang Y., Zong T., Rehman M.U., Wang X., Huang S. The fungicide thiram perturbs gut microbiota community and causes lipid metabolism disorder in chickens. Ecotoxicol Environ Saf. 2020;206 doi: 10.1016/j.ecoenv.2020.111400. [DOI] [PubMed] [Google Scholar]
- Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.J., Hase K. Gut microbiota-generated metabolites in animal health and disease. Nat. Chem. Biol. 2014;10:416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- Ley R.E., Peterson D.A., Gordon J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Li K., Dan Z., Gesang L., Wang H., Zhou Y., Du Y., Ren Y., Shi Y., Nie Y. Comparative analysis of gut microbiota of native tibetan and han populations living at different altitudes. PLoS One. 2016;11 doi: 10.1371/journal.pone.0155863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhao X. Comparative analyses of fecal microbiota in Tibetan and Chinese Han living at low or high altitude by barcoded 454 pyrosequencing. Sci. Rep. 2015;5:14682. doi: 10.1038/srep14682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Tian S., Jin L., Zhou G., Li Y., Zhang Y., Wang T., Yeung C.K., Chen L., Ma J., Zhang J., Jiang A., Li J., Zhou C., Zhang J., Liu Y., Sun X., Zhao H., Niu Z., Lou P., Xian L., Shen X., Liu S., Zhang S., Zhang M., Zhu L., Shuai S., Bai L., Tang G., Liu H., Jiang Y., Mai M., Xiao J., Wang X., Zhou Q., Wang Z., Stothard P., Xue M., Gao X., Luo Z., Gu Y., Zhu H., Hu X., Zhao Y., Plastow G.S., Wang J., Jiang Z., Li K., Li N., Li X., Li R. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 2013;45:1431–1438. doi: 10.1038/ng.2811. [DOI] [PubMed] [Google Scholar]
- Lindström E.S., Kamst M.P., Zwart G. Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water retention time. Appl. Environ. Microbiol. 2005;71:8201–8206. doi: 10.1128/AEM.71.12.8201-8206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Z., Jiang C., Duan X., Yang X., Qin H., Dan L. Incubation effect on eggs of tibetan chicken under low altitude condition. Anim. Husbandry Feed Sci. 2017;04:206–208. [Google Scholar]
- Magoč T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity C., Adak A., Pathak T.K., Pati B.R., Chandra M.K. Study of the cultivable microflora of the large intestine of the rat under varied environmental hyperbaric pressures. J. Microbiol. Immunol. Infect. 2012;45:281–286. doi: 10.1016/j.jmii.2011.12.002. [DOI] [PubMed] [Google Scholar]
- McLelland J. Anatomy of the avian cecum. J. Exp. Zool. Suppl. 1989;3:2–9. doi: 10.1002/jez.1402520503. [DOI] [PubMed] [Google Scholar]
- Mountzouris K.C., Tsirtsikos P., Kalamara E., Nitsch S., Schatzmayr G., Fegeros K. Evaluation of the efficacy of a probiotic containing Lactobacillus, Bifidobacterium, Enterococcus, and Pediococcus strains in promoting broiler performance and modulating cecal microflora composition and metabolic activities. Poult. Sci. 2007;86:309–317. doi: 10.1093/ps/86.2.309. [DOI] [PubMed] [Google Scholar]
- Murphy E.F., Cotter P.D., Healy S., Marques T.M., O'Sullivan O., Fouhy F., Clarke S.F., O'Toole P.W., Quigley E.M., Stanton C., Ross P.R., O'Doherty R.M., Shanahan F. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59:1635–1642. doi: 10.1136/gut.2010.215665. [DOI] [PubMed] [Google Scholar]
- Parks D.H., Tyson G.W., Hugenholtz P., Beiko R.G. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge I.G. Studies on digestion and absorption in the intestines of growing pigs: 3. Net movements of mineral nutrients in the digestive tract. Br. J. Nutr. 1978;39:527–537. doi: 10.1079/bjn19780068. [DOI] [PubMed] [Google Scholar]
- Qiu Q., Zhang G., Ma T., Qian W., Wang J., Ye Z., Cao C., Hu Q., Kim J., Larkin D.M., Auvil L., Capitanu B., Ma J., Lewin H.A., Qian X., Lang Y., Zhou R., Wang L., Wang K., Xia J., Liao S., Pan S., Lu X., Hou H., Wang Y., Zang X., Yin Y., Ma H., Zhang J., Wang Z., Zhang Y., Zhang D., Yonezawa T., Hasegawa M., Zhong Y., Liu W., Zhang Y., Huang Z., Zhang S., Long R., Yang H., Wang J., Lenstra J.A., Cooper D.N., Wu Y., Wang J., Shi P., Wang J., Liu J. The yak genome and adaptation to life at high altitude. Nat. Genet. 2012;44:946–949. doi: 10.1038/ng.2343. [DOI] [PubMed] [Google Scholar]
- Reuter G. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2001;2:43–53. [PubMed] [Google Scholar]
- Rojo-Bezares B., Sáenz Y., Poeta P., Zarazaga M., Ruiz-Larrea F., Torres C. Assessment of antibiotic susceptibility within lactic acid bacteria strains isolated from wine. Int. J. Food Microbiol. 2006;111:234–240. doi: 10.1016/j.ijfoodmicro.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Sakaki T., Takeshima T., Tominaga M., Hashimoto H., Kawaguchi S. Recurrence of ICA-PCoA aneurysms after neck clipping. J. Neurosurg. 1994;80:58–63. doi: 10.3171/jns.1994.80.1.0058. [DOI] [PubMed] [Google Scholar]
- Sanders M.E., Morelli L., Tompkins T.A. Sporeformers as human probiotics: bacillus, sporolactobacillus, and brevibacillus. Compr. Rev. Food Sci. Food Saf. 2003;2:101–110. doi: 10.1111/j.1541-4337.2003.tb00017.x. [DOI] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome. Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Von der Weid I., Paiva E., Nóbrega A., Van Elsas J.D., Seldin L. Diversity of Paenibacillus polymyxa strains isolated from the rhizosphere of maize planted in Cerrado soil. Res. Microbiol. 2000;151:369–381. doi: 10.1016/s0923-2508(00)00160-1. [DOI] [PubMed] [Google Scholar]
- Waite D.W., Taylor M.W. Characterizing the avian gut microbiota: membership, driving influences, and potential function. Front. Microbiol. 2014;5:223. doi: 10.3389/fmicb.2014.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.S., Li Y., Peng M.S., Zhong L., Wang Z.J., Li Q.Y., Tu X.L., Dong Y., Zhu C.L., Wang L., Yang M.M., Wu S.F., Miao Y.W., Liu J.P., M.Irwin D., Wang W., Wu D.D., Zhang Y.P. Genomic analyses reveal potential independent adaptation to high altitude in Tibetan chickens. Mol. Biol. Evol. 2015;32:1880–1889. doi: 10.1093/molbev/msv071. [DOI] [PubMed] [Google Scholar]
- Wen C., Yan W., Mai C., Duan Z., Zheng J., Sun C., Yang N. Joint contributions of the gut microbiota and host genetics to feed efficiency in chickens. Microbiome. 2021;9:126. doi: 10.1186/s40168-021-01040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing P., Hahn M.W., Wu Q.L. Low taxon richness of bacterioplankton in high-altitude lakes of the eastern tibetan plateau, with a predominance of Bacteroidetes and Synechococcus spp. Appl. Environ. Microbiol. 2009;75:7017–7025. doi: 10.1128/AEM.01544-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Xu W., Li J., Dai L., Xiong C., Tang X., Yang Y., Mu Y., Zhou J., Ding J., Wu Q., Huang Z. Metagenomic analysis of the Rhinopithecus bieti fecal microbiome reveals a broad diversity of bacterial and glycoside hydrolase profiles related to lignocellulose degradation. BMC Genom. 2015;16:174. doi: 10.1186/s12864-015-1378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Yang H., Zhang L., Su Y., Shi D., Xiao H., Tian Y. High-throughput sequencing technology to reveal the composition and function of cecal microbiota in Dagu chicken. BMC Microbiol. 2016;16:259. doi: 10.1186/s12866-016-0877-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Sun C., Yuan J., Yang N. Gut metagenomic analysis reveals prominent roles of Lactobacillus and cecal microbiota in chicken feed efficiency. Sci. Rep. 2017;7:45308. doi: 10.1038/srep45308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.M., Cao G.T., Ferket P.R., Liu T.T., Zhou L., Zhang L., Xiao Y.P., G.Chen A. Effects of probiotic, Clostridium butyricum, on growth performance, immune function, and cecal microflora in broiler chickens. Poult. Sci. 2012;91:2121–2129. doi: 10.3382/ps.2011-02131. [DOI] [PubMed] [Google Scholar]
- Yılmazdikmen B., Şahan Ü., İpek A., Aydın C., Kederli E. Effect of oxygen supplementation in a hatchery at high altitude and growth performance of broilers reared at low altitude. South African J. Anim. Sci. 2014;44:350–359. [Google Scholar]
- Yin C., B.Xia S.Tang, Cao A., Liu L., Zhong R., Chen L., Zhang H. The effect of exogenous bile acids on antioxidant status and gut microbiota in heat-stressed broiler chickens. Front. Nutr. 2021;8 doi: 10.3389/fnut.2021.747136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Lei F., Zhu L., Li S., Wu Z., Zhang R., Gao G.F., Zhu B., Wang X. Exposure of different bacterial inocula to newborn chicken affects gut microbiota development and ileum gene expression. ISME J. 2010;4:367–376. doi: 10.1038/ismej.2009.128. [DOI] [PubMed] [Google Scholar]
- Yuan Z., Yan W., Wen C., Zheng J., Yang N., Sun C. Enterotype identification and its influence on regulating the duodenum metabolism in chickens. Poult. Sci. 2020;99:1515–1527. doi: 10.1016/j.psj.2019.10.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng B., Zhao J., Guo W., Zhang S., Hua Y., Tang J., Kong F., Yang X., Fu L., Liao K., Yu X., Chen G., Jin L., Shuai S., Yang J., Si X., Ning R., Mishra S., Li Y. High-altitude living shapes the skin microbiome in humans and pigs. Front. Microbiol. 2017;8:1929. doi: 10.3389/fmicb.2017.01929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Su W., Zhang B., Ling Y., Kim W.K., Zhang H. Comprehensive analysis of coding and non-coding RNA transcriptomes related to hypoxic adaptation in Tibetan chickens. J. Anim. Sci. Biotechnol. 2021;12:60. doi: 10.1186/s40104-021-00582-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wu C.X., Chamba Y., Ling Y. Blood characteristics for high altitude adaptation in Tibetan chickens. Poult. Sci. 2007;86:1384–1389. doi: 10.1093/ps/86.7.1384. [DOI] [PubMed] [Google Scholar]
- Zhang T., Ding H., Chen L., Yin L., Gong Y., Pan Z., Zhang G., Xie K., Dai G., Wang J. Antibiotic-Induced dysbiosis of microbiota promotes Chicken lipogenesis by altering metabolomics in the cecum. Metabolites. 2021;11:487. doi: 10.3390/metabo11080487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Gou W., Wang X., Zhang Y., Ma J., Zhang H., Zhang Y., Zhang H. Genome Resequencing Identifies Unique Adaptations of Tibetan Chickens to Hypoxia and High-Dose Ultraviolet Radiation in High-Altitude Environments. Genome. Biol. Evol. 2016;8:765–776. doi: 10.1093/gbe/evw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Li N., X.Tang N.Liu, Zhao W. Changes in intestinal microbiota across an altitudinal gradient in the lizard Phrynocephalus vlangalii. Ecol. Evol. 2018;8:4695–4703. doi: 10.1002/ece3.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Yao Y., Li D., Xu H., Wu J., Wen A., et al. Characterization of the gut microbiota in six geographical populations of Chinese rhesus macaques (Macaca mulatta), implying an adaptation to high-altitude environment. Microb. Ecol. 2018;76:565–577. doi: 10.1007/s00248-018-1146-8. [DOI] [PubMed] [Google Scholar]
- Zhou X., Jiang X., Yang C., Ma B., Lei C., Xu C., Zhang A., Yang X., Xiong Q., Zhang P., Men S., Xiang R., Wang H. Cecal microbiota of Tibetan chickens from five geographic regions were determined by 16S rRNA sequencing. Microbiologyopen. 2016;5:753–762. doi: 10.1002/mbo3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.Y., Zhong T., Pandya Y., Joerger R.D. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 2002;68:124–137. doi: 10.1128/AEM.68.1.124-137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]