Abstract
The structures of prion protein (PrP)–based mammalian prions have long been elusive. However, cryo-EM has begun to reveal the near-atomic resolution structures of fully infectious ex vivo mammalian prion fibrils as well as relatively innocuous synthetic PrP amyloids. Comparisons of these various types of PrP fibrils are now providing initial clues to structural features that correlate with pathogenicity. As first indicated by electron paramagnetic resonance and solid-state NMR studies of synthetic amyloids, all sufficiently resolved PrP fibrils of any sort (n > 10) have parallel in-register intermolecular β-stack architectures. Cryo-EM has shown that infectious brain-derived prion fibrils of the rodent-adapted 263K and RML scrapie strains have much larger ordered cores than the synthetic fibrils. These bona fide prion strains share major structural motifs, but the conformational details and the overall shape of the fibril cross sections differ markedly. Such motif variations, as well as differences in sequence within the ordered polypeptide cores, likely contribute to strain-dependent templating. When present, N-linked glycans and glycophosphatidylinositol (GPI) anchors project outward from the fibril surface. For the mouse RML strain, these posttranslational modifications have little effect on the core structure. In the GPI-anchored prion structures, a linear array of GPI anchors along the twisting fibril axis appears likely to bind membranes in vivo, and as such, may account for pathognomonic membrane distortions seen in prion diseases. In this review, we focus on these infectious prion structures and their implications regarding prion replication mechanisms, strains, transmission barriers, and molecular pathogenesis.
Keywords: prion, prion disease, cryo-EM, amyloid, protein aggregation, protein structure, neurodegenerative disease, infectious disease, N-linked glycosylation, glycosylphosphatidylinositol anchor
Abbreviations: 4RβS, 4-rung β-solenoid; aRML, anchorless RML; GPI, glycophosphatidylinositol; PIRIBS, parallel in-register intermolecular β-sheet/stack; ssNMR, solid state NMR
Self-propagating prions cause fatal and transmissible spongiform encephalopathies in mammals. Classical transmissible spongiform encephalopathy or prion diseases involve the conversion of the host’s normal cellular PrP (PrPC) to aberrant infectious aggregates that have generically been called PrPSc (1). However, multiple abnormal PrP structures have been observed both in vivo and in vitro, and a key challenge is to determine which are relevant to disease, that is, infectious and/or toxic (2). Here, we will use the term PrPd to refer to any disease-associated form of PrP regardless of its infectivity or pathogenicity (3). When histologically stained in infected brain tissue, PrPd deposits can range from small, diffuse deposits to much larger extracellular amyloid plaques, with the relative amounts and distributions of such deposits being dependent on prion strain and host type (reviewed in (3)). For example, some prion diseases result in predominantly diffuse PrPd deposits with little, if any, amyloid plaque while others have mixtures of both diffuse and plaque deposits. Still other types of prion disease have predominant amyloid plaques (4, 5, 6). Ultrastructural analyses using immunogold labeling have shown that PrPd can accumulate not only as fibrils and amyloid plaques but also in forms that are not discernably fibrillar (reviewed in (3, 7)). Such forms are often closely associated with membranes that obscure visualization of PrPd morphology. When PrPd is extracted from tissues, it is typically found in the form of amyloid fibrils (8, 9); however, two-dimensional crystalline arrays and other nonfibrillar or subfibrillar ultrastructures have also been observed (10, 11, 12, 13, 14, 15, 16). Both fibrillar and subfibrillar structures have been linked to prion infectivity (14), but the structural relationships and pathophysiological roles of these various prion ultrastructures remain unclear.
The normal precursor to PrPSc, PrPC, has substantial sequence homology among mammals (17) but its general physiological function(s) are uncertain. PrPC has multiple ligands and plays roles in processes such as cellular differentiation, signaling, adhesion, stress tolerance, and redox activities (reviewed in (18, 19, 20)), suggesting a cell surface scaffolding function that organizes and mediates a variety of physiological functions (21). Unlike most proteins that are converted to pathologic oligomers and amyloids, PrPC usually contains N-linked glycans (22, 23) and a glycophosphatidylinositol (GPI) anchor (24). In its native monomeric state, PrPC has predominant intrinsically disordered and α-helical domains (reviewed in (25)). In contrast, PrPSc is multimeric (14, 26, 27), rich in β-sheet content (28, 29, 30), detergent-insoluble, and partially protease-resistant (31, 32) (a.k.a. PrPRes). PrPSc multimers propagate by binding and refolding PrPC as they elongate (26, 33). This type of propagation mechanism, whereby seeding overcomes a kinetic barrier between the native PrPC state and the formation of ordered aggregates, is characteristic of PrPSc-based and other prion-like neurodegenerative diseases. A small subset (10%–15%) of human prion disease cases involve expression of PrPC mutants with potentially altered folding and/or stability (34, 35), but most cases (i.e., those with sporadic or acquired prion disease) express WT PrPC. Although the steps are poorly understood, the exposure of PrPC to the PrPSc template perturbs the canonical PrPC folding pathway to allow complete refolding of the polypeptide chain (29, 30, 33, 36, 37). Detailed characterization of the structures involved is crucial in understanding the molecular pathogenesis of prion diseases and designing and/or screening for potentially therapeutic vaccines and conversion inhibitors.
We recently reported the first high-resolution structure of a fully infectious ex vivo prion (hamster 263K scrapie) (38, 39), and since then, structures of WT (40) and GPI-deficient (anchorless) (41) forms of another prion strain (mouse RML) have also been reported by Manka et al. and us. Here, we compare the available infectious prion structures to each other and to previously determined near-atomic structures for synthetic noninfectious fibrils. We also discuss implications for the molecular bases for prion pathogenesis, strain diversity, and transmission barriers.
Experimental underpinnings of initial parallel in-register and 4-rung β-solenoid models
Several experimental barriers have impeded attempts to solve the atomic structure of the bona fide prions. The tendency of tissue-derived prions to clump without forming regular three-dimensional crystalline arrays has prevented the determination of their detailed structures by X-ray or electron diffraction techniques. However, fiber diffraction data have provided clear evidence that infectious brain derived PrPSc fibrils have a regular ∼4.9 Å spacing of polypeptide strands perpendicular to the fibril axis (reviewed in (42)). This is a characteristic feature of amyloid fibrils formed by stacked β-strands. Methodological difficulties in the preparation of highly infectious synthetic prions using isotopically labeled recombinant PrP substrates hindered the solution of high-resolution structures by solid state NMR (ssNMR) spectroscopy. Nonetheless, ground-breaking electron paramagnetic resonance (EPR) and ssNMR studies of the protease-resistant cores of synthetic recombinant PrP fibrils first demonstrated that various recombinant PrP molecules can form fibrils in which the monomers are stacked parallel and in-register, that is, with residues of one monomer aligned immediately adjacent to the corresponding residues of the adjacent monomers (43, 44, 45, 46, 47, 48, 49). Based on these and other types of empirically derived lower-resolution data, parallel in-register intermolecular β-sheet/stack (PIRIBS) (Fig. 1) models of prion fibrils and strain permutations were proposed (45).
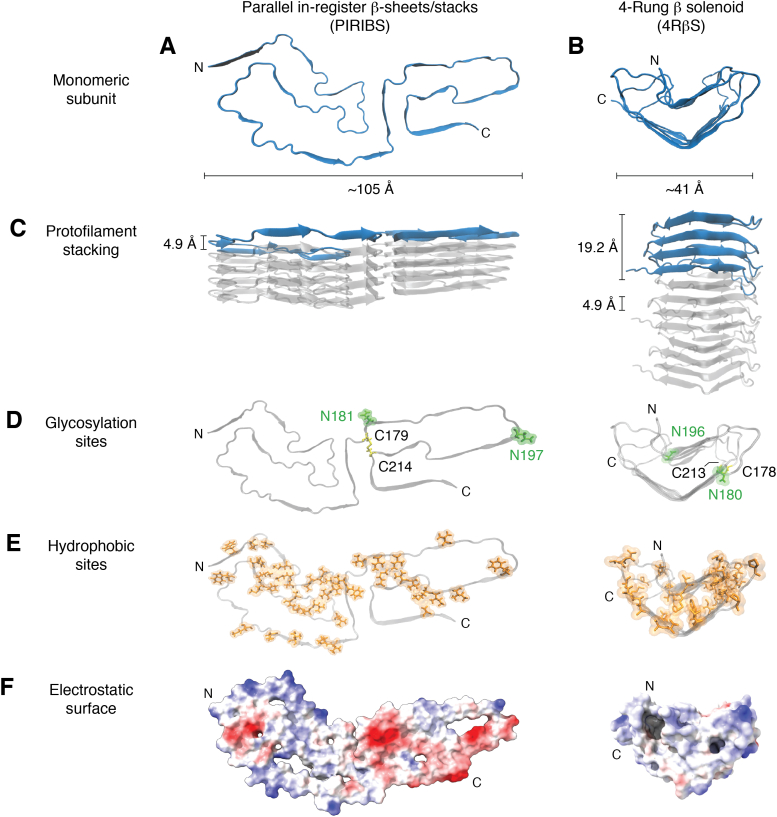
Figure 1.
Recent models for prion fibrils.A, monomeric subunit of the 263K prion within its parallel in-register intermolecular β-sheet or stack (PIRIBS) architecture (38, 39). B, monomeric subunit within a 4-rung β-solenoid (4RβS) architecture hypothesized initially for the anchorless RML (aRML) prion (50), but see contrary data demonstrating PIRIBS architecture for this strain (41). Spacing of cross-β strands in both models is ∼4.9 Å (C). This is also the spacing of each monomer in the PIRIBS structure, whereas in the 4RβS model the monomers span 19.2 Å along the fibril axis (C). Glycosylation sites (D) at positions N181 and N197 (hamster numbering, PIRIBS) and N180 and N196 (mouse numbering, 4RβS) are labeled in green and a disulfide bond between C179 and C214 (PIRIBS) and C178 and C213 (4RβS) is colored in yellow. Note: an extra glycine residue at position 53 in the hamster PrP sequence shifts the hamster-mouse sequence alignment for subsequent residues. The lower panels show the distribution of hydrophobic residues in orange (E) and the electrostatic surface (F) (red and blue indicate negative and positive charges, respectively).
To accommodate the ∼135 to 150 residue protease-resistant core that is common in multiple PrPSc strains, such PIRIBS models require serpentine windings of the polypeptide chain to allow it to fit within the cross sectional dimensions of prion fibril cores (45). An alternative 4-rung β-solenoid (4RβS) model (Fig. 1) (42, 50, 51) for the GPI-anchorless RML (aRML) prion fibril was proposed based on observations of brain-derived prion fiber diffraction patterns, low-resolution cryo-EM, and other data. Key among those observations was evidence of repeating features along the fibril axis with a spacing of 19.2 Å in addition to the ∼4.9 Å spacing between strands within β-sheets (as is seen in the PIRIBS model). The 19.2 Å spacing along the axis of a protofilament was suggested to correspond to interfaces between monomers if each provided four successive rungs, as opposed to one in a PIRIBS architecture. The resulting 4RβS protofilament would be much narrower cross sectionally than the PIRIBS structure. Accordingly, whereas PIRIBS modeling posited that ex vivo prion fibrils were comprised of a single protofilament, the 4RβS fibril model suggested two intertwined protofilaments. Thus, the hypothesized PIRIBS and 4RβS fibril architectures were fundamentally different; however, prior to the last year and a half, no empirical data unequivocally discriminated between these architectures for bona fide infectious prions.
Recent advances in the field of cryo-EM have allowed for the atomic and near-atomic resolution of many macromolecular complexes (reviewed in (52, 53)). Conformational and compositional heterogeneity often precludes conventional methods of structure determination (such as NMR and X-ray crystallography) from resolving large biomolecular assemblies, membrane proteins, and systems with polymorphism (reviewed in (53)). When imaging with cryo-EM, careful sample preparation, preservation of important scaffolding molecules and cofactors, and proper vitrification may decrease haphazard clumping of amyloids and help produce grids with an adequate distribution of fibrillar assemblies (54). Advanced neural network algorithms can aid in particle picking, noise reduction, and image class averaging (55, 56). Helical reconstruction capabilities in analysis software such as RELION 3.1 (https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page) (57, 58, 59) allow for the de novo modeling of fibrils from 3D reconstructions, and the acceleration of data processing with the usage of graphical processing units can further achieve higher resolution of structures (60).
Structures of synthetic PrP fibrils by cryo-EM
Significant insights into how PrP molecules can assemble into amyloid fibrils have come from cryo-EM studies of synthetic recombinant PrP fibrils. Such structures include fibrils of recombinant human PrP94 to 178 (rhu PrP94–178) (61) (Fig. 2A), full-length human PrP23 to 231 (rhu PrP23–231) (62) (Fig. 2B), mutant full-length human E196K PrP23 to 231 (rhu PrP23–231 E196 K) (63) (Fig. 2C), human PrP23 to 144 (rhu PrP23–144) (64), and a much shorter synthetic peptide (residues 168–176) of bank vole PrP (65). Importantly, the infectivity of these synthetic fibrils has not been documented to our knowledge. The protease-resistant cores are much smaller than those found in bona fide tissue-derived infectious PrPSc fibrils making it likely that these synthetic fibrils are either noninfectious or at least many orders of magnitude less infectious per unit protein (66, 67, 68). It is notable, however, that the murine homolog of the rhu PrP23–144 peptide has been shown to cause prion disease with a 100% attack rate in transgenic PrP-overexpressing mice (69). This murine PrP sample was not titered, but the long incubation periods were consistent with the titer being rather low relative to ex vivo prions.
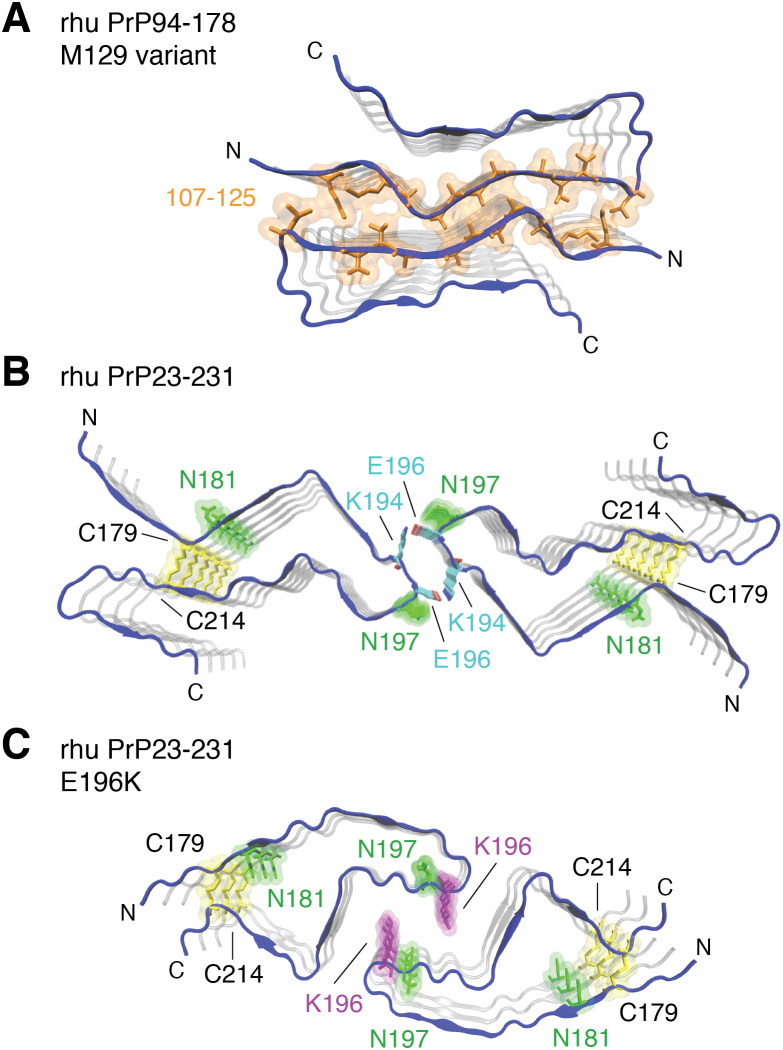
Figure 2.
Ordered core structures of synthetic prion fibrils derived from cryo-EM.A, the two protofilaments of the rhu PrP94–178 (M129 variant) (61) are traced in blue. Hydrophobic residues at the interface of the two protofilaments are shown in orange. B, the two protofilaments of the rhu PrP23–231 fibril (62) with highlighted glycosylated sidechains (green) and disulfide bond (yellow). Salt-bridges formed between K194 and E196 at the interface of opposing protofibrils are colored by heteroatom (carbon, cyan; oxygen, red; nitrogen, blue). C, protofilaments of the E196K mutant (63) with highlighted as in glycosylated sidechains (green; N181 and N197), the disulfide bond (yellow; C179 and C214), and the E196K mutation (purple).
These various synthetic PrP fibrils all have PIRIBS architectures but with different sequences constituting their ordered fibrillar cores. In the rhu PrP94–178 fibrils, the core is formed by two closely packed, symmetrical protofilaments, with each protofilament core comprised of a β-arch of residues 106 to 145 (61). In this context, β-arch has been used to mean a hairpin-shaped motif in an amyloid cross section with a loop at its tip and intermolecular β-sheets on its flanks (61). Residues 106 to 145 also form the well-resolved core of fibrils of rhu PrP23–144 but assume a markedly distinct conformation and a four-protofilament assembly. This sequence corresponds to that expressed in humans with a form of Gerstmann–Sträussler–Scheinker syndrome linked to expression of PrP with the rare Y145Stop mutation (4, 70).
Rhu PrP23–231-derived fibrils also have two protofilaments but with ordered cores formed by C-terminal residues 170 to 229 (62). In contrast to the rhu PrP94–178 fibrils, the main feature of these protofilament cores is a β-arch linked at the base by the natural disulfide bond formed between Cys179 and Cys214. Importantly, this intramolecular disulfide bond is native to PrPC and is known to be retained in the conversion to PrPSc (71, 72, 73). This disulfide β-arch is related to disulfide β-arches suggested previously by multiple EPR and ssNMR analyses of analogous synthetic human and rodent PrP fibrils (43, 44, 46, 48, 61, 62, 74). Rhu PrP23–231 E196K fibrils formed from PrP containing the familial human prion disease-linked E196K mutation have a PIRIBS architecture with an ordered core dominated by a disulfide β-arch (63). However, in this case the arch has a distinct conformation, providing evidence that the disulfide β-arch motif can vary between types of PrP fibrils.
Collectively, these studies of synthetic PrP fibrils demonstrated two major structural features. Firstly, recombinant C-terminally truncated PrP molecules tend to form PIRIBS-based amyloid fibrils with ordered cores spanning the central hydrophobic domain of residues ∼113 to 131 (46, 47, 49, 61, 75). Secondly, both full-length and N-terminally truncated PrP molecules can form fibrils with PIRIBS-based cores spanning residues ∼170 to 229, which are centered on disulfide-linked β-arches (43, 44, 45, 48, 62, 63). Fibrils of the latter type have been obtained in vitro with either spontaneous nucleation or initial seeding by tissue-derived PrPSc (45).
High-resolution cryo-EM analyses of infectious tissue-derived prion fibrils
Three high-resolution cryo-EM structures of fully infectious, brain-derived prion fibrils have now been reported (38, 39, 40, 41). These include the hamster-adapted scrapie strain 263K (Fig. 3A) (38, 39) and both WT (40) and GPI-anchorless (Fig. 3B) (41) forms of the mouse RML scrapie strain. Animal bioassays demonstrated that each of these bona fide PrPSc (PrPRes) preparations contained roughly a billion 50% lethal doses (LD50) per milligram protein. The WT 263K and RML prions also have GPI anchors and abundant N-linked glycans. In contrast to the synthetic fibrils reviewed above, these ex vivo prions have much larger proteinase K–resistant cores spanning from residues ∼81–90 to ∼231. Indeed, this span of core residues is also larger than those of most, if not all, other neuropathologic PIRIBS-based protein amyloids, such as those composed of Aβ (76) and tau (77).
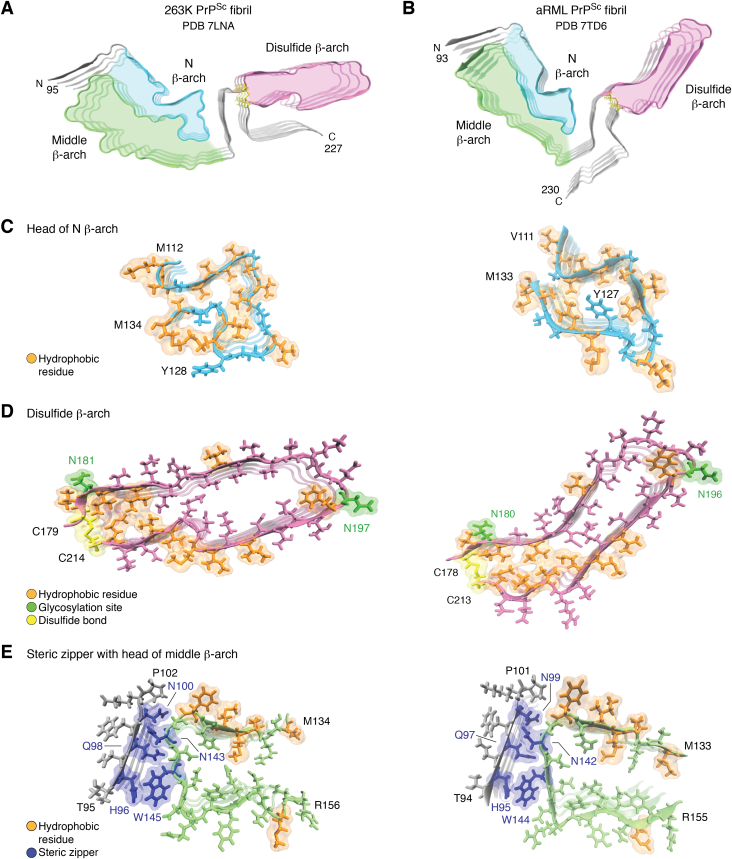
Figure 3.
Comparison of 263K and aRML prion structures. Cross-sections of the (A) 263K and (B) aRML fibril (4-monomer segments) highlighting analogous β-arch motifs (N β-arch, cyan; middle β-arch, green; disulfide β-arch, pink). C, tips of N β-arches of 263K (left) and aRML (right) with hydrophobic residues in orange. D, disulfide β-arches (pink) with respective disulfide bonds (yellow), glycosylation sites (green), and hydrophobic residues (orange). E, steric zippers (blue) formed between tips middle β-arches and respective N-terminal residues. The 263 K structure illustrated in this figure includes residues 194 to 196 which were not specified in PDB ID 7LNA. aRML, anchorless RML; PDB, Protein Data Bank.
Like the synthetic PrP amyloids, each of these ex vivo prion fibril structures has PIRIBS cores. However, commensurate with the latter’s much larger protease-resistant cores, the span of residues that form their highly resolved PIRIBS cores is more than twice that seen in the synthetic fibrils. Specifically, the 263K, wtRML, and aRML fibril cores span residues 95 to 227 (38), 94 to 225 (40), and 93 to 230 (41), respectively. With these much larger PIRIBS cores, single monomers span the entire fibril cross section (Fig. 3, A and B). Morphologic analyses of the wtRML (40) and aRML (41) preparations detected a small proportion of paired fibrils (e.g., ∼10% of wtRML), but such duplexes were not regular enough to be resolved as a discrete subpopulation by single particle analysis of any of the brain-derived prion preparations. While the aRML and wtRML fibril cross sections are quite similar, they are both more acutely V-shaped than the 263K cross section (compare Fig. 3, A and B). It had long been assumed, based primarily on negatively stained transmission electron microscopy images, that prion fibrils or rods were comprised of two protofilaments. However, we now suspect that what appeared to be separation between two protofilaments was actually deposition of stain in the trough of the “V” between the N- and C-terminal lobes of the fibril structure as we suggested earlier (45).
The two types of β-arch motifs that are seen in synthetic fibrils, namely those spanning approximately residues 113 to 131 and 170 to 229, are also found in the 263K (38, 39) and RML prions (40, 41) (Fig. 3, A–C); however, whereas the synthetic fibrils have either one or the other of these N- or C-proximal β-arches, the much larger cores of ex vivo prions have both arches at once. We provisionally refer to the more N-terminal of these β-arches as the “N β-arch” (Fig. 3, A–C) and the C-terminal one as the “disulfide β-arch” (Fig. 3, A and B, D) with reference to the disulfide bond at its base. Another more central β-arch, which shares its N-terminal flank with the N β-arch, is called the middle β-arch (Fig. 3, A and B, E). Still another key feature of the 263K and RML prions is a steric zipper (Fig. 3E) that holds the extreme N-terminal residues of the core against the head of the middle β-arch. This steric zipper involves the tight interdigitation of the sidechains of alternating residues (blue), which are homologous in the hamster and mouse PrP sequences. Steric zippers have been observed in the spines of multiple amyloid fibrils (78, 79, 80).
Although the 263K and RML prion structures share structural motifs that, in the cases of the N β-arches and disulfide β-arches, overlap those seen previously in various synthetic PrP fibrils, their conformational details are substantially different from the latter and each other (38, 39, 40, 41). For example, the heads of the N β-arches of 263K (Fig. 3A) and RML (Fig. 3B) fibrils have strikingly different anvil- or mushroom-shaped heads despite having identical glycine and hydrophobic amino acid sequences spanning residues 113 to 138 (hamster numbering) (Fig. 3C). Perhaps sequence differences that are immediately N- and C-terminal to this head region differentially influence the folding of the intervening or proximal residues.
Strain-dependent conformational differences are also striking in the C-terminal half of the prion fibril cores (38, 39, 40, 41). Specifically, the disulfide β-arches (Fig. 3D) of the RML strains are almost perpendicular to the N β-arch, while in 263K these two β-arches are nearly aligned. The extreme C termini are also quite distinct. In 263K, the C-terminal linkage to the GPI anchor closely flanks the disulfide β-arch while in the RML structures, these residues project in the opposite direction (Fig. 3, A and B). Comparison of the aRML and wtRML structures also indicates that, whereas the ordered core of aRML spans residues 93 to 230, the wtRML core extends only to residue 225. The less-ordered extreme C terminus of wtRML likely relates to the presence of the structurally heterogeneous GPI anchors, which are absent on aRML.
Overall, comparison of the 263K and the RML strains reveals that their fibrillar cross sections, and hence, growth templates, are distinct. Given that the hamster and mouse PrP sequences differ at eight positions within the fibril core, some of the difference between the 263K and RML prion conformation may be influenced by sequence as aforementioned. Clearer demonstrations of the purely conformational determinants of prion strain will require high-resolution analyses of strains isolated from hosts of the same genotype.
Finally, we note that a PIRIBS fibril architecture has recently been posted for highly infectious synthetic prions formed in vitro from bank vole PrP (109I) 23 to 231 based on PITHIRDS solid-state NMR analyses (74). This work adds significantly to the small, but growing, list of PIRIBS-based fully infectious prion structures.
Conversion of PrPC to PrPSc
Presumably, the conformational conversion and incorporation of incoming PrP monomers or, perhaps, preformed oligomeric intermediates occurs on the ends of the prion fibrils (38, 39). The sequence of events in the refolding of PrPC and the extent to which that each of those events is guided directly by interactions with the PrPSc template are far from clear. However, comparison of the conformations of the hamster PrPC (e.g. (81)) and PrPSc (38) structures indicates that an entire restructuring of PrPC is required (Fig. 4), including elimination of all original tertiary structure, unfurling of the three α-helices, and dismantling of the small intermolecular β1-β2 sheet (38, 39). Formed in their place are extensive intermolecular β-sheets running parallel to the fibril axis with interspersed loops to form β-arches as well as other features of the tightly packed fibril core. Key among the required conversion steps would be the dissociation of PrPC’s β1-helix 1–β2 loop from the hairpin formed by helices 2 and 3 (Fig. 4, “Speculative intermediate”), a process that has been predicted and described as a “banana-peeling model” (82). As noted previously, the disulfide bond remains intact throughout the conversion process (71, 72, 73), consistent with conversion occurring in oxidizing environments such as the cell surface, endosomes, or extracellular spaces (6, 83, 84, 85, 86, 87).
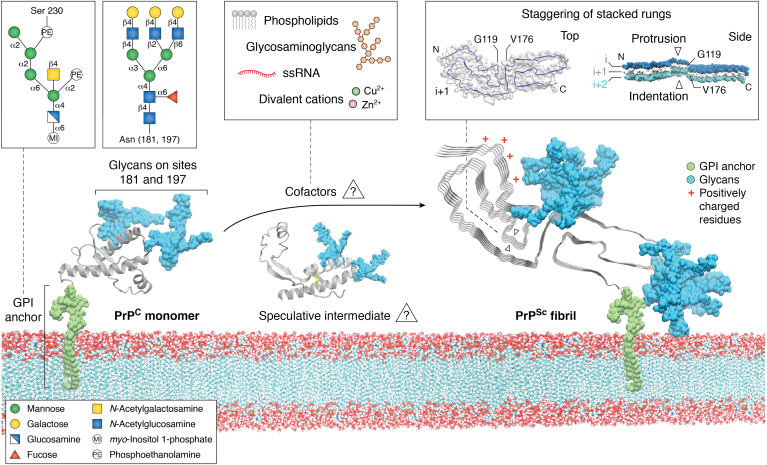
Figure 4.
Conversion of PrPCto PrPSc. Schematic of membrane bound PrPC (only the ordered C-terminal domain determined by solution NMR (81) is shown) with GPI-anchor (green) and N-linked glycan moieties (blue) in POPC lipid bilayer. Insets depict representative GPI anchors and glycans. Membrane-bound 263K pentamer is shown with N-linked glycans (blue) and GPI anchor (green) determined by cryo-EM (39). A cluster of positively charged residues near the N terminus is marked with red asterisks. The inset on the right shows top view of a single polypeptide chain with arrows at positions G119 and V179 (left) and a side view of three stacked monomers (silver, blue, and purple) exhibiting fibril staggering and the diagonal orientation of G119 and V179 of the silver monomer. Also shown is a speculative conversion intermediate with PrPC’s β1–helix 1-β2 loop peeled away from helices 2 & 3 (113) as well as an inset listing types of cofactors that can affect prion propagation in vitro (88, 89, 91, 92, 93) and might therefore be peripherally associated with prion fibrils. GPI, glycophosphatidylinositol.
Lateral views of the templating surfaces at each end of the 263K and RML fibrils show that polypeptide chains of the monomers are not entirely coplanar, with, for example, the hydrophobic heads of the respective N β-arch motifs protruding at one end and receding at the other (Fig. 4, upper right inset) (38, 39, 40, 41). It is possible that at the protruding end, an interaction between this sticky head and the analogous residues of PrPC, or a partially unfolded intermediate, might initiate refolding. Thereafter, sequential refolding and intermolecular backbone hydrogen bonding might follow along the “track” of the templating polypeptide backbone toward the N and C termini. We anticipate that PrPC-to-PrPSc conversion process will also be influenced crucially by interactions with polyanionic cofactors (Fig. 4, middle inset), which have been shown to be important (88, 89, 90, 91, 92, 93, 94, 95), and membranes, with the latter presumably being orchestrated primarily by GPI anchoring (96, 97, 98, 99).
PrPSc glycans and GPI anchors—implications
Many previous studies have shown that PrPRes glycoform patterns can vary systematically between prion strains within a given type of host and that such characteristic variations can be used to differentiate strains. For example, different subtypes of human sporadic Creutzfeldt–Jacob disease are discriminated in part based on glycoform patterns on immunoblots of brain-derived PrPRes (reviewed in (100)). As noted above, the N-linked glycans and GPI anchors are displayed on the surfaces of 263K and wtRML fibrils, with potentially far-reaching implications in prion pathogenesis (38, 39). Studies of several different prion strains in WT mice versus transgenic mice expressing only GPI-anchorless PrPC have also demonstrated profound roles of the GPI group on disease phenotypes (5, 6, 101, 102, 103). Without GPI anchors, as in prion-infected anchorless PrP transgenic mice or humans expressing anchorless PrP mutants, such as Y145X 163X, Y226X, Q227X, and G131V (104), PrPSc is free to accumulate in the large extracellular amyloid plaques that are prominent in such hosts. However, these posttranslational modifications do not seem to substantially alter the core structures of at least three murine prions strains, as probed by infrared spectroscopy (105). Also, the RML strain has been shown to maintain its fundamental strain characteristics such as incubation period and neuropathological lesion profile when passaged from WT mice to anchorless PrP mice and back again. However, two studies have described more subtle effects of passage of RML prions through anchorless mice with respect to their sensitivity to inhibitors in a cell panel assay (106) and the exposure of histidines 176 and 186 to H/D exchange with solvent (107). Nonetheless, the fact that the aRML and wtRML cryo-EM–based structures are similar to one another, yet different from the 263K structure, is consistent with prion core structures enciphering the primary properties of strains. However, clearly, the phenotypes of those strains can be further modulated markedly by the availability, or lack thereof, of GPI anchors and glycans in a given type of host or tissue (5, 6, 101, 104, 108, 109, 110, 111, 112). Importantly, both anchorless and GPI-anchored PrPSc can be infectious and capable of killing the host with equal speed (107).
The outward display of glycans and GPI anchors on the 263K (Fig. 4) and wtRML fibrils suggests that they could strongly modulate the interactions of prions with their environments in vivo. In particular, blanketing of at least the C-terminal half of the fibril cores with glycans and tethered membranes should severely restrict the access of other macromolecules (38). Macromolecules of particular significance might include proteostatic or innate immune factors involved in prion recognition and clearance. We also have proposed that pathognomonic PrPd-associated membrane distortions that are observed in prion diseases might be due to attachment of membranes to PrPSc fibrils via the array of GPI anchors as they follow the twist of the fibril. Such distortions include invaginations, protrusions, and spiral membrane inclusions, any of which might disrupt cellular functions. The spreading of prions via membranous particles such as exosomes and tunneling nanotubes (7, 113, 114, 115) might also be facilitated by GPI-meditated tethering. A direct comparison of spreading mechanisms in WT and anchorless PrP mouse models after microinjections of 22L prions into the brain demonstrated profound differences. In WT mice spreading primarily followed neuronal circuitry, whereas in anchorless PrP mice, spread was slower and involved the brain interstitial fluid drainage system (102, 116). Moreover, neuroinvasion and neural spread of prion infection from the periphery are highly dependent on the presence of GPI-anchored PrP (101).
Species/transmission barriers mechanisms
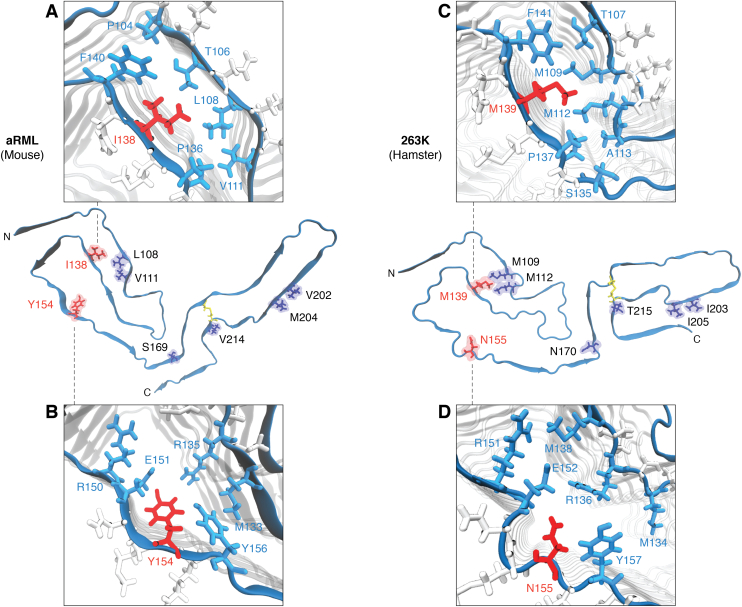
An important issue in coping with prion diseases is the extent to which prion disease strain in one host species might infect other hosts with different PrP amino acid sequences. A most troubling example of interspecies transmission was the bovine spongiform encephalopathy epidemic in cattle that gave rise to variant Creutzfeldt–Jacob disease in humans (reviewed in (117)). There are analogous concerns about the possibility of chronic wasting disease causing prion disease in humans who hunt, eat, and/or are otherwise exposed to infected cervids (117, 118, 119). Also of concern are transmissions between nonhuman animal species, particularly, those of significance in agriculture and wildlife management. Many studies have shown that differences in PrP sequence between the infecting prion and the host’s PrPC can range from being inconsequential to being responsible for profound transmission barriers (120, 121, 122, 123, 124, 125, 126). Even a single mismatch may make a host highly resistant to infection with certain prion strains (127, 128, 129, 130). The availability of the 263K cryo-EM structure has allowed us to suggest a plausible mechanism for one such species barrier, that is, between hamsters and mice (38). When considered together, prior studies in transgenic mice (126) and cell cultures (131) have indicated that most important sequence mismatch in this transmission barrier was at residue 155 (hamster numbering), which is N in hamsters and Y in mice (Fig. 5). In silico modeling based on the 263K structure indicated that insertion of the bulkier Y sidechain into the space occupied by the N in the 263K structure would lead to steric clashes and require longer–range adjustments in polypeptide backbone of a hybrid prion structure (38). Although the PrPC conversion mechanism at the PrPSc templating surface has not been defined, clashes at residue 155 appear likely to affect the kinetics of conversion and/or the stability of the product in a way that strongly restricts the ability of hamster 263K prions to grow and propagate when inoculated into mice. However, other less direct factors that have not yet been explored might also play roles in this species barrier mechanism.
Figure 5.
Potential transmission barrier mechanisms. Sidechain representations (purple, red) of the eight residue variations between mouse and hamster sequences are displayed on the monomer chains (aRML and 263K, respectively). Two residues (mouse: I138, Y154; hamster: M139, N155) implicated in cross species barrier mechanisms are displayed in red. Insets show mouse residues I138 (A) and Y154 (B) and corresponding hamster resides M139 (C) and N155 (D) and their sidechain orientations relative to those of neighboring residues (cyan) and give insight into the surrounding chemical environment. aRML, anchorless RML.
Interestingly, many of the other sequence mismatches between the hamster and mouse PrP sequences are much less inhibitory (126, 131). Tolerance of these mismatches can be rationalized as being the consequence of those residues being outside the fibril core, on the surface, or in positions within the core where their sidechains would not subject to steric clashes or other destabilizing forces. More generally, considering the known diversity in the PrP sequence mismatches that influence the relative interspecies transmissibilities of prions and the conformational diversity between prion strains, it seems likely that the mechanisms involved will be manifold.
Conclusions
At this early stage, all of the available near-atomic structures of both highly infectious prions and much less pathogenic synthetic PrP fibrils are fibrillar with PIRIBS architectures. Many types of PrP-based prions exist in mammals, and further studies will be needed to determine the full range of prion structures. The increasing availability of such structures allows for more insight into the outstanding questions of the prion field. Already, we can see a tendency of PrP molecules in PIRIBS fibrils to form β-arch structural motifs that involve analogous sequences but vary in their detailed conformations. Variations in the conformations and relative positioning of these common β-arch motifs and other structural features can create specific conformational templates for the refolding and incorporation of PrPC molecules, providing a molecular basis for the faithful propagation of distinct mammalian prion strains. The tight packing observed in certain regions of the 263K prion cores, together with molecular modeling, suggest a molecular basis for the 263K prion transmission barrier between hamsters and mice. Finally, the availability of near-atomic authentic prion structures should aid in the rational design and screening of drugs or vaccines that can interfere with pathological conversions of PrPC to PrPd, as well as strategies to promote detoxification and/or clearance of prions in infected individuals.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowlegments
This work was supported in part by the Intramural Research Program of the NIAID, Mary Hilderman Smith, Zoë Smith Jaye, and Jenny Smith Unruh in memory of Jeffrey Smith; and the Britton Fund, and CWRU School of Medicine.
Author contributions
B. C., E. A., and A. K. conceptualization; B. C. writing–original draft; B. C., E. A., and A. K. writing–review & editing; E. A. visualization.
Edited by Mike Shipston
Contributor Information
Allison Kraus, Email: alk127@case.edu.
Byron Caughey, Email: bcaughey@nih.gov.
References
- 1.Prusiner S.B. Prions. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caughey B., Kraus A. Transmissibility versus pathogenicity of Self-propagating protein aggregates. Viruses. 2019;11:1044. doi: 10.3390/v11111044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeffrey M., McGovern G., Siso S., Gonzalez L. Cellular and sub-cellular pathology of animal prion diseases: Relationship between morphological changes, accumulation of abnormal prion protein and clinical disease. Acta Neuropathol. 2011;121:113–134. doi: 10.1007/s00401-010-0700-3. [DOI] [PubMed] [Google Scholar]
- 4.Ghetti B., Piccardo P., Spillantini M.G., Ichimiya Y., Porro M., Perini F., et al. Vascular variant of prion protein cerebral amyloidosis with tau-positive neurofibrillary tangles: The phenotype of the stop codon 145 mutation in PRNP. Proc. Natl. Acad. Sci. U. S. A. 1996;93:744–748. doi: 10.1073/pnas.93.2.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chesebro B., Race B., Meade-White K., LaCasse R., Race R., Klingeborn M., et al. Fatal transmissible amyloid encephalopathy: a new type of prion disease associated with lack of prion protein membrane anchoring. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chesebro B., Trifilo M., Race R., Meade-White K., Teng C., LaCasse R., et al. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science. 2005;308:1435–1439. doi: 10.1126/science.1110837. [DOI] [PubMed] [Google Scholar]
- 7.Caughey B., Baron G.S., Chesebro B., Jeffrey M. Getting a grip on prions: oligomers, amyloids, anchors and pathological membrane interactions. Annu. Rev. Biochem. 2009;78:177–204. doi: 10.1146/annurev.biochem.78.082907.145410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merz P.A., Somerville R.A., Wisniewski H.M., Iqbal K. Abnormal fibrils from scrapie-infected brain. Acta Neuropathol. 1981;54:63–74. doi: 10.1007/BF00691333. [DOI] [PubMed] [Google Scholar]
- 9.Diringer H., Gelderblom H., Hilmert H., Özel M., Edelbluth C., Kimberlin R. Scrapie infectivity, fibrils and low molecular weight protein. Nature. 1983;306:476–478. doi: 10.1038/306476a0. [DOI] [PubMed] [Google Scholar]
- 10.Wille H., Govaerts C., Borovinskiy A., Latawiec D., Downing K.H., Cohen F.E., et al. Electron crystallography of the scrapie prion protein complexed with heavy metals. Arch. Biochem. Biophys. 2007;467:239–248. doi: 10.1016/j.abb.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Govaerts C., Wille H., Prusiner S.B., Cohen F.E. Evidence for assembly of prions with left-handed beta-helices into trimers. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wille H., Michelitsch M.D., Guenebaut V., Supattapone S., Serban A., Cohen F.E., et al. Structural studies of the scrapie prion protein by electron crystallography. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3563–3568. doi: 10.1073/pnas.052703499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wille H., Prusiner S.B. Ultrastructural studies on scrapie prion protein crystals obtained from reverse micellar solutions. Biophys.J. 1999;76:1048–1062. doi: 10.1016/S0006-3495(99)77270-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silveira J.R., Raymond G.J., Hughson A.G., Race R.E., Sim V.L., Hayes S.F., et al. The most infectious prion protein particles. Nature. 2005;437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanni I., Pirisinu L., Acevedo-Morantes C., Kamali-Jamil R., Rathod V., Di Bari M.A., et al. Isolation of infectious, non-fibrillar and oligomeric prions from a genetic prion disease. Brain. 2020;143:1512–1524. doi: 10.1093/brain/awaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortez L.M., Nemani S.K., Duque Velásquez C., Sriraman A., Wang Y., Wille H., et al. Asymmetric-flow field-flow fractionation of prions reveals a strain-specific continuum of quaternary structures with protease resistance developing at a hydrodynamic radius of 15 nm. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watts J.C., Westaway D. The prion protein family: diversity, rivalry, and dysfunction. Biochim. Biophys. Acta. 2007;1772:654–672. doi: 10.1016/j.bbadis.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Caughey B., Baron G.S. Prions and their partners in crime. Nature. 2006;443:803–810. doi: 10.1038/nature05294. [DOI] [PubMed] [Google Scholar]
- 19.Sigurdson C.J., Bartz J.C., Glatzel M. Cellular and molecular mechanisms of prion disease. Annu. Rev. Pathol. Mech. Dis. 2019;14:497–516. doi: 10.1146/annurev-pathmechdis-012418-013109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biasini E., Turnbaugh J.A., Unterberger U., Harris D.A. Prion protein at the crossroads of physiology and disease. Trends Neurosci. 2012;35:92–103. doi: 10.1016/j.tins.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linden R. The biological function of the prion protein: a cell surface scaffold of signaling modules. Front. Mol. Neurosci. 2017;10:77. doi: 10.3389/fnmol.2017.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolton D.C., Meyer R.K., Prusiner S.B. Scrapie PrP 27-30 is a sialoglycoprotein. J. Virol. 1985;53:596–606. doi: 10.1128/jvi.53.2.596-606.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Multhaup G., Diringer H., Hilmert H., Prinz H., Heukeshoven J., Beyreuther K. The protein component of scrapie-associated fibrils is a glycosylated low molecular weight protein. EMBO J. 1985;4:1495–1501. doi: 10.1002/j.1460-2075.1985.tb03808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stahl N., Borchelt D.R., Hsiao K., Prusiner S.B. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51:229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 25.Wüthrich K., Riek R. Three-dimensional structures of prion proteins. Adv. Protein Chem. 2001;57:55–82. doi: 10.1016/s0065-3233(01)57018-7. [DOI] [PubMed] [Google Scholar]
- 26.Caughey B., Kocisko D.A., Raymond G.J., Lansbury P.T., Jr. Aggregates of scrapie-associated prion protein induce the cell-free conversion of protease-sensitive prion protein to the protease-resistant state. Chem. Biol. 1995;2:807–817. doi: 10.1016/1074-5521(95)90087-x. [DOI] [PubMed] [Google Scholar]
- 27.Caughey B., Raymond G.J., Kocisko D.A., Lansbury P.T., Jr. Scrapie infectivity correlates with converting activity, protease resistance, and aggregation of scrapie-associated prion protein in guanidine denaturation studies. J. Virol. 1997;71:4107–4110. doi: 10.1128/jvi.71.5.4107-4110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caughey B.W., Dong A., Bhat K.S., Ernst D., Hayes S.F., Caughey W.S. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 29.Safar J., Roller P., Gajdusek D., Gibbs C., Jr. Conformational transitions, dissociation, and unfolding of scrapie amyloid (prion) protein. J. Biol. Chem. 1993;268:20276–20284. [PubMed] [Google Scholar]
- 30.Pan K.-M., Baldwin M., Nguyen J., Gasset M., Serban A., Groth D., et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. U. S. A. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKinley M.P., Bolton D.C., Prusiner S.B. A protease-resistant protein is a structural component of the scrapie prion. Cell. 1983;35:57–62. doi: 10.1016/0092-8674(83)90207-6. [DOI] [PubMed] [Google Scholar]
- 32.Caughey B., Neary K., Buller R., Ernst D., Perry L., Chesebro B., et al. Normal and scrapie-associated forms of prion protein differ in their sensitivities to phospholipase and proteases in intact neuroblastoma cells. J. Virol. 1990;64:1093–1101. doi: 10.1128/jvi.64.3.1093-1101.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kocisko D.A., Come J.H., Priola S.A., Chesebro B., Raymond G.J., Lansbury P.T., et al. Cell-free formation of protease-resistant prion protein. Nature. 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 34.Vanik D.L., Surewicz W.K. Disease-associated F198S mutation increases the propensity of the recombinant prion protein for conformational conversion to scrapie-like form. J. Biol. Chem. 2002;277:49065–49070. doi: 10.1074/jbc.M207511200. [DOI] [PubMed] [Google Scholar]
- 35.Apetri A.C., Vanik D.L., Surewicz W.K. Polymorphism at residue 129 modulates the conformational conversion of the D178N variant of human prion protein 90− 231. Biochemistry. 2005;44:15880–15888. doi: 10.1021/bi051455+. [DOI] [PubMed] [Google Scholar]
- 36.Bessen R.A., Kocisko D.A., Raymond G.J., Nandan S., Lansbury P.T., Jr., Caughey B. Nongenetic propagation of strain-specific phenotypes of scrapie prion protein. Nature. 1995;375:698–700. doi: 10.1038/375698a0. [DOI] [PubMed] [Google Scholar]
- 37.Horiuchi M., Chabry J., Caughey B. Specific binding of normal prion protein to the scrapie form via a localized domain initiates its conversion to the protease-resistant state. EMBO J. 1999;18:3193–3203. doi: 10.1093/emboj/18.12.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraus A., Hoyt F., Schwartz C.L., Hansen B., Artikis E., Hughson A.G., et al. High-resolution structure and strain comparison of infectious mammalian prions. Mol. Cell. 2021;81:4540–4551.e6. doi: 10.1016/j.molcel.2021.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Kraus A., Hoyt F., Schwartz C.L., Hansen B., Hughson A.G., Artikis E., et al. Structure of an infectious mammalian prion. bioRxiv. 2021 doi: 10.1016/j.molcel.2021.08.011. preprint. [DOI] [PubMed] [Google Scholar]
- 40.Manka S.W., Zhang W., Wenborn A., Betts J., Joiner S., Saibil H.R. 2.7 Å cryo-EM structure of ex vivo RML prion fibrils. Nat. Commun. 2022 doi: 10.1038/s41467-022-30457-7. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoyt F., Standke H.G., Artikis E., Schwartz C.L., Hansen B., Li K., et al. Structure of anchorless RML prion reveals motif variation between strains. Nat. Commun. 2022 doi: 10.1038/s41467-022-30458-6. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wille H., Requena J.R. The structure of PrP(Sc) prions. Pathogens. 2018;7:20. doi: 10.3390/pathogens7010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cobb N.J., Sonnichsen F.D., McHaourab H., Surewicz W.K. Molecular architecture of human prion protein amyloid: a parallel, in-register beta-structure. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18946–18951. doi: 10.1073/pnas.0706522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cobb N.J., Apetri A.C., Surewicz W.K. Prion protein amyloid formation under native-like conditions involves refolding of the C-terminal alpha-helical domain. J. Biol. Chem. 2008;283:34704–34711. doi: 10.1074/jbc.M806701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Groveman B.R., Dolan M.A., Taubner L.M., Kraus A., Wickner R.B., Caughey B. Parallel in-register intermolecular beta-sheet architectures for prion-seeded prion protein (PrP) amyloids. J. Biol. Chem. 2014;289:24129–24142. doi: 10.1074/jbc.M114.578344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theint T., Nadaud P.S., Aucoin D., Helmus J.J., Pondaven S.P., Surewicz K., et al. Species-dependent structural polymorphism of Y145Stop prion protein amyloid revealed by solid-state NMR spectroscopy. Nat. Commun. 2017;8:753. doi: 10.1038/s41467-017-00794-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theint T., Xia Y., Nadaud P.S., Mukhopadhyay D., Schwieters C.D., Surewicz K., et al. Structural studies of amyloid fibrils by paramagnetic solid-state nuclear magnetic resonance spectroscopy. J. Am. Chem. Soc. 2018;140:13161–13166. doi: 10.1021/jacs.8b06758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tycko R., Savtchenko R., Ostapchenko V.G., Makarava N., Baskakov I.V. The alpha-helical C-terminal domain of full-length recombinant PrP converts to an in-register parallel beta-sheet structure in PrP fibrils: evidence from solid state nuclear magnetic resonance. Biochemistry. 2010;49:9488–9497. doi: 10.1021/bi1013134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shannon M.D., Theint T., Mukhopadhyay D., Surewicz K., Surewicz W.K., Marion D., et al. Conformational dynamics in the core of human Y145Stop prion protein amyloid probed by relaxation dispersion NMR. Chemphyschem. 2019;20:311–317. doi: 10.1002/cphc.201800779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spagnolli G., Rigoli M., Orioli S., Sevillano A.M., Faccioli P., Wille H., et al. Full atomistic model of prion structure and conversion. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vazquez-Fernandez E., Vos M.R., Afanasyev P., Cebey L., Sevillano A.M., Vidal E., et al. The structural architecture of an infectious mammalian prion using electron cryomicroscopy. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saibil H.R. Cryo-EM in molecular and cellular biology. Mol. Cell. 2022;82:274–284. doi: 10.1016/j.molcel.2021.12.016. [DOI] [PubMed] [Google Scholar]
- 53.Bai X.-C., McMullan G., Scheres S.H. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 2015;40:49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 54.Zielinski M., Röder C., Schröder G.F. Challenges in sample preparation and structure determination of amyloids by cryo-EM. J. Biol. Chem. 2021;297:100938. doi: 10.1016/j.jbc.2021.100938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu Y., Ouyang Q., Mao Y. A deep convolutional neural network approach to single-particle recognition in cryo-electron microscopy. BMC Bioinformatics. 2017;18:1–10. doi: 10.1186/s12859-017-1757-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kimanius D., Dong L., Sharov G., Nakane T., Scheres S.H. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 2021;478:4169–4185. doi: 10.1042/BCJ20210708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thurber K.R., Yin Y., Tycko R. Automated picking of amyloid fibrils from cryo-EM images for helical reconstruction with RELION. J. Struct. Biol. 2021;213 doi: 10.1016/j.jsb.2021.107736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He S., Scheres S.H. Helical reconstruction in RELION. J. Struct. Biol. 2017;198:163–176. doi: 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kimanius D., Forsberg B.O., Scheres S.H., Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016;5 doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Glynn C., Sawaya M.R., Ge P., Gallagher-Jones M., Short C.W., Bowman R., et al. Cryo-EM structure of a human prion fibril with a hydrophobic, protease-resistant core. Nat. Struct. Mol. Biol. 2020;27:417–423. doi: 10.1038/s41594-020-0403-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L.Q., Zhao K., Yuan H.Y., Wang Q., Guan Z., Tao J., et al. Cryo-EM structure of an amyloid fibril formed by full-length human prion protein. Nat. Struct. Mol. Biol. 2020;27:598–602. doi: 10.1038/s41594-020-0441-5. [DOI] [PubMed] [Google Scholar]
- 63.Wang L.Q., Zhao K., Yuan H.Y., Li X.N., Dang H.B., Ma Y., et al. Genetic prion disease-related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abg9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Q., Jaroniec C.P., Surewicz W.K. Cryo-EM structure of disease-related prion fibrils provides insights into seeding barriers. bioRxiv. 2021 doi: 10.1101/2021.08.10.455830. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gallagher-Jones M., Glynn C., Boyer D.R., Martynowycz M.W., Hernandez E., Miao J., et al. Sub-angstrom cryo-EM structure of a prion protofibril reveals a polar clasp. Nat. Struct. Mol. Biol. 2018;25:131–134. doi: 10.1038/s41594-017-0018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Groveman B.R., Raymond G.J., Campbell K.J., Race B., Raymond L.D., Hughson A.G., et al. Role of the central lysine cluster and scrapie templating in the transmissibility of synthetic prion protein aggregates. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kraus A., Raymond G.J., Race B., Campbell K.J., Hughson A.G., Anson K.J., et al. PrP P102L and nearby lysine mutations promote spontaneous in vitro formation of transmissible prions. J. Virol. 2017;91 doi: 10.1128/JVI.01276-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Q., Wang F., Xiao X., Kim C., Bohon J., Kiselar J., et al. Structural attributes of mammalian prion infectivity: insights from studies with synthetic prions. J. Biol. Chem. 2018;293:18494–18503. doi: 10.1074/jbc.RA118.005622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi J.-K., Cali I., Surewicz K., Kong Q., Gambetti P., Surewicz W.K. Amyloid fibrils from the N-terminal prion protein fragment are infectious. Proc. Natl. Acad. Sci. U. S. A. 2016;113:13851–13856. doi: 10.1073/pnas.1610716113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghetti B., Piccardo P., Frangione B., Bugiani O., Giaccone G., Young K., et al. Prion protein amyloidosis. Brain Pathol. 1996;6:127–145. doi: 10.1111/j.1750-3639.1996.tb00796.x. [DOI] [PubMed] [Google Scholar]
- 71.Turk E., Teplow D.B., Hood L.E., Pruisner S.B. Purification and properties of the cellular and scrapie hamster prion proteins. Eur. J. Biochem. 1988;176:21–30. doi: 10.1111/j.1432-1033.1988.tb14246.x. [DOI] [PubMed] [Google Scholar]
- 72.Herrmann L.M., Caughey B. The importance of the disulfide bond in prion protein conversion. Neuroreport. 1998;9:2457–2461. doi: 10.1097/00001756-199808030-00006. [DOI] [PubMed] [Google Scholar]
- 73.Welker E., Raymond L.D., Scheraga H.A., Caughey B. Intramolecular Versus intermolecular disulfide bonds in prion proteins. J. Biol. Chem. 2002;277:33477–33481. doi: 10.1074/jbc.M204273200. [DOI] [PubMed] [Google Scholar]
- 74.Martín-Pastor M., Codeseira Y.B., Spagnolli G., Eraña H., Fernández L.C., Martin D., et al. Solid state NMR reveals a parallel in register architecture for an infectious recombinant prion. bioRxiv. 2021 doi: 10.1101/2021.07.20.453078. preprint. [DOI] [Google Scholar]
- 75.Aucoin D., Xia Y., Theint T., Nadaud P.S., Surewicz K., Surewicz W.K., et al. Protein-solvent interfaces in human Y145Stop prion protein amyloid fibrils probed by paramagnetic solid-state NMR spectroscopy. J. Struct. Biol. 2019;206:36–42. doi: 10.1016/j.jsb.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang Y., Arseni D., Zhang W., Huang M., Lövestam S., Schweighauser M., et al. Cryo-EM structures of amyloid-β 42 filaments from human brains. Science. 2022;375:167–172. doi: 10.1126/science.abm7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shi Y., Zhang W., Yang Y., Murzin A.G., Falcon B., Kotecha A., et al. Structure-based classification of tauopathies. Nature. 2021;598:359–363. doi: 10.1038/s41586-021-03911-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sawaya M.R., Hughes M.P., Rodriguez J.A., Riek R., Eisenberg D.S. The expanding amyloid family: structure, stability, function, and pathogenesis. Cell. 2021;184:4857–4873. doi: 10.1016/j.cell.2021.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chuang E., Hori A.M., Hesketh C.D., Shorter J. Amyloid assembly and disassembly. J. Cell Sci. 2018;131 doi: 10.1242/jcs.189928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chatani E., Yuzu K., Ohhashi Y., Goto Y. Current understanding of the structure, stability and dynamic properties of amyloid fibrils. Int. J. Mol. Sci. 2021;22:4349. doi: 10.3390/ijms22094349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.James T.L., Liu H., Ulyanov N.B., Farr-Jones S., Zhang H., Donne D.G., et al. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adrover M., Pauwels K., Prigent S., de Chiara C., Xu Z., Chapuis C., et al. Prion Fibrillization is mediated by a native structural element that comprises helices H2 and H3. J. Biol. Chem. 2010;285:21004–21012. doi: 10.1074/jbc.M110.111815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Caughey B., Raymond G.J. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease-and phospholipase-sensitive. J. Biol. Chem. 1991;266:18217–18223. [PubMed] [Google Scholar]
- 84.Caughey B., Raymond G.J., Ernst D., Race R.E. N-Terminal truncation of the scrapie-associated form of PrP by lysosomal protease (s): Implications regarding the site of conversion of PrP to the protease-resistant state. J. Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Borchelt D., Taraboulos A., Prusiner S. Evidence for synthesis of scrapie prion proteins in the endocytic pathway. J. Biol. Chem. 1992;267:16188–16199. [PubMed] [Google Scholar]
- 86.Yim Y.-I., Park B.-C., Yadavalli R., Zhao X., Eisenberg E., Greene L.E. The multivesicular body is the major internal site of prion conversion. J. Cell Sci. 2015;128:1434–1443. doi: 10.1242/jcs.165472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Austin C.D., Wen X., Gazzard L., Nelson C., Scheller R.H., Scales S.J. Oxidizing potential of endosomes and lysosomes limits intracellular cleavage of disulfide-based antibody–drug conjugates. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17987–17992. doi: 10.1073/pnas.0509035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Deleault N.R., Lucassen R.W., Supattapone S. RNA molecules stimulate prion protein conversion. Nature. 2003;425:717–720. doi: 10.1038/nature01979. [DOI] [PubMed] [Google Scholar]
- 89.Deleault N.R., Geoghegan J.C., Nishina K., Kascsak R., Williamson R.A., Supattapone S. Protease-resistant prion protein amplification reconstituted with partially purified substrates and synthetic polyanions. J. Biol. Chem. 2005;280:26873–26879. doi: 10.1074/jbc.M503973200. [DOI] [PubMed] [Google Scholar]
- 90.Deleault N.R., Harris B.T., Rees J.R., Supattapone S. Formation of native prions from minimal components in vitro. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Deleault N.R., Kascsak R., Geoghegan J.C., Supattapone S. Species-dependent differences in cofactor utilization for formation of the protease-resistant prion protein in vitro. Biochemistry. 2010;49:3928–3934. doi: 10.1021/bi100370b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Deleault N.R., Piro J.R., Walsh D.J., Wang F., Ma J., Geoghegan J.C., et al. Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8546–8551. doi: 10.1073/pnas.1204498109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Deleault N.R., Walsh D.J., Piro J.R., Wang F., Wang X., Ma J., et al. Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E1938–E1946. doi: 10.1073/pnas.1206999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Miller M.B., Wang D.W., Wang F., Noble G.P., Ma J., Woods V.L., Jr., et al. Cofactor molecules induce structural transformation during infectious prion formation. Structure. 2013;21:2061–2068. doi: 10.1016/j.str.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang F., Wang X., Yuan C.G., Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science. 2010;327:1132–1135. doi: 10.1126/science.1183748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Baron G.S., Wehrly K., Dorward D.W., Chesebro B., Caughey B. Conversion of raft associated prion protein to the protease-resistant state requires insertion of PrP-res (PrP(Sc)) into contiguous membranes. EMBO J. 2002;21:1031–1040. doi: 10.1093/emboj/21.5.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baron G.S., Caughey B. Effect of glycosylphosphatidylinositol anchor-dependent and - independent prion protein association with model raft membranes on conversion to the protease-resistant Isoform. J. Biol. Chem. 2003;278:14883–14892. doi: 10.1074/jbc.M210840200. [DOI] [PubMed] [Google Scholar]
- 98.Rouvinski A., Karniely S., Kounin M., Moussa S., Goldberg M.D., Warburg G., et al. Live imaging of prions reveals nascent PrPSc in cell-surface, raft-associated amyloid strings and webs. J. Cell Biol. 2014;204:423–441. doi: 10.1083/jcb.201308028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wegmann S., Miesbauer M., Winklhofer K.F., Tatzelt J., Muller D.J. Observing fibrillar assemblies on scrapie-infected cells. Pflugers Arch. 2008;456:83–93. doi: 10.1007/s00424-007-0433-x. [DOI] [PubMed] [Google Scholar]
- 100.Rossi M., Baiardi S., Parchi P. Understanding prion strains: evidence from studies of the disease forms affecting humans. Viruses. 2019;11:309. doi: 10.3390/v11040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Klingeborn M., Race B., Meade-White K.D., Rosenke R., Striebel J.F., Chesebro B. Crucial role for prion protein membrane anchoring in the neuroinvasion and neural spread of prion infection. J. Virol. 2011;85:1484–1494. doi: 10.1128/JVI.02167-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rangel A., Race B., Klingeborn M., Striebel J., Chesebro B. Unusual cerebral vascular prion protein amyloid distribution in scrapie-infected transgenic mice expressing anchorless prion protein. Acta Neuropathol. Commun. 2013;1:25. doi: 10.1186/2051-5960-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Raymond G.J., Race B., Hollister J.R., Offerdahl D.K., Moore R.A., Kodali R., et al. Isolation of novel synthetic prion strains by amplification in transgenic mice coexpressing wild-type and anchorless prion proteins. J.Virol. 2012;86:11763–11778. doi: 10.1128/JVI.01353-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Race B., Williams K., Hughson A.G., Jansen C., Parchi P., Rozemuller A.J.M., et al. Familial human prion diseases associated with prion protein mutations Y226X and G131V are transmissible to transgenic mice expressing human prion protein. Acta Neuropathol. Commun. 2018;6:13. doi: 10.1186/s40478-018-0516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Baron G.S., Hughson A.G., Raymond G.J., Offerdahl D.K., Barton K.A., Raymond L.D., et al. Effect of glycans and the glycophosphatidylinositol anchor on strain dependent conformations of scrapie prion protein: Improved purifications and infrared spectra. Biochemistry. 2011;50:4479–4490. doi: 10.1021/bi2003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mahal S.P., Jablonski J., Suponitsky-Kroyter I., Oelschlegel A.M., Herva M.E., Oldstone M., et al. Propagation of RML prions in mice expressing PrP devoid of GPI anchor leads to formation of a novel, stable prion strain. PLoS.Pathog. 2012;8 doi: 10.1371/journal.ppat.1002746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Aguilar-Calvo P., Xiao X., Bett C., Eraña H., Soldau K., Castilla J., et al. Post-translational modifications in PrP expand the conformational diversity of prions in vivo. Sci. Rep. 2017;7:1–15. doi: 10.1038/srep43295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sevillano A.M., Aguilar-Calvo P., Kurt T.D., Lawrence J.A., Soldau K., Nam T.H., et al. Prion protein glycans reduce intracerebral fibril formation and spongiosis in prion disease. J. Clin. Invest. 2020;130:1350–1362. doi: 10.1172/JCI131564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bett C., Kurt T.D., Lucero M., Trejo M., Rozemuller A.J., Kong Q., et al. Defining the conformational features of anchorless, poorly neuroinvasive prions. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cancellotti E., Bradford B.M., Tuzi N.L., Hickey R.D., Brown D., Brown K.L., et al. Glycosylation of PrPC determines timing of neuroinvasion and targeting in the brain following transmissible spongiform encephalopathy infection by a peripheral route. J. Virol. 2010;84:3464–3475. doi: 10.1128/JVI.02374-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wiseman F.K., Cancellotti E., Piccardo P., Iremonger K., Boyle A., Brown D., et al. The glycosylation status of PrPC is a key factor in determining transmissible spongiform encephalopathy transmission between species. J. Virol. 2015;89:4738–4747. doi: 10.1128/JVI.02296-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Makarava N., Chang J.C., Molesworth K., Baskakov I.V. Posttranslational modifications define course of prion strain adaptation and disease phenotype. J. Clin. Invest. 2020;130:4382–4395. doi: 10.1172/JCI138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kraus A., Groveman B.R., Caughey B. Prions and the potential transmissibility of protein misfolding diseases. Annu. Rev. Microbiol. 2013;67:543–564. doi: 10.1146/annurev-micro-092412-155735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gousset K., Schiff E., Langevin C., Marijanovic Z., Caputo A., Browman D.T., et al. Prions hijack tunnelling nanotubes for intercellular spread. Nat. Cell Biol. 2009;11:328–336. doi: 10.1038/ncb1841. [DOI] [PubMed] [Google Scholar]
- 115.Vassileff N., Cheng L., Hill A.F. Extracellular vesicles - propagators of neuropathology and sources of potential biomarkers and therapeutics for neurodegenerative diseases. J. Cell Sci. 2020;133 doi: 10.1242/jcs.243139. [DOI] [PubMed] [Google Scholar]
- 116.Rangel A., Race B., Phillips K., Striebel J., Kurtz N., Chesebro B. Distinct patterns of spread of prion infection in brains of mice expressing anchorless or anchored forms of prion protein. Acta Neuropathol. Commun. 2014;2:1–14. doi: 10.1186/2051-5960-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Houston F., Andreoletti O. Animal prion diseases: the risks to human health. Brain Pathol. 2019;29:248–262. doi: 10.1111/bpa.12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Race B., Williams K., Orru C.D., Hughson A.G., Lubke L., Chesebro B. Lack of transmission of chronic wasting disease to cynomolgus macaques. J. Virol. 2018;92 doi: 10.1128/JVI.00550-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang Z., Qin K., Camacho M.V., Cali I., Yuan J., Shen P., et al. Generation of human chronic wasting disease in transgenic mice. Acta Neuropathol. Commun. 2021;9:158. doi: 10.1186/s40478-021-01262-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bossers A., Belt P.B.G.M., Raymond G.J., Caughey B., de Vries R., Smits M.A. Scrapie susceptibility-linked polymorphisms modulate the in vitro conversion of sheep prion protein to protease-resistant forms. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4931–4936. doi: 10.1073/pnas.94.10.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kocisko D.A., Priola S.A., Raymond G.J., Chesebro B., Lansbury P.T., Jr., Caughey B. Species specificity in the cell-free conversion of prion protein to protease-resistant forms: A model for the scrapie species barrier. Proc. Natl. Acad. Sci. U. S. A. 1995;92:3923–3927. doi: 10.1073/pnas.92.9.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Priola S.A., Caughey B., Race R.E., Chesebro B. Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J.Virol. 1994;68:4873–4878. doi: 10.1128/jvi.68.8.4873-4878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Prusiner S.B., Scott M., Foster D., Pan K.M., Groth D., Mirenda C., et al. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 124.Raymond G.J., Bossers A., Raymond L.D., O'Rourke K.I., McHolland L.E., Bryant P.K., III, et al. Evidence of a molecular barrier limiting susceptibility of humans, cattle and sheep to chronic wasting disease. EMBO J. 2000;19:4425–4430. doi: 10.1093/emboj/19.17.4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Raymond G.J., Hope J., Kocisko D.A., Priola S.A., Raymond L.D., Bossers A., et al. Molecular assessment of the transmissibilities of BSE and scrapie to humans. Nature. 1997;388:285–288. doi: 10.1038/40876. [DOI] [PubMed] [Google Scholar]
- 126.Scott M., Groth D., Foster D., Torchia M., Yang S.L., DeArmond S.J., et al. Propagation of prions with artificial properties in transgenic mice expressing chimeric PrP genes. Cell. 1993;73:979–988. doi: 10.1016/0092-8674(93)90275-u. [DOI] [PubMed] [Google Scholar]
- 127.Goldmann W., Hunter N., Smith G., Foster J., Hope J. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J. Gen. Virol. 1994;75:989–995. doi: 10.1099/0022-1317-75-5-989. [DOI] [PubMed] [Google Scholar]
- 128.Mead S., Whitfield J., Poulter M., Shah P., Uphill J., Campbell T., et al. A novel protective prion protein variant that colocalizes with kuru exposure. New Engl. J. Med. 2009;361:2056–2065. doi: 10.1056/NEJMoa0809716. [DOI] [PubMed] [Google Scholar]
- 129.Asante E.A., Smidak M., Grimshaw A., Houghton R., Tomlinson A., Jeelani A., et al. A naturally occurring variant of the human prion protein completely prevents prion disease. Nature. 2015;522:478–481. doi: 10.1038/nature14510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Priola S.A., Chesebro B. A single hamster amino acid blocks conversion to protease-resistant PrP in scrapie-infected mouse neuroblastoma cells. J. Virol. 1995;69:7754–7758. doi: 10.1128/jvi.69.12.7754-7758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Priola S.A., Chabry J.l., Chan K. Efficient conversion of normal prion protein (PrP) by abnormal hamster PrP is determined by homology at amino acid residue 155. J. Virol. 2001;75:4673–4680. doi: 10.1128/JVI.75.10.4673-4680.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]