Abstract
Since antibiotic resistance is a global health issues, the use of antibiotics in animal feed for growth promotion has been restricted in many countries. Bacillus licheniformis probiotic is a potential alternative to antibiotics for increasing poultry performance. Through metagenomic sequencing, this study investigated the effects of B. licheniformis–fermented products (BLFPs) and enramycin on the microbial community composition and antibiotic resistance gene (ARG) distribution in the cecal digesta of broilers at the age of 35 d. In total, 144 one-day-old male broiler chicks (Ross 308) were randomly assigned to 4 dietary treatments as follows: basal diet (control [C] group), basal diet plus 10 mg/kg enramycin (E group), basal diet plus 1 g/kg BLFPs (L group), and basal diet plus 3 g/kg BLFPs (H group), with 6 replicate cages per treatment group and 6 birds per cage. The results indicated that the cecal alpha diversity (richness and evenness) of bacterial species was higher in the H group than in the C group. Principal coordinate analysis of microbiota and the ARG composition indicated clear differences among the cecal samples of the groups. In the cecal digesta, the abundance of active bacteria associated with probiotic properties, such as Lactobacillus crispatus and Akkermansia muciniphila, was higher in the H group than in the other groups. Enramycin treatment promoted the expression of peptide (bcrA), glycopeptide (vanRI), and lincosamide (lsaE) resistance genes but inhibited the expression of aminocoumarin (parY) and pleuromutilin (TaeA) resistance genes. BLFP (1 and 3 g/kg) treatment suppressed the expression of aminoglycoside (ANT(6)-Ib), streptogramin (vatB), and peptide (ugd) resistance genes but enhanced the expression of macrolide (efrA) and aminocoumarin (novA) resistance genes. The abundance of peptide resistance genes in Bacteroides spp. was lower in the H group than in the C group. The abundance of lincosamide resistance genes in Lactobacillus spp. was higher in the E group than in the other groups. These results demonstrated that differential changes in the structure of 3 g/kg BLFPs and enramycin-induced cecal microbial communities accompany changes in the abundance of bacterial hosts carrying specific ARGs in the cecal microbiota of broilers.
Key words: antibiotic resistance gene, Bacillus licheniformis, broiler, enramycin, microbiota
INTRODUCTION
Broilers raised in intensive industrial farming systems are exposed to frequent pathogenic challenges and exhibit a low growth rate and poor feed conversion ratio, resulting in large economic losses to the poultry industry worldwide (Estevez, 2007). In broilers, antibiotics are commonly administered at low doses for infectious disease prevention, thereby improving their health and growth. However, the misuse and overuse of antibiotics as growth promoters may lead to the evolution or selection of antibiotic-resistant bacteria among broilers (Marshall and Levy, 2011). Therefore, the use of antibiotic growth promoters in poultry feed has been banned in many countries.
Numerous feed additives, such as probiotics, have been developed as replacements for antibiotic growth promoters. Of these probiotics, Bacillus licheniformis has been recognized as being safe for animal production, with applicability as alternative antibiotic growth promoters (EFSA, 2007). Dietary supplementation with B. licheniformis has been reported to promote growth performance and prevent pathogenic infection in broilers (Knap et al., 2010; Liu et al., 2012; Gong et al., 2018; Musa et al., 2019; Abudabos et al., 2020). We previously demonstrated that B. licheniformis–fermented products (BLFPs) reduce enteric disease risk, improve growth performance, and modulate gut microbiota in broilers (Chen and Yu, 2020; Cheng et al., 2021a,b; Yu et al., 2021). The antimicrobial lipopeptides derived from BLFPs exhibit activity against Clostridium perfringens and Eimeria tenella (Horng et al., 2019; Yu et al., 2021).
Bacterial community shifts are strongly correlated with the occurrence of antibiotic resistance alterations in broilers (Xiong et al., 2018). Through 16S rRNA amplicon sequencing, we previously found that BLFP supplementation increased the abundance of Lactobacillus spp. in the feces and cecal digesta of broilers (Chen and Yu, 2020; Cheng et al., 2021a). Moreover, the BLFP-induced alteration of gut microbiota is positively correlated with growth performance in broilers (Chen and Yu, 2020; Cheng et al., 2021a). However, the effects of BLFPs on antibiotic resistance gene (ARG) distribution and diversity and their bacterial hosts in the broiler gut have not been characterized. Quantitative and qualitative analyses of ARG-harboring bacteria through metagenomic sequencing facilitates the evaluation of the feasibility of BLFPs as an alternative to antibiotics.
Enramycin, a polypeptide antibiotic, is an inhibitor of N-acetylglucosaminyl transferase (Fang et al., 2006). It is widely used as a feed additive for poultry to prevent necrotic enteritis induced by Gram-positive gut pathogens (Benno et al., 1988). BLFPs contain B. licheniformis spores and their derived antimicrobial lipopeptides. Bacillus spp. have been proposed to inhibit gut pathogens through various mechanisms, such as competitive exclusion, competition for substrates and limiting resources, and production of antimicrobial substances (Bahaddad et al., 2022). These findings indicate that enramycin and BLFPs exhibit distinct antibacterial mechanisms in the gut. Therefore, in this paper, we hypothesize that BLFPs and enramycin differentially modulate the bacterial community composition in the cecal digesta of broilers. Alteration of the cecal microbial community structure accompanies changes in the abundance of bacterial hosts carrying specific ARGs.
To test this hypothesis, we investigated the effects of BLFPs and enramycin on the microbiota and antibiotic resistome in the cecal digesta of broilers through metagenomic sequencing. Our previous study demonstrated that similar to enramycin, 3 g/kg BLFPs could improve broiler growth performance (Chen and Yu, 2020). BLFPs and enramycin differentially regulate fecal microbiota of broilers (Chen and Yu, 2020). On the basis of these findings, the microbial community composition and ARG distribution in the cecal digesta of broilers were examined further in the present study.
MATERIALS AND METHODS
Experimental Design
The fermented products produced in our previous study were used in this study, and the concentrations of B. licheniformis spores and B. licheniformis-derived antimicrobial cyclic lipopeptides in the fermented products were 3 × 1012 colony-forming unit/g and 4.7 mg/g, respectively (Chen and Yu, 2020). All experiments were performed in accordance with approved guidelines. The animal protocol was approved by the Institutional Animal Care and Use Committee of National Ilan University. In total, 144 one-day-old male broiler chicks (Ross 308) were obtained from a local commercial hatchery, reared in temperature-controlled stainless-steel cages (190 × 50 × 35 cm3), and randomly assigned to 1 of 4 treatments, with 6 replicate cages per treatment group and 6 birds per cage. The treatments were as follows: 1) basal diet (control [C] group), 2) basal diet plus 10 mg/kg enramycin (E group), 3) basal diet plus 1 g/kg BLFPs (L group), and 4) basal diet plus 3 g/kg BLFPs (H group). The experimental diets were formulated to meet all the minimum nutrient requirements for birds (NRC, 1994). The detailed composition of the basal diet has been described in our previous study (Chen and Yu, 2020). Enramycin and BLFPs were added to the diets in powder form. The broilers were fed the test diets from 1 to 35 d of age; moreover, the broilers aged 1 to 14 d and 15 to 35 d were considered to be in the starter and growth phases, respectively. The birds were provided with drinking water and feed ad libitum, and a 20/4-h light–darkness cycle was applied. The ambient temperature on d 1 to 3 was set at 33°C, and it was gradually reduced to 30°C on d 4 to 7, 27°C on d 8 to 14, and 24°C on d 15 to 35. The Newcastle disease and infectious bronchitis vaccines were administered through nose drops on d 4 and 14.
DNA Extraction, Library Construction, Sequencing, and Annotation
At the end of the experiment, the broilers were humanely killed through carbon dioxide inhalation. The cecal digesta from 2 broilers per replicate were freshly collected and pooled. Four replicates were used for cecal microbiota and ARG analyses. Total genomic DNA from the cecal digesta was extracted using a ZymoBIOMICS DNA Miniprep Kit (Zymo Research, Irvine, CA) according to the manufacturer's instructions. A paired-end sequencing library was prepared from the extracted DNA on an Illumina Nextera XTLibrary Preparation Kit (Illumina, San Diego, CA). Library quality was assessed using a Qubit 2.0 Fluorometer (Thermo Scientific, Waltham, MA) and an Bioanalyzer 2100 system (Agilent, Santa Clara, CA). The library was then sequenced on an Illumina NovaSeq 6000 platform (Illumina) to generate 150-bp paired-end reads. Quality control and filtering of sequenced raw reads were performed using Trimmomatic (version 0.38). A mean quality lower than Q20 in a 100-bp sliding window was considered the criterion. The reads that mapped to chicken, human, maize, soybean, wheat, and fish genomes on Bowtie2 (version 2.3.4.1) were filtered out. The clean reads were assembled using MEGAHIT (version 1.1.3) under the pair-end mode. All contigs of size >500 bp were applied to predict genes by using Prodigal (version 2.6.3) with the parameter “-p meta.” The low-identity and unaligned sequences obtained from all the assemblies were combined, and any redundancies were removed using CD-HIT (version 4.6.6) with the parameter “-c 0.95 -n 10 -G 0 -aS 0.9.” Taxonomic assignment of sequences was conducted through DIAMOND (version 0.9.22.123) alignment against the NR database of NCBI. The functional assignment of protein sequences was performed through DIAMOND alignment against the Comprehensive Antibiotic Resistance Database.
Microbiota and ARG Analysis
Bioinformatics analysis was performed using MicrobiomeAnalyst (Dhariwal et al., 2017). Fisher's alpha index (species richness) and Shannon index (species evenness) were used to evaluate the alpha diversity of the microbial and ARG compositions. The overall differences in the bacterial community and ARG structure were analyzed through heatmap and principal coordinate analysis (PCoA) on QIIME 2 (version 2017.4). Correlation analysis was performed using Spearman's correlation coefficient, and visualized using the R package “corrplot” (version 0.84).
Statistical Analysis
Individual cages were considered to be replicates defined as experimental units. The Shapiro–Wilk test was used for testing data distribution normality. The differences among the dietary treatment groups were analyzed through one-way ANOVA followed by Tukey's honestly significant difference test conducted using SAS (version 9.4, 2012; SAS Institute, Cary, NC). The results are expressed as means ± SEMs. All P values were adjusted through the Benjamini and Hochberg method, and significance was established at P ≤ 0.05. The Bray–Curtis dissimilarity measure was used for calculating the distance matrix for PCoA. The correlations among bacteria were measured using Spearman's correlation coefficient. Spearman's correlation coefficient in the ranges of 0.1 to 0.3, 0.3 to 0.6, and 0.6 to 1.0 were considered indicate weak, moderate, and strong correlations, respectively.
RESULTS
Effect of BLFPs and Enramycin on the Cecal Microbial Community
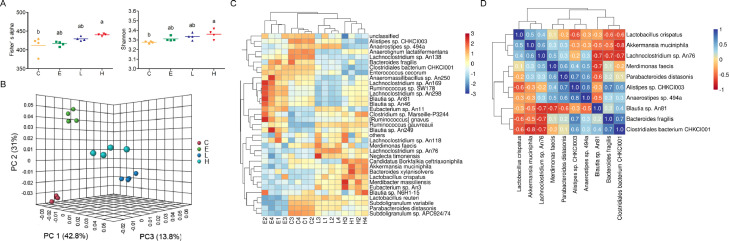
After stringent quality trimming of raw data, the averages of high-quality reads from the cecal digesta of broilers fed only a basal diet, enramycin, 1 g/kg BLFPs, and 3 g/kg BLFPs were 36,325,241, 41,542,243, 44,340,865, and 43,536,929, respectively. The species richness (Fisher's alpha index) and species evenness (Shannon index) were higher in the cecal digesta of the groups fed 3 g/kg BLFPs than in the C group (P < 0.05; Figure 1A). PCoA revealed significant intergroup differences in the bacterial community composition in the cecal digesta (Figure 1B). Figure 1C presents an overview of the species-level taxonomy and a heat map of the 35 most abundant bacteria in the cecal digesta. Specific bacterial clusters, such as those including Alistipes sp. CHKCI003, Anaerotignum lactatifermentans, and Lachnoclostridium sp. An138, were identified in the C group. Some bacterial clusters, such as those including C. bacterium CHKCI001, Enterococcus cecorum, and Anaeromassilibacillus sp. An250, partially overlapped between the C and E groups. Specific bacterial clusters, such as those including Ruminococcus sp. SW178, Lachnoclostridium sp. An298, Blautia sp. An81, and Blautia sp. An46, were identified only in the E group. Similar bacterial clusters, such as those including L. reuteri, S. variabile, P. distasonis, and S. sp. APC924/74, were identified in the C, L, and H groups. Specific bacterial clusters including Candidatus Borkfalkia ceftriaxoniphila, A. muciniphila, and Bact. xylanisolvens were identified only in the H group.
Figure 1.
Comparison of the microbial community structure in the cecal digesta through advanced analysis. (A) Scatter plot of bacterial alpha diversity in group C (basal diet), E (basal diet plus 10 mg/kg enramycin), L (basal diet plus 1 g/kg B. licheniformis–fermented products [BLFPs]), and H (basal diet plus 3 g/kg BLFPs) cecal digesta. Each bar represents mean ± SEM (n = 4). Different superscripts indicate significant differences between groups. (B) Principal coordinate analysis of the cecal bacterial communities in the C, E, L, and H groups (n = 4). (C) Heat map of species abundance of microbiota in cecal digesta. Abundance distribution of 35 dominant species (Y-axis) across all samples (X-axis) is displayed (n = 4). The values are normalized using Z-score. (D) Correlation analysis of bacterial abundance among the dominant 10 species.
Effect of BLFPs and Enramycin on the Cecal Bacterial Taxonomic Composition
The effects of BLFPs and enramycin on the bacterial taxonomy in the cecal digesta of broilers are summarized in Table 1. At the phylum level, the abundance of the phylum Firmicutes was lower in the C group than in the other groups (P < 0.001). The abundance of the phylum Bacteroidetes was higher in the C group than in the other groups (P < 0.001). The Firmicutes-to-Bacteroidetes ratio was lower in the C group than in the other groups (P < 0.001). At the genus level, the abundance of unclassified species was lower in the H group than in the other groups (P = 0.003). The abundance of Lactobacillus spp. was higher in the H group than in the C and E groups (P = 0.009). The abundance of Blautia spp. was higher in the E group than in the C and L groups (P = 0.006). The abundance of Alistipes spp. was lower in the H group than in the other groups (P < 0.001). The abundance of Lachnoclostridium spp. was lower in the C group than in the E and L groups (P < 0.001). Supplementation with enramycin led to the lowest abundance of Parabacteroides spp., whereas the C group demonstrated the highest abundance of Parabacteroides spp. (P < 0.001). The abundance of Faecalibacterium spp. was lower in the E and H groups than in the C and L groups (P < 0.001). The abundance of Clostridium and Ruminococcus spp. was lower in the C group than in the other groups (P < 0.001). At the species level, the abundance of unclassified species was lower in the H group than in the C group (P < 0.001). The abundance of Alistipes sp. CHKCI003, Parabacteroides distasonis, and Subdoligranulum variabile was higher in the C group than in the other groups (P < 0.001). Supplementation with 3 g/kg BLFPs led to the lowest abundance of Alistipes sp. CHKCI003 (P < 0.001), whereas supplementation with enramycin led to the lowest abundance of P. distasonis and S. variabile (P < 0.001). The abundance of Bacteroides fragilis was lower in the H group than in the C group (P = 0.017). The abundance of Lachnoclostridium sp. An76 was higher in the L group than in the other groups (P < 0.001). The abundance of Lactobacillus crispatus and Akkermansia muciniphila was higher in the H group than in the C and E groups (P < 0.01). The abundance of Anaerostipes sp. 494a was lower in the E and H groups than in the C and L groups (P < 0.001). The abundance of Clostridiales bacterium CHKCI001 was lower in the L and H groups than that in the C and E groups (P < 0.001). Supplementation with 3 g/kg BLFPs led to the lowest abundance of the C. bacterium CHKCI001 (P < 0.001). Moreover, the abundance of Blautia sp. An81, Lachnoclostridium sp. An169, Ruminococcus sp. SW178, and Lachnoclostridium sp. An298 was higher in the E group than in the other groups (P < 0.01). The abundance of Merdimonas faecis was higher in the L group than in the E and H groups (P = 0.004). The abundance of L. reuteri was lower in the E group than in the other groups (P < 0.001). The abundance of Ruminococcus gauvreauii was higher in the L and H groups than that in the C and E groups (P < 0.001). The C group had the lowest abundance of R. gauvreauii (P < 0.001). The abundance of Blautia sp. An249 and Blautia sp. An46 was higher in the E group than in the C and L groups (P < 0.01). Supplementation with 1 g/kg BLFPs led to the lowest abundance of Blautia sp. An46 (P < 0.001). The abundance of Eubacterium sp. An11 was higher in the E and L groups than in the other groups (P < 0.001). Figure 1D presents the correlation among the 10 most abundant bacteria in the cecal digesta. The abundance of L. crispatus was positively associated with that of A. muciniphila and Lachnoclostridium sp. An76. By contrast, the abundance of the L. crispatus was negatively correlated with that of the C. bacterium CHKCI001, Bact. fragilis, and Alistipes sp. CHKCI003. The abundance of M. faecis, P. distasonis, Alistipes sp. CHKCI003, and Anaerostipes sp. 494a was positively correlated (Figure 3A). The abundance of Bact. fragilis was positively associated with that of C. bacterium CHKCI001. The abundance of Bact. fragilis and C. bacterium CHKCI001 was negatively correlated with that of the L. crispatus, A. muciniphila, and Lachnoclostridium sp. An76.
Table 1.
Effect of B. licheniformis–fermented products and enramycin on bacterial taxonomy in cecal digesta of broilers.
| Relative abundance (%) |
||||||
|---|---|---|---|---|---|---|
| C1 | E | L | H | SEM | P value | |
| Phylum | ||||||
| Firmicutes | 70.73b | 77.35a | 75.70a | 74.99a | 0.72 | < 0.001 |
| Bacteroidetes | 23.23a | 17.14b | 18.48b | 17.85b | 0.68 | < 0.001 |
| Firmicutes/Bacteroidetes | 3.05b | 4.55a | 4.10a | 4.23a | 0.17 | < 0.001 |
| Genus | ||||||
| unclassified | 32.50a | 32.88a | 32.19a | 29.92b | 0.36 | 0.003 |
| Bacteroides | 11.67 | 11.57 | 10.42 | 11.83 | 0.29 | 0.310 |
| Lactobacillus | 7.81b | 8.60b | 9.58ab | 11.78a | 0.49 | 0.009 |
| Blautia | 6.62b | 7.97a | 7.02b | 7.43ab | 0.16 | 0.006 |
| Alistipes | 5.30a | 3.51b | 3.82b | 2.93c | 0.23 | < 0.001 |
| Lachnoclostridium | 4.99b | 5.77a | 5.67a | 5.39ab | 0.09 | < 0.001 |
| Parabacteroides | 2.73a | 0.71d | 1.78b | 1.19c | 0.20 | < 0.001 |
| Faecalibacterium | 2.73a | 1.73b | 2.62a | 1.88b | 0.13 | < 0.001 |
| Clostridium | 1.81b | 1.95a | 2.03a | 1.99a | 0.03 | < 0.001 |
| Ruminococcus | 1.72b | 3.22a | 2.97a | 3.16a | 0.16 | < 0.001 |
| Species | ||||||
| unclassified | 51.40a | 50.99ab | 50.68ab | 50.24b | 0.14 | 0.015 |
| Alistipes sp. CHKCI003 | 4.21a | 2.78b | 3.03b | 2.22c | 0.19 | < 0.001 |
| Bacteroides fragilis | 3.18a | 3.07ab | 2.59ab | 2.51b | 0.10 | 0.017 |
| Lachnoclostridium sp. An76 | 1.40c | 1.35c | 2.10a | 1.72b | 0.08 | < 0.001 |
| Lactobacillus crispatus | 1.25b | 1.46b | 1.68ab | 2.02a | 0.09 | 0.006 |
| Akkermansia muciniphila | 1.07b | 0.54b | 1.14b | 2.39a | 0.19 | < 0.001 |
| Anaerostipes sp. 494a | 1.04a | 0.65b | 1.12a | 0.53b | 0.07 | < 0.001 |
| Clostridiales bacterium CHKCI001 | 0.88a | 0.84a | 0.50b | 0.34c | 0.06 | < 0.001 |
| Blautia sp. An81 | 0.75b | 0.95a | 0.59b | 0.75b | 0.04 | < 0.001 |
| Parabacteroides distasonis | 0.60a | 0.15d | 0.39b | 0.26c | 0.04 | < 0.001 |
| Merdimonas faecis | 0.58ab | 0.52c | 0.60a | 0.55bc | 0.01 | 0.004 |
| Lachnoclostridium sp. An169 | 0.59b | 0.66a | 0.53b | 0.57b | 0.01 | 0.001 |
| Lactobacillus reuteri | 0.55a | 0.12c | 0.60a | 0.37b | 0.05 | < 0.001 |
| Ruminococcus gauvreauii | 0.58c | 1.35b | 1.78a | 1.86a | 0.13 | < 0.001 |
| Ruminococcus sp. SW178 | 0.57b | 1.08a | 0.42b | 0.64b | 0.07 | < 0.001 |
| Subdoligranulum variabile | 0.57a | 0.21c | 0.44b | 0.44b | 0.03 | < 0.001 |
| Blautia sp. An249 | 0.55b | 0.74a | 0.62b | 0.66ab | 0.07 | 0.002 |
| Lachnoclostridium sp. An298 | 0.53bc | 1.10a | 0.39c | 0.63b | 0.07 | < 0.001 |
| Blautia sp. An46 | 0.48b | 0.65a | 0.30c | 0.47b | 0.03 | < 0.001 |
| Eubacterium sp. An11 | 0.44b | 0.79a | 0.70a | 0.46b | 0.04 | < 0.001 |
C = Basal diet; E = Basal diet plus 10 mg/kg enramycin; L = Basal diet plus 1 g/kg B. licheniformis–fermented products; H = Basal diet plus 3 g/kg B. licheniformis–fermented products.
Means in a row with no common superscript are significantly different (P ≤ 0.05).
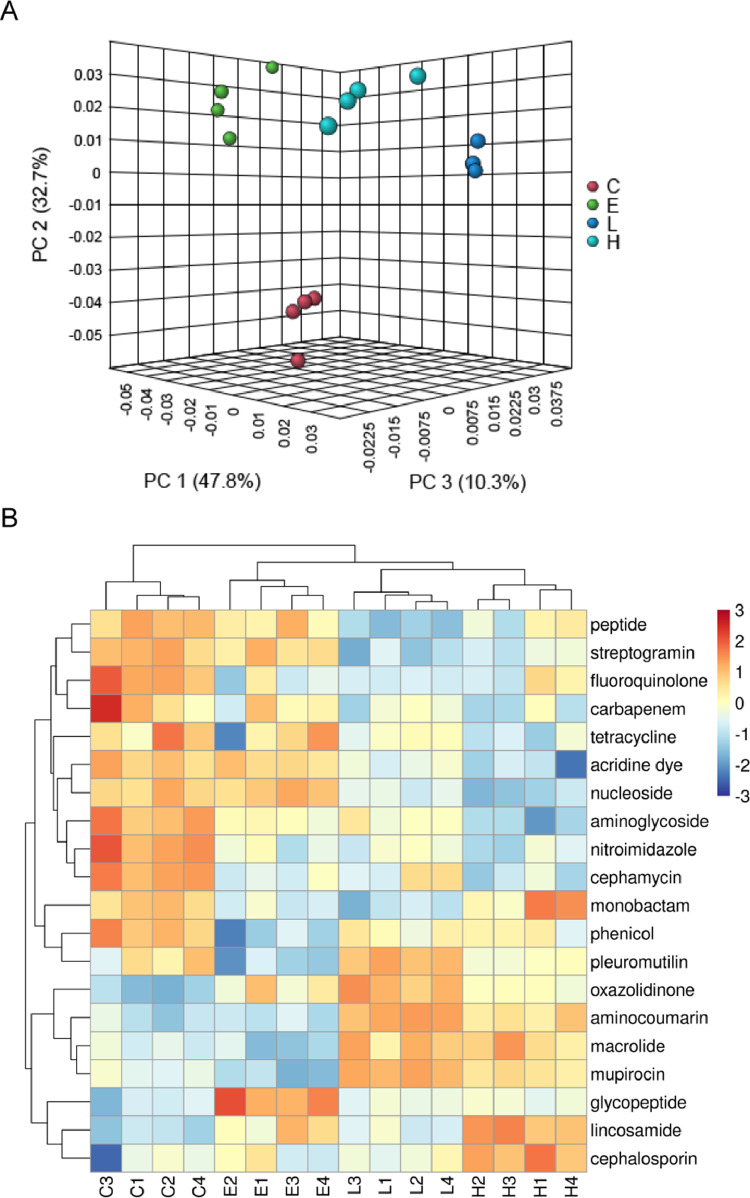
Figure 3.
Comparison of the antibiotic resistance class structure in cecal digesta through advanced analysis. (A) Principal coordinate analysis of the antibiotic resistance classes of basal diet in group C (basal diet), E (basal diet plus 10 mg/kg enramycin), L (basal diet plus 1 g/kg B. licheniformis–fermented products [BLFPs]), and H (basal diet plus 3 g/kg BLFPs) cecal digesta (n = 4). (B) Heat map of antibiotic resistance class abundance from cecal digesta. Abundance distribution of 20 dominant antibiotic resistance classes (Y-axis) across all samples (X-axis) is displayed (n = 4).
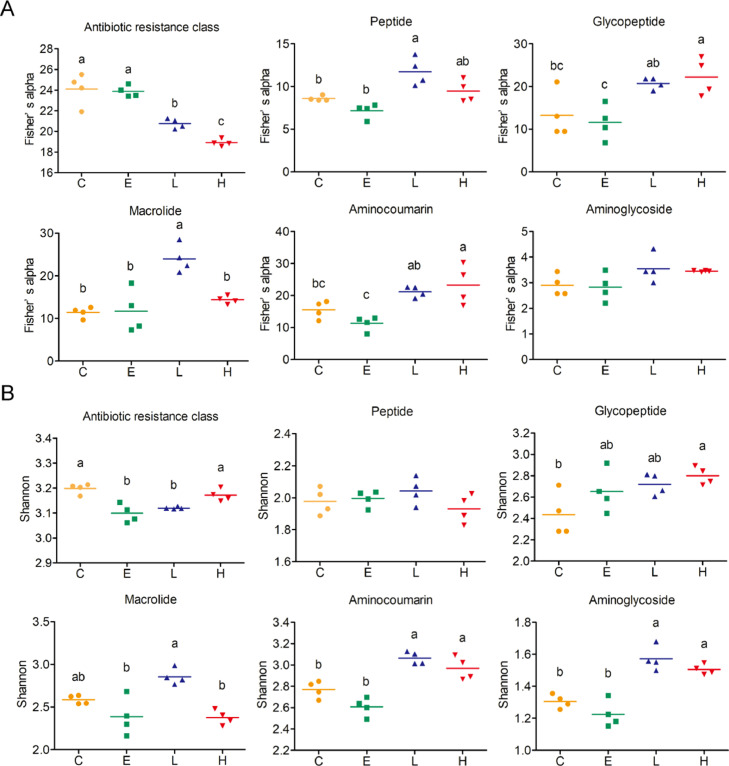
Effect of BLFPs and Enramycin on Alpha and Beta Diversities of Antibiotic Resistance Classes
Figure 2 presents the alpha diversity of antibiotic resistance classes in the cecal digesta of broilers. The richness (Fisher's alpha) of total antibiotic resistance classes was lower in the groups fed 1 and 3 g/kg BLFPs than in the C and E groups (P < 0.05; Figure 2A). Supplementation with 3 g/kg BLFPs led to the lowest richness of antibiotic resistance classes (P < 0.05; Figure 2A). The richness of peptide resistance genes was higher in the group fed 1 g/kg BLFPs than in the C and E groups (P < 0.05; Figure 2A). The richness of glycopeptide and aminocoumarin resistance genes was higher in the group fed 3 g/kg BLFPs than in the C and E groups (P < 0.05; Figure 2A). The richness of macrolide resistance genes was higher in the group fed 1 g/kg BLFPs than in the other groups (P < 0.05; Figure 2A). The evenness (Shannon index) of total antibiotic resistance classes was lower in the groups fed enramycin and 1 g/kg BLFPs than in the C and H groups (P < 0.05; Figure 2B). The evenness of glycopeptide resistance genes was higher in the group fed 3 g/kg BLFPs than in the C group (P < 0.05; Figure 2B). The evenness of macrolide resistance genes was higher in the group fed 1 g/kg BLFPs than in the E and H groups (P < 0.05; Figure 2B). The evenness of aminocoumarin and aminoglycoside resistance genes was higher in the groups fed 1 and 3 g/kg BLFPs than in the C and E groups (P < 0.05; Figure 2B). Figure 3 presents the beta diversity of antibiotic resistance classes in the cecal digesta of broilers. Our PCoA results revealed significant intergroup differences in the total antibiotic resistance class composition (Figure 3A). Figure 3B presents a heat map of the 20 most abundant antibiotic resistance classes in the cecal digesta of broilers. Specific antibiotic resistance classes, such as aminoglycosides, nitroimidazoles, and cephamycins, were identified in the C group. Some antibiotic resistance classes, such as streptogramins, acridine dyes, and nucleosides, partially overlapped between the C and E groups. Glycopeptide and cephalosporin resistance genes were specifically identified in the E and H groups, respectively. Similar antibiotic resistance classes, such as aminocoumarins, macrolides, and mupirocins, were identified in the L and H groups. The effects of BLFPs and enramycin on the antibiotic resistance classes in the cecal digesta of broilers are summarized in Table 2. Total ARG abundance per sample from the cecal digesta of broilers fed only a basal diet, enramycin, 1 g/kg BLFPs, and 3 g/kg BLFPs was 18,270, 22,122, 23,451, and 22,625, respectively. Peptide resistance gene abundance was lower in the L and H groups than in the C group (P < 0.001). Supplementation with 1 g/kg BLFPs led to the lowest peptide resistance gene abundance (P < 0.001). The abundance of macrolide and aminocoumarin resistance genes was higher in the L and H groups than in the C and E groups (P < 0.001). Supplementation with 1 g/kg BLFPs led to the highest aminocoumarin resistance gene abundance (P < 0.001). The abundance of glycopeptide resistance genes was higher in the E group than in the other groups (P < 0.001). The abundance of lincosamide resistance genes was higher in the E and H group than in the C and L groups (P < 0.001). The abundance of aminoglycoside, phenicol, fluoroquinolone, and nitroimidazole resistance genes was higher in the C group than in the other groups (P < 0.001). Supplementation with 3 g/kg BLFPs led to the lowest aminoglycoside resistance gene abundance in the cecal digesta (P < 0.001), and supplementation with enramycin led to the lowest phenicol resistance gene abundance in the cecal digesta (P < 0.001). The abundance of mupirocin resistance genes was higher in the L and H groups than in the C and E groups (P < 0.001). Supplementation with 1 g/kg BLFPs led to the highest mupirocin resistance gene abundance, whereas enramycin led to the lowest mupirocin resistance gene abundance (P < 0.001). The abundance of streptogramin resistance genes was lower in the L and H groups than in the C and E groups (P < 0.001). The abundance of pleuromutilin resistance genes was lower in the enramycin groups than in the other groups (P < 0.001). The abundance of oxazolidinone resistance genes was lower in the C group than in the other groups (P < 0.001). Supplementation with 1 g/kg BLFPs led to the highest oxazolidinone resistance gene abundance (P < 0.001). Finally, the abundance of carbapenem resistance genes was lower in the H group than in the C group (P = 0.03).
Figure 2.
Alpha diversity of different antibiotic resistance classes measured using antibiotic resistance genes in cecal digesta of broilers. Scatter plot of the alpha diversity of antibiotic resistance classes in group C (basal diet), E (basal diet plus 10 mg/kg enramycin), L (basal diet plus 1 g/kg B. licheniformis–fermented products [BLFPs]), and H (basal diet plus 3 g/kg BLFPs) cecal digesta. Each bar represents mean ± SEM (n = 4). Different superscripts indicate significant differences between groups.
Table 2.
Effect of B. licheniformis–fermented products and enramycin on antibiotic resistance classes in cecal digesta of broilers.
| Relative abundance (%) |
||||||
|---|---|---|---|---|---|---|
| C1 | E | L | H | SEM | P value | |
| Total ARGs abundance per sample (CPM)2 | 18,270.49 | 22,121.52 | 23,451.25 | 22,624.68 | 1,159.58 | 0.436 |
| Peptide | 20.48a | 20.22ab | 19.35c | 19.91b | 0.12 | < 0.001 |
| Macrolide | 15.11b | 14.71b | 16.1a | 16.01a | 0.17 | < 0.001 |
| Glycopeptide | 10.77b | 14.14a | 11.44b | 11.54b | 0.35 | < 0.001 |
| Lincosamide | 9.73b | 10.52a | 10.02b | 10.99a | 0.13 | < 0.001 |
| Aminocoumarin | 7.84c | 7.87c | 8.90a | 8.54b | 0.12 | < 0.001 |
| Aminoglycoside | 7.20a | 6.70b | 6.71b | 6.18c | 0.10 | < 0.001 |
| Tetracycline | 6.32 | 6.24 | 6.22 | 6.14 | 0.03 | 0.164 |
| Mupirocin | 3.82c | 3.57d | 4.28a | 4.07b | 0.07 | < 0.001 |
| Phenicol | 3.69a | 2.66c | 3.25b | 3.21b | 0.10 | < 0.001 |
| Streptogramin | 3.26a | 3.16a | 2.67b | 2.83b | 0.07 | < 0.001 |
| Fluoroquinolone | 2.96a | 2.20b | 2.14b | 2.33b | 0.10 | < 0.001 |
| Pleuromutilin | 2.79ab | 2.26c | 3.05a | 2.63b | 0.08 | < 0.001 |
| Oxazolidinone | 2.38c | 2.73b | 2.93a | 2.66b | 0.05 | < 0.001 |
| Nitroimidazole | 2.18a | 1.80b | 1.82b | 1.74b | 0.05 | < 0.001 |
| Carbapenem | 0.46a | 0.39ab | 0.36ab | 0.33b | 0.02 | 0.030 |
C = Basal diet; E = Basal diet plus 10 mg/kg enramycin; L = Basal diet plus 1 g/kg B. licheniformis–fermented products; H = Basal diet plus 3 g/kg B. licheniformis–fermented products.
Abbreviations: ARG, antibiotic resistance gene; CPM, counts per million.
Means in a row with no common superscript are significantly different (P ≤ 0.05).
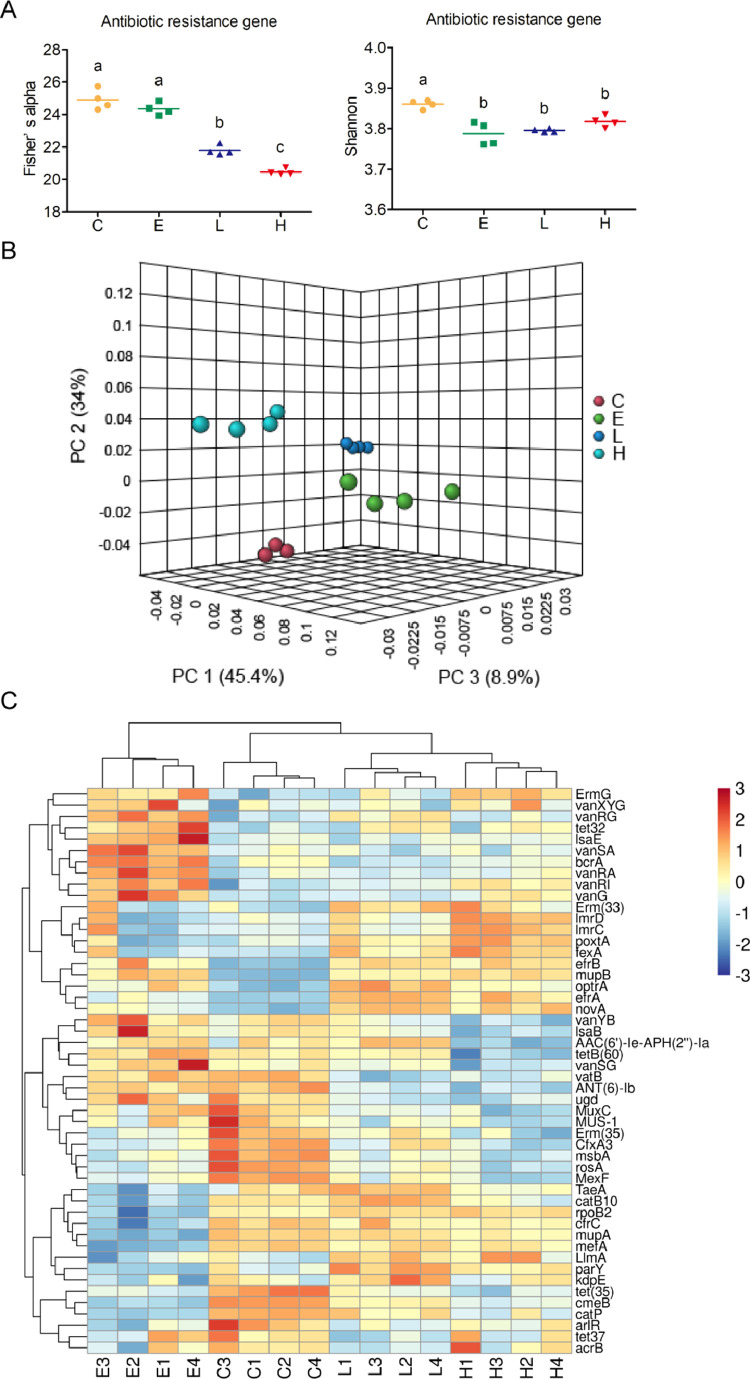
Effect of BLFPs and Enramycin on Alpha and Beta Diversities of ARGs
The richness (Fisher's alpha) of total ARGs was lower in the groups fed 1 and 3 g/kg BLFPs than in the C and E groups (P < 0.05; Figure 4A). Moreover, 3 g/kg BLFPs led to the lowest ARG richness (P < 0.05; Figure 4A). The evenness (Shannon index) of total ARGs was higher in the C group than in the other groups (P < 0.05; Figure 4A). Our PCoA results revealed significant intergroup differences in the total ARG composition (Figure 4B). Figure 4C illustrates a heat map of the 50 most abundant ARGs in the cecal digesta of broilers. Specific ARGs such as Erm(35), CfxA3, msbA, rosA, MexF, and tet(35) were identified in the C group, whereas ARGs such as vatB, ANT(b)-lb, and ugd partially overlapped between the C and E groups. Specific ARGs such as lsaE, vanSA, bcrA, and vanRA were identified in the E group. Moreover, ARGs such as efrB, mupB, and optrA partially overlapped in the E, L, and H groups, whereas ARGs such as TaeA, rpoB2, cfrC, mupA, and mefA overlapped in the C, L, and H groups. efrA and novA were specifically identified in the L and H groups. The effects of BLFPs and enramycin on ARGs in the cecal digesta of broilers are summarized in Table 3. bcrA, vanRI, and lsaE abundance was higher in the E group than in the other groups (P < 0.001), but rpoB2 and parY abundance was lower in the E group than in the other groups (P < 0.001). LlmA and TaeA abundance was higher in the L and H groups than in the E group (P < 0.001), but efrB abundance lower in the C group than in the other groups (P < 0.001). Moreover, efrA and novA abundance was higher in the L and H groups than in the other groups (P < 0.001). efrA and novA abundance was lower in the C group than in the other groups (P < 0.001). ANT(6)-Ib, vatB, and ugd abundance was higher in the C and E groups than in the other groups (P < 0.01). Supplementation with 1 g/kg BLFPs led to the highest optrA abundance, whereas the C group had the lowest optrA abundance (P < 0.001). Finally, tet32 abundance was higher in the E group than in the C group (P < 0.01).
Figure 4.
Comparison of the antibiotic resistance genes (ARGs) in cecal digesta through advanced analysis. (A) Scatter plot of alpha diversity of ARGs in group C (basal diet), E (basal diet plus 10 mg/kg enramycin), L (basal diet plus 1 g/kg B. licheniformis–fermented products [BLFPs]), and H (basal diet plus 3 g/kg BLFPs) cecal digesta. Each bar represents mean ± SEM (n = 4). Different superscripts indicate significant differences between groups. (B) Principal coordinate analysis of the cecal ARGs in the C, E, L, and H groups (n = 4). (C) Heat map of ARG abundance of microbiota from cecal digesta. Abundance distribution of 50 dominant ARGs (Y-axis) across all samples (X-axis) is displayed (n = 4). The values are normalized using Z-score.
Table 3.
Effect of B. licheniformis–fermented products and enramycin on antibiotic resistance genes in cecal digesta of broilers.
| Relative abundance (%) |
||||||
|---|---|---|---|---|---|---|
| C1 | E | L | H | SEM | P value | |
| bcrA | 9.54b | 11.12a | 9.00b | 9.49b | 0.22 | < 0.001 |
| rpoB2 | 5.99a | 4.77b | 6.38a | 6.38a | 0.18 | < 0.001 |
| parY | 5.08b | 4.80c | 5.53a | 5.26b | 0.07 | < 0.001 |
| LlmA | 4.71ab | 4.62b | 5.01a | 4.98a | 0.06 | 0.012 |
| efrB | 4.16b | 4.87a | 4.95a | 4.91a | 0.09 | < 0.001 |
| efrA | 4.09c | 4.49b | 5.17a | 4.91a | 0.11 | < 0.001 |
| ANT(6)-Ib | 4.12a | 3.98a | 3.53b | 3.37b | 0.08 | < 0.001 |
| vatB | 3.13a | 3.06a | 2.55b | 2.69b | 0.07 | < 0.001 |
| ugd | 3.22a | 3.22a | 2.79b | 2.75b | 0.07 | 0.002 |
| TaeA | 2.79ab | 2.26c | 3.05a | 2.63b | 0.08 | < 0.001 |
| vanRI | 2.61c | 3.67a | 2.86c | 3.23b | 0.11 | < 0.001 |
| novA | 2.74c | 3.05b | 3.37a | 3.28a | 0.07 | < 0.001 |
| optrA | 2.38c | 2.73b | 2.93a | 2.66b | 0.05 | < 0.001 |
| tet32 | 2.25b | 2.53a | 2.36ab | 2.33ab | 0.03 | 0.014 |
| lsaE | 2.12b | 2.59a | 2.08b | 2.16b | 0.06 | < 0.001 |
C = Basal diet; E = Basal diet plus 10 mg/kg enramycin; L = Basal diet plus 1 g/kg B. licheniformis–fermented products; H = Basal diet plus 3 g/kg B. licheniformis–fermented products.
Means in a row with no common superscript are significantly different (P ≤ 0.05).
Effect of BLFPs and Enramycin on Taxonomic Assignment of ARGs
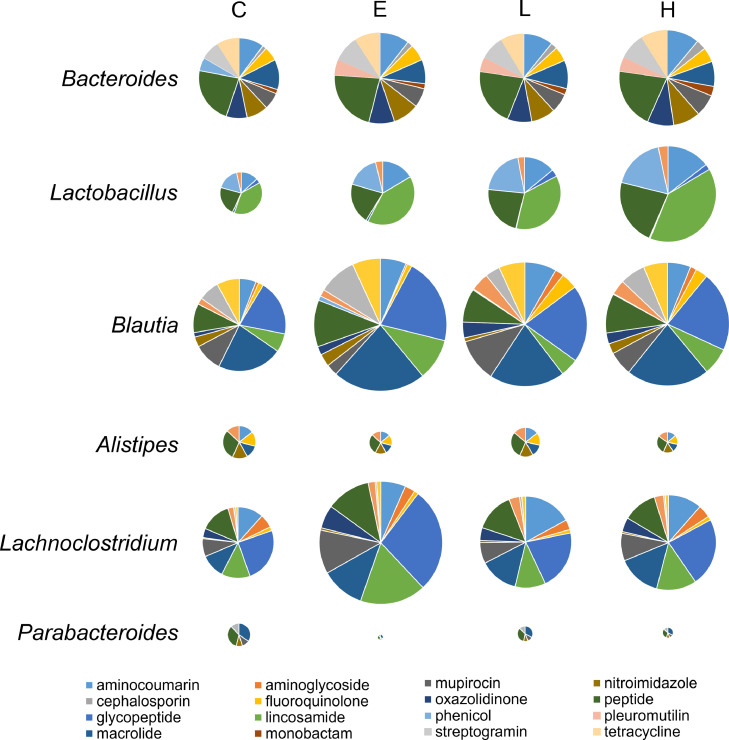
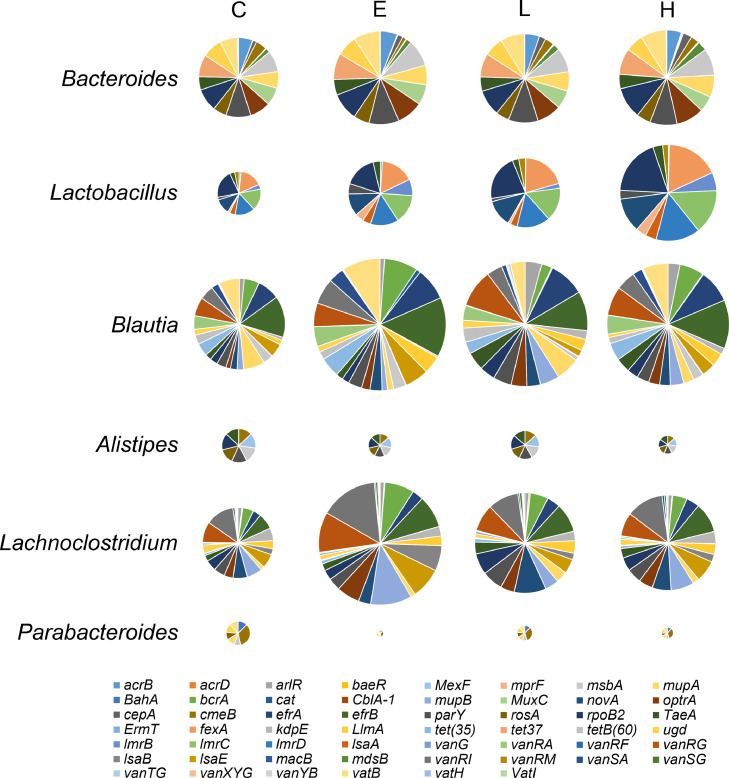
The detected ARGs were predicted to belong to 132 different bacterial genera, with Bacteroides, Lactobacillus, Blautia, Alistipes, Lachnoclostridium, and Parabacteroides accounting for 55.8% of the total ARGs abundance. Figure 5 presents the most abundant genera with the assigned ARGs from the main antibiotic resistance classes in the cecal digesta of broilers. In Bacteroides and Alistipes spp., ARGs from peptide antibiotic resistance classes were predominant, whereas those from the macrolide antibiotic resistance classes were the most abundant in Blautia and Parabacteroides spp. In Lactobacillus spp., ARGs from lincosamide antibiotic resistance classes were predominant, whereas those from glycopeptide antibiotic resistance classes were the most abundant in Lachnoclostridium spp. The effects of BLFPs and enramycin on the dominant genera with the assigned ARGs from the main antibiotic resistance classes in the cecal digesta of broilers are summarized in Table 4. Peptide resistance gene abundance in Bacteroides spp. was lower in the H group than in the C group (P = 0.016). The abundance of lincosamide resistance genes in Lactobacillus spp. was lower in the E group than in the C group (P < 0.01). Supplementation with 1 g/kg BLFPs led to the lowest lincosamide resistance gene abundance in Lactobacillus spp. (P < 0.001). The abundance of macrolide resistance genes in Blautia spp. and that of glycopeptide resistance genes in genus Lachnoclostridium spp. were lower in the L group than in the C and E groups (P < 0.05). Figure 6 presents the most abundant genera with the assigned ARGs in the cecal digesta. The aminocoumarin resistance gene parY was found to be predominantly associated with Bacteroides spp., whereas the peptide resistance gene rpoB2 was the most abundant in Lactobacillus and Alistipes spp. The macrolide resistance gene efrB was predominantly associated with Blautia spp. Moreover, the glycopeptide resistance gene vanRI was predominantly associated with Lachnoclostridium spp., whereas the macrolide resistance gene cmeB was the most abundant in Parabacteroides spp. The effects of BLFPs and enramycin on the dominant genera with the assigned ARGs in the cecal digesta of broilers are presented in Table 4. parY abundance in Bacteroides spp. was lower in the H group than in the other groups (P < 0.001). Supplementation with enramycin led to the lowest rpoB2 gene abundance in Lactobacillus spp. (P < 0.001). efrB abundance in Blautia spp. and vanRI abundance in Lachnoclostridium spp. were lower in the L group than in the other groups (P < 0.001).
Figure 5.
Pie chart for the distribution of most abundant genera with assigned antibiotic resistance genes from the main antibiotic resistance classes. The size of each pie chart is proportional to the antibiotic resistance class abundance within each group (n = 4).
Table 4.
Effect of B. licheniformis–fermented products and enramycin on dominant genera with assigned antibiotic resistance class and gene in cecal digesta of broilers.
| Relative abundance (%) |
|||||||
|---|---|---|---|---|---|---|---|
| C1 | E | L | H | SEM | P value | ||
| Genus | Antibiotic resistance class | ||||||
| Bacteroides | Peptide | 22.89a | 22.17ab | 21.38ab | 20.62b | 0.29 | 0.016 |
| Lactobacillus | Lincosamide | 38.62b | 41.30a | 36.49c | 39.69b | 0.48 | < 0.001 |
| Blautia | Macrolide | 22.71a | 22.64a | 19.61b | 21.77ab | 0.42 | 0.010 |
| Alistipes | Peptide | 30.28 | 29.22 | 29.75 | 28.78 | 0.31 | 0.389 |
| Lachnoclostridium | glycopeptide | 25.04ab | 27.69a | 21.20c | 23.28bc | 0.71 | < 0.001 |
| Parabacteroides | macrolide | 34.45 | 32.91 | 33.17 | 33.20 | 0.42 | 0.603 |
| Genus | Antibiotic resistance gene | ||||||
| Bacteroides | parY | 10.10b | 10.41ab | 10.79a | 9.24c | 0.16 | < 0.001 |
| Lactobacillus | rpoB2 | 20.96a | 16.37c | 21.52a | 18.96b | 0.54 | < 0.001 |
| Blautia | efrB | 14.75a | 14.54a | 10.24b | 12.92a | 0.53 | < 0.001 |
| Alistipes | rpoB2 | 16.02 | 15.76 | 16.08 | 14.60 | 0.22 | 0.055 |
| Lachnoclostridium | vanRI | 13.00ab | 15.19a | 9.92c | 12.64b | 0.55 | < 0.001 |
| Parabacteroides | cmeB | 34.45 | 32.91 | 33.17 | 33.20 | 0.42 | 0.603 |
C = Basal diet; E = Basal diet plus 10 mg/kg enramycin; L = Basal diet plus 1 g/kg B. licheniformis–fermented products; H = Basal diet plus 3 g/kg B. licheniformis–fermented products.
Means in a row with no common superscript are significantly different (P ≤ 0.05).
Figure 6.
Pie chart for the distribution of most abundant genera with assigned antibiotic resistance genes (ARGs). The size of each pie chart is proportional to the ARG abundance within each group (n = 4).
DISCUSSION
Dietary supplementation with BLFPs in broilers modulates gut morphology and microbiota, leading to improved health and growth performance under C. perfringens and E. tenella challenge (Cheng et al., 2021a,b; Yu et al., 2021). We previously found that supplementation with 3 g/kg BLFPs led to body weight improvement in broilers, increasing their weight by 12.9% on d 35 compared with that in control broilers (Chen and Yu, 2020). Furthermore, the performance of broilers fed 3 g/kg BLFPs did not differ from those in the enramycin-treated group (Chen and Yu, 2020). The abundance of Lactobacillus spp. in the feces is strongly associated with the feed conversion ratio of broilers (Singh et al., 2012). We previously found that the abundance of Lactobacillus spp. in the feces of broilers increased after treatment with 3 g/kg BLFPs (Chen and Yu, 2020). Moreover, the abundance of Lactobacillus spp. in feces was positively correlated with the body weight (r = 0.78), average daily gain (r = 0.78), and average daily feed intake (r = 0.24) of broilers but was negatively associated with their feed conversion ratio (r = −0.75). Similarly, in the present study, we observed that supplementation with 3 g/kg BLFPs increased the abundance of Lactobacillus spp. in the cecal digesta of broilers. The abundance of the genus Lactobacillus in the cecal digesta was also positively correlated with the body weight (r = 0.26) and average daily gain (r = 0.26) of broilers but was negatively associated with their feed conversion ratio (r = −0.45; data were analyzed on the basis of the previous study). The correlation between Lactobacillus spp. abundance and broiler growth performance was found to weaken in the present study—consistent with the results of Chen and Yu (2020). These results also corroborated those of Stanley et al. (2015), who found that cecal microbiota is qualitatively similar to fecal microbiota but quantitatively different in broilers. In a previous study, we demonstrated that the abundance of Lactobacillus spp. was high in the feces of enramycin-fed broilers (Chen and Yu, 2020). However, in the present study, supplementation with enramycin did not affect the abundance of Lactobacillus spp. in the cecal digesta of broilers. Chlortetracycline has been found to differentially modulate the abundance of Lactobacillus spp. in broiler ceca and ilea (She et al., 2018). These results imply that enramycin results in the rapid repopulation of bacteria and restores the dominance of the genus Lactobacillus in the distal gut of broilers. BLFPs at 3 g/kg can increase the cecal and fecal abundance of Lactobacillus spp. in broilers, and this increased abundance may have a beneficial effect on broiler performance. In summary, BLFPs and enramycin differentially modulate Lactobacillus spp. abundance in the cecal and fecal contents of broilers.
Accumulating evidence suggests that antibiotics modulate gut microbiota, and that the antibiotic-mediated alteration of the bacterial community structure affects ARG-harboring bacterial hosts (Costa et al., 2017; Xiong et al., 2018). Bacillus-based probiotics and Bacillus species–fermented products can modulate the intestinal microbiota of broilers (Chen and Yu, 2020; Bilal et al., 2021; Chen and Yu, 2021). However, the relationship between the Bacillus species–mediated alteration of microbiota and ARG-harboring bacterial hosts remains unclear. In the present study, ARGs in the cecal digesta of broilers were mainly harbored by diverse bacterial genera including Bacteroides, Lactobacillus, Blautia, Alistipes, Lachnoclostridium, and Parabacteroides. Bacteroides species in the human and poultry gut have been reported to be ARG reservoirs (Xiong et al., 2018; Yan et al., 2022). Among all antibiotic resistance classes, peptide resistance genes were dominant in the cecal digesta of broilers in the present study. This finding is not consistent with the observation of Xiong et al. (2018), who reported that multidrug resistance genes are dominant class in the feces of broilers. The differences in these findings may be due to the differences in sample source (cecal digesta vs. feces). Peptide antibiotics inhibit pathogen growth by blocking cell wall synthesis; however, bacteria carrying peptide resistance genes have been noted in the gut of broilers receiving growth-promoting doses of peptide antibiotics (Thibodeau et al., 2008). In the present study, we confirmed that peptide resistance genes represent the predominant antibiotic resistance class in Bacteroides spp. in the cecal digesta of broilers. The abundance of Bacteroides spp. in the cecal digesta did not change among the groups; nevertheless, 3 g/kg BLFPs reduced peptide resistance gene abundance in Bacteroides spp. In addition, the total number of peptide resistance genes in the cecal digesta decreased in response to 3 g/kg BLFP treatment. These findings suggest that treatment with 3 g/kg BLFPs induces a qualitative change in the ARGs of Bacteroides spp. in the cecal digesta of broilers. A study demonstrated that cloacal Lactobacillus spp. from broilers are resistant to macrolide and lincosamide antibiotics (Cauwerts et al., 2006). Macrolides and lincosamides prevent bacterial replication through a bacteriostatic mechanism by interfering with protein synthesis. In this study, we found that lincosamide and macrolide resistance genes were the dominant antibiotic resistance classes in Lactobacillus and Blautia spp., respectively. Although supplementation with 1 g/kg BLFPs did not alter Lactobacillus and Blautia spp. abundance in the cecal digesta, lincosamide resistance genes in Lactobacillus spp. and macrolide resistance genes in Blautia spp. decreased in broilers treated with 1 g/kg BLFPs. The abundance of Lactobacillus spp. in the cecal digesta increased in response to 3 g/kg BLFP treatment; however, lincosamide resistance gene abundance in Lactobacillus spp. did not increase accordingly. By contrast, enramycin supplementation did not increase Lactobacillus spp. abundance in the cecal digesta; however, lincosamide resistance gene abundance in Lactobacillus spp. was high in response to enramycin treatment. Taken together, these results suggest that BLFP supplementation can inhibit ARGs at the antibiotic resistance class level. In this study, 1 and 3 g/kg BLFP supplementation provided differential inhibition of antimicrobial classes, and BLFP and enramycin differentially regulated antimicrobial class abundance in the cecal digesta of broilers.
A recent study indicated that the most abundant ARGs in chicken cecal microbial commensals are tetracycline resistance genes, such as tetW, tet32, and tetO (Juricova et al., 2021). By contrast, the dominant ARGs in the cecal digesta of broilers were bcrA, rpoB2, and parY in the present study. bcrA encodes ABC transporter in Bacillus spp. and confers peptide antibiotic resistance (Podlesek et al., 1995). We found that bcrA gene abundance increased in the cecal digesta of broilers in response to enramycin treatment, indicating that peptide antibiotics added to the feed at low subtherapeutic levels could increase peptide resistance gene abundance. parY, which encodes an aminocoumarin-resistant topoisomerase, has also been identified in fecal microbiota of birds (Cao et al., 2020). In this study, supplementation with 1 g/kg BLFPs promoted parY abundance in Bacteroides spp., whereas supplementation with 3 g/kg BLFPs inhibited parY abundance. rpoB2 encodes the RNA polymerase beta subunit in Nocardia species and confers rifampin antibiotic resistance (Ishikawa et al., 2006). In the present study, supplementation with enramycin and 3 g/kg BLFPs inhibited rpoB2 abundance in Lactobacillus spp. efrB encodes ABC transporter and confers multidrug resistance (Lee et al., 2003), and vanRI encodes a regulatory protein associated with glycopeptide antibiotic resistance (Kruse et al., 2014). In this study, supplementation with 1 g/kg BLFPs inhibited efrB abundance in Blautia spp. and vanRI in Lachnoclostridium spp. in the cecal digesta of broilers, whereas supplementation with enramycin promoted vanRI abundance in Lachnoclostridium spp. Taken together, these results indicate that BLFPs and enramycin differentially modulate the abundance of individual ARGs at the genus level in the cecal and fecal contents of broilers.
Enramycin, a polypeptide antibiotic, inhibits MurG, which is involved in peptidoglycan synthesis in Gram-positive bacteria (Fang et al., 2006). Peptidoglycan synthesis inhibition compromises cell wall integrity, consequently leading to cell lysis and death. BLFPs contain B. licheniformis spores and their derived antimicrobial lipopeptides. The main antibacterial mechanisms of Bacillus spp. in the gut include competitive exclusion, competition for substrates and limiting resources, and antimicrobial substance synthesis (Bahaddad et al., 2022). We previously demonstrated that the antimicrobial lipopeptides isolated from BLFPs exhibit antibacterial activity against Gram-positive and Gram-negative bacteria (Horng et al., 2019). Antimicrobial lipopeptides may target and bind to the bacterial membrane directly, causing rapid depolarization of the antibacterial membrane (Avrahami and Shai, 2004). Because of the differences in the antibacterial mechanisms of action of BLFPs and enramycin, the cecal microbial community of broilers may be differentially regulated under their action. In the present study, we also found that supplementation with 1 g/kg BLFPs specifically increased L. crispatus and A. muciniphila abundance but reduced that of B. fragilis and C. bacterium CHKCI001. Our correlation results further demonstrated that L. crispatus and A. muciniphila abundance was negatively associated with B. fragilis and C. bacterium CHKCI001 abundance. These results indicate that L. crispatus and A. muciniphila may suppress B. fragilis and C. bacterium CHKCI001 growth in the cecal digesta of broilers. L. crispatus and A. muciniphila have been shown to exhibit beneficial effects in broilers and mice (Taheri et al., 2010; Zhao et al., 2017). Supplementation with enramycin alone can increase Blautia sp. An81 abundance but can reduce that of P. distasonis in the cecal digesta of broilers. Although the function of Blautia sp. An81 in the gut remains unclear, Blautia spp. may be involved in propionate biosynthesis in chickens (Polansky et al., 2015). P. distasonis has been shown to alleviate obesity and metabolic dysfunction in a diet-induced obesity mouse model (Wang et al., 2019). We also observed that Blautia sp. An81 and P. distasonis were not positively correlated with L. crispatus and A. muciniphila. These findings thus demonstrate that 3 g/kg BLFPs and enramycin differentially modulate the cecal bacterial community, even though both of them lead to similar growth performance in broilers.
In conclusion, the current study, for the first time, performed the integral analysis of the bacterial community composition and ARG distribution in the cecal digesta of broilers in response to BLFP and enramycin treatment. Differential ARG distribution was observed between BLFPs and enramycin because of the dominance of different bacterial taxa. Our results provide valuable insights into how BLFPs and enramycin differentially modulate the bacterial community composition to promote healthier growth in broilers. The results related to ARG may facilitate the evaluation of the feasibility of BLFPs as an alternative to antibiotics.
ACKNOWLEDGMENTS
This work was supported by the Ministry of Science and Technology [MOST 110-2313-B-197-005-MY3] in Taiwan.
DISCLOSURES
The authors have no conflicts of interest to declare.
References
- Abudabos A.M., Aljumaah M.R., Alkhulaifi M.M., Alabdullatif A., Suliman G.M., Al Sulaiman A.R. Comparative effects of Bacillus subtilis and Bacillus licheniformis on live performance, blood metabolites and intestinal features in broiler inoculated with Salmonella infection during the finisher phase. Microb. Pathog. 2020;139 doi: 10.1016/j.micpath.2019.103870. [DOI] [PubMed] [Google Scholar]
- Avrahami D., Shai Y. A new group of antifungal and antibacterial lipopeptides derived from non-membrane active peptides conjugated to palmitic acid. J. Biol. Chem. 2004;279:12277–12285. doi: 10.1074/jbc.M312260200. [DOI] [PubMed] [Google Scholar]
- Bahaddad S.A., Almalki M.H.K., Alghamdi O.A., Sohrab S.S., Yasir M., Azhar E.I., Chouayekh H. Bacillus species as direct-fed microbial antibiotic alternatives for monogastric production. Probiotics Antimicrob. Proteins. 2022;29:1–16. doi: 10.1007/s12602-022-09909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benno Y., Endo K., Mitsuoka T. Isolation of fecal Clostridium perfringens from broiler chickens and their susceptibility to eight antimicrobial agents for growth promotion. Nihon Juigaku Zasshi. 1988;50:832–834. doi: 10.1292/jvms1939.50.832. [DOI] [PubMed] [Google Scholar]
- Bilal M., Achard C., Barbe F., Chevaux E., Ronholm J., Zhao X. Bacillus pumilus and Bacillus subtilis promote early maturation of cecal microbiota in broiler chickens. Microorganisms. 2021;9:1899. doi: 10.3390/microorganisms9091899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Hu Y., Liu F., Wang Y., Bi Y., Lv N., Li J., Zhu B., Gao G.F. Metagenomic analysis reveals the microbiome and resistome in migratory birds. Microbiome. 2020;8:26. doi: 10.1186/s40168-019-0781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauwerts K., Pasmans F., Devriese L.A., Martel A., Haesebrouck F., Decostere A. Cloacal Lactobacillus isolates from broilers show high prevalence of resistance towards macrolide and lincosamide antibiotics. Avian Pathol. 2006;35:160–164. doi: 10.1080/03079450600598137. [DOI] [PubMed] [Google Scholar]
- Chen J.Y., Yu Y.H. Bacillus subtilis-fermented products ameliorate the growth performance and alter cecal microbiota community in broilers under lipopolysaccharide challenge. Poult. Sci. 2021;100:875–886. doi: 10.1016/j.psj.2020.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.C., Yu Y.H. Bacillus licheniformis-fermented products improve growth performance and the fecal microbiota community in broilers. Poult. Sci. 2020;99:1432–1443. doi: 10.1016/j.psj.2019.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.H., Horng Y.B., Chen W.J., Hua K.F., Dybus A., Yu Y.H. Effect of fermented products produced by Bacillus licheniformis on the growth performance and cecal microbial community of broilers under coccidial challenge. Animals. 2021;11:1245. doi: 10.3390/ani11051245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.H., Horng Y.B., Dybus A., Yu Y.H. Bacillus licheniformis-fermented products improve growth performance and intestinal gut morphology in broilers under Clostridium perfringens challenge. J. Poult. Sci. 2021;58:30–39. doi: 10.2141/jpsa.0200010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M.C., Bessegatto J.A., Alfieri A.A., Weese J.S., Filho J.A., Oba A. Different antibiotic growth promoters induce specific changes in the cecal microbiota membership of broiler chicken. PLoS One. 2017;12 doi: 10.1371/journal.pone.0171642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhariwal A., Chong J., Habib S., King I.L., Agellon L.B., Xia J. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45:W180–W188. doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA Introduction of a qualified presumption of safety (QSP) approach for assessment of selected microorganisms referred to EFSA - opinion of the scientific committee. EFSA J. 2007;587:1–16. [Google Scholar]
- Estevez I. Density allowances for broilers: where to set the limits? Poult. Sci. 2007;86:1265–1272. doi: 10.1093/ps/86.6.1265. [DOI] [PubMed] [Google Scholar]
- Fang X., Tiyanont K., Zhang Y., Wanner J., Boger D., Walker S. The mechanism of action of ramoplanin and enduracidin. Mol. Biosyst. 2006;2:69–76. doi: 10.1039/b515328j. [DOI] [PubMed] [Google Scholar]
- Gong L., Wang B., Mei X., Xu H., Qin Y., Li W., Zhou Y. Effects of three probiotic Bacillus on growth performance, digestive enzyme activities, antioxidative capacity, serum immunity, and biochemical parameters in broilers. Anim. Sci. J. 2018;89:1561–1571. doi: 10.1111/asj.13089. [DOI] [PubMed] [Google Scholar]
- Horng Y.B., Yu Y.H., Dybus A., Hsiao F.S.H., Cheng Y.H. Antibacterial activity of Bacillus species-derived surfactin on Brachyspira hyodysenteriae and Clostridium perfringens. AMB Express. 2019;9:188. doi: 10.1186/s13568-019-0914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa J., Chiba K., Kurita H., Satoh H. Contribution of rpoB2 RNA polymerase beta subunit gene to rifampin resistance in Nocardia species. Antimicrob. Agents Chemother. 2006;50:1342–1346. doi: 10.1128/AAC.50.4.1342-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juricova H., Matiasovicova J., Kubasova T., Cejkova D., Rychlik I. The distribution of antibiotic resistance genes in chicken gut microbiota commensals. Sci. Rep. 2021;11:3290. doi: 10.1038/s41598-021-82640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knap I., Lund B., Kehlet A.B., Hofacre C., Mathis G. Bacillus licheniformis prevents necrotic enteritis in broiler chickens. Avian Dis. 2010;54:931–935. doi: 10.1637/9106-101509-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Kruse T., Levisson M., de Vos W.M., Smidt H. vanI: a novel D-Ala-D-Lac vancomycin resistance gene cluster found in Desulfitobacterium hafniense. Microb. Biotechnol. 2014;7:456–466. doi: 10.1111/1751-7915.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.W., Huda M.N., Kuroda T., Mizushima T., Tsuchiya T. EfrAB, an ABC multidrug efflux pump in Enterococcus faecalis. Antimicrob. Agents Chemother. 2003;47:3733–3738. doi: 10.1128/AAC.47.12.3733-3738.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yan H., Lv L., Xu Q., Yin C., Zhang K., Wang P., Hu J. Growth performance and meat quality of broiler chickens supplemented with Bacillus licheniformis in drinking water. Asian Australas. J. Anim. Sci. 2012;25:682–689. doi: 10.5713/ajas.2011.11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall B.M., Levy S.B. Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 2011;24:718–733. doi: 10.1128/CMR.00002-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa B.B., Duan Y., Khawar H., Sun Q., Ren Z., Elsiddig Mohamed M.A., Abbasi I.H.R., Yang X. Bacillus subtilis B21 and Bacillus licheniformis B26 improve intestinal health and performance of broiler chickens with Clostridium perfringens-induced necrotic enteritis. J. Anim. Physiol. Anim. Nutr. 2019;103:1039–1049. doi: 10.1111/jpn.13082. [DOI] [PubMed] [Google Scholar]
- NRC . Nutrient Requirements of Poultry. 9th rev. ed. National Research Council, National Academy Press; Washington, DC: 1994. [Google Scholar]
- Podlesek Z., Comino A., Herzog-Velikonja B., Zgur-Bertok D., Komel R., Grabnar M. Bacillus licheniformis bacitracin-resistance ABC transporter: relationship to mammalian multidrug resistance. Mol. Microbiol. 1995;16:969–976. doi: 10.1111/j.1365-2958.1995.tb02322.x. [DOI] [PubMed] [Google Scholar]
- Polansky O., Sekelova Z., Faldynova M., Sebkova A., Sisak F., Rychlik I. Important metabolic pathways and biological processes expressed by chicken cecal microbiota. Appl. Environ. Microbiol. 2015;82:1569–1576. doi: 10.1128/AEM.03473-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Y., Cai H., Liu G. Effects of antibiotic on microflora in ileum and cecum for broilers by 16S rRNA sequence analysis. Anim. Sci. J. 2018;89:1680–1691. doi: 10.1111/asj.13113. [DOI] [PubMed] [Google Scholar]
- Singh K.M., Shah T., Deshpande S., Jakhesara S.J., Koringa P.G., Rank D.N., Joshi C.G. High through put 16S rRNA gene-based pyrosequencing analysis of the fecal microbiota of high FCR and low FCR broiler growers. Mol. Biol. Rep. 2012;39:10595–10602. doi: 10.1007/s11033-012-1947-7. [DOI] [PubMed] [Google Scholar]
- Stanley D., Geier M.S., Chen H., Hughes R.J., Moore R.J. Comparison of fecal and cecal microbiotas reveals qualitative similarities but quantitative differences. BMC Microbiol. 2015;15:51. doi: 10.1186/s12866-015-0388-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri H.R., Moravej H., Tabandeh F., Zaghari M., Shivazad M. Efficacy of combined or single use of Lactobacillus crispatus LT116 and L. johnsonii LT171 on broiler performance. Br. Poult. Sci. 2010;51:580–585. doi: 10.1080/00071668.2010.508491. [DOI] [PubMed] [Google Scholar]
- Thibodeau A., Quessy S., Guévremont E., Houde A., Topp E., Diarra M.S., Letellier A. Antibiotic resistance in Escherichia coll and Enterococcus spp. isolates from commercial broiler chickens receiving growth-promoting doses of bacitracin or virginiamycin. Can. J. Vet. Res. 2008;72:129–136. [PMC free article] [PubMed] [Google Scholar]
- Wang K., Liao M., Zhou N., Bao L., Ma K., Zheng Z., Wang Y., Liu C., Wang W., Wang J., Liu S.J., Liu H. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 2019;26:222–235. doi: 10.1016/j.celrep.2018.12.028. [DOI] [PubMed] [Google Scholar]
- Xiong W., Wang Y., Sun Y., Ma L., Zeng Q., Jiang X., Li A., Zeng Z., Zhang T. Antibiotic-mediated changes in the fecal microbiome of broiler chickens define the incidence of antibiotic resistance genes. Microbiome. 2018;6:34. doi: 10.1186/s40168-018-0419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Hall A.B., Jiang X. Bacteroidales species in the human gut are a reservoir of antibiotic resistance genes regulated by invertible promoters. NPJ Biofilms Microbiomes. 2022;8:1. doi: 10.1038/s41522-021-00260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.H., Wu C.M., Chen W.J., Hua K.F., Liu J.R., Cheng Y.H. Effectiveness of Bacillus licheniformis-fermented products and its derived antimicrobial lipopeptides in controlling coccidiosis in broilers. Animals. 2021;11:3576. doi: 10.3390/ani11123576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Liu W., Wang J., Shi J., Sun Y., Wang W., Ning G., Liu R., Hong J. Akkermansia muciniphila improves metabolic profiles by reducing inflammation in chow diet-fed mice. J. Mol. Endocrinol. 2017;58:1–14. doi: 10.1530/JME-16-0054. [DOI] [PubMed] [Google Scholar]